Abstract
Understanding the cellular and molecular mechanisms underlying the formation and maintenance of memories is a central goal of the neuroscience community. It is well regarded that an organism's ability to lastingly adapt its behavior in response to a transient environmental stimulus relies on the central nervous system's capability for structural and functional plasticity. This plasticity is dependent on a well-regulated program of neurotransmitter release, post-synaptic receptor activation, intracellular signaling cascades, gene transcription, and subsequent protein synthesis. In the last decade, epigenetic markers like DNA methylation and post-translational modifications of histone tails have emerged as important regulators of the memory process. Their ability to regulate gene transcription dynamically in response to neuronal activation supports the consolidation of long-term memory. Furthermore, the persistent and self-propagating nature of these mechanisms, particularly DNA methylation, suggests a molecular mechanism for memory maintenance. In this review, we will examine the evidence that supports a role of epigenetic mechanisms in learning and memory. In doing so, we hope to emphasize (1) the widespread involvement of these mechanisms across different behavioral paradigms and distinct brain regions, (2) the temporal and genetic specificity of these mechanisms in response to upstream signaling cascades, and (3) the functional outcome these mechanisms may have on structural and functional plasticity. Finally, we consider the future directions of neuroepigenetic research as it relates to neuronal storage of information.
Learning and memory can be broadly defined as lasting alterations of a behavioral output produced in response to a transient environmental input (Sweatt 2010). In order for a transient stimulus to induce a lasting change in behavior, cells must undergo a complex set of stimulus-specific cellular and molecular changes that will consolidate a memory into an everlasting trace. Since the 1960s, memory researchers have recognized the importance of gene transcription and protein synthesis in long-term memory formation in a variety of experimental memory paradigms (Agranoff 1965; Agranoff et al. 1965, 1966; Squire et al. 1980). However, given that most proteins turn over on a timescale of hours, these findings raised important conceptual questions regarding the molecular basis for lifelong memory maintenance. It soon became evident that a self-perpetuating biochemical reaction would be required to preserve the molecular changes induced by short-lived environmental stimuli. With this necessity in mind, Crick (1984) and Holliday (1999) put forth the proposition that epigenetic mechanisms, particularly DNA methylation, possess the biochemical properties necessary to propagate memories over a lifetime. DNA methylation has long been appreciated as a stable and self-perpetuating regulator of cellular identity through the establishment and propagation of persistent, heritable changes in gene expression across cell divisions (Bird 2002). This mnemogenic quality suggested that epigenetic mechanisms may provide a suitable molecular basis for memory formation and maintenance.
As a result, the last decade has seen a series of studies demonstrating that epigenetic markers are actively and transiently regulated in post-mitotic neurons of adult rodents, honeybees, aplysia, and drosophila during the normal process of learning and memory (e.g., Levenson et al. 2004; Chwang et al. 2006; Lubin et al. 2008; Miller et al. 2008; Gupta et al. 2010; Lockett et al. 2010; Miller et al. 2010; Kramer et al. 2011; Maddox and Schafe 2011; Monsey et al. 2011; Biergans et al. 2012). Considering the stable nature of DNA methylation during development, it was surprising to find both dynamic and activity-induced changes in DNA methylation in the adult central nervous system. This unconventional operation of epigenetic mechanisms led to the formulation of a subfield of epigenetics, termed neuroepigenetics (also referred to as behavioral epigenetics [Lester et al. 2011]). Neuroepigenetics encompasses “the unique mechanisms and processes allowing dynamic experience-dependent regulation of the epigenome in nondividing cells of the nervous system” (Day and Sweatt 2011). A growing number of studies under this umbrella have demonstrated a critical role for epigenetic mechanisms in a wide range of learning and memory tasks and across diverse brain regions. Thus, epigenetic mechanisms seem to play a ubiquitous role in the establishment of lasting neural and behavioral modifications in response to environmental stimuli.
In this review, we will primarily discuss the role of epigenetic mechanisms in the formation and maintenance of fear memory. We also hope to demonstrate the universal role these mechanisms have in other learning and memory behavioral paradigms. In doing so, we will review the evidence that transient epigenetic modifications mediate memory consolidation by regulating gene expression within the first few hours after learning, whereas sustained changes in epigenetic modifications in cortical brain regions underlie memory maintenance over prolonged periods of time. Finally, we will examine the current understanding of the basic principles that regulate the establishment of specific patterns of epigenetic modifications and speculate on some possible mechanisms through which these modifications translate into cellular changes that support memory formation and maintenance.
Defining epigenetics
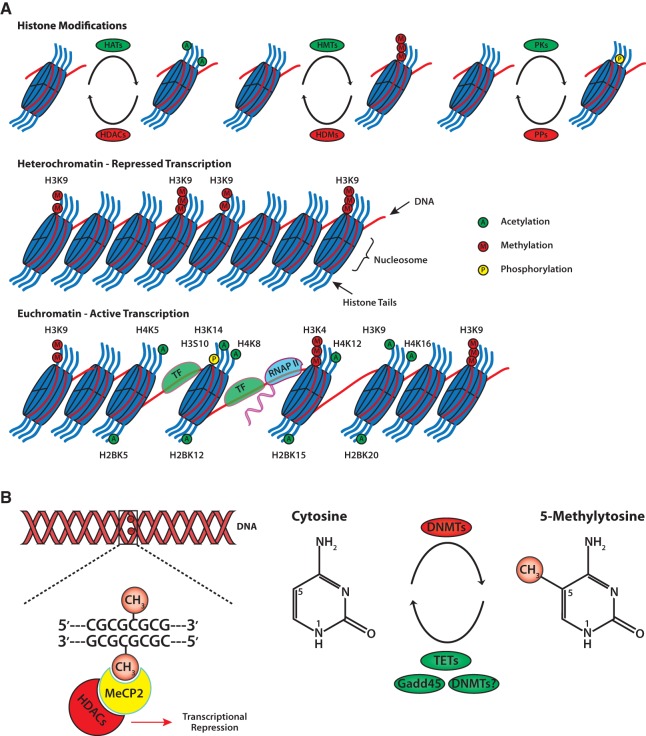
Epigenetic mechanisms are key regulators of DNA compaction and transcription. To allow long stretches of DNA to fit inside the cell nucleus, 146 bp sections of DNA are coiled around an octamer of histone proteins, which contains two pairs of histone H2A-H2B dimers and an H3-H4 histone tetramer (Quina et al. 2006). Adjacent nucleosomes join with one another via the linker histone H1 to form chromatin (Happel and Doenecke 2009), which can exist either as heterochromatin, characterized by a closed, highly compacted state restrictive to transcription, or as euchromatin, characterized by an open state amenable to transcription (Arney and Fisher 2004). The switching between the opposing chromatin states and the assembly of transcriptional machinery at gene promoters is mediated by epigenetic modifications, primarily DNA methylation and post-translational modifications of histones (see Fig. 1). DNA methylation preferentially occurs on cytosines positioned adjacent to guanine nucleobases (CpG) and is established via DNA methyltransferase (DNMT) enzymes, which catalyze the covalent binding of a methyl group from the methyl donor S-adenosyl-methionine (SAM) to the 5′ position on the cytosine-pyrimidine ring (Chiang et al. 1996; Turker 1999; Bird 2002; Price 2010). Distinct subgroups of DNMTs carry out distinct functions, whereby de novo DNMTs 3a and 3b establish novel methylation marks and the maintenance DNMT1 maintains previously established methylation marks (Cheng et al. 2010). Methylation of a cytosine on one strand prompts maintenance DNMTs to methylate the corresponding cytosine on the opposite strand, which allows for the self-perpetuation and persistence of this mark throughout cell division and in the face of DNA damage (Santos et al. 2005). Although DNA methylation primarily represses transcription by interfering with the binding of transcriptional machinery to regulatory sites on DNA (Iguchi-Ariga and Schaffner 1989) and by promoting closed chromatin states via the recruitment of transcriptional repressors (Karymov et al. 2001; Drewell et al. 2002; Fuks et al. 2003), recent evidence suggests that methyl-CpG-binding protein 2 (MeCP2) can also activate transcription through interactions with CREB (Chahrour et al. 2008). Such duality of function was recently reported for de novo DNMTs as well, whereby DNMT3a1 and DNMT3a2 isoforms are associated with heterochromatin and euchromatin, respectively (Chen et al. 2002; Kotini et al. 2011).
Figure 1.
General schematic of epigenetic modifications. (A) Packaging of DNA into chromatin is achieved through the wrapping of 146 bp of DNA around octamers of histone proteins. Chromatin-modifying enzymes dynamically regulate the addition and removal of post-translational modifications on histone N-terminal tails. Modifications associated with learning and memory include histone acetylation, phosphorylation, and methylation. The specific combination of histone tail modifications dictate whether or not the chromatin exists as heterochromatin or euchromatin. Heterochromatin is characterized by condensed chromatin and subsequent transcriptional repression. Euchromatin is characterized by a relaxed chromatin state that allows transcriptional machinery access to DNA for gene expression. (B) Methylation of DNA involves covalent addition of a methyl group to the 5′ position of the cytosine pyrimidine ring by DNMTs. DNA methylation commonly occurs at genes enriched with cytosine-guanine nucleotides (CpG islands). Proteins with methyl-binding domains, like MeCP2, bind to methylated DNA and recruit repressor complexes containing HDACs. Recent evidence suggests that active DNA demethylation can occur via several mechanisms involving members of the Gadd45 family, TET family, and DNMTs themselves. (DNMTs) DNA methyltransferases, (Gadd45) growth arrest and DNA damage 45, (HATs) histone acetyltransferases, (HDACs) histone deacetylases, (HDMs) histone demethylases, (HMTs) histone methyltransferases, (MeCP2) methyl CpG binding protein 2, (PKs) protein kinases, (PPs) protein phosphatases, (TETs) ten eleven translocation, (TF) transcription factor, (RNAP II) RNA polymerase II.
Post-translational modifications (PTMs) of histones are another critically important regulator of chromatin compaction and gene expression. The positive charge of unmodified histone proteins facilitates interactions with negatively charged DNA and promotes closed chromatin states (Muhlbacher et al. 2006). Histones can undergo a number of modifications, including acetylation, phosphorylation, and methylation, which alter their charge and binding properties (Muhlbacher et al. 2006; Sanchez Mde and Gutierrez 2009). Histone acetylation is the most widely studied modification and involves the transfer of an acetyl group from acetyl coenzyme A to lysine residues of histone tails via histone acetyltransferase (HAT) enzymes (Hebbes et al. 1988). In contrast to DNA methylation, histone acetylation is associated with transcriptional activation, which is largely attributed to acetylated histones acting as recognition sites for chromatin-remodeling proteins, transcriptional regulators, and RNA polymerase II (Mujtaba et al. 2007). Whereas acetylation and phosphorylation are primarily associated with transcriptional activation, histone methylation can either promote or repress transcription, depending on which residue is modified and with how many methyl groups (Nakayama et al. 2001; Peters and Schubeler 2005). For example, methylation of H3 lysine 4 (H3K4) is associated with transcriptional activation regardless of the number of methyl groups, whereas di- or tri-methylation of H3K9 is associated with transcriptional repression (Binda et al. 2010). Different types of modifications are not independent, in that specific modifications tend to co-occur, based largely on their role as transcriptional activators or repressors (Strahl and Allis 2000). In addition, DNA methylation interferes with histone acetylation through the recruitment of complexes that include histone deacetylase (HDAC) enzymes, which remove acetyl groups from histones (Wade 2001a,b). The opposite may also hold true, as a recent study in plants has implicated HAT enzymes in active DNA demethylation though a yet unknown mechanism (Qian et al. 2012). It is important to note that the roles of specific modifications in either activation or repression of transcription are based on generalizations of predominant instead of absolute associations with transcriptional outcomes. For example, bidirectional promoters are silenced on one side and active on the other in spite of similar chromatin states (Lin et al. 2007; Vastenhouw and Schier 2012), and histone H3K9me2 is considered to be a repressive mark even though it does not strictly correlate with transcriptional activation or repression on a genome-wide scale (He and Lehming 2003; Barski et al. 2007). Thus, caution is warranted when extrapolating transcriptional outcomes from observations of histone modifications in isolation. Even more important, the imperfect congruence between individual epigenetic modifications and transcription emphasizes the importance of investigating the overall pattern of epigenetic modifications on transcriptional outcomes, particularly when drawing inferences regarding complex outcomes, such as learning and memory.
