Abstract
The protein kinase catalytic domain contains several conserved residues of unknown functions. Here, using a combination of computational and experimental approaches, we show that the function of some of these residues is to maintain the backbone geometry of the active site in a strained conformation. Specifically, we find that the backbone geometry of the catalytically important HRD motif deviates from ideality in high-resolution structures and the strained geometry results in favorable hydrogen bonds with conserved noncatalytic residues in the active site. In particular, a conserved aspartate in the F-helix hydrogen bonds to the strained HRD backbone in diverse eukaryotic and eukaryotic-like protein kinase crystal structures. Mutations that alter this hydrogen-bonding interaction impair catalytic activity in Aurora kinase. Although the backbone strain is present in most active conformations, several inactive conformations lack the strain because of a peptide flip in the HRD backbone. The peptide flip is correlated with loss of hydrogen bonds with the F-helix aspartate as well as with other interactions associated with kinase regulation. Within protein kinases that are regulated by activation loop phosphorylation, the strained residue is an arginine, which coordinates with the activation loop phosphate. Based on analysis of strain across the protein kinase superfamily, we propose a model in which backbone strain co-evolved with conserved residues for allosteric control of catalytic activity. Our studies provide new clues for the design of allosteric protein kinase inhibitors.
Keywords: allostery, conformational strain, DFG flip, hydrophobic spine, Ramachandran plot
Protein phosphorylation represents a fundamental mechanism by which our cells respond to environmental signals (1, 2). In eukaryotes, protein phosphorylation is carried out by a large and diverse family of protein kinases (3), which share a conserved catalytic domain. The catalytic domain is a dynamic scaffold that toggles between various conformational states, including a catalytically “on” and “off” state, to faithfully propagate cellular signals. In the “on” state, the catalytic domain is capable of catalyzing the transfer of γ-phosphate from ATP to a receiving hydroxyl group in a protein or small molecule substrate, whereas in the “off” state it is incapable of phosphoryl-transfer. Because a constitutively “on” or “off” kinase can be catastrophic, the protein kinase “switch” is tightly controlled by a diverse array of regulatory mechanisms. Allosteric regulation is one such mode of control (4–9) in which conformational changes in the catalytic domain, upon ligand or regulatory protein binding, alters catalytic activity. Understanding the conformational control of protein kinase activity is essential for the development of new therapies for human diseases such as cancer, which is associated with abnormal regulation of protein kinase activity (10, 11).
Crystal structures of protein kinases in various conformational states and NMR studies on protein kinase A (PKA) have provided important insights into the conformational changes associated with catalysis and regulation. These studies have shown that the catalytic domain cycles through various conformational states, including an apo (open state), ATP-bound (closed binary complex), and ATP/substrate-bound (closed ternary complex) state, as part of its catalytic cycle. ATP and substrate binding promotes synchronous motions in the active site and establishes a conformational equilibrium between an “open” inactive conformation and a “closed” active conformation (12, 13). The transition from inactive to active state also involves large conformational changes in key structural elements of the catalytic core. In the Abl tyrosine kinase, for example, a conserved DFG motif in the active site switches from an inactive (DFG-out) conformation to an active (DFG-in) conformation (14). This conformational change involves rotation of the DGF aspartate (DFG-Asp) backbone torsion angles from a disfavored region of the Ramachandran plot to a fully allowed region (14). This conformational change is believed to be modulated by the protonation state of the DFG-Asp (15). More recently, a network of hydrophobic interactions connecting the ATP and substrate binding lobes of the kinase domain, called the regulatory spine, has been suggested to play an important role in kinase activation (16, 17). According to the spine model, the assembly of the hydrophobic interactions linking the ATP and substrate binding regions leads to kinase activation (16, 17).
