Abstract
The control of normal cell growth is a balance between stimulatory and inhibitory signals. MYC is a pleiotropic transcription factor that both activates and represses a broad range of target genes and is indispensable for cell growth. While much is known about gene activation by MYC, there is no established mechanism for the majority of MYC repressed genes. We report that MYC transcriptionally activates the PTEN tumor suppressor in normal cells to inactivate the PI3K pathway, thus suppressing AKT activation. Suppression of AKT enhances the activity of the EZH2 histone methyltransferase, a subunit of the epigenetic repressor Polycomb Repressive Complex 2 (PRC2), while simultaneously stabilizing the protein. MYC mediated enhancement in EZH2 protein level and activity results in local and genome-wide elevation in the repressive H3K27me3 histone modification, leading to widespread gene repression including feedback autoregulation of the MYC gene itself. Depletion of either PTEN or EZH2 and inhibition of the PI3K/AKT pathway leads to gene derepression. Importantly, expression of a phospho-defective EZH2 mutant is sufficient to recapitulate nearly half of all MYC-mediated gene repression. We present a novel epigenetic model for MYC-mediated gene repression and propose that PTEN and MYC exist in homeostatic balance to control normal growth which is disrupted in cancer cells.
Keywords: Myc, PTEN, Polycomb Repressive Complex 2, Ezh2, epigenetic
INTRODUCTION
Cancer is driven by the activation of oncogenes and the loss of tumor suppressors. One tumor suppressor that is frequently inactivated in diverse cancers is the phosphatase and tensin homolog gene (PTEN) (reviewed in (1)). PTEN acts as a negative regulator of the PI3K pathway by converting PIP3 to PIP2, which in turn prevents activation of the Akt pathway downstream of PI3K. The PI3K/Akt pathway is an important component of cell signaling that regulates a myriad of biological processes including growth, proliferation and apoptosis (1). Cancer cells often exhibit mutations in the PI3K/Akt pathway, including loss of PTEN, ultimately resulting in activation of the pathway and its downstream effectors (2).
The MYC gene is the most frequently amplified gene in human cancer, and deregulated expression of MYC is a hallmark of 70% of all cancers (3). Myc is a well-established pleiotropic transcription factor and significant progress has been made in understanding the role of Myc as a transcriptional activator (4). In addition to activating target genes, Myc also represses an almost equal number of genes (3), and this repression is important for Myc mediated cell proliferation and transformation (5, 6). However, despite major advances in the Myc field, there is no uniform mechanism of repression that satisfactorily accounts for the majority of Myc repressed genes. In fact, the first Myc regulated gene ever described was the MYC gene itself, through what is generally considered an autoregulatory feedback loop (7, 8). Autoregulation is postulated to be important in fine-tuning the amount of Myc in a cell since small changes in Myc expression are sufficient to shift the balance from normal to aberrant growth. Notably, many cancer cells have lost the ability to autoregulate (9, 10), and more Myc is advantageous for growth and proliferation of cancer.
One pathway implicated in autoregulation of the MYC ortholog in Drosophila (dmyc) is the Polycomb group of proteins (11). The Polycomb Repressive Complex 2 (PRC2) epigenetically silences genes by trimethylating lysine 27 on histone H3 (H3K27me3), a function carried out by the methyltransferase Enhancer of Zeste 2 (Ezh2) which is an integral component of the complex (12). The Ezh2 gene is frequently amplified or overexpressed in many tumors and was described as an E2F responsive oncogene (13).
In this study, we show that Myc suppresses the PI3K/Akt pathway via transcriptional upregulation of the PTEN tumor suppressor. Significantly, suppression of Akt results in Ezh2-mediated gene repression in two mammalian systems, including autoregulation. Activation of Ezh2 is both necessary and sufficient to account for nearly half of all Myc repressed genes. We propose a general mechanism for Myc-mediated repression and autoregulation linked to an important tumor suppressor pathway.
MATERIAL AND METHODS
Cell lines, drug treatments and Western blotting
c-myc−/− (HO 15.19) and Phoenix cells were maintained in DMEM supplemented with 10% fetal bovine serum. Immortalized mammary epithelial cells (IMECs) (14) were cultured in DMEM:F12 50:50 media supplemented with epidermal growth factor, insulin, hydrocortisone and 5% FBS. To generate stable cell lines, retroviral vectors were used to create polyclonal populations. For LY294002 (Cayman Chemical; #70920) treatments, cells were plated subconfluently 12–16 hours prior to treatment. Treatments lasted for 2 hours followed by RIPA lysis for immunoblotting. Antibody information used for immunoblotting and ChIPs can be found in Supplemental Information.
Plasmids and RNA interference
To generate the myc promoter-luciferase reporter, a 2.5 kb fragment containing the human MYC promoters (P1 and P2) and upstream regulatory elements was cloned into the pGL3 Basic vector. Ezh2/S21A and Ezh2/S21E mutants were generated using the QuikChange II Site Directed Mutagenesis Kit (Stratagene) as per manufacturers protocol. pUSE MYR-AKT was a gift from Dr. Lienhard (Dartmouth). Silencer Select Pre-designed siRNAs were obtained from Ambion (Applied Biosystems) and transfected using Lipofectamine RNAi Max (Invitrogen; 10nM siRNA). At the indicated time point, cells were harvested for protein or RNA. siRNA sequences are listed in Supplemental Information (Supplemental Table S2). Silencer Select Negative Control siRNA was used as a transfection control and to account for non-specific effects.
Real-time RT-PCR
Total RNA was harvested from log phase cultures with Trizol (Invitrogen) and cDNA was synthesized using the Super Script kit from Invitrogen. Two-step real-time polymerase chain reaction (PCR) was performed using the SYBR Green Mix (BioRad) on a BioRad C1000 Thermal cycler. The expression of GAPDH or ACTIN was used for normalization. Primer sequences are available upon request.
Chromatin immunoprecipitation
Log phase cultures were fixed with 1% formaldehyde for 10 min and subjected to ChIP assays with minor modifications as previously described (15). Briefly, cells were lysed in Lysis Buffer (1% SDS, 10mM EDTA, 50mM TRIS pH8.1) and sonicated to generate DNA fragments between 200–1000bps. Cleared lysates were then diluted and incubated with the following antibodies overnight at 4°C. IPs were washed with RIPA buffer and precipitated DNA was recovered. Real-time PCR amplification was then performed using specific primers. Primer sequences are available upon request. The data is presented as % binding compared to input for each sample. All experiments were performed three to four times and error bars represent S.D.
Luciferase assay
Luciferase assays were performed with the Dual luciferase Reporter Kit (Promega). The pRL vector constitutively expressing Renilla luciferase was used to normalize for transfection efficiency. 2×105 IMECs were plated in 12 well dishes 24 hours prior to transfections. On the day of transfection, each well was transfected with pRL, pGL3 Basic (to asses basal reporter activity) or MYC promoter-GL3 and the indicated plasmids. 24 hours later, luciferase activity was measured using the Wallac 1450 MicroBeta TriLux system (Perkin Elmer). Experiments were carried out three times in triplicates and error bars represent S.D.
Soft agar assay
In 6 well plates, 10,000 cells (+/− siRNA treatment for 48 hours) were plated in 0.3% agar, layered over a 0.6% agar base layer. Wells were re-fed with 200ul complete media every other day. 14 days after plating, colonies were counted and imaged.