There is now growing evidence that DNA methylation is dynamically and bidirectionally regulated in response to a variety of experience-induced events, including neural activity in the brain, estrogen treatment in human cells, and exercise in muscle (Kangaspeska et al. 2008; Metivier et al. 2008; Guo et al. 2011a,b; Barres et al. 2012). Tremendous gains have been rapidly made in identifying the mechanisms of active DNA demethylation, which include base excision repair in response to deamination of a methylated cytosine by Gadd45 or oxidation by TET proteins, which convert methylated cytosines (5mC) into hydroxyl-methyl-cytosines (5hmC) (Gehring et al. 2009; Kriaucionis and Heintz 2009; Ma et al. 2009; Tahiliani et al. 2009; Wu and Sun 2009; Guo et al. 2011b; Niehrs and Schafer 2012). In addition, the same DNMT enzymes that methylate DNA have been implicated in DNA demethylation (Metivier et al. 2008), but this mechanism has not yet been investigated in the brain. Thus, active regulation of epigenetic marks is a critical regulator of gene expression in a variety of tissue types.
Achieving signaling- and task-specificity at the level of epigenetic regulation
For epigenetic mechanisms to support the formation of distinct and diverse memories, epigenetic modifications must be responsive to signaling cascades induced by environmental stimuli. Resulting modifications of chromatin structure must then regulate the expression of memory-associated genes within the appropriate neural networks. In other words, epigenetic modifications must be actively and selectively induced by specific signaling cascades at specific genes in the cells and brain regions that support specific types of memory. Indeed, many studies have now shown that epigenetic changes that support memory formation and maintenance involve task-, region-, gene-, time-, and signaling-cascade specific changes in the epigenetic regulation of gene expression.
Epigenetic mechanisms regulate memory across tasks and brain regions
The majority of evidence supporting an epigenetic basis for memory formation and maintenance has been found using hippocampus-dependent tasks that include contextual fear conditioning, Morris water maze (MWM), novel object recognition (NOR), and object-location memory. In general, hippocampus-dependent tasks have been associated with global increases of euchromatin-related post-translational modifications of histones and with positive regulation of gene expression. For example, contextual fear conditioning produced increased levels of acetylation at H3 lysine 14 (H3K14), phosphorylation at H3 serine 10 (H3S10), and trimethylation at H3 lysine 4 (H3K4me3) in the hippocampus (Levenson et al. 2004; Chwang et al. 2006), although a recent study also found increased levels of the heterochromatin-related dimethylation at H3 lysine 9 (H3K9me2) (Gupta et al. 2010; Gupta-Agarwal et al. 2012). Similarly, training on the Morris water maze induced increased acetylation at H4 lysine 12 (H4K12) and pan-acetylation of H2B (tetra-acetylated-H2BK5K12K15K20) (Bousiges et al. 2010).
Memory consolidation for a particular task involves the establishment of distinct epigenetic modifications in individual components of memory-supportive networks. For example, contextual fear conditioning induced distinct H3K9me2 and H3K4me3 patterns in the hippocampus compared with those of the entorhinal cortex, and the inhibition of H3K9me2 in the entorhinal cortex, but not in the hippocampus, enhanced memory formation (Gupta-Agarwal et al. 2012). Similarly, cued fear conditioning as well as BDNF-induced plasticity in cell culture resulted in different patterns of histone PTMs at the homer1 promoter in hippocampal and amygdala neurons (Mahan et al. 2012), indicating that the same gene can be differentially regulated by identical stimuli in different regions of the brain. Epigenetic modifications can be further localized to discrete subregions of the hippocampus. For example, Castellano and colleagues (2012) found that training in a one-day redundant place/cue version of the MWM induced increased pan-acetylation of H3 and H4 and decreased acetylation of H3 lysine 9 (H3K9) in the CA1, whereas only H3 pan-acetylation was increased in area CA3. In addition to increased pan-acetylation of H3, the dentate gyrus (DG) was also characterized by sparse immunolabeling for H3S10. These data highlight the subregion specificity of histone modifications and exemplify the need to further refine our analysis to distinct subregions and, ideally, to specific genes and cell populations to better understand the sort of information encoded by these modifications.
In contrast to studies involving contextual fear conditioning and MWM that directly measured histone PTMs induced by learning, the role of histone modifications in object-recognition and object-location memory have been substantiated primarily via pharmacological and genetic manipulations of HATs and HDACs (Vecsey et al. 2007; Stefanko et al. 2009; Roozendaal et al. 2010; Haettig et al. 2011; McQuown et al. 2011). Given the role of histone acetylation in long-term memory formation, many groups have targeted different HATs such as CREB-binding protein (CBP), E1A-binding protein (p300), and p300/CBP-associated factor (PCAF) (Oike et al. 1999; Bourtchouladze et al. 2003; Alarcon et al. 2004; Korzus et al. 2004; Wood et al. 2005, 2006; Maurice et al. 2008; Chen et al. 2010; Barrett et al. 2011; Oliveira et al. 2011; Valor et al. 2011). In a comprehensive review, Barrett and Wood (2008) examined the numerous memory deficits associated with mouse models containing mutations in one of the three previously mentioned HATs. Interestingly, a deficit in NOR memory was the most common impairment among all the different genetically modified mice, suggesting the relative importance of HAT activity and histone acetylation for NOR memory.
Studies using a variety of behavioral paradigms in brain regions outside of the hippocampus, particularly in the amygdala, the prefrontal cortex, the insular cortex, and the striatum, have begun to investigate the role of epigenetic mechanisms in relation to learning and memory paradigms associated with those brain regions, including cued fear conditioning, memory extinction, conditioned taste aversion, and reward learning, respectively (Swank and Sweatt 2001; Bredy et al. 2007; Kwon and Houpt 2010; e.g., Barros et al. 2011; Bayerlein et al. 2011; Monsey et al. 2011). For example, increased expression of euchromatin-associated modifications of histones was reported during the consolidation of cued fear conditioning in the amygdala (Monsey et al. 2011). In addition, histone modifications may be important for the consolidation of conditioned taste aversion (CTA) memories. Studies in the mollusk Helix lucorum show unilateral increases in H3K14 acetylation in premotor interneurons that initiate withdrawal behavior in response to a food item previously paired with an aversive electric shock (Danilova et al. 2010). In rodents, a role for histone acetylation in CTA memory is indirectly supported by increased levels of lysine-HAT activity in the insular cortex after training (Swank and Sweatt 2001). There is also a rapidly growing literature on the role of epigenetic modifications in the striatum in the development of drug addiction and a number of excellent reviews are available on the subject (e.g., Maze and Nestler 2011; Robison and Nestler 2011). The breadth of epigenetic involvement in various learning and memory tasks is indicative of the seemingly universal requirement for epigenetic mechanisms in producing lasting changes in neural function and behavior in response to a variety of transient environmental stimuli.
Fewer studies have been conducted on the role of DNA methylation in learning and memory and this role is best characterized for fear conditioning. In 2007, Miller and Sweatt reported the first evidence for increased de novo DNMT expression in the hippocampus in response to contextual fear conditioning. This change in enzyme levels was accompanied by increased DNA methylation at the promoter of the memory-suppressor gene protein phosphatase I (PP1) and decreased methylation at the promoter of the plasticity-associated gene reelin. Later studies identified learning-induced changes in DNA methylation of BDNF, arc, and calcineurin genes (Lubin et al. 2008; Miller et al. 2010; Munoz et al. 2010; Penner et al. 2011), which play critical roles in memory formation and maintenance. Recent studies from Glen Schafe's group demonstrated that DNA methylation is important for the consolidation and re-consolidation of cued fear conditioning in the amygdala using DNMT inhibitors to block DNA methylation (Maddox and Schafe 2011; Monsey et al. 2011), although that group has not directly investigated DNA methylation changes at specific genes. A number of studies have also found DNA methylation to be associated with drug responses and drug-related reward learning in striatal structures (e.g., Barros et al. 2011; Bayerlein et al. 2011; Nielsen et al. 2012) and with the establishment of lasting behavioral modification across species, including Aplysia and the honey bee (Lockett et al. 2010; Biergans et al. 2012; Rajasethupathy et al. 2012). Although the specific nature of the link between DNA methylation and behavioral modification is not clear, the available evidence suggests that DNA methylation is critical for coordinating appropriate patterns of gene silencing and activation.
DNA methylation and the balance between memory activators and memory suppressors
An interesting feature of DNA methylation is the gene-specificity and the directionality of observed changes, with some genes exhibiting increased and others exhibiting decreased DNA methylation after learning (Miller and Sweatt 2007; Lubin et al. 2008; Feng et al. 2010; Gupta et al. 2010; Miller et al. 2010). Given this observation, it is not clear whether memory deficits observed after DNMT inhibition reflect a greater functional relevance of methylation over demethylation, or a disrupted balance between these opposing modifications. There is ample evidence to support the need for a balance between memory activators and inhibitors, including the opposing actions of proteases and phosphatases in the cytoplasm and of transcriptional activators and repressors at gene promoters (Blitzer et al. 1995; Wang and Kelly 1997; Wang et al. 1997; Koshibu et al. 2009; Lee and Silva 2009; Rajasethupathy et al. 2012). In fact, it has been suggested that a similar need for balance may be required at the epigenetic level (Koshibu et al. 2009), where DNA methylation must remain in balance with demethylation, and histone acetylation must be balanced with histone deacetylation, and so on. A requirement for an epigenetic balance is consistent with the observation that increased expression of plasticity genes reelin and BDNF in response to DNMT inhibition is not sufficient to support memory consolidation in the face of increased expression of PP1 (Miller and Sweatt 2007; Lubin et al. 2008).
The available evidence suggests that DNA methylation may tip the balance to favor the expression of plasticity-associated genes by inhibiting the activity of memory-suppressor genes (Miller and Sweatt 2007; Miller et al. 2010; Rajasethupathy et al. 2012). Indeed, such a mechanism has been described for SIRT1, a class III HDAC that promotes memory formation by inhibiting microRNA134-mediated degradation of CREB (Gao et al. 2010). Memory-suppressor genes PP1 (protein phosphatase 1) and calcineurin (a.k.a. Ca2+/calmodulin dependent protein phosphatase, PP2) in rodents, and CREB2 in Aplysia provide a powerful constraint on memory, such that the silencing of any one of these genes reduces the threshold for memory formation and improves memory retention (Bartsch et al. 1995; Malleret et al. 2001; Genoux et al. 2002; Koshibu et al. 2009). Cytoplasmic PP1 promotes memory suppression through dephosphorylation of signaling molecules critical for memory formation, including CaMKII and GluR1 (Genoux et al. 2002), whereas nuclear PP1 promotes memory suppression through dephosphorylation of serine 10 on histone H3 (Koshibu et al. 2009, 2011). In addition, PP1 interacts with HDACs and histone demethylases to increase their activity, thus promoting transcriptional silencing through histone deacetylation and demethylation (Koshibu et al. 2009). Calcineurin enhances PP1 activity by dephosphorylating a key PP1 inhibitor (Malleret et al. 2001), whereas CREB2 mediates memory suppression through inhibition of CREB1, a transcriptional activator critical for memory formation in Aplysia (Bartsch et al. 1995). DNA methylation relieves the repressive effects of these genes to allow for memory consolidation and the expression of plasticity-promoting genes in rodents and in Aplysia. As mentioned previously, fear conditioning is associated with increased methylation and decreased expression of hippocampal PP1 1 h after training (Miller and Sweatt 2007) and with increased methylation and decreased expression of cortical calcineurin 30 d after training (Miller et al. 2010). Similarly, treatment with the memory modulator serotonin induces DNA methylation and transcriptional repression of CREB2, the major memory suppressor in Aplysia (Rajasethupathy et al. 2012). These studies provide correlational evidence to support the hypothesis that methylation of memory-suppressor genes may prove to be a key mechanism for supporting memory consolidation, but additional studies are required to test this hypothesis directly.