Catalytic activation of protein kinases also involves precise positioning of the conserved HRD motif (3, 18, 19) in the active site. The HRD-Asp orients the substrate for catalysis (20) and has also been suggested to function as a catalytic base (21). Indeed, the HRD-Asp is one of the most highly conserved residues in the protein kinase superfamily and mutation of the HRD-Asp abolishes catalytic activity in many protein kinases (20, 22, 23). Unlike the HRD-Asp, the arginine (Arg) within the HRD motif is conserved in most but not all kinases, and plays a largely regulatory role (9, 22, 24, 25). In kinases that are regulated by phosphorylation of the substrate binding activation loop (Fig. 1), the HRD-Arg coordinates with the phosphorylated residue in the activation loop (7, 26). This coordination allows allosteric coupling between the regulatory site (phosphorylation site) and the active site (7, 27). Unlike the HRD-Asp and HRD-Arg, the precise role of the HRD-His in kinase functions is not as well understood.
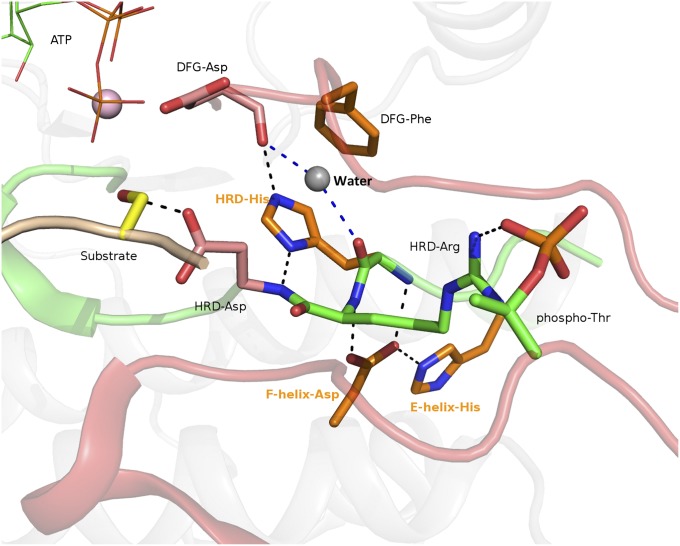
Fig. 1.
Protein kinase active site showing structural interactions associated with catalytic and EPK-ELK component residues. Catalytic residues are colored pink and EPK-ELK component residues are colored orange. The HRD-Arg and phospho-threonine unique to EPKs are shown in green. All nitrogen atoms are colored blue and oxygen atoms are colored red. ATP is shown in a “lines” representation and the substrate serine is shown in yellow. A conserved water molecule seen in all active kinase conformations between HRD and DFG motifs is shown in gray, and the hydrogen bonds made by this water are shown as blue dashed lines. The figure was generated based on a CDK-substrate complex (PDB ID 1QMZ) using PyMOL. The EPK-ELK component residues in Aurora kinase are: E-helix-His (H248), F-helix-Asp (D311), and HRD-His (H254). The corresponding residues in PKA are: E-helix-His (H158), F-helix-Asp (D220), and HRD-His (Y164).
Quantitative comparisons of protein kinase sequences and structures from diverse organisms have shown that the HRD-His is part of an ancient structural component that distinguishes eukaryotic protein kinases (EPKs) and eukaryotic-like kinases (ELKs) from distantly related atypical kinases (APKs) (18). The HRD-His, along with co-conserved residues in the substrate-binding lobe (F-helix-Asp and E-helix-His in Fig. 1) form a contiguous network of hydrogen bonds that couple the backbone of the HRD motif to ATP and substrate-binding regions. This network, called the EPK-ELK structural component (18), is conserved in EPKs and ELKs, but absent in APKs. The ability of APKs to adopt the protein kinase-fold without the hydrogen-bonding network indicated that the EPK-ELK structural component is not required for maintaining protein kinase structure or fold, but rather for as-yet-unidentified functions (18, 28).