RESULTS
Myc suppresses AKT through activation of PTEN expression
The PTEN tumor suppressor was previously shown to be a direct Myc target gene with an E-box that is occupied by Myc in vivo (16, 17). We analyzed PTEN expression and found that it is Myc-activated at both the protein and mRNA levels in human mammary epithelial cells (IMECs) and myc −/− rat fibroblast cells (Figures 1A and 1B). Therefore, we decided to explore the functional consequences of PTEN activation by Myc.
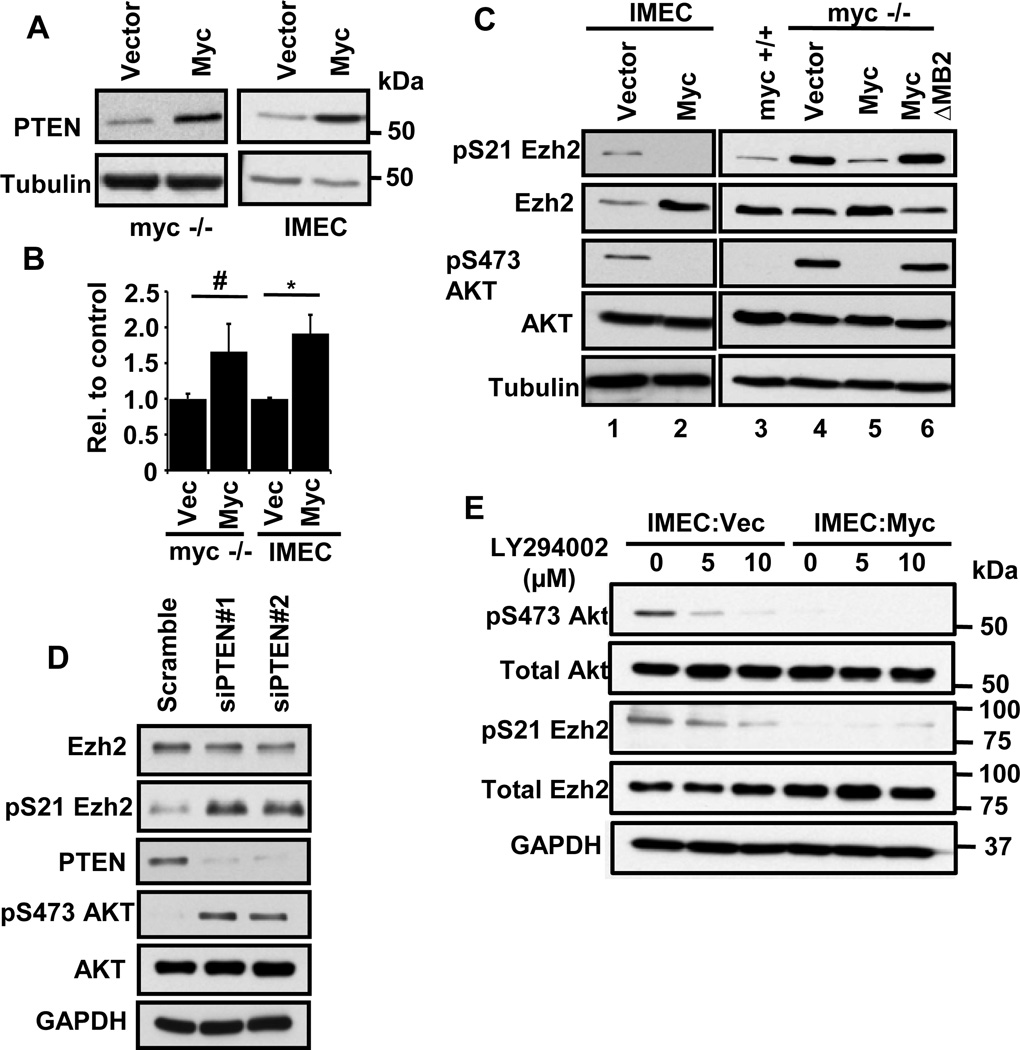
Figure 1. Myc induces PTEN expression and inhibition of the PI3K/Akt pathway resulting in increased active Ezh2.
(A) Immunoblot of PTEN protein levels in rat fibroblasts with or without Myc and in IMECs with or without exogenous Myc.
(B) RT-PCR of PTEN RNA levels in rat fibroblasts with or without Myc and in IMECs with or without exogenous Myc. * denotes p-value < 0.05, # denotes p-value < 0.01
(C) Immunoblot analysis of the indicated proteins in myc−/− fibroblasts and IMECs expressing empty vector or exogenous Myc. Experiments were performed three or more times and representative blots are shown.
(D) siRNA-mediated depletion of PTEN leads to enhanced pEzh2 and pAKT in IMECs.
(E) IMECs as in (C) were treated with LY294002 for 2 hours or left untreated. Cells were harvested and lysates were immunoblotted with the indicated antibodies. Inhibition of the PI3K/Akt axis suppresses pAkt (P-S473) and pEzh2 (P-S21 Ezh2).
PTEN is a dual lipid and protein phosphatase that acts as a negative regulator of the PI3K/Akt pathway (1). Since PTEN dephosphorylates PIP3 to PIP2, thus attenuating Akt activation, we tested if PTEN activation had an effect on Akt signaling. Activation of Akt involves phosphorylation of Serine 473 (pS473) so we assessed the levels of pS473 in response to exogenous Myc expression. Notably, we found that Myc inhibits Akt S473 phosphorylation in both IMECs and myc−/− rat fibroblasts (Figure 1C). In IMECs, there was a basal level of pS473 in the parental/vector cells that becomes undetectable with Myc overexpression. In rat fibroblasts, there is no detectable pS473 in myc+/+ cells which express native levels of Myc. In contrast, myc−/− cells derived by genetic knockout have highly elevated pS473 which is subsequently suppressed to undetectable levels by reconstitution of Myc. Repression of Akt pS473 is not evident with a Myc mutant lacking the conserved Myc Box 2 transactivation domain (ΔMB2) (Figure 1C). MB2, an evolutionarily conserved motif in the Myc transactivation domain, is essential for most of Myc’s biological activities including oncogenic transformation, transactivation and gene repression (4). To determine if PTEN is responsible for regulating Akt phosphorylation, we depleted PTEN with siRNA and analyzed the level of Akt pS473. Consistent with the established signaling pathway, depleting PTEN in IMECs expressing exogenous Myc restored active Akt (pS473) (Figure 1D). Taken together, this data demonstrates that Myc induces PTEN to inhibit AKT activation, highlighting the biological consequence of a relationship that has only been alluded to previously.
To ensure we were not observing artifacts of overexpressing Myc beyond physiological levels, we monitored the amount of exogenous Myc in these systems. The level of Myc overexpression in IMECs is within the range observed in breast cancer cell lines, while the amount of Myc reconstituted in myc−/− fibroblasts is similar to that of endogenous Myc in parental myc+/+ fibroblasts (Fig S1A) (18). Other than PTEN, several other phosphatases have been reported to inactivate Akt via dephosphorylation but depletion had no effect on pAKT levels (Fig S1B, C).
EZH2 is activated by Myc-mediated suppression of AKT kinase activity
It was reported previously that Akt suppresses the activity of the Ezh2 histone methyltransferase by phosphorylation on Serine 21 (pS21) (23). Given the previous link in Drosophila between Myc-mediated gene repression and Polycomb complexes (11), we decided to examine the status of this modification using phospho-specific antibodies. We observed a loss of phospho-Ezh2 (pS21) in all cells with exogenous Myc expression and suppression of pAkt (Figure 1C) along with an increase in total Ezh2, suggesting an increase in the pool of active Ezh2 upon increased Myc expression. Additionally we found that Myc stabilizes Ezh2, without altering its mRNA levels, and that this stability is negatively correlated with its phosphorylation at Serine 21 (Fig S2). To test more directly if PTEN activation was responsible for reduced pEzh2, we used two independent siRNAs to deplete PTEN in IMECs and observed a significant increase in phospho-Ezh2 upon loss of PTEN (Figure 1D).