A somewhat peculiar observation regarding the balance between opposing epigenetic modifications is that memory deficits produced by DNMT inhibitors can be reversed by treatment with HDAC inhibitors (Miller et al. 2008; Maddox and Schafe 2011; Monsey et al. 2011). In other models, including cancer, DNMT and HDAC inhibitors tend to have synergistic effects (Zhu and Otterson 2003; Fraczek et al. 2012), an outcome that is consistent with the transcriptionally repressive role of both enzymes. We speculate that the opposing action of DNMT and HDAC inhibitors in the hippocampus may be at least partly explained by increased expression of PP1 in response to DNMT inhibition (Miller and Sweatt 2007). Inhibition of nuclear PP1 reduces HDAC activity (Koshibu et al. 2009) and HDAC inhibitors disrupt the interaction between HDAC and PP1 complexes (Brush et al. 2004). Thus, by increasing PP1 expression, DNMT inhibitors may also result in increased HDAC activity, which would account for the reversal of memory deficits by treatment with HDAC inhibitors. Although this hypothesis has not been investigated directly, a recent study has shown that intra-cortical administration of DNMT inhibitors reduced histone acetyltransferase expression and produced a concomitant decrease in H3 and H4 acetylation (Sui et al. 2012). Based on these observations, we hypothesize that epigenetic modifications may regulate a fine balance between the activity of memory suppressors and promoters at the level of individual genes in a fashion consistent with the required balance between memory suppressors and activators at all levels of signaling (Blitzer et al. 1995; Wang and Kelly 1997; Wang et al. 1997; Koshibu et al. 2009; Lee and Silva 2009; Rajasethupathy et al. 2012). Direct tests of this hypothesis will provide a critical contribution to our understanding of epigenetic mechanisms in memory formation.
Additional explanations for memory impairment produced by DNMT inhibitors are also possible and are not mutually exclusive. For example, some evidence suggests that DNA methylation may not always repress transcription. In fact, the methyl CpG binding protein MeCP2 is capable of acting as a transcriptional repressor when bound to HDACs, or an activator when bound to CREB (Chahrour et al. 2008). Such mechanisms appear relevant in the brain, as regulation of the gene encoding the norepinephrine transporter was associated with changes in MeCP2 binding, but not with changes in DNA methylation in mouse cortical cells (Harikrishnan et al. 2010). Nonrepressive methylation mechanisms may also be relevant for contextual fear conditioning, as zif268 expression was enhanced 30 min after training even though promoter methylation was increased at that time point (Gupta et al. 2010). Another possibility is that DNMTs may be involved in both DNA methylation and demethylation (Metivier et al. 2008), such that blanket inhibition of these enzymes with DNMT inhibitors would disrupt both processes, although this hypothesis has not been tested in neural tissue. However, a recent study showed that a novel DNMT isoform, DNMT3a2, is associated with transcriptional activation rather than repression (Chen et al. 2002) and is positively associated with memory formation (Oliveira et al. 2012), although the mechanism that mediates this positive association is not clear. Currently, there are no techniques available to manipulate epigenetic modifications at individual genes selectively, but the available techniques can, nevertheless, provide important insights into potential ways in which these mechanisms interact to regulate memory formation and maintenance.
Time-course specificity
Temporal specificity of gene expression and protein synthesis in appropriate brain regions is essential for the formation and persistence of a lasting memory trace (e.g., Katche et al. 2010). Numerous studies show that learning induces temporally distinct waves of gene expression and protein synthesis in the hippocampus (Igaz et al. 2002, 2004a,b; Bekinschtein et al. 2007; Katche et al. 2010; Lonergan et al. 2010). Specifically, early changes occurring within 3 h of training are critical for the initial memory formation, whereas delayed changes occurring between 12 and 24 h after training are required for memory persistence over time (Bekinschtein et al. 2007). Changes in protein expression that support memory persistence at later time points are driven by transcriptional events at earlier time points. This is illustrated in a study by Bekinschtein and colleagues (2007), who found that blocking hippocampal BDNF 12 h after training impaired c-fos expression at 24 h and impaired memory recall at 7 d (Bekinschtein et al. 2007; Katche et al. 2010), indicating that the timing of gene expression within the hippocampus is a critical regulator of memory formation and stabilization. The delayed changes in hippocampal gene expression may reflect the process of systems consolidation, in which the hippocampus has a temporally restricted role in memory formation and undergoes a process of “downloading” the memory to the cortex for maintenance over prolonged periods of time (Frankland et al. 2004, 2006; Frankland and Bontempi 2005; Teixeira et al. 2006; Ding et al. 2008; Wang et al. 2009; Lesburgueres et al. 2011).
The majority of available studies on epigenetic mechanisms in memory have found that epigenetic markers are dynamically and specifically regulated during the initial consolidation window in the hippocampus. Using fear conditioning as a model of associative learning, Miller and Sweatt (2007) found that DNA methylation was rapidly altered 1 h after training and that the changes in DNA methylation returned to baseline within 24 h. Also using fear conditioning, it was found that histone acetylation, phosphorylation, and methylation followed a similar temporal pattern (Levenson et al. 2004; Chwang et al. 2006; Miller et al. 2008; Gupta et al. 2010; Gupta-Agarwal et al. 2012). Consistent with a role for epigenetic mechanisms in initial memory consolidation, the administration of DNMT inhibitors impaired fear memory and HDAC inhibitors improved fear memory (Miller and Sweatt 2007; Lubin et al. 2008; Miller et al. 2008; Monsey et al. 2011; Fass et al. 2013) only if administered during the restricted consolidation window shortly after training. Neither drug was effective at reversing memory if administered 6 h after training (Miller et al. 2008; Monsey et al. 2011). However, more recent studies have reported protracted epigenetic changes during the hippocampus-dependent process of consolidation. For example, the transcriptionally repressive H3K9me2 mark was increased 1 h and reduced 24 h after fear conditioning in the hippocampus (Gupta et al. 2010), whereas the transcriptionally permissive H3K4me3 mark was increased 1 h and decreased 24 h after fear conditioning in the entorhinal cortex (Gupta-Agarwal et al. 2012) compared to untrained controls. These results indicate that epigenetic changes may also occur in waves that contribute to temporally distinct patterns of gene expression that are involved in establishing transient and persistent memory traces. However, temporal dynamics of epigenetic changes in the hippocampus have not been extensively studied and much more work is required to test directly a potential epigenetic basis for regulating distinct waves of gene expression at different stages of consolidation.
The finding that epigenetic modifications in the hippocampus are transient challenged the initial hypothesis that persistent changes in DNA methylation support long-lasting memories (Miller and Sweatt 2007). The systems consolidation theory of memory maintenance (Frankland and Bontempi 2005; Frankland et al. 2006) suggests that the transient changes in epigenetic markers in the hippocampus parallel the transient involvement of the hippocampus in memory formation. According to the theory, memory consolidation is dependent on the hippocampus immediately after and for ∼7 d after training, whereas older memories (≥7 d, approximately) are downloaded to the cortex for maintenance over prolonged periods of time. Accordingly, a number of studies have shown that newly acquired memories are associated with transiently increased gene expression and spine density in the hippocampus, but that older memories are associated with altered gene expression and increased spine density in the cortex (Maviel et al. 2004; Restivo et al. 2009). Indeed, in contrast to transient changes (<24 h) in DNA methylation observed in the hippocampus (Miller and Sweatt 2007; Gupta et al. 2010), increased DNA methylation in the medial prefrontal cortex becomes evident 1 d after training, increases thereafter, and persists for at least 30 d (Miller et al. 2010). Altered DNA methylation at 30 d is critical for memory maintenance, as interference with DNA methylation by the administration of DNMT inhibitors into the medial prefrontal cortex blocks the recall of memory, suggesting that stable changes in cortical DNA methylation may support the maintenance of memory over time. Gräff et al. (2012) recently implicated dynamic histone modifications during systems-wide consolidation of hippocampus-dependent memories. As with DNA methylation, hippocampal H3S10 phosphorylation and H3K14 and H4K5 acetylation were transiently induced after object-recognition learning, whereas modifications in the cortex were delayed and persisted for up to 7 d. These biochemical changes were measured after memory recall and may thus reflect retrieval-induced epigenetic modifications in the cortex, a hypothesis that is indirectly supported by the absence of hippocampal histone modifications at 24 h in the absence of recall. Although fascinating in its own right, additional studies are required to confirm the spatiotemporal dynamics of histone modifications during memory consolidation and in the absence of memory recall.
Studies of cortical DNA methylation in remote memory indicate that distinct memory-associated genes exhibit temporally distinct patterns of methylation (Miller et al. 2010). Of the candidate genes examined, only calcineurin was persistently methylated, whereas reelin was transiently methylated and downregulated during the period of transition (approximately encompassing the first 7 d after training) from the hippocampus to the medial prefrontal cortex (Miller et al. 2010). We speculate that cortical DNA methylation at earlier time points (1–7 d) may direct sustained changes in DNA methylation at later time points (≥7 d) to regulate the timing and the duration of gene expression required for different stages of memory formation and maintenance. Although this notion is largely speculative at this point, there is some evidence to suggest that DNA methylation and histone modifications may delay or prolong the inhibition of memory-suppressor genes to allow for sufficient relief from the mechanisms that promote forgetting. For example, CREB2, a memory suppressor that inhibits activation of the memory promoter CREB1 in Aplysia (Bartsch et al. 1995), exhibits the highest levels of methylation and inhibition between 12 and 24 h after serotonin application, which corresponds with the period of increased activity of CREB1 and memory consolidation (Rajasethupathy et al. 2012). Importantly, these data point to a dual role of epigenetic modifications as dynamic regulators of gene expression in the hippocampus and as persistent maintainers of remote memories in the cortex. More work is required to investigate the role of temporally and region-specific changes in DNA methylation in memory formation and maintenance, as well as the distinct ways in which these temporally distinct epigenetic modifications translate into functional memories.
Epigenetic modifications are regulated in response to specific signaling cascades
Intertwined with the requirement for temporal specificity is the need for epigenetic marks to be laid down in a regulated fashion to allow for precise changes in gene expression by appropriate upstream signaling events. For example, NMDA receptor activation initiates the extracellular signal-regulated kinase/mitogen-activated protein kinase (ERK/MAPK) signaling cascade, which, via nuclear kinases such as mitogen- and stress-activated protein kinase 1 (MSK1), is involved in downstream histone H3 acetylation and phosphorylation in the hippocampus (Levenson et al. 2004; Chwang et al. 2006, 2007). Importantly, stimulation of ERK signaling (Levenson et al. 2004) and treatment with HDAC inhibitors (Graff and Tsai 2011; Graff et al. 2011) produced gene- and histone-specific changes in PTMs, indicating that distinct signaling cascades may establish precise histone codes that correspond to particular types of memory (Graff et al. 2011). Moreover, BDNF activity regulates the consolidation of contextual fear conditioning through alterations of histone- and residue-specific post-translational modifications at the homer1 promoter in the hippocampus and the amygdala (Mahan et al. 2012). In addition, nitric oxide (NO) has been implicated in histone acetylation by regulating the dissociation of HDAC2 from CREB-regulated gene promoters (Nott et al. 2008; Nott and Riccio 2009) and, most recently, HDAC inhibitors were shown to reverse fear conditioning deficits in NO knockout mice through increased H3 acetylation in the hippocampus and the amygdala (Itzhak et al. 2012). Similarly, the enhancement of object-recognition memory by HDAC inhibitors can be blocked with antagonism of the glucocorticoid receptor and the downstream activation of PKA (Roozendaal et al. 2010). DNA methylation and histone acetylation also appear to be regulated by overlapping signaling cascades, as evidenced by impaired DNA methylation in response to NMDA receptor antagonist treatment in the hippocampus (Lubin et al. 2008; Miller et al. 2010) and reduced DNMT3a expression in response to ERK/MAPK inhibition in the amygdala (Monsey et al. 2011). Similarly, DNA methylation of the memory-suppressor gene CREB2 in Aplysia is dependent on serotonin signaling, in that DNMT inhibitors blocked serotonin-induced CREB2 silencing and the associated enhancement of cellular activation, and the inhibition of serotonin signaling blocked DNMT-inhibitor mediated alterations of cell activity (Rajasethupathy et al. 2012).
Many of the post-translational modifications of histones discussed above are specifically induced at particular residues on specific histones, while others are not affected (e.g., Chwang et al. 2006; Gupta et al. 2010; Gupta-Agarwal et al. 2012; Mahan et al. 2012), indicating that upstream signaling cascades may differentially regulate the activity of multiple HATs, HDACs, histone methylases, and histone demethylases that regulate specific modifications at distinct residues. For example, specific HATs, including HPA2 and Gcn, specifically acetylate H3K14 (Angus-Hill et al. 1999; P Cheung et al. 2000a,b; WL Cheung et al. 2000), and the histone methyltransferase Mll specifically methylates H3K4 (Milne et al. 2002), whereas the G9a/G9a-like protein (GLP) lysine dimethyltransferase complex catalyzes methylation of K3K9 (Kubicek et al. 2007; Leung et al. 2011; Shinkai and Tachibana 2011).