Here, we present evidence that the remarkable conservation of the EPK-ELK structural component is because of its role in a conformational strain switch in the active site of protein kinases. Specifically, we find that the hydrogen bonds mediated by the EPK-ELK component residues maintain the backbone torsion angles of the HRD motif in a “strained” conformation in most active structures, and release of the backbone strain via a peptide flip correlates with conformational changes in the EPK-ELK component residues. Furthermore, we show that mutation of the EPK-ELK component residues in Aurora kinase impairs catalytic activity. We propose that the EPK-ELK component is an ancient allosteric network that has co-evolved with the catalytic loop strain to regulate activity in EPKs and ELKs. EPKs have elaborated on this core network through the addition of EPK-specific regulatory features, such as the HRD-Arg and phosphorylatable activation loop, to provide additional layers of allosteric control.
Results
Identification of a Hidden Backbone Strain in the Catalytic Loop of Protein Kinases.
As shown in Fig. 1, the EPK-ELK component residues, such as the HRD-His, and the F-helix-Asp form precise hydrogen bonds with the backbone of the HRD motif. To understand why these interactions are maintained across diverse EPK and ELK structures, we performed detailed conformational analysis of the HRD motif backbone in available crystal structures. We analyzed the backbone torsion angles, and the bond-angle values of the HRD motif residues in 103 high-resolution (<1.7 Å) structures from diverse protein kinase families (Materials and Methods). This process revealed that the torsion angles of the HRD-Arg occurs in disfavored regions of the Ramachandran plot in 89 of the 103 structures (Fig. 2A and Table S1). In these 89 structures, the bond angle (N-Ca-C) of the HRD-Arg also deviates from ideal values (Fig. 2B and Table S1). We define these unusual conformations of the HRD-Arg backbone as “conformational strain” (Materials and Methods). Conformation strain in the HRD-Arg backbone also persists in a larger dataset of kinase structures at medium resolution (<2.5 Å) (Fig. S1A).
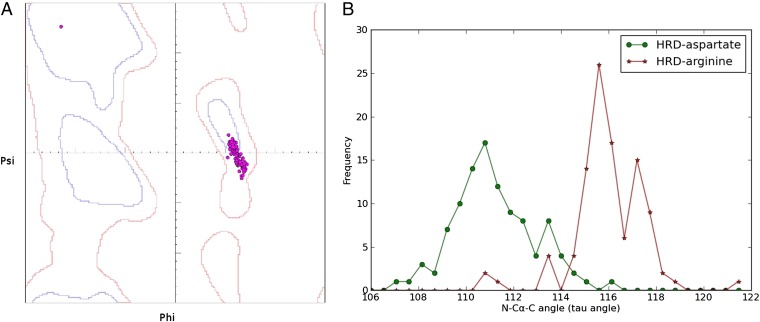
Fig. 2.
Conformational strain in the HRD-Arg backbone. (A) Ramachandran plot showing the torsion angle values (ϕ/ψ) of the HRD-Arg residue in 103 high-resolution structures (<1.7 Å) from diverse families. Contours in the Ramachandran plot are based on the definition of Molprobity (35, 36). The blue faint contour indicates the “favored” regions (enclosing 98% of observed conformations) and the red outer contour indicates the “disfavored but allowed” regions (enclosing 99.8% of observed conformations). Torsion-angle values of the HRD-Arg are indicated by dots. In 89 of the 103 high-resolution structures, the torsion-angle values of the HRD-Arg occur in the disfavored region and are defined as strained. (B) Conformational strain in the HRD-Arg backbone is supported by nonideal bond-angle values. Distribution of the N-Cα-C angle bond-angle values for the HRD-Asp and HRD-Arg in high-resolution structures. Shift in the HRD-Arg bond-angle values (116.33 Å ± 1.6) from the HRD-Asp suggests energetic strain because of bond bending. The average deviation from ideal bond-angle values (111 Å) is estimated to be 2 SDs (111 Å ± 2.8).