The PI3K/Akt pathway is frequently hyperactivated in a variety of tumors and thus many drugs are available to inhibit it. We used the compound LY294002 that inactivates PI3K to further investigate the role of the PI3K/Akt pathway in Ezh2 phosphorylation (24). We found that inhibiting the PI3K/Akt pathway with LY294002 in parental IMECs prevents Akt activation and phosphorylation at Serine 473 (Figure 1E). In addition, loss of pS473 Akt induces loss of phospho-Ezh2 (pS21) and a modest increase in total Ezh2.
MYC represses genes via Ezh2 and Polycomb Repressive Complex 2
To test the consequence of activating Ezh2 through loss of phosphorylation, we assessed whole cell levels of the Ezh2-associated H3K27me3 modification that is indicative of repressed chromatin. Consistent with an increase in the active fraction of Ezh2, we observed Myc dependent elevation in global levels of H3K27me3 in both cell lines (Figure 2A). This genome-wide increase in H3K27me3 could contribute to the large number of genes that are Myc repressed. This led us to hypothesize that Myc mediated inhibition of the PI3K pathway via PTEN upregulation results in increased genome-wide gene repression elicited by PRC2. It is important to note that because we analyzed histones from whole cell extracts, we cannot distinguish between chromatin bound and non-chromatin bound histones.
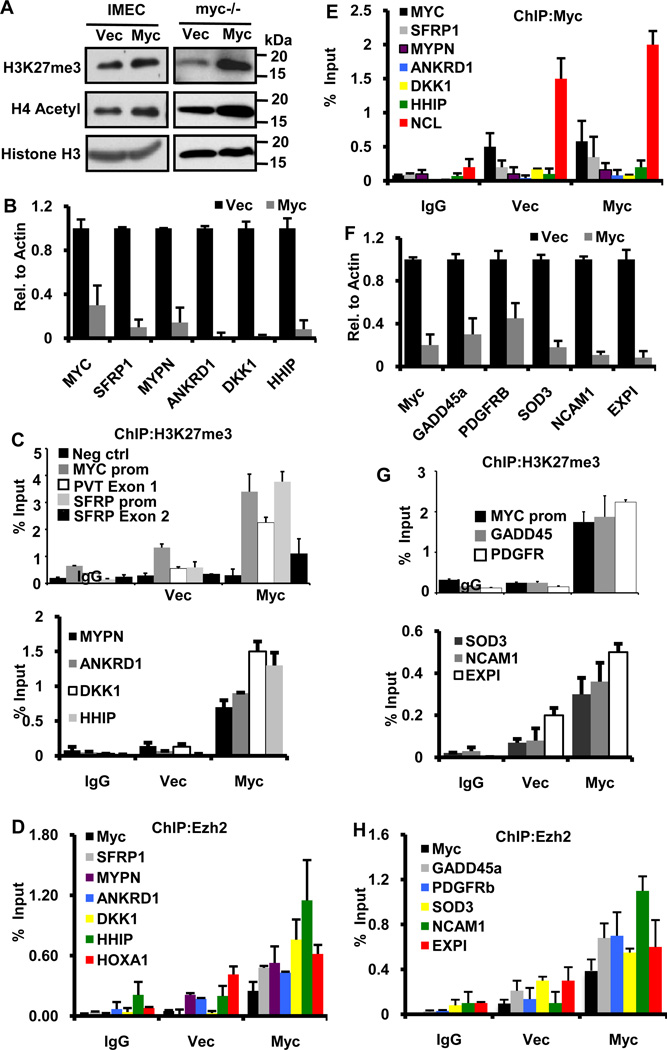
Figure 2. Overexpression of Myc enhances global H3K27 methylation and specific H3K27 methylation at the MYC promoter and Myc-repressed genes.
(A) Immunoblot analysis of H3K27me3 levels in IMECs +/− Myc overexpression (left panel) or in myc−/− rat fibroblasts reconstituted with Myc (right panel). Myc also enhances acetylated H4 as previously described (47).
(B) qRT-PCR for Myc repressed targets in IMECs transduced with empty vector or exogenous Myc. Primers specific to the indicated genes were used in analyzing transcriptional repression by Myc.
(C) Chromatin immunoprecipitation (ChIP) with anti-H3K27me3 was performed using IMECs transduced with empty vector or exogenous Myc. IP-ed DNA was assessed for H3K27me3 at the MYC promoter, PVT exon 1, SFRP promoter, SFRP exon 2, MYPN promoter, ANKRD1 promoter, DKK1 promoter and HHIP promoter. IgG served as a control for nonspecific binding to beads (IgG) and NUP214 served as a negative control (Neg ctrl) for the H3K27me3 mark.
(D) ChIP with anti-Ezh2 antibody. In IMECs, the same gene promoters as in (C) were analyzed for Ezh2 binding, and increased binding of Ezh2 was observed with exogenous Myc.
(E) ChIP with anti-MYC was performed in IMECs and immunoprecipitated DNA was analyzed with primers specific for all promoters as in (B). There was no significant binding over background, even with added exogenous Myc (Vec and Myc). A known Myc binding site in the nucleolin (NCL) gene served as a positive control.
(F) Quantitative RT-PCR of the indicated genes in myc−/− rat fibroblasts before and after reconstitution with Myc.
(G) ChIP with anti-H3K27me3 in myc−/− fibroblasts shows enhanced H3K27me3 at the endogenous MYC promoter and Myc-repressed genes GADD45, PDGFRb SOD3, NCAM1 and EXPI.
(H) Repressed gene promoters as in (G) were analyzed for Ezh2 binding in myc−/− fibroblasts, showing increased binding with exogenous Myc. ChIP and qRT-PCR data is presented as an average of three independent experiments with error bars denoting S.D.
To assess a role for Ezh2 activity and H3K27me3 in gene repression, we studied several Myc-repressed genes in two different cell systems. As discussed earlier, the first Myc repressed gene described was MYC itself so we tested if the autoregulatory feedback loop involved repression by H3K27me3. In IMECs, we analyzed the Myc-repressed genes MYC, SFRP1, DKK1, MYPN, ANKRD1 and HHIP (Figure 2B). It was previously shown that repression of SFRP1 and DKK1 is functionally important for Myc-mediated transformation (6). MYPN, ANKRD1 and HHIP are additional Myc repressed genes obtained from microarray data in IMECs (25). We observed an enrichment of the H3K27me3 modification at the endogenous human MYC promoter in response to autorepression by ectopic Myc (Figure 2C). A similar enrichment of H3K27me3 was also observed at exon 3 of MYC and at a region 30kb downstream of the MYC gene encompassing the transcriptional start site of the noncoding PVT RNA (Figure 2C and Figure S3B), which is regulated in parallel with MYC (26). These data suggest that the H3K27me3 mark may extend over a large area, consistent with spreading of H3K27me3 observed in Drosophila (27). As with the MYC gene itself, we found significant enrichment of the PRC2-mediated H3K27me3 modification in response to exogenous Myc expression at the promoters of all repressed genes tested (Figure 2C). In line with the propagation of H3K27me3 along gene bodies, we observed an enrichment of H3K27me3 at exon 2 of SFRP1, approximately 10 kb downstream of the promoter (Figure 2C). In contrast, two well-studied Myc activated target genes, nucleolin (NCL) and fibrillarin (FBL), did not show an enhancement of H3K27me3 in response to exogenous Myc (Fig. S3B), and Myc activated ribosomal protein targets were unaffected by overexpression of Ezh2 in myc−/− fibroblasts (Supplemental Table S1). We also found no binding of Myc protein itself at any of the Myc repressed promoters (Figure 2E).