Different enzymes that catalyze the addition and removal of post-translational modifications appear to have at least partially independent effects on memory formation. For example, knocking out different HATs, CBP or p300, produces distinct patterns of memory deficits in mice (Korzus et al. 2004; Wood et al. 2005), and HDAC2 (and not HDAC1) is negatively associated with spatial memory (Guan et al. 2009). In contrast, class III HDACs (also called sirtuins) are positively associated with memory formation (Kim et al. 2007; Gao et al. 2010), supporting the idea that different HATs and HDACs may regulate different types of memory (Graff and Tsai 2011). However, it is not entirely clear whether relevant modifications occur only at specific residues, or whether general enhancement or reduction of a particular modification is critical for memory formation. The best example of residue-specific modifications in memory has been described in a recent study of H3K9 methylation in Drosophila. Kramer et al. (2011) isolated H3K9me by knocking out an H3K9-specific euchromatin histone methyltransferase (EHMT) that resulted in deficient learning and memory, and impaired dendrite development. The deficits were associated with changes in H3K9 dimethylation on neural plasticity-related genes and were reversed with the induction of EHMT expression in adulthood. This study provides an excellent example of isolating a single modification that is dynamically regulated on a subset of genes, many of which are involved in memory formation, although the modification remained stable on other genes. This observation provides further evidence for the dual role of epigenetic modifications in the maintenance of stable patterns of gene expression and in regulating dynamic changes in gene expression in response to environmental signals.
In addition to subcategories of methyltransferases, HDACs, HATs, and histone methylases, DNMTs can also be classified into either de novo or maintenance subcategories and recent studies have identified different subtypes of DNMT3a, wherein DNMT3a1 is associated with gene repression and DNMT3a2 is associated with gene activation (Chen et al. 2002; Kotini et al. 2011). The latter is selectively and positively associated with memory for trace fear conditioning and novel object recognition (Oliveira et al. 2012). Overall, these findings indicate that a precise pattern of chromatin modifications in the nucleus is established in response to upstream signaling cascades, although the role of specific enzymes and modifications of specific sites in learning and memory need to be better elucidated. Based on their position downstream of environmental stimuli and the associated signaling cascades, epigenetic mechanisms are well suited to integrate the upstream signaling information and translate it into gene-specific transcriptional regulation.
Translation of epigenetic mechanisms into a functional memory trace
Traditionally, studies of memory formation have focused on activity-dependent changes at the synapse, particularly long-term potentiation (LTP) and the formation of new synaptic contacts indexed by changes in spine density (Bliss and Coolingridge 1993; Restivo et al. 2009). However, it has been difficult to reconcile such a synapse-specific basis of memory with the presumably cell-wide changes produced by epigenetic modifications in the nucleus. One of the most obvious links between epigenetic mechanisms and synaptic function is the epigenetic regulation of genes that have a known role in the establishment of LTP and memory formation. Reelin and BDNF are epigenetically regulated (Miller and Sweatt 2007; Lubin et al. 2008) and have an established role in LTP induction, synapse maturation, and spine development (Weeber et al. 2002; Beffert et al. 2005; Qiu and Weeber 2007; Niu et al. 2008; Mei et al. 2011; Amaral and Pozzo-Miller 2012; Vigers et al. 2012). A number of studies have found evidence supporting a role for HDACs in synapse formation and plasticity (Kim et al. 2007; Guan et al. 2009; Gao et al. 2010; Calfa et al. 2012). Effects appear to be HDAC specific, wherein HDAC2 is associated with reduced synaptic plasticity, synapse number, and spine density (Guan et al. 2009), and the HDAC SIRT1 is associated with enhanced synaptic plasticity (Gao et al. 2010) and greater dendritic complexity (Michan et al. 2010). Moreover, DNMT inhibitors reduce synaptic plasticity and impair LTP induction (Levenson et al. 2006; Nelson et al. 2008), thus highlighting the importance of epigenetic modifications in regulating traditional mechanisms of memory. Although the mechanism underlying the effect of epigenetic modifications on synaptic structure and function is not clear, recent studies have found that homer1 and TrkB (a receptor for BDNF) may serve as activity-regulated synaptic tags that could localize BDNF and other plasticity-associated proteins to recently activated synapses (Okada et al. 2009; Lu et al. 2011), implicating synaptic tagging as a potential mechanism for targeting epigenetically regulated genes to appropriate synaptic sites. Eric Kandel's group recently put forth another interesting idea regarding the role of DNA methylation as a regulator of memory allocation (Rajasethupathy et al. 2012). Memory formation occurs in a subset of cells that exhibit higher levels of CREB1, such that the memory trace is preferentially “allocated” to neurons expressing higher CREB1 levels (Han et al. 2009; Zhou et al. 2009). Kandel's group observed widespread inter-cell variation in CREB2 methylation, which is positively associated with CREB1 expression. This led the group to propose that CREB2 inhibition may distinguish neurons that are currently involved in memory formation from those that are not, thereby implicating DNA methylation in regulating the sequence of cellular involvement in particular forms of memory.
Euchromatin-associated modifications enhance memory
So far in our discussion, we have focused primarily on the ability of epigenetic mechanisms to promote memory by selectively responding to specific stimuli in the environment. However, many studies have found that, in addition to the specific epigenetic modifications that occur only in response to associative learning (e.g., Levenson et al. 2004; Chwang et al. 2006; Miller et al. 2008; Gupta et al. 2010; Gupta-Agarwal et al. 2012), exposure to nonassociative control treatments also induces epigenetic modifications. In fact, exposure to a novel environment produced increased levels of ERK1/2 dependent H3 phosphorylation and acetylation that was abolished by environmental habituation (Sarantis et al. 2012). Similarly, context exposure alone produced increased H3K9me2 in the CA1 (Gupta-Agarwal et al. 2012) and exposure to a visible-platform control condition produced similar changes in H3 acetylation to those seen after MWM training (Castellano et al. 2012). With respect to DNA methylation, the specificity of the changes observed in response to the context and shock pairing is dependent on the gene of interest, with some genes, including egr1/zif268 and bdnf exon 1, exhibiting similar modifications when shock and context are presented individually as when they are presented in combination (Lubin et al. 2008; Miller et al. 2010). Divergent patterns of specificity can even be observed at the single cytosine level, as evidenced by a training-specific methylation at only one cytosine in the promoter region of bdnf exon IV among the multiple modifications that occurred in response to nonassociative context exposure (Lubin et al. 2008). Such findings suggest that certain epigenetic modifications may be induced by specific sets of environmental stimuli, some of which reflect associative memory whereas others are induced by novel environmental inputs that are independent of associative learning.
The nonspecific sensitivity of epigenetic mechanisms to diverse inputs from the environment suggests that exposure to new stimuli can promote neural plasticity irrespective of associative learning. By extension, the nonspecific epigenetic modifications produced by environmental exploration and novelty may serve a function that alters the epigenome in a way that may either promote or impair future learning. This is exactly the argument that has been made to account for the effects of environmental enrichment on learning and memory, as evidenced by heightened levels of histone acetylation and open chromatin states in rodents exposed to enriched environments (for review, see Graff and Tsai 2011). Indeed, Graff and Tsai (2011) have argued for beneficial effects of such an “increased dose” of euchromatin-associated epigenetic modifications based on evidence that manipulations that increase euchromatin-associated PTMs, such as PP1 inhibition, estrogen treatment, or the activation of glucocorticoid receptors (Genoux et al. 2002; Koshibu et al. 2009; Roozendaal et al. 2010; Zhao et al. 2010; Koshibu et al. 2011) also enhance memory formation. This link is further supported by evidence that HDAC inhibitors enhance learning in response to weak stimuli that do not induce memory on their own (e.g., Fass et al. 2012; Stafford et al. 2012). In addition, variations in maternal behavior in early life, which result in different patterns of hippocampal and cortical DNA methylation in adult offspring, are associated with altered patterns of learning and memory (Caldji et al. 1998; Bredy et al. 2003; Champagne et al. 2008; Roth et al. 2009), indicating that preexisting differences in epigenetic modifications can reduce the threshold for learning and memory. Similarly, one study has found that post-training individual differences in DNA methylation of the BDNF gene in the hippocampus correlate with performance on a spontaneous object-recognition task (Munoz et al. 2010), implying that preexisting changes in epigenetic marks may mediate the responsivity of epigenetic marks to memory-inducing stimuli. Ultimately, these findings point to a bidirectional relationship between epigenetic mechanisms and learning and memory, whereby learning induces the formation of novel epigenetic marks and preexisting levels of epigenetic marks regulate the threshold for learning and memory.
General considerations and limitations
Within the last decade, increased interest in neuroepigenetics has resulted in exciting methodological and conceptual advances in the field. As with any new field of study, however, it is important to consider the inherent technical limitations and their impact on the interpretation of data. For example, many studies of DNA methylation used bisulfite sequencing to obtain single-nucleotide resolution of cytosine methylation, which is unable to discriminate between 5mc and 5hmc (Huang et al. 2010; Jin et al. 2010; Nestor et al. 2010), suggesting that the observed changes in DNA methylation in previous studies will have to be reexamined for the presence of hydroxyl methylation. Recently, several variants of bisulfite sequencing, including oxidative bisulfite sequencing (oxBS-Seq) and Tet-assisted bisulfite sequencing (TAB-Seq), have been developed in order to address such limitations (Booth et al. 2012; Yu et al. 2012). Furthermore, for mapping studies that do not require single base resolution, immunoprecipitation techniques that employ 5mc and 5hmc specific antibodies or proteins (e.g., hydroxylated/methylated DNA immunoprecipitation [h/MeDIP] and methylated-CpG island recovery assay [MIRA]) are available as reasonable alternatives if a DNA fragment does not contain both modified cytosines (Jin et al. 2010). As the field continues to grow, it is imperative that such techniques become the standard in the field to ensure accurate 5mc and 5hmc measurements.
On a similar note, certain commercially available antibodies used to evaluate histone PTMs may, in fact, also lack the necessary specificity to distinguish one modification from another. Using peptide array technology, Castellano et al. (2012) showed that several of their purchased antibodies recognized various histone modifications in addition to the ones supposedly targeted by their antibodies. This lack of antibody specificity has important implications for both past and future studies that examine how combinatorial patterns of histone modifications work together to regulate gene expression. Furthermore, a lack of studies that examine antibody cross-reactivity complicates cross-study comparisons and the proper resolution of potential discrepancies (Castellano et al. 2012). Use of pharmacological approaches also has inherent limitations. For example, a causal link between histone modifications and behavior is often inferred on the basis of pharmacological enhancement of memory with HDAC inhibitors, many of which have widespread effects that are not specific to histones, HDAC subtypes, or to modifications of specific histone residues (for review, see Zovkic and Sweatt 2013), although important advances have been made in understanding the role of specific HATs and HDACs from studies that used genomic tools to interfere with subtype-specific expression (e.g., Wood et al. 2005, 2006; McQuown et al. 2011; Gräff et al. 2012). Although the results of the latter studies are generally in agreement with those obtained with pharmacological HDAC inhibitors, they nevertheless point to a need to develop additional HDAC-specific compounds that mimic effects observed with genetic manipulations, as reported for the pharmacological and genetic inhibition of HDAC3 (e.g., McQuown et al. 2011).