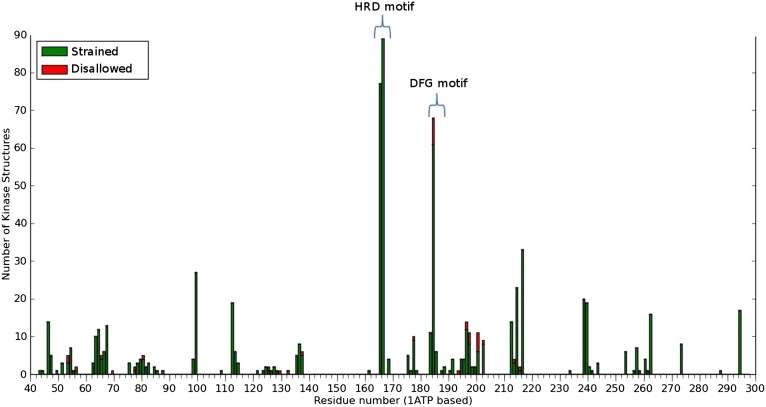
To determine if regions other than the HRD motif display backbone strain, we plotted the frequency of strained and disallowed conformations at each of the 295 residue positions that define the protein kinase domain. This process revealed that in addition to the HRD-Arg, the backbone of the DFG-Asp also displays backbone strain (Fig. 3). The strain in the DFG-Asp has been noted previously (14); however, the strain in the catalytic loop (HRD-Arg) has not been noted before.
Fig. 3.
Frequency of disfavored (strained) and disallowed conformations observed at each residue position in the protein kinase domain. The residues are numbered according to the cAMP dependent kinase (PDB ID 1ATP).
Catalytic Loop Strain Is Conserved in EPKs and ELKs That Are Not Regulated by Activation Loop Phosphorylation.
In EPKs, the side-chain of the HRD-Arg typically coordinates with the phosphate group of a phosphorylated Thr/Ser/Tyr residue in the activation loop. To determine if the HRD-Arg backbone strain is a consequence of activation loop phosphorylation, we analyzed the phosphorylation status of the activation loop in all of the strained conformations. We found no correlation between the occurrence of strain at the HRD-Arg backbone and the phosphorylation status of the activation loop. Furthermore, families that do not conserve an arginine at the HRD-Arg position also display strain at the equivalent position. For example, DAPK kinase, which conserve a phenylalanine at the HRD-Arg position display the catalytic loop strain (PDB ID 2W4J) (Fig. S1). Likewise, ELKs, which are not regulated by activation loop phosphorylation, also display the strain (Fig. S1). However, the strain was not detected in distantly related APKs, such as actin-fragmin kinase (29).
Association and Coevolution of the Catalytic Loop Strain with the EPK-ELK Structural Component.
We hypothesized that the hydrogen bonds mediated by the EPK-ELK conserved residues is essential to maintain the HRD-Arg backbone strain (Fig. 1). To test this hypothesis, we analyzed all the crystal structures that display the strain (2,269 structures) (Materials and Methods). We found that in a significant number of structures where the HRD-Arg backbone is conformationally strained, the hydrogen bonding network mediated by the EPK-ELK component residues (Fig. 1) are present (1,357 of 2,269, P value 1.41 × 10−52) (Table S2). In particular, the F-helix-Asp hydrogen bonds to the backbone amides of the HRD-His and HRD-Arg in nearly all the structures where the strain is present (2,198 of 2,269 structures, P value 2.348 × 10−42) (Table S2). In addition, conserved water-mediated interactions (30) between the HRD and DFG motif backbone atoms are observed in the strained conformations (Fig. 1). We also found that the strained conformation occurs predominantly in the active state (Table S1). The strain status in other functional states of the kinase, such as the inactive, liganded, and unliganded states, is provided in Table S3.
To obtain insights into how the strain may have evolved during the course of kinase evolution, we analyzed the phylogenetic distribution of the various amino acids conserved at the HRD-Arg position in EPKs and ELKs. This analysis revealed that although an arginine within the HRD motif is distinctive of EPKs, ELKs generally conserve a glycine, alanine, or asparagine at the HRD-Arg position (Fig. S1). Rio kinases, in particular, are noteworthy in this regard because they conserve a glycine at the HRD-Arg position that can adopt the active conformation without being strained.