Since Ezh2 is the methyltransferase responsible for the H3K27me3 mark, we analyzed genes with enriched H3K27me3 for Ezh2 binding by chromatin immunoprecipitation. As expected, Ezh2 was detected at repressed genes with the H3K27me3 mark (Figure 2D). Surprisingly, we did not observe enrichment of Ring1b, which is a component of PRC1, at Myc-repressed genes in IMEC cells (Figure S3A). This is consistent with a recent report showing the presence of PRC2, independent of PRC1, at bivalent domains in ES cells (28).
To further validate the role of PRC2 in Myc-mediated repression, we used the myc−/− rat fibroblast cell line in which two different selectable markers, neo and his, have been placed under control of the endogenous MYC promoters (29). Reconstituting Myc expression into these cells represses the endogenous MYC promoter similar to autorepression (30). We observed a strong induction of the H3K27me3 mark in response to reconstituted mouse Myc expression at the endogenous MYC promoter, at previously reported Myc repressed genes GADD45 and PDGFRb, and at additional Myc repressed genes SOD3, NCAM1 and EXPI which were selected from unpublished microarrays (Figures 2F and 2G). Additionally, we detected an accumulation of H3K27me3 on the HOXA1 promoter, a known Polycomb repressed target, but not at the promoter of a Myc activated target, Nol5a (Fig S3C). Ezh2 was also detected in these repressed genes in accordance with the IMEC data (Figure 2H). Thus, enhanced levels of H3K27me3 appear to be a general feature of Myc-repressed genes, including the MYC promoter itself.
PTEN and Ezh2 are indispensable components of Myc-mediated repression
To test if PTEN and Ezh2 are required for Myc-mediated repression, we depleted each with specific siRNAs. PTEN depletion dramatically reversed the repression of the Myc-repressed genes and prevented the accumulation of the H3K27me3 modification (Figure 3A–B and FigS4B). Similarly, depletion of Ezh2 (Figure 3C) restored expression of endogenous human MYC, SFRP1, HHIP, MYPN, ANKRD1 and DKK1 genes (Figure 3D and FigS4A) and induced a loss of H3K27me3 at these promoters (Figure 3E). Thus both PTEN and Ezh2 are required for sustained repression and accumulation of H3K27me3. Unfortunately, myc−/− fibroblasts do not permit transient transfection of siRNA to perform the same experiment.
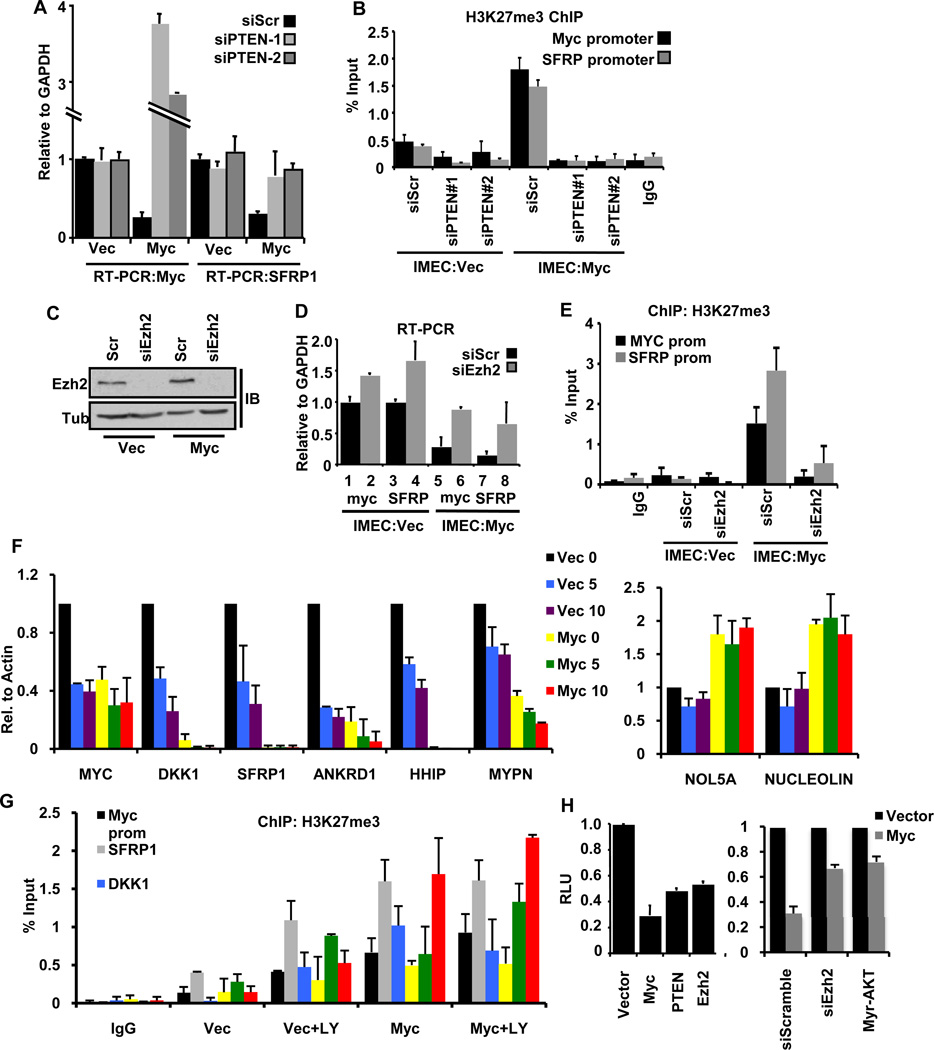
Figure 3. PTEN and Ezh2 are required for Myc-mediated gene repression.
(A) siRNA-mediated depletion of PTEN in IMECs leads to derepression of both endogenous MYC and SFRP expression.
(B) Depletion of PTEN leads to loss of H3K27me3 at the MYC and SFRP promoters that are repressed by exogenous Myc.
(C) Immunoblot analysis of Ezh2 upon siRNA depletion of Ezh2 in IMECs transduced with either empty vector or exogenous Myc.
(D) qRT-PCR for endogenous MYC and SFRP mRNAs. Expression of both genes is suppressed by ectopic Myc overexpression (compare lanes 1 and 5, lanes 3 and 7). Depletion of Ezh2 derepresses both MYC and SFRP (compare lanes 5 and 6, lanes 7 and 8).
(E) ChIP with anti-H3K27me3 after siRNA treatment of Ezh2 in IMECs. Depletion of Ezh2 abolishes the H3K27me3 mark at the endogenous MYC and SFRP1 promoters in IMEC:Myc cells.
(F) qRT-PCR with RNA obtained from cells treated with LY294002 as in Figure 1F. Left panel: Inhibition of Akt activation induces repression of Myc-repressed genes in Vector cells, mimicking the effect of exogenous Myc (compare Vec + LY to Myc for each target gene). Right panel: Myc activated target genes, Nol5a and Nucleolin were unaffected by LY treatment.