Conclusions and future directions
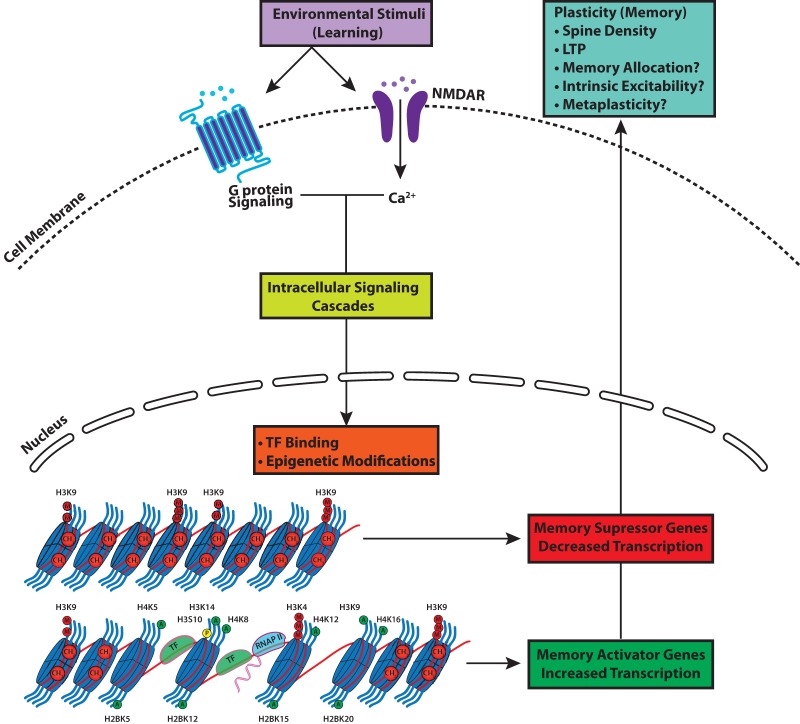
In this review, we hope to have underscored the notion that epigenetic regulation of gene expression is a ubiquitous mechanism across learning paradigms and brain regions. It is now evident that integration and regulation of epigenetic modifications allows for complex control of gene expression necessary for long-term memory formation and maintenance. Dynamic changes in DNA methylation and chromatin structure are the result of well-established intracellular signaling cascades that converge on the nucleus to adjust the precise equilibrium of gene repression and activation. It is through this altered transcriptional profile that cells are then able to modulate the plasticity that underlies memory formation, as depicted in Figure 2. Together, these findings usher an exciting era of neuroepigenetics that will certainly continue to grow.
Figure 2.
A model depicting the role of epigenetic mechanisms in memory formation and maintenance. Environmental stimuli, which consist primarily of associative learning tasks in animal models, initiate cellular communication by activating specific post-synaptic receptors. Receptor activation stimulates specific intracellular signaling cascades that lead to particular patterns of epigenetic modifications, which in turn regulate the access of transcription factors (TF) and RNA polymerase II (RNA P II) to gene promoters. These regulatory processes result in an increased transcription of memory activator genes and decreased transcription of memory-suppressor genes, which ultimately promote memory formation and maintenance through effects on long-term potentiation (LTP), spine density, memory allocation, cell excitability, and metaplasticity.
As the field expands, several mechanistic questions remain to be answered. Specifically, although tremendous progress has been made in recent years, more research is required to better understand the mechanisms by which epigenetic modifications are generated, maintained, and removed. For example, the processes that direct DNA methylation to specific sequences are largely unknown. Some studies indicate that transcription factors may act as “docking stations” for DNMT enzymes, which exhibit minimal sequence specificity on their own (Brenner et al. 2005; Cheng et al. 2010). Other evidence suggests that factors intrinsic to the DNA sequence are relevant, including spacing of CpGs in CpG islands (Cokus et al. 2008; Zhang et al. 2009; Cheng et al. 2010). More work is also needed to investigate how the epigenetic code manifests functional change within specific cells and neural circuits. It is clear that epigenetic modifications alter LTP and synaptic plasticity (Levenson et al. 2006; Guan et al. 2009; Gao et al. 2010), but the mechanism through which this relationship occurs is not clear. In addition, a potential role for epigenetic modifications in regulating different types of plasticity is not well defined. For example, little is known about the involvement of epigenetic mechanisms in regulating intrinsic plasticity, defined as the efficiency of coupling between excitatory potentials and spikes in the post-synaptic neuron, compared to synaptic plasticity, defined as the efficiency of synaptic connections between neurons (Benito and Barco 2010). Although indirect evidence implicates epigenetic mechanisms in both cell excitability and synaptic plasticity (Guan et al. 2002; Levenson et al. 2004, 2006; Yeh et al. 2004; Miller et al. 2008; Nelson et al. 2008; Feng et al. 2010), the nature of this link is still not clear. Further, a potential role for epigenetic mechanisms in memory allocation and the related concept of metaplasticity also remain unexplored, although some studies suggest a possible involvement of DNA methylation in both. As mentioned earlier, memory allocation refers to the distribution of memory to specific cells (Silva et al. 2009), whereas metaplasticity refers to the ability of a neuron's activation history to prime it for future encoding (Abraham 2008). The ability of epigenetic modifications to be selectively induced and to persist under appropriate conditions makes them perfectly positioned to regulate the likelihood of a particular cell to be activated in the future. Indeed, just such a mechanism was recently proposed by Kandel's group, as discussed above (Rajasethupathy et al. 2012).
To address these questions, future studies will benefit from methodologies that increase the cellular and molecular resolution of our investigations. A higher degree of cellular resolution will allow researchers to restrict their analysis to specific cell populations, ideally focusing on cells that make up the memory trace for a particular behavioral paradigm. To fulfill this need, techniques that allocate memories to specific cell populations within a neural circuit (Han et al. 2009; Zhou et al. 2009) or that tag cells activated during memory acquisition (Reijmers et al. 2007; Tayler et al. 2011) could be combined with techniques such as laser capture microscopy and fluorescence-associated cell sorting. Additionally, these techniques in combination with reporter rodent models could also be used to address how epigenetic mechanisms may be differentially regulated in distinct cell populations (excitatory neurons vs. inhibitory neurons vs. glia) and how these mechanisms may integrate to regulate the function of an entire memory circuit. Recent evidence suggests that there is cell-type specific expression of different HDAC isoforms (Baltan et al. 2011), again underscoring the importance of developing and using targeted manipulations of epigenetic modifying enzymes.
In addition to increased cellular resolution, a higher degree of molecular resolution will allow researchers to better understand how signaling cascades and nuclear protein complexes interact to generate specific epigenetic modifications and how these modifications integrate to regulate overall neuronal function and synaptic plasticity. The advent of high-throughput sequencing has already increased the molecular resolution available to researchers by allowing genome-wide analysis of DNA methylation and post-translational modifications of histones. Although the studies conducted thus far have focused on candidate genes that have a known role in memory, genome-wide studies may identify additional epigenetically regulated genes that are critical for memory formation and maintenance. Determining whether these genes regulate synaptic or intrinsic plasticity will help elucidate how exactly these epigenetic marks contribute to learning and memory. Presumably, regulation of certain genes will lead to consolidation of synaptic plasticity via the regulation of synaptic effector molecules, whereas other genes might be better positioned to regulate the intrinsic excitability of a cell via modulation of Na+ and K+ channel functions. With these considerations in mind, future research will undoubtedly enrich our understanding of the cellular and molecular underpinnings of learning and memory at large, as well as further elucidate the role epigenetic mechanisms have in this process.
Acknowledgments
The authors' work is supported by NIH grants MH091122, MH57014, NS 057098, and AG031722, as well as NR012686 (J.D.S.) and NSERC-PDF (I.B.Z.).
References
- Abraham WC 2008. Metaplasticity: Tuning synapses and networks for plasticity. Nat Rev Neurosci 9: 387–399 [DOI] [PubMed] [Google Scholar]
- Agranoff BW 1965. Molecules and memories. Perspect Biol Med 9: 13–22 [DOI] [PubMed] [Google Scholar]
- Agranoff BW, Davis RE, Brink JJ 1965. Memory fixation in the goldfish. Proc Natl Acad Sci 54: 788–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agranoff BW, Davis RE, Brink JJ 1966. Chemical studies on memory fixation in goldfish. Brain Res 1: 303–309 [DOI] [PubMed] [Google Scholar]
- Alarcon JM, Malleret G, Touzani K, Vronskaya S, Ishii S, Kandel ER, Barco A 2004. Chromatin acetylation, memory, and LTP are impaired in CBP+/− mice: A model for the cognitive deficit in Rubinstein-Taybi syndrome and its amelioration. Neuron 42: 947–959 [DOI] [PubMed] [Google Scholar]
- Amaral MD, Pozzo-Miller L 2012. Intracellular Ca2+ stores and Ca2+ influx are both required for BDNF to rapidly increase quantal vesicular transmitter release. Neural Plast 2012: 203536 10.1155/2012/203536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angus-Hill ML, Dutnall RN, Tafrov ST, Sternglanz R, Ramakrishnan V 1999. Crystal structure of the histone acetyltransferase Hpa2: A tetrameric member of the Gcn5-related N-acetyltransferase superfamily. J Mol Biol 294: 1311–1325 [DOI] [PubMed] [Google Scholar]
- Arney KL, Fisher AG 2004. Epigenetic aspects of differentiation. J Cell Sci 117: 4355–4363 [DOI] [PubMed] [Google Scholar]
- Baltan S, Bachleda A, Morrison RS, Murphy SP 2011. Expression of histone deacetylases in cellular compartments of the mouse brain and the effects of ischemia. Transl Stroke Res 2: 411–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barres R, Yan J, Egan B, Treebak JT, Rasmussen M, Fritz T, Caidahl K, Krook A, O'Gorman DJ, Zierath JR 2012. Acute exercise remodels promoter methylation in human skeletal muscle. Cell Metab 15: 405–411 [DOI] [PubMed] [Google Scholar]
- Barrett RM, Wood MA 2008. Beyond transcription factors: The role of chromatin modifying enzymes in regulating transcription required for memory. Learn Mem 15: 460–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett RM, Malvaez M, Kramar E, Matheos DP, Arrizon A, Cabrera SM, Lynch G, Greene RW, Wood MA 2011. Hippocampal focal knockout of CBP affects specific histone modifications, long-term potentiation, and long-term memory. Neuropsychopharmacology 36: 1545–1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barros M, Dempster EL, Illott N, Chabrawi S, Maior RS, Tomaz C, De Souza Silva MA, Huston JP, Mill J, Muller CP 2011. Decreased methylation of the NK3 receptor coding gene (TACR3) after cocaine-induced place preference in marmoset monkeys. Addict Biol 16: 10.1111/j.1369-1600.2011.00409 [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K 2007. High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 [DOI] [PubMed] [Google Scholar]
- Bartsch D, Ghirardi M, Skehel PA, Karl KA, Herder SP, Chen M, Bailey CH, Kandel ER 1995. Aplysia CREB2 represses long-term facilitation: Relief of repression converts transient facilitation into long-term functional and structural change. Cell 83: 979–992 [DOI] [PubMed] [Google Scholar]
- Bayerlein K, Kraus T, Leinonen I, Pilniok D, Rotter A, Hofner B, Schwitulla J, Sperling W, Kornhuber J, Biermann T 2011. Orexin A expression and promoter methylation in patients with alcohol dependence comparing acute and protracted withdrawal. Alcohol 45: 541–547 [DOI] [PubMed] [Google Scholar]
- Beffert U, Weeber EJ, Durudas A, Qiu S, Masiulis I, Sweatt JD, Li WP, Adelmann G, Frotscher M, Hammer RE, et al. 2005. Modulation of synaptic plasticity and memory by Reelin involves differential splicing of the lipoprotein receptor Apoer2. Neuron 47: 567–579 [DOI] [PubMed] [Google Scholar]
- Bekinschtein P, Cammarota M, Igaz LM, Bevilaqua LR, Izquierdo I, Medina JH 2007. Persistence of long-term memory storage requires a late protein synthesis- and BDNF-dependent phase in the hippocampus. Neuron 53: 261–277 [DOI] [PubMed] [Google Scholar]
- Benito E, Barco A 2010. CREB's control of intrinsic and synaptic plasticity: Implications for CREB-dependent memory models. Trends Neurosci 33: 230–240 [DOI] [PubMed] [Google Scholar]
- Biergans SD, Jones JC, Treiber N, Galizia CG, Szyszka P 2012. DNA methylation mediates the discriminatory power of associative long-term memory in honeybees. PLoS One 7: e39349 10.1371/journal.pone.0039349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binda O, LeRoy G, Bua DJ, Garcia BA, Gozani O, Richard S 2010. Trimethylation of histone H3 lysine 4 impairs methylation of histone H3 lysine 9: Regulation of lysine methyltransferases by physical interaction with their substrates. Epigenetics 5: 767–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A 2002. DNA methylation patterns and epigenetic memory. Genes Dev 16: 6–21 [DOI] [PubMed] [Google Scholar]