(G) Myc repressed promoters were assessed for H3K27me3 accumulation via ChIP in samples treated with 5µM LY. Treatment of IMEC:Vec cells with LY enhances H3K27me3 mark at Myc repressed promoters in line with reduced transcription (F).
(H) Left panel: Myc, PTEN and Ezh2 all repress a MYC promoter-luciferase reporter in transient assays in IMECs. Right panel: Cells were treated with RNAi against Ezh2 or transfected with Myr-Akt 24 hours prior to transfection with the appropriate plasmids for the luciferase assay as stated in Materials and Methods. Graphs represent an average of at three independent replicates and error bars indicate S.D.
To provide additional evidence that the PI3K pathway was linked to gene repression, we treated parental IMECs with LY294002. Chemical inhibition of the PI3K pathway (Figure 1D) resulted in repression of Myc repressed genes without altering the transcriptional activity of Myc induced targets (Figure 3F). Along with reduced expression, Myc repressed promoters showed an accumulation of the repressive H3K27me3 mark upon treatment with LY294002 (Figure 3G). Altogether, these data provide strong evidence for the importance of Myc-mediated inhibition of the PI3K/Akt pathway to initiate and maintain repression of genes.
To test more directly if the levels of PTEN and Ezh2 can suppress the MYC promoter analogous to autoregulation, we performed a transient assay with a MYC promoter-luciferase reporter. Ectopic expression of Myc, PTEN and Ezh2 all suppressed luciferase expression to a similar extent (Figure 3H, left panel). Depletion of Ezh2 with RNAi or expression of a constitutively active form of AKT (Myr-AKT) (31) is sufficient to alleviate repression of the MYC promoter (Figure 3H, right panel). These data support a model where Myc and PTEN exist in homeostatic balance to regulate Myc mediated gene repression and autoregulation, which is supported by a previous report that small changes in PTEN levels is sufficient to alter PI3K/Akt signaling and promote tumorigenesis (32).
Ezh2-mediated repression is independent of Miz-1
While we observed a strong dependence on Ezh2 for Myc-mediated gene repression, neither cell system exhibited a similar dependence on Miz-1 (ZBTB17), a zinc finger transcription factor previously reported to play a role in Myc-mediated repression (33, 34) (Figure S4 C–G). We also did not observe Miz-1 occupancy at Myc-repressed gene promoters in IMECs (Figure S4C) or a gain of repression in control (IMEC:Vec) cells upon loss of Miz-1 (Figure S4E, left panel). Similar conclusions could be drawn from myc−/− fibroblasts stably expressing a Myc mutant (V394D) defective in Miz-1 binding (35). myc−/− fibroblasts stably expressing exogenous mouse WT-Myc or V394E were able to repress the transcription of genes similarly (Figures S4F and S4G). These data suggest an alternate mechanism of Myc-mediated repression that is Miz-1 independent.
Expression of ectopic Ezh2 mimics Myc-mediated repression
Since overexpression of PTEN could affect multiple signaling pathways, we wanted to determine if altered activity of Ezh2 alone could account for Myc-mediated repression and autoregulation. To this end, we analyzed gene expression in myc−/− cell lines stably expressing Ezh2WT, Ezh2-S21A (phospho-defective) and Ezh2-S21E (phospho-mimetic), and compared it to repression in response to Myc over-expression. Exogenous Ezh2 expression was comparable to endogenous levels for all three constructs (Figure 4A), which can be resolved because exogenous Ezh2 has a tag that alters its size. Notably, expression of Ezh2-S21A in myc−/− cells is sufficient to repress the MYC promoter similar to autorepression, and similar repression was also observed for other Myc-repressed genes (Figure 4B). Expression of Ezh2-WT induced a modest repression, whereas the Ezh2-S21A mutant repressed as strongly as Myc. In contrast, there was no repression at all with the Ezh2-S21E mutant. Ezh2-WT enhanced H3K27me3 levels at all three genes (Figure 4C). The phospho-defective mutant (S21A) induced even higher H3K27me3 levels, which correlated with stronger repression, whereas the phospho-mimetic Ezh2-S21E induced barely detectable changes (Figure 4C). Additionally, we analyzed the level of a different histone modification (H3K4me3) which is associated with actively transcribed promoters. Interestingly, we found that overexpression of either Myc or any form of Ezh2 led to a complete loss of H3K4me3 at Myc-repressed promoters, confirming the established inverse relationship between these modifications (Figure 4D) (36). In addition to Myc WT and Ezh2, we also analyzed Myc mutants with defects in either the transactivation domain (ΔMB2) or DNA binding domain (ΔC). Neither mutant had any effect on H3K27me3 or H3K4me3 levels (Figures 4C and 4D), consistent with their defect in gene repression (Figure 4B).
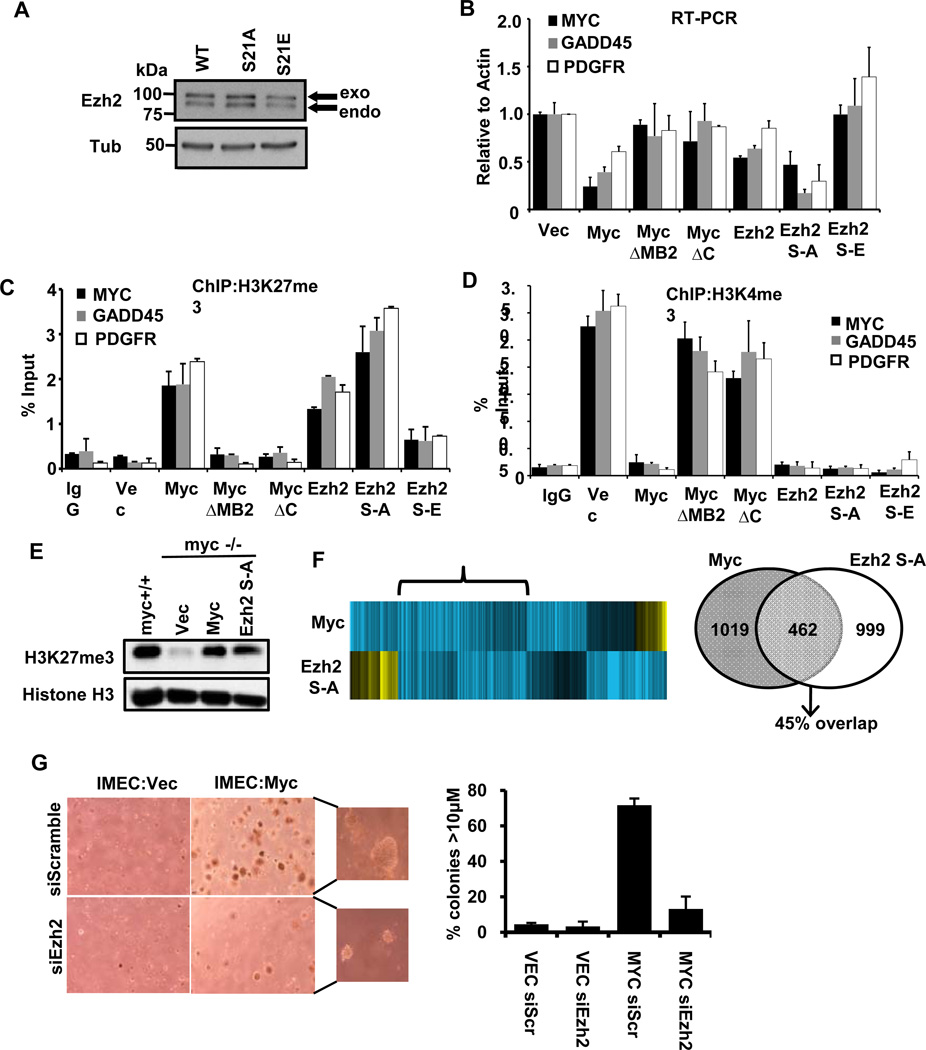
Figure 4. Ezh2 alone can repress MYC and Myc-repressed genes.