- Bliss TV, Collingridge GL 1993. A synaptic model of memory: Long-term potentiation in the hippocampus. Nature 361: 31–39 [DOI] [PubMed] [Google Scholar]
- Blitzer RD, Wong T, Nouranifar R, Iyengar R, Landau EM 1995. Postsynaptic cAMP pathway gates early LTP in hippocampal CA1 region. Neuron 15: 1403–1414 [DOI] [PubMed] [Google Scholar]
- Booth MJ, Branco MR, Ficz G, Oxley D, Krueger F, Reik W, Balasubramanian S 2012. Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science 336: 934–937 [DOI] [PubMed] [Google Scholar]
- Bourtchouladze R, Lidge R, Catapano R, Stanley J, Gossweiler S, Romashko D, Scott R, Tully T 2003. A mouse model of Rubinstein-Taybi syndrome: Defective long-term memory is ameliorated by inhibitors of phosphodiesterase 4. Proc Natl Acad Sci 100: 10518–10522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousiges O, Vasconcelos AP, Neidl R, Cosquer B, Herbeaux K, Panteleeva I, Loeffler JP, Cassel JC, Boutillier AL 2010. Spatial memory consolidation is associated with induction of several lysine-acetyltransferase (histone acetyltransferase) expression levels and H2B/H4 acetylation-dependent transcriptional events in the rat hippocampus. Neuropsychopharmacology 35: 2521–2537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredy TW, Grant RJ, Champagne DL, Meaney MJ 2003. Maternal care influences neuronal survival in the hippocampus of the rat. Eur J Neurosci 18: 2903–2909 [DOI] [PubMed] [Google Scholar]
- Bredy TW, Wu H, Crego C, Zellhoefer J, Sun YE, Barad M 2007. Histone modifications around individual BDNF gene promoters in prefrontal cortex are associated with extinction of conditioned fear. Learn Mem 14: 268–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner C, Deplus R, Didelot C, Loriot A, Vire E, De Smet C, Gutierrez A, Danovi D, Bernard D, Boon T, et al. 2005. Myc represses transcription through recruitment of DNA methyltransferase corepressor. EMBO J 24: 336–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brush MH, Guardiola A, Connor JH, Yao TP, Shenolikar S 2004. Deacteylase inhibitors disrupt cellular complexes containing protein phosphatases and deacetylases. J Biol Chem 279: 7685–7691 [DOI] [PubMed] [Google Scholar]
- Caldji C, Tannenbaum B, Sharma S, Francis D, Plotsky PM, Meaney MJ 1998. Maternal care during infancy regulates the development of neural systems mediating the expression of fearfulness in the rat. Proc Natl Acad Sci 95: 5335–5340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calfa G, Chapleau CA, Campbell S, Inoue T, Morse SJ, Lubin FD, Pozzo-Miller L 2012. HDAC activity is required for BDNF to increase quantal neurotransmitter release and dendritic spine density in CA1 pyramidal neurons. Hippocampus 22: 1493–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano JF, Fletcher BR, Kelley-Bell B, Kim DH, Gallagher M, Rapp PR 2012. Age-related memory impairment is associated with disrupted multivariate epigenetic coordination in the hippocampus. PLoS One 7: e33249 10.1371/journal.pone.0033249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chahrour M, Jung SY, Shaw C, Zhou X, Wong ST, Qin J, Zoghbi HY 2008. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science 320: 1224–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champagne DL, Bagot RC, van Hasselt F, Ramakers G, Meaney MJ, de Kloet ER, Joels M, Krugers H 2008. Maternal care and hippocampal plasticity: Evidence for experience-dependent structural plasticity, altered synaptic functioning, and differential responsiveness to glucocorticoids and stress. J Neurosci 28: 6037–6045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Ueda Y, Xie S, Li E 2002. A novel Dnmt3a isoform produced from an alternative promoter localizes to euchromatin and its expression correlates with active de novo methylation. J Biol Chem 277: 38746–38754 [DOI] [PubMed] [Google Scholar]
- Chen G, Zou X, Watanabe H, van Deursen JM, Shen J 2010. CREB binding protein is required for both short-term and long-term memory formation. J Neurosci 30: 13066–13077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Hashimoto H, Horton JR, Zhang X 2010. Mechanisms of DNA methylation, methyl-CpG recognition, and demethylation in mammals. In Handbook of epigenetics (ed. Tollefsbol TO), pp. 9–24 Academic Press, Oxford, UK [Google Scholar]
- Cheung P, Allis CD, Sassone-Corsi P 2000a. Signaling to chromatin through histone modifications. Cell 103: 263–271 [DOI] [PubMed] [Google Scholar]
- Cheung P, Tanner KG, Cheung WL, Sassone-Corsi P, Denu JM, Allis CD 2000b. Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol Cell 5: 905–915 [DOI] [PubMed] [Google Scholar]
- Cheung WL, Briggs SD, Allis CD 2000. Acetylation and chromosomal functions. Curr Opin Cell Biol 12: 326–333 [DOI] [PubMed] [Google Scholar]
- Chiang PK, Gordon RK, Tal J, Zeng GC, Doctor BP, Pardhasaradhi K, McCann PP 1996. S-Adenosylmethionine and methylation. FASEB J 10: 471–480 [PubMed] [Google Scholar]
- Chwang WB, O'Riordan KJ, Levenson JM, Sweatt JD 2006. ERK/MAPK regulates hippocampal histone phosphorylation following contextual fear conditioning. Learn Mem 13: 322–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chwang WB, Arthur JS, Schumacher A, Sweatt JD 2007. The nuclear kinase mitogen- and stress-activated protein kinase 1 regulates hippocampal chromatin remodeling in memory formation. J Neurosci 27: 12732–12742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD, Pradhan S, Nelson SF, Pellegrini M, Jacobsen SE 2008. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 452: 215–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick F 1984. Memory and molecular turnover. Nature 312: 101. [DOI] [PubMed] [Google Scholar]
- Danilova AB, Kharchenko OA, Shevchenko KG, Grinkevich LN 2010. Histone H3 Acetylation is asymmetrically induced upon learning in identified neurons of the food aversion network in the mollusk Helix lucorum. Front Behav Neurosci 4: 180 10.3389/fnbeh.2010.00180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day JJ, Sweatt JD 2011. Cognitive neuroepigenetics: A role for epigenetic mechanisms in learning and memory. Neurobiol Learn Mem 96: 2–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding HK, Teixeira CM, Frankland PW 2008. Inactivation of the anterior cingulate cortex blocks expression of remote, but not recent, conditioned taste aversion memory. Learn Mem 15: 290–293 [DOI] [PubMed] [Google Scholar]
- Drewell RA, Goddard CJ, Thomas JO, Surani MA 2002. Methylation-dependent silencing at the H19 imprinting control region by MeCP2. Nucleic Acids Res 30: 1139–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fass DM, Reis SA, Ghosh B, Hennig KM, Joseph NF, Zhao WN, Nieland TJ, Guan JS, Groves Kuhnle CE, Tang W, et al. 2013. Crebinostat: A novel cognitive enhancer that inhibits histone deacetylase activity and modulates chromatin-mediated neuroplasticity. Neuropharmacology 64: 81–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Zhou Y, Campbell SL, Le T, Li E, Sweatt JD, Silva AJ, Fan G 2010. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat Neurosci 13: 423–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraczek JE, Vinken M, Tourwe D, Vanhaecke T, Rogiers V 2012. Synergetic effects of DNA demethylation and histone deacetylase inhibition in primary rat hepatocytes. Invest New Drugs 30: 1715–1724 [DOI] [PubMed] [Google Scholar]
- Frankland PW, Bontempi B 2005. The organization of recent and remote memories. Nat Rev Neurosci 6: 119–130 [DOI] [PubMed] [Google Scholar]
- Frankland PW, Bontempi B, Talton LE, Kaczmarek L, Silva AJ 2004. The involvement of the anterior cingulate cortex in remote contextual fear memory. Science 304: 881–883 [DOI] [PubMed] [Google Scholar]
- Frankland PW, Ding HK, Takahashi E, Suzuki A, Kida S, Silva AJ 2006. Stability of recent and remote contextual fear memory. Learn Mem 13: 451–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuks F, Hurd PJ, Wolf D, Nan X, Bird AP, Kouzarides T 2003. The methyl-CpG-binding protein MeCP2 links DNA methylation to histone methylation. J Biol Chem 278: 4035–4040 [DOI] [PubMed] [Google Scholar]
- Gao J, Wang WY, Mao YW, Graff J, Guan JS, Pan L, Mak G, Kim D, Su SC, Tsai LH 2010. A novel pathway regulates memory and plasticity via SIRT1 and miR-134. Nature 466: 1105–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring M, Reik W, Henikoff S 2009. DNA demethylation by DNA repair. Trends Genet 25: 82–90 [DOI] [PubMed] [Google Scholar]
- Genoux D, Haditsch U, Knobloch M, Michalon A, Storm D, Mansuy IM 2002. Protein phosphatase 1 is a molecular constraint on learning and memory. Nature 418: 970–975 [DOI] [PubMed] [Google Scholar]
- Graff J, Tsai LH 2011. Cognitive enhancement: A molecular memory booster. Nature 469: 474–475 [DOI] [PubMed] [Google Scholar]
- Graff J, Kim D, Dobbin MM, Tsai LH 2011. Epigenetic regulation of gene expression in physiological and pathological brain processes. Physiol Rev 91: 603–649 [DOI] [PubMed] [Google Scholar]
- Gräff J, Woldemichael BT, Berchtold D, Dewarrat G, Mansuy IM 2012. Dynamic histone marks in the hippocampus and cortex facilitate memory consolidation. Nat Commun 3: 991. [DOI] [PubMed] [Google Scholar]
- Guan Z, Giustetto M, Lomvardas S, Kim JH, Miniaci MC, Schwartz JH, Thanos D, Kandel ER 2002. Integration of long-term-memory-related synaptic plasticity involves bidirectional regulation of gene expression and chromatin structure. Cell 111: 483–493 [DOI] [PubMed] [Google Scholar]
- Guan JS, Haggarty SJ, Giacometti E, Dannenberg JH, Joseph N, Gao J, Nieland TJ, Zhou Y, Wang X, Mazitschek R, et al. 2009. HDAC2 negatively regulates memory formation and synaptic plasticity. Nature 459: 55–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JU, Ma DK, Mo H, Ball MP, Jang MH, Bonaguidi MA, Balazer JA, Eaves HL, Xie B, Ford E, et al. 2011a. Neuronal activity modifies the DNA methylation landscape in the adult brain. Nat Neurosci 14: 1345–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JU, Su Y, Zhong C, Ming GL, Song H 2011b. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell 145: 423–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Kim SY, Artis S, Molfese DL, Schumacher A, Sweatt JD, Paylor RE, Lubin FD 2010. Histone methylation regulates memory formation. J Neurosci 30: 3589–3599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta-Agarwal S, Franklin AV, Deramus T, Wheelock M, Davis RL, McMahon LL, Lubin FD 2012. G9a/GLP histone lysine dimethyltransferase complex activity in the hippocampus and the entorhinal cortex is required for gene activation and silencing during memory consolidation. J Neurosci 32: 5440–5453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haettig J, Stefanko DP, Multani ML, Figueroa DX, McQuown SC, Wood MA 2011. HDAC inhibition modulates hippocampus-dependent long-term memory for object location in a CBP-dependent manner. Learn Mem 18: 71–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han JH, Kushner SA, Yiu AP, Hsiang HL, Buch T, Waisman A, Bontempi B, Neve RL, Frankland PW, Josselyn SA 2009. Selective erasure of a fear memory. Science 323: 1492–1496 [DOI] [PubMed] [Google Scholar]
- Happel N, Doenecke D 2009. Histone H1 and its isoforms: Contribution to chromatin structure and function. Gene 431: 1–12 [DOI] [PubMed] [Google Scholar]
- Harikrishnan KN, Bayles R, Ciccotosto GD, Maxwell S, Cappai R, Pelka GJ, Tam PP, Christodoulou J, El-Osta A 2010. Alleviating transcriptional inhibition of the norepinephrine slc6a2 transporter gene in depolarized neurons. J Neurosci 30: 1494–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H, Lehming N 2003. Global effects of histone modifications. Brief Funct Genomics 2: 234–243 [DOI] [PubMed] [Google Scholar]