(A) Ezh2 and Ezh2 mutants were stably expressed in myc−/− fibroblasts.
(B) Expression of the MYC promoter, GADD45 and PDGFRb were assessed by RT-PCR.
(C) ChIP for H3K27me3 levels at the MYC, GADD45 and PDGFRb promoters in the same lines from B.
(D) ChIP for H3K4me3 levels at the MYC, GADD45 and PDGFR promoters in the same lines from B.
(E) Immunoblot analysis of total cell lysates from myc+/+ and myc−/− samples expressing empty Vector (Vec), exogenous Myc (Myc) or Ezh2 S-A. Myc and Ezh2 increase global levels of H3K27me3.
(F) Heat map of repressed genes from microarray data. Blue (downregulated) and yellow (upregulated). The bracket marks genes that are repressed by both Myc and Ezh2S21A. Venn diagram demonstrates the overlap of genes repressed by 2-fold or more in both samples.
(G) Soft agar transformation assay with IMEC cells expressing empty vector (Vec) or exogenous Myc (Myc) upon depletion of Ezh2 with RNAi. Graph represents mean number of colonies that were larger than 10µM per 100 colonies counted (48). Data presented is average of three independent replicates and error bars indicate S.D.
We were particularly interested in determining the amount of Myc-mediated repression that can be accounted for by enhanced Ezh2 activity and elevated H3K27me3 levels. We analyzed whole genome expression by microarray using RNA from myc−/− fibroblasts expressing Myc-WT or Ezh2-S21A. We chose the hyperactive, phospho-defective form of Ezh2 to avoid phosphorylation of Ezh2-WT by active Akt (Figure 1C, lane 4) in myc/− cells. Comparable levels of H3K27me3 were present in myc+/+ parental fibroblasts, Myc-reconstituted myc−/− fibroblasts and myc−/− fibroblasts expressing ectopic Ezh2-S21A (Figure 4E). After normalizing to empty vector, we selected all genes that were repressed 2-fold or more by Myc WT and assessed their response to Ezh2-S21A. There were 1802 probes (1019 unique genes) repressed >2-fold by Myc, and 1724 probes (999 genes) repressed by Ezh2 S21A. Of the Myc-repressed genes, 814 probes (462 genes) overlapped with Ezh2-S21A repression, or a 45% overlap of repressed genes (Fig 4F). These data strongly support an integral role for Ezh2 in Myc-mediated gene repression and autoregulation.
Additionally, in our experimental setting, we observed that the transforming activity of Myc is directly tied to functional PRC2, and thus gene repression (Fig 4G). Depletion of Ezh2 significantly impairs the ability of IMEC cells expressing exogenous Myc to form colonies in soft agar. These data corroborate previous reports that have linked Myc-mediated gene repression to its ability to transform cells (5, 37). Altogether, these data provide evidence that Myc-mediated repression is an important component of Myc biology and its function as a potent oncogene.
DISCUSSION
This study presents a new model for the mechanism of Myc-mediated repression based on activation of PTEN transcription and the modulation of Ezh2 activity and repressive histone modifications. While complete loss of the PTEN tumor suppressor is common in cancer, even modest changes in PTEN levels are sufficient to promote oncogenic transformation (32, 38). In parallel, it has been established that incremental changes in Myc expression are a common driving force in cancer and can also contribute to inherited cancer predispositions (3, 15, 39, 40). Given these findings along with the data presented here, we propose that controlling the amount of Myc in a cell by autoregulation via modulation of PTEN expression is pivotal in maintaining the delicate balance of normal growth. Our data are consistent with recent studies showing that elevated PTEN suppresses MYC expression (38).
The regulation of Ezh2 by Myc occurs through the Akt pathway, which was previously shown to directly modify Ezh2 activity by phosphorylation (23). Suppression of Akt activity by PTEN reduces Ezh2 phosphorylation, increasing methyltransferase activity and simultaneously increasing Ezh2 protein levels by protein stabilization. Suppression of pAkt, pEzh2, and Ezh2 methyltransferase activity are dependent on both the MB2 and C-terminal DNA binding domains of Myc, consistent with previous mapping of Myc domains required for gene repression and autoregulation. The dependence on the Myc transactivation domain for gene repression stems from a requirement to induce the expression of PTEN. A recent study shows consistent activation of the Akt pathway in Burkitt’s lymphomas (41), but it is difficult to interpret these findings in relation to the model presented here because tumors may acquire complex mutational profiles that drive oncogenic growth independent of the MYC pathway or gene repression.
We show that the MYC gene itself as well as numerous Myc-repressed genes acquire high levels of H3K27me3 which is necessary and largely sufficient to suppress transcription (Figure 5). Repression by H3K27me3 involves a number of mechanisms such as the recruitment of PRC1 to the H3K27me3 deposited by PRC2 (42), but we find no enrichment of PRC1 components at Myc-repressed promoters. However, since PRC2 can be recruited to the histone modification that it creates (43), a small increase in Ezh2 activity could amplify the regional H3K27me3 modification through positive feedback at responsive genes (43, 44). Stable association of PRC2 complexes with repressed genes could also block the activating H3K4me3 modification as we observe. We show that Myc and Ezh2 share 45% of their repressed genes and that overexpression of Ezh2 alone can recapitulate Myc-mediated gene repression and autoregulation. These data suggest that the modulation of Ezh2 is a major mediator of Myc gene repression.
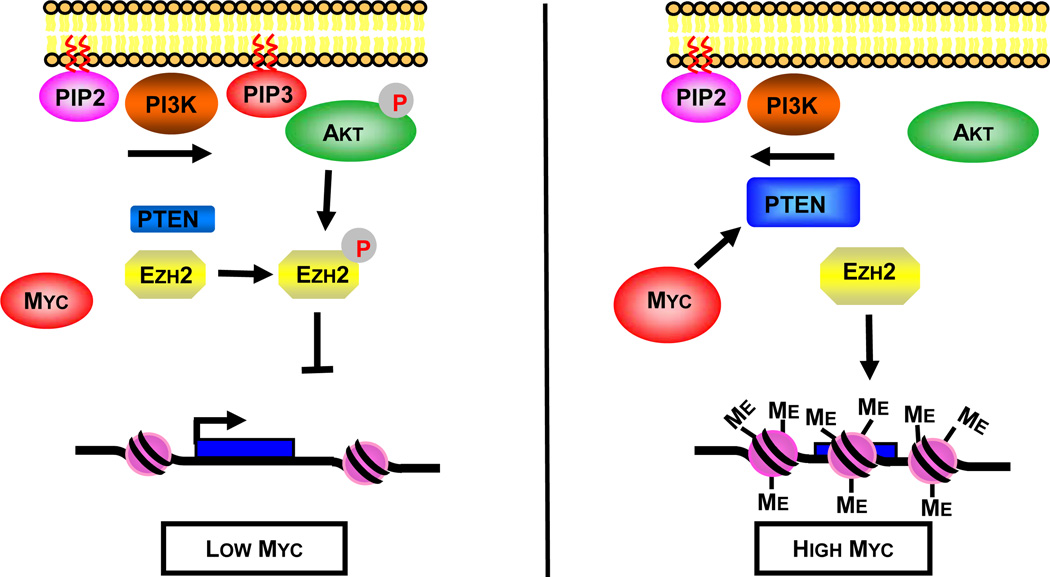
Figure 5. An epigenetic model for Myc-mediated gene repression.