- Hebbes TR, Thorne AW, Crane-Robinson C 1988. A direct link between core histone acetylation and transcriptionally active chromatin. EMBO J 7: 1395–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday R 1999. Is there an epigenetic component in long-term memory?. J Theor Biol 200: 339–341 [DOI] [PubMed] [Google Scholar]
- Huang Y, Pastor WA, Shen Y, Tahiliani M, Liu DR, Rao A 2010. The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing. PLos One 5: e8888 10.1371/journal.pone/0008888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igaz LM, Vianna MR, Medina JH, Izquierdo I 2002. Two time periods of hippocampal mRNA synthesis are required for memory consolidation of fear-motivated learning. J Neurosci 22: 6781–6789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igaz LM, Bekinschtein P, Izquierdo I, Medina JH 2004a. One-trial aversive learning induces late changes in hippocampal CaMKIIα, Homer 1a, Syntaxin 1a and ERK2 protein levels. Brain Res Mol Brain Res 132: 1–12 [DOI] [PubMed] [Google Scholar]
- Igaz LM, Bekinschtein P, Vianna MM, Izquierdo I, Medina JH 2004b. Gene expression during memory formation. Neurotox Res 6: 189–204 [DOI] [PubMed] [Google Scholar]
- Iguchi-Ariga SM, Schaffner W 1989. CpG methylation of the cAMP-responsive enhancer/promoter sequence TGACGTCA abolishes specific factor binding as well as transcriptional activation. Genes Dev 3: 612–619 [DOI] [PubMed] [Google Scholar]
- Itzhak Y, Anderson KL, Kelley JB, Petkov M 2012. Histone acetylation rescues contextual fear conditioning in nNOS KO mice and accelerates extinction of cued fear conditioning in wild type mice. Neurobiol Learn Mem 97: 409–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin SG, Kadam S, Pfeifer GP 2010. Examination of the specificity of DNA methylation profiling techniques towards 5-methylcytosine and 5-hydroxymethylcytosine. Nucleic Acids Res 38: e125 10.1093/nar/gkq223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kangaspeska S, Stride B, Metivier R, Polycarpou-Schwarz M, Ibberson D, Carmouche RP, Benes V, Gannon F, Reid G 2008. Transient cyclical methylation of promoter DNA. Nature 452: 112–115 [DOI] [PubMed] [Google Scholar]
- Karymov MA, Tomschik M, Leuba SH, Caiafa P, Zlatanova J 2001. DNA methylation-dependent chromatin fiber compaction in vivo and in vitro: Requirement for linker histone. FASEB J 15: 2631–2641 [DOI] [PubMed] [Google Scholar]
- Katche C, Bekinschtein P, Slipczuk L, Goldin A, Izquierdo IA, Cammarota M, Medina JH 2010. Delayed wave of c-Fos expression in the dorsal hippocampus involved specifically in persistence of long-term memory storage. Proc Natl Acad Sci 107: 349–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Nguyen MD, Dobbin MM, Fischer A, Sananbenesi F, Rodgers JT, Delalle I, Baur JA, Sui G, Armour SM, et al. 2007. SIRT1 deacetylase protects against neurodegeneration in models for Alzheimer's disease and amyotrophic lateral sclerosis. EMBO J 26: 3169–3179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korzus E, Rosenfeld MG, Mayford M 2004. CBP histone acetyltransferase activity is a critical component of memory consolidation. Neuron 42: 961–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshibu K, Graff J, Beullens M, Heitz FD, Berchtold D, Russig H, Farinelli M, Bollen M, Mansuy IM 2009. Protein phosphatase 1 regulates the histone code for long-term memory. J Neurosci 29: 13079–13089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshibu K, Graff J, Mansuy IM 2011. Nuclear protein phosphatase-1: An epigenetic regulator of fear memory and amygdala long-term potentiation. Neuroscience 173: 30–36 [DOI] [PubMed] [Google Scholar]
- Kotini AG, Mpakali A, Agalioti T 2011. Dnmt3a1 upregulates transcription of distinct genes and targets chromosomal gene clusters for epigenetic silencing in mouse embryonic stem cells. Mol Cell Biol 31: 1577–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer JM, Kochinke K, Oortveld MA, Marks H, Kramer D, de Jong EK, Asztalos Z, Westwood JT, Stunnenberg HG, Sokolowski MB, et al. 2011. Epigenetic regulation of learning and memory by Drosophila EHMT/G9a. PLoS Biol 9: e1000569 10.1371/image.pbio.v09.i03 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriaucionis S, Heintz N 2009. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 324: 929–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubicek S, O'Sullivan RJ, August EM, Hickey ER, Zhang Q, Teodoro ML, Rea S, Mechtler K, Kowalski JA, Homon CA, et al. 2007. Reversal of H3K9me2 by a small-molecule inhibitor for the G9a histone methyltransferase. Mol Cell 25: 473–481 [DOI] [PubMed] [Google Scholar]
- Kwon B, Houpt TA 2010. Phospho-acetylation of histone H3 in the amygdala after acute lithium chloride. Brain Res 1333: 36–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Silva AJ 2009. The molecular and cellular biology of enhanced cognition. Nat Rev Neurosci 10: 126–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesburgueres E, Gobbo OL, Alaux-Cantin S, Hambucken A, Trifilieff P, Bontempi B 2011. Early tagging of cortical networks is required for the formation of enduring associative memory. Science 331: 924–928 [DOI] [PubMed] [Google Scholar]
- Lester BM, Tronick E, Nestler E, Abel T, Kosofsky B, Kuzawa CW, Marsit CJ, Maze I, Meaney MJ, Monteggia LM, et al. 2011. Behavioral epigenetics. Ann N Y Acad Sci 1226: 14–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung DC, Dong KB, Maksakova IA, Goyal P, Appanah R, Lee S, Tachibana M, Shinkai Y, Lehnertz B, Mager DL, et al. 2011. Lysine methyltransferase G9a is required for de novo DNA methylation and the establishment, but not the maintenance, of proviral silencing. Proc Natl Acad Sci 108: 5718–5723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levenson JM, O'Riordan KJ, Brown KD, Trinh MA, Molfese DL, Sweatt JD 2004. Regulation of histone acetylation during memory formation in the hippocampus. J Biol Chem 279: 40545–40559 [DOI] [PubMed] [Google Scholar]
- Levenson JM, Roth TL, Lubin FD, Miller CA, Huang IC, Desai P, Malone LM, Sweatt JD 2006. Evidence that DNA (cytosine-5) methyltransferase regulates synaptic plasticity in the hippocampus. J Biol Chem 281: 15763–15773 [DOI] [PubMed] [Google Scholar]
- Lin PJ, Collins PJ, Trinklein ND, Fu Y, Xi H, Myers RM, Weng Z 2007. Transcription factor binding and modified histones in human bidirectional promoters. Genome Res 17: 818–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockett GA, Helliwell P, Maleszka R 2010. Involvement of DNA methylation in memory processing in the honey bee. Neuroreport 21: 812–816 [DOI] [PubMed] [Google Scholar]
- Lonergan ME, Gafford GM, Jarome TJ, Helmstetter FJ 2010. Time-dependent expression of Arc and zif268 after acquisition of fear conditioning. Neural Plast 2010: 139891 10.1155/2010/139891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Ji Y, Ganesan S, Schloesser R, Martinowich K, Sun M, Mei F, Chao MV, Lu B 2011. TrkB as a potential synaptic and behavioral tag. J Neurosci 31: 11762–11771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubin FD, Roth TL, Sweatt JD 2008. Epigenetic regulation of BDNF gene transcription in the consolidation of fear memory. J Neurosci 28: 10576–10586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma DK, Jang MH, Guo JU, Kitabatake Y, Chang ML, Pow-Anpongkul N, Flavell RA, Lu B, Ming GL, Song H 2009. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science 323: 1074–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox SA, Schafe GE 2011. Epigenetic alterations in the lateral amygdala are required for reconsolidation of a Pavlovian fear memory. Learn Mem 18: 579–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahan AL, Mou L, Shah N, Hu JH, Worley PF, Ressler KJ 2012. Epigenetic modulation of Homer1a transcription regulation in amygdala and hippocampus with Pavlovian fear conditioning. J Neurosci 32: 4651–4659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malleret G, Haditsch U, Genoux D, Jones MW, Bliss TV, Vanhoose AM, Weitlauf C, Kandel ER, Winder DG, Mansuy IM 2001. Inducible and reversible enhancement of learning, memory, and long-term potentiation by genetic inhibition of calcineurin. Cell 104: 675–686 [DOI] [PubMed] [Google Scholar]
- Maurice T, Duclot F, Meunier J, Naert G, Givalois L, Meffre J, Celerier A, Jacquet C, Copois V, Mechti N, et al. 2008. Altered memory capacities and response to stress in p300/CBP-associated factor (PCAF) histone acetylase knockout mice. Neuropsychopharmacology 33: 1584–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maviel T, Durkin TP, Menzaghi F, Bontempi B 2004. Sites of neocortical reorganization critical for remote spatial memory. Science 305: 96–99 [DOI] [PubMed] [Google Scholar]
- Maze I, Nestler EJ 2011. The epigenetic landscape of addiction. Ann N Y Acad Sci 1216: 99–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuown SC, Barrett RM, Matheos DP, Post RJ, Rogge GA, Alenghat T, Mullican SE, Jones S, Rusche JR, Lazar MA, et al. 2011. HDAC3 is a critical negative regulator of long-term memory formation. J Neurosci 31: 764–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei F, Nagappan G, Ke Y, Sacktor TC, Lu B 2011. BDNF facilitates L-LTP maintenance in the absence of protein synthesis through ΠKMζ. PLoS One 6: e21568 10.1371/journal.pone.0021568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metivier R, Gallais R, Tiffoche C, Le Peron C, Jurkowska RZ, Carmouche RP, Ibberson D, Barath P, Demay F, Reid G, et al. 2008. Cyclical DNA methylation of a transcriptionally active promoter. Nature 452: 45–50 [DOI] [PubMed] [Google Scholar]
- Michan S, Li Y, Chou MM, Parrella E, Ge H, Long JM, Allard JS, Lewis K, Miller M, Xu W, et al. 2010. SIRT1 is essential for normal cognitive function and synaptic plasticity. J Neurosci 30: 9695–9707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CA, Sweatt JD 2007. Covalent modification of DNA regulates memory formation. Neuron 53: 857–869 [DOI] [PubMed] [Google Scholar]
- Miller CA, Campbell SL, Sweatt JD 2008. DNA methylation and histone acetylation work in concert to regulate memory formation and synaptic plasticity. Neurobiol Learn Mem 89: 599–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CA, Gavin CF, White JA, Parrish RR, Honasoge A, Yancey CR, Rivera IM, Rubio MD, Rumbaugh G, Sweatt JD 2010. Cortical DNA methylation maintains remote memory. Nat Neurosci 13: 664–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne TA, Briggs SD, Brock HW, Martin ME, Gibbs D, Allis CD, Hess JL 2002. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol Cell 10: 1107–1117 [DOI] [PubMed] [Google Scholar]
- Monsey MS, Ota KT, Akingbade IF, Hong ES, Schafe GE 2011. Epigenetic alterations are critical for fear memory consolidation and synaptic plasticity in the lateral amygdala. PLoS One 6: e19958 10.1371/journal.pone.0019958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlbacher F, Schiessel H, Holm C 2006. Tail-induced attraction between nucleosome core particles. Phys Rev E Stat Nonlin Soft Matter Phys 74: 031919 10.1103/PhysRevE.74.031919 [DOI] [PubMed] [Google Scholar]
- Mujtaba S, Zeng L, Zhou MM 2007. Structure and acetyl-lysine recognition of the bromodomain. Oncogene 26: 5521–5527 [DOI] [PubMed] [Google Scholar]
- Munoz PC, Aspe MA, Contreras LS, Palacios AG 2010. Correlations of recognition memory performance with expression and methylation of brain-derived neurotrophic factor in rats. Biol Res 43: 251–258 [PubMed] [Google Scholar]
- Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI 2001. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292: 110–113 [DOI] [PubMed] [Google Scholar]
- Nelson ED, Kavalali ET, Monteggia LM 2008. Activity-dependent suppression of miniature neurotransmission through the regulation of DNA methylation. J Neurosci 28: 395–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestor CE, Ottaviano R, Reddington J, Sproul D, Reinhardt D, Dunican D, Katz E, Dixon JM, Harrison DJ, Meehan RR 2012. Tissue-type is a major modifier of the 5-hydroxymethylcytosine content of human genes. Genome Res 22: 467–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niehrs C, Schafer A 2012. Active DNA demethylation by Gadd45 and DNA repair. Trends Cell Biol 22: 220–227 [DOI] [PubMed] [Google Scholar]