(left) Cells with low levels of Myc have lower levels of PTEN, allowing more active AKT (pAKT), suppression of Ezh2 activity by phosphorylation (pEzh2), and reduced H3K27me3 levels in the cell.
(right) Elevated levels of Myc transcriptionally activate expression of the PTEN tumor suppressor which suppresses AKT activity. Suppression of AKT activity in turn suppresses phosphorylation of Ezh2, elevating its methyltransferase activity and also its protein level through reduced turnover. Higher Ezh2 activity promotes repressive H3K27me3 histone modifications on the MYC promoter and Myc-repressed genes.
One aspect of our proposed mechanism that remains unclear is why particular promoters are responsive to Myc-mediated repression. No common motif has been associated with Myc-repressed genes other than the core initiator element in the promoter (33, 34, 45). One possibility is that certain promoters are poised to respond to a variation in H3K27me3 levels because they exist at a threshold in the balance between repressive and activating chromatin configurations. A small shift toward elevated repressive histone modifications could nucleate a localized expansion in repressive chromatin and downregulate gene expression. The balance of repressive and activating chromatin could be highly variable between cell types for individual genes, which could explain why Myc-repressed genes are so variable. An alternate possibility that is not mutually exclusive is that PRC2 complexes are guided to specific genes by noncoding RNAs that may vary in different cell types (46) or that may even be Myc induced. Nevertheless, our study presents a novel feedback pathway linking the potent tumor suppressor PTEN to MYC regulation and global changes in gene expression and chromatin modification.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Carol Ringelberg for help with microarray data analysis. We thank members of the Cole lab for helpful discussions and feedback. We also thank Dr. M.C. Hung for reagents. This work was supported by a grant from the National Cancer Institute (CA055248).
Footnotes
The authors disclose no potential conflicts of interest
REFERENCES
- 1.Chalhoub N, Baker SJ. PTEN and the PI3-kinase pathway in cancer. Annu Rev Pathol. 2009;4:127–150. doi: 10.1146/annurev.pathol.4.110807.092311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yuan TL, Cantley LC. PI3K pathway alterations in cancer: variations on a theme. Oncogene. 2008;27:5497–5510. doi: 10.1038/onc.2008.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dang CV, O'Donnell KA, Zeller KI, Nguyen T, Osthus RC, Li F. The c-Myc target gene network. Seminars in cancer biology. 2006;16:253–264. doi: 10.1016/j.semcancer.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 4.Cowling VH, Cole MD. Mechanism of transcriptional activation by the Myc oncoproteins. Seminars in cancer biology. 2006;16:242–252. doi: 10.1016/j.semcancer.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 5.Xiao Q, Claassen G, Shi J, Adachi S, Sedivy J, Hann SR. Transactivation-defective c-MycS retains the ability to regulate proliferation and apoptosis. Genes Dev. 1998;12:3803–3808. doi: 10.1101/gad.12.24.3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cowling VH, D'Cruz CM, Chodosh LA, Cole MD. c-Myc transforms human mammary epithelial cells through repression of the Wnt inhibitors DKK1 and SFRP1. Mol Cell Biol. 2007;27:5135–5146. doi: 10.1128/MCB.02282-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nishikura K, ar-Rushdi A, Erikson J, Watt R, Rovera G, Croce CM. Differential expression of the normal and of the translocated human c-myc oncogenes in B cells. Proceedings of the National Academy of Sciences of the United States of America. 1983;80:4822–4826. doi: 10.1073/pnas.80.15.4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Penn LJ, Brooks MW, Laufer EM, Land H. Negative autoregulation of c-myc transcription. The EMBO journal. 1990;9:1113–1121. doi: 10.1002/j.1460-2075.1990.tb08217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grignani F, Lombardi L, Inghirami G, Sternas L, Cechova K, Dalla-Favera R. Negative autoregulation of c-myc gene expression is inactivated in transformed cells. The EMBO journal. 1990;9:3913–3922. doi: 10.1002/j.1460-2075.1990.tb07612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Penn LJZ, Brooks MW, Laufer EM, Littlewood TD, Morgenstern JP, Evan GI, et al. Domains of human c-Myc protein required for autosuppression and cooperation with ras oncogenes are overlapping. Mol Cell Biol. 1990;10:4961–4966. doi: 10.1128/mcb.10.9.4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goodliffe JM, Wieschaus E, Cole MD. Polycomb mediates Myc autorepression and its transcriptional control of many loci in Drosophila. Genes Dev. 2005;19:2941–2946. doi: 10.1101/gad.1352305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morey L, Helin K. Polycomb group protein-mediated repression of transcription. Trends Biochem Sci. 2010;35:323–332. doi: 10.1016/j.tibs.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 13.Bracken AP, Pasini D, Capra M, Prosperini E, Colli E, Helin K. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. The EMBO journal. 2003;22:5323–5335. doi: 10.1093/emboj/cdg542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DiRenzo J, Signoretti S, Nakamura N, Rivera-Gonzalez R, Sellers W, Loda M, et al. Growth factor requirements and basal phenotype of an immortalized mammary epithelial cell line. Cancer Res. 2002;62:89–98. [PubMed] [Google Scholar]
- 15.Wright JB, Brown SJ, Cole MD. Upregulation of c-MYC in cis through a large chromatin loop linked to a cancer risk-associated single-nucleotide polymorphism in colorectal cancer cells. Mol Cell Biol. 2010;30:1411–1420. doi: 10.1128/MCB.01384-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neiman PE, Ruddell A, Jasoni C, Loring G, Thomas SJ, Brandvold KA, et al. Analysis of gene expression during myc oncogene-induced lymphomagenesis in the bursa of Fabricius. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:6378–6383. doi: 10.1073/pnas.111144898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fernandez PC, Frank SR, Wang L, Schroeder M, Liu S, Greene J, et al. Genomic targets of the human c-Myc protein. Genes Dev. 2003;17:1115–1129. doi: 10.1101/gad.1067003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cowling VH, Cole MD. The Myc Transactivation Domain Promotes Global Phosphorylation of the RNA Polymerase II Carboxy-Terminal Domain Independently of Direct DNA Binding. Mol Cell Biol. 2007;27:2059–2073. doi: 10.1128/MCB.01828-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao T, Furnari F, Newton AC. PHLPP: a phosphatase that directly dephosphorylates Akt, promotes apoptosis, and suppresses tumor growth. Mol Cell. 2005;18:13–24. doi: 10.1016/j.molcel.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 20.Brognard J, Sierecki E, Gao T, Newton AC. PHLPP and a second isoform, PHLPP2, differentially attenuate the amplitude of Akt signaling by regulating distinct Akt isoforms. Mol Cell. 2007;25:917–931. doi: 10.1016/j.molcel.2007.02.017. [DOI] [PubMed] [Google Scholar]
- 21.Kuo YC, Huang KY, Yang CH, Yang YS, Lee WY, Chiang CW. Regulation of phosphorylation of Thr-308 of Akt, cell proliferation, and survival by the B55alpha regulatory subunit targeting of the protein phosphatase 2A holoenzyme to Akt. J Biol Chem. 2008;283:1882–1892. doi: 10.1074/jbc.M709585200. [DOI] [PubMed] [Google Scholar]
- 22.Resjo S, Goransson O, Harndahl L, Zolnierowicz S, Manganiello V, Degerman E. Protein phosphatase 2A is the main phosphatase involved in the regulation of protein kinase B in rat adipocytes. Cell Signal. 2002;14:231–238. doi: 10.1016/s0898-6568(01)00238-8. [DOI] [PubMed] [Google Scholar]
- 23.Cha TL, Zhou BP, Xia W, Wu Y, Yang CC, Chen CT, et al. Akt-mediated phosphorylation of EZH2 suppresses methylation of lysine 27 in histone H3. Science. 2005;310:306–310. doi: 10.1126/science.1118947. [DOI] [PubMed] [Google Scholar]
- 24.Vlahos CJ, Matter WF, Hui KY, Brown RF. A specific inhibitor of phosphatidylinositol 3-kinase, 2-(4-morpholinyl)-8-phenyl-4H-1-benzopyran-4-one (LY294002) J Biol Chem. 1994;269:5241–5248. [PubMed] [Google Scholar]
- 25.Doe MR, Ascano JM, Kaur M, Cole MD. Myc posttranscriptionally induces HIF1 protein and target gene expression in normal and cancer cells. Cancer Res. 2012;72:949–957. doi: 10.1158/0008-5472.CAN-11-2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carramusa L, Contino F, Ferro A, Minafra L, Perconti G, Giallongo A, et al. The PVT-1 oncogene is a Myc protein target that is overexpressed in transformed cells. J Cell Physiol. 2007;213:511–518. doi: 10.1002/jcp.21133. [DOI] [PubMed] [Google Scholar]
- 27.Schwartz YB, Kahn TG, Nix DA, Li XY, Bourgon R, Biggin M, et al. Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nature genetics. 2006;38:700–705. doi: 10.1038/ng1817. [DOI] [PubMed] [Google Scholar]
- 28.Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, et al. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 2008;4 doi: 10.1371/journal.pgen.1000242. e1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mateyak MK, Obaya AJ, Adachi S, Sedivy JM. Phenotypes of c-Myc-deficient rat fibroblasts isolated by targeted homologous recombination. Cell Growth Differ. 1997;8:1039–1048. [PubMed] [Google Scholar]
- 30.Bush A, Mateyak M, Dugan K, Obaya A, Adachi S, Sedivy J, et al. c-myc null cells misregulate cad and gadd45 but not other proposed c-Myc targets. Genes Dev. 1998;12:3797–3802. doi: 10.1101/gad.12.24.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Watanabe S, Umehara H, Murayama K, Okabe M, Kimura T, Nakano T. Activation of Akt signaling is sufficient to maintain pluripotency in mouse and primate embryonic stem cells. Oncogene. 2006;25:2697–2707. doi: 10.1038/sj.onc.1209307. [DOI] [PubMed] [Google Scholar]
- 32.Alimonti A, Carracedo A, Clohessy JG, Trotman LC, Nardella C, Egia A, et al. Subtle variations in Pten dose determine cancer susceptibility. Nature genetics. 2010;42:454–458. doi: 10.1038/ng.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Staller P, Peukert K, Kiermaier A, Seoane J, Lukas J, Karsunky H, et al. Repression of p15INK4b expression by Myc through association with Miz-1. Nat Cell Biol. 2001;3:392–399. doi: 10.1038/35070076. [DOI] [PubMed] [Google Scholar]
- 34.Seoane J, Pouponnot C, Staller P, Schader M, Eilers M, Massague J. TGFbeta influences Myc, Miz-1 and Smad to control the CDK inhibitor p15INK4b. Nat Cell Biol. 2001;3:400–408. doi: 10.1038/35070086. [DOI] [PubMed] [Google Scholar]
- 35.Herold S, Wanzel M, Beuger V, Frohme C, Beul D, Hillukkala T, et al. Negative regulation of the mammalian UV response by Myc through association with Miz-1. Mol Cell. 2002;10:509–521. doi: 10.1016/s1097-2765(02)00633-0. [DOI] [PubMed] [Google Scholar]
- 36.Mills AA. Throwing the cancer switch: reciprocal roles of polycomb and trithorax proteins. Nat Rev Cancer. 2010;10:669–682. doi: 10.1038/nrc2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cowling VH, Cole MD. An N-Myc truncation analogous to c-Myc-S induces cell proliferation independently of transactivation but dependent on Myc homology box II. Oncogene. 2008;27:1327–1332. doi: 10.1038/sj.onc.1210734. [DOI] [PubMed] [Google Scholar]
- 38.Garcia-Cao I, Song MS, Hobbs RM, Laurent G, Giorgi C, de Boer VC, et al. Systemic elevation of PTEN induces a tumor-suppressive metabolic state. Cell. 2012;149:49–62. doi: 10.1016/j.cell.2012.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pomerantz MM, Ahmadiyeh N, Jia L, Herman P, Verzi MP, Doddapaneni H, et al. The 8q24 cancer risk variant rs6983267 shows long-range interaction with MYC in colorectal cancer. Nature genetics. 2009;41:882–884. doi: 10.1038/ng.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tuupanen S, Turunen M, Lehtonen R, Hallikas O, Vanharanta S, Kivioja T, et al. The common colorectal cancer predisposition SNP rs6983267 at chromosome 8q24 confers potential to enhanced Wnt signaling. Nature genetics. 2009;41:885–890. doi: 10.1038/ng.406. [DOI] [PubMed] [Google Scholar]
- 41.Schmitz R, Young RM, Ceribelli M, Jhavar S, Xiao W, Zhang M, et al. Burkitt lymphoma pathogenesis and therapeutic targets from structural and functional genomics. Nature. 2012 doi: 10.1038/nature11378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bracken AP, Helin K. Polycomb group proteins: navigators of lineage pathways led astray in cancer. Nat Rev Cancer. 2009;9:773–784. doi: 10.1038/nrc2736. [DOI] [PubMed] [Google Scholar]
- 43.Hansen KH, Helin K. Epigenetic inheritance through self-recruitment of the polycomb repressive complex 2. Epigenetics. 2009;4:133–138. doi: 10.4161/epi.4.3.8483. [DOI] [PubMed] [Google Scholar]
- 44.Margueron R, Li G, Sarma K, Blais A, Zavadil J, Woodcock CL, et al. Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol Cell. 2008;32:503–518. doi: 10.1016/j.molcel.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Facchini LM, Chen S, Marhin WW, Lear JN, Penn LZ. The Myc negative autoregulation mechanism requires Myc-Max association and involves the c-Myc P2 minimal. Mol Cell Biol. 1997;17:100–114. doi: 10.1128/mcb.17.1.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith E, Shilatifard A. The chromatin signaling pathway: diverse mechanisms of recruitment of histone-modifying enzymes and varied biological outcomes. Mol Cell. 2010;40:689–701. doi: 10.1016/j.molcel.2010.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Knoepfler PS, Zhang XY, Cheng PF, Gafken PR, McMahon SB, Eisenman RN. Myc influences global chromatin structure. The EMBO journal. 2006;25:2723–2734. doi: 10.1038/sj.emboj.7601152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cowling VH, Cole MD. E-cadherin repression contributes to c-Myc-induced epithelial cell transformation. Oncogene. 2007;26:3582–3586. doi: 10.1038/sj.onc.1210132. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.