- Nielsen DA, Huang W, Hamon SC, Maili L, Witkin BM, Fox RG, Cunningham KA, Moeller FG 2012. Forced abstinence from cocaine self-administration is associated with DNA methylation changes in myelin genes in the corpus callosum: A preliminary study. Front Psychiatry 3: 60 10.3389/fpsyt.2012.00060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu S, Yabut O, D'Arcangelo G 2008. The Reelin signaling pathway promotes dendritic spine development in hippocampal neurons. J Neurosci 28: 10339–10348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nott A, Riccio A 2009. Nitric oxide-mediated epigenetic mechanisms in developing neurons. Cell Cycle 8: 725–730 [DOI] [PubMed] [Google Scholar]
- Nott A, Watson PM, Robinson JD, Crepaldi L, Riccio A 2008. S-Nitrosylation of histone deacetylase 2 induces chromatin remodelling in neurons. Nature 455: 411–415 [DOI] [PubMed] [Google Scholar]
- Oike Y, Hata A, Mamiya T, Kaname T, Noda Y, Suzuki M, Yasue H, Nabeshima T, Araki K, Yamamura K 1999. Truncated CBP protein leads to classical Rubinstein-Taybi syndrome phenotypes in mice: Implications for a dominant-negative mechanism. Hum Mol Genet 8: 387–396 [DOI] [PubMed] [Google Scholar]
- Okada D, Ozawa F, Inokuchi K 2009. Input-specific spine entry of soma-derived Vesl-IS protein conforms to synaptic tagging. Science 324: 904–909 [DOI] [PubMed] [Google Scholar]
- Oliveira AM, Estevez MA, Hawk JD, Grimes S, Brindle PK, Abel T 2011. Subregion-specific p300 conditional knock-out mice exhibit long-term memory impairments. Learn Mem 18: 161–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira AM, Hemstedt TJ, Bading H 2012. Rescue of aging-associated decline in Dnmt3a2 expression restores cognitive abilities. Nat Neurosci 15: 1111–1113 [DOI] [PubMed] [Google Scholar]
- Penner MR, Roth TL, Chawla MK, Hoang LT, Roth ED, Lubin FD, Sweatt JD, Worley PF, Barnes CA 2011. Age-related changes in Arc transcription and DNA methylation within the hippocampus. Neurobiol Aging 32: 2198–2210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters AH, Schubeler D 2005. Methylation of histones: Playing memory with DNA. Curr Opin Cell Biol 17: 230–238 [DOI] [PubMed] [Google Scholar]
- Price JC, Guan S, Burlingame A, Prusiner SB, Ghaemmaghami S 2010. Analysis of proteome dynamics in the mouse brain. Proc Natl Acad Sci 107: 14508–14513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian W, Miki D, Zhang H, Liu Y, Zhang X, Tang K, Kan Y, La H, Li X, Li S, et al. 2012. A histone acetyltransferase regulates active DNA demethylation in Arabidopsis. Science 336: 1445–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu S, Weeber EJ 2007. Reelin signaling facilitates maturation of CA1 glutamatergic synapses. J Neurophysiol 97: 2312–2321 [DOI] [PubMed] [Google Scholar]
- Quina AS, Buschbeck M, Di Croce L 2006. Chromatin structure and epigenetics. Biochem Pharmacol 72: 1563–1569 [DOI] [PubMed] [Google Scholar]
- Rajasethupathy P, Antonov I, Sheridan R, Frey S, Sander C, Tuschl T, Kandel ER 2012. A role for neuronal piRNAs in the epigenetic control of memory-related synaptic plasticity. Cell 149: 693–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reijmers LG, Perkins BL, Matsuo N, Mayford M 2007. Localization of a !stable neural correlate of associative memory. Science 317: 1230–1233 [DOI] [PubMed] [Google Scholar]
- Restivo L, Vetere G, Bontempi B, Ammassari-Teule M 2009. The formation of recent and remote memory is associated with time-dependent formation of dendritic spines in the hippocampus and anterior cingulate cortex. J Neurosci 29: 8206–8214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robison AJ, Nestler EJ 2011. Transcriptional and epigenetic mechanisms of addiction. Nat Rev Neurosci 12: 623–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roozendaal B, Hernandez A, Cabrera SM, Hagewoud R, Malvaez M, Stefanko DP, Haettig J, Wood MA 2010. Membrane-associated glucocorticoid activity is necessary for modulation of long-term memory via chromatin modification. J Neurosci 30: 5037–5046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth TL, Lubin FD, Funk AJ, Sweatt JD 2009. Lasting epigenetic influence of early-life adversity on the BDNF gene. Biol Psychiatry 65: 760–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez Mde L, Gutierrez C 2009. Novel insights into the plant histone code: Lessons from ORC1. Epigenetics 4: 205–208 [DOI] [PubMed] [Google Scholar]
- Santos KF, Mazzola TN, Carvalho HF 2005. The prima donna of epigenetics: The regulation of gene expression by DNA methylation. Braz J Med Biol Res 38: 1531–1541 [DOI] [PubMed] [Google Scholar]
- Sarantis K, Antoniou K, Matsokis N, Angelatou F 2012. Exposure to novel environment is characterized by an interaction of D1/NMDA receptors underlined by phosphorylation of the NMDA and AMPA receptor subunits and activation of ERK1/2 signaling, leading to epigenetic changes and gene expression in rat hippocampus. Neurochem Int 60: 55–67 [DOI] [PubMed] [Google Scholar]
- Shinkai Y, Tachibana M 2011. H3K9 methyltransferase G9a and the related molecule GLP. Genes Dev 25: 781–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva AJ, Zhou Y, Rogerson T, Shobe J, Balaji J 2009. Molecular and cellular approaches to memory allocation in neural circuits. Science 326: 391–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squire LR, Davis HP, Spanis CW 1980. Neurobiology of amnesia. Science 209: 836–837 [DOI] [PubMed] [Google Scholar]
- Stafford JM, Raybuck JD, Ryabinin AE, Lattal KM 2012. Increasing histone acetylation in the hippocampus-infralimbic network enhances fear extinction. Biol Psychiatry 72: 25–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanko DP, Barrett RM, Ly AR, Reolon GK, Wood MA 2009. Modulation of long-term memory for object recognition via HDAC inhibition. Proc Natl Acad Sci 106: 9447–9452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl BD, Allis CD 2000. The language of covalent histone modifications. Nature 403: 41–45 [DOI] [PubMed] [Google Scholar]
- Sui L, Wang Y, Ju LH, Chen M 2012. Epigenetic regulation of reelin and brain-derived neurotrophic factor genes in long-term potentiation in rat medial prefrontal cortex. Neurobiol Learn Mem 97: 425–440 [DOI] [PubMed] [Google Scholar]
- Swank MW, Sweatt JD 2001. Increased histone acetyltransferase and lysine acetyltransferase activity and biphasic activation of the ERK/RSK cascade in insular cortex during novel taste learning. J Neurosci 21: 3383–3391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweatt JD 2010. Mechanisms of memory Elsevier, Burlington, MA [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. 2009. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324: 930–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tayler KK, Lowry E, Tanaka K, Levy B, Reijmers L, Mayford M, Wiltgen BJ 2011. Characterization of NMDAR-independent learning in the hippocampus. Front Behav Neurosci 5: 28 10.3389/fnbeh.2011.00028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira CM, Pomedli SR, Maei HR, Kee N, Frankland PW 2006. Involvement of the anterior cingulate cortex in the expression of remote spatial memory. J Neurosci 26: 7555–7564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turker MS 1999. The establishment and maintenance of DNA methylation patterns in mouse somatic cells. Semin Cancer Biol 9: 329–337 [DOI] [PubMed] [Google Scholar]
- Valor LM, Pulopulos MM, Jimenez-Minchan M, Olivares R, Lutz B, Barco A 2011. Ablation of CBP in forebrain principal neurons causes modest memory and transcriptional defects and a dramatic reduction of histone acetylation but does not affect cell viability. J Neurosci 31: 1652–1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vastenhouw NL, Schier AF 2012. Bivalent histone modifications in early embryogenesis. Curr Opin Cell Biol 24: 374–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vecsey CG, Hawk JD, Lattal KM, Stein JM, Fabian SA, Attner MA, Cabrera SM, McDonough CB, Brindle PK, Abel T, et al. 2007. Histone deacetylase inhibitors enhance memory and synaptic plasticity via CREB:CBP-dependent transcriptional activation. J Neurosci 27: 6128–6140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigers AJ, Amin DS, Talley-Farnham T, Gorski JA, Xu B, Jones KR 2012. Sustained expression of brain-derived neurotrophic factor is required for maintenance of dendritic spines and normal behavior. Neuroscience 212: 1–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade PA 2001a. Methyl CpG-binding proteins and transcriptional repression. Bioessays 23: 1131–1137 [DOI] [PubMed] [Google Scholar]
- Wade PA 2001b. Methyl CpG binding proteins: Coupling chromatin architecture to gene regulation. Oncogene 20: 3166–3173 [DOI] [PubMed] [Google Scholar]
- Wang JH, Kelly PT 1997. Postsynaptic calcineurin activity downregulates synaptic transmission by weakening intracellular Ca2+ signaling mechanisms in hippocampal CA1 neurons. J Neurosci 17: 4600–4611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JH, Ko GY, Kelly PT 1997. Cellular and molecular bases of memory: Synaptic and neuronal plasticity. J Clin Neurophysiol 14: 264–293 [DOI] [PubMed] [Google Scholar]
- Wang SH, Teixeira CM, Wheeler AL, Frankland PW 2009. The precision of remote context memories does not require the hippocampus. Nat Neurosci 12: 253–255 [DOI] [PubMed] [Google Scholar]
- Weeber EJ, Beffert U, Jones C, Christian JM, Forster E, Sweatt JD, Herz J 2002. Reelin and ApoE receptors cooperate to enhance hippocampal synaptic plasticity and learning. J Biol Chem 277: 39944–39952 [DOI] [PubMed] [Google Scholar]
- Wood MA, Kaplan MP, Park A, Blanchard EJ, Oliveira AM, Lombardi TL, Abel T 2005. Transgenic mice expressing a truncated form of CREB-binding protein (CBP) exhibit deficits in hippocampal synaptic plasticity and memory storage. Learn Mem 12: 111–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood MA, Attner MA, Oliveira AM, Brindle PK, Abel T 2006. A transcription factor-binding domain of the coactivator CBP is essential for long-term memory and the expression of specific target genes. Learn Mem 13: 609–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Sun YE 2009. Reversing DNA methylation: New insights from neuronal activity-induced Gadd45b in adult neurogenesis. Sci Signal 2: e17 10.1126/scisignal.264pe17 [DOI] [PubMed] [Google Scholar]
- Yeh SH, Lin CH, Gean PW 2004. Acetylation of nuclear factor-κB in rat amygdala improves long-term but not short-term retention of fear memory. Mol Pharmacol 65: 1286–1292 [DOI] [PubMed] [Google Scholar]
- Yu M, Hon GC, Szulwach KE, Song CX, Zhang L, Kim A, Li X, Dai Q, Shen Y, Park B, et al. 2012. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell 149: 1368–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Rohde C, Tierling S, Jurkowski TP, Bock C, Santacruz D, Ragozin S, Reinhardt R, Groth M, Walter J, et al. 2009. DNA methylation analysis of chromosome 21 gene promoters at single base pair and single allele resolution. PLoS Genet 5: e1000438 10.1371/journal.pgen.1000438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Fan L, Frick KM 2010. Epigenetic alterations regulate estradiol-induced enhancement of memory consolidation. Proc Natl Acad Sci 107: 5605–5610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Won J, Karlsson MG, Zhou M, Rogerson T, Balaji J, Neve R, Poirazi P, Silva AJ 2009. CREB regulates excitability and the allocation of memory to subsets of neurons in the amygdala. Nat Neurosci 12: 1438–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu WG, Otterson GA 2003. The interaction of histone deacetylase inhibitors and DNA methyltransferase inhibitors in the treatment of human cancer cells. Curr Med Chem Anticancer Agents 3: 187–199 [DOI] [PubMed] [Google Scholar]
- Zovkic IB, Sweatt JD 2013. Epigenetic mechanisms in learned fear: Implications for PTSD. Neuropsychopharmacology 38: 77–93 [DOI] [PMC free article] [PubMed] [Google Scholar]