Catalytic Loop Strain Switch Is Correlated with Conformational Changes in the EPK-ELK Structural Component, the DFG Motif, and the Hydrophobic Spine.
Switching of HRD-Arg backbone from a strained to relaxed conformation (observed in 173 structures) is accompanied by a concerted conformational change in the EPK-ELK structural component residues. This relaxation breaks the two canonical hydrogen bonds between the F-helix-Asp side-chain and the HRD-Arg and HRD-His backbone in all of the structures where the catalytic loop strain is lost. Similarly, the hydrogen bonds between the F-helix-Asp and E-helix-His are also lost in structures where the HRD-Arg torsion angle is in the favored region of Ramachandran plot (169 of 173, P value 1.23 × 10−4). The loss of the catalytic loop strain is also correlated with the loss of HRD-His–mediated hydrogen bonds to the backbone of DFG-Asp (128 of 173 structures P value 5.73 × 10−14). In addition, the water-mediated interactions between the HRD and DFG motifs, described in the previous section, are not observed in structures where the strain is lost.
The loss of the catalytic loop strain is also correlated with conformational changes in the DFG motif and the hydrophobic spine. In a significant number of structures where the HRD-Arg backbone occurs in the relaxed conformation, the DFG motif occurs in the “DFG-out” conformation (16 structures, P value 0.039) (Table S2). An intermediate state to the DFG-out conformation occurs in many protein kinase families, such as Aurora (PDB ID 2J50) and CDK (PDB ID 3MTL), and these structures lack the catalytic loop strain. In addition to conformational changes in the DFG motif, we note that the hydrophobic spine that connects the DFG and HRD motifs is also disassembled in structures where the strain is lost (16 structures, P value 0.039).
Mutation of the EPK-ELK Component Residues Alter Aurora Kinase Activity.
Because the EPK-ELK component residues are directly associated with the strain, we hypothesized that mutation of the EPK-ELK component residues should alter the strain status, and consequently protein kinase activity. To test this hypothesis, we performed mutational analysis of EPK-ELK component residues in human Aurora kinase A. Aurora kinase conserves all of the EPK-ELK component residues and phosphorylates its physiological substrate, Histone H3 (at Ser-10), upon auto-phosphorylation of the activation loop (31). Throughout the experimental analysis, a catalytically inactive mutant K162A in the N lobe that impairs ATP binding, a catalytically inactive aspartate mutant (D256A), and a wild-type kinase reaction without ATP was used as negative controls.
Mutational analysis of F-helix-Asp.
We mutated the F-helix-Asp to a leucine, asparagine, alanine, and glutamate. The aspartate to glutamate mutant showed activity similar to or greater than that of the wild-type (Fig. 4A). However, the other three mutants (D311A, D311L, and D311N) did not show any detectable catalytic activity compared to the control wild-type assay without ATP and the catalytically inactive mutants. In addition to impaired phosphorylation of Histone H3, the three mutants (D311A, D311L, and D311N) also prevent autophosphorylation of the activation loop threonine (T288) (Fig. 4A).
Fig. 4.
Mutational analysis of EPK-ELK component residues. (A) F-helix-Asp mutations. (Left) The controls (with two known kinase dead mutants, D256A and K162A) and the F-helix-Asp single mutants (D311A to D311N). For each mutant, the status of the autophosphorylation, total amount of Aurora protein, substrate phosphorylation status (using phospho-Histone H3 antibody), and total Histone H3 in each lane is given. (Right) The data for double mutants in a background of activating T288E (activation loop phospho-mimic) mutant. (B) HRD-His and E-helix-His in the T288E background.
We also generated an activating T288E mutation that mimics constitutive autophosphorylation, and performed our mutational analysis of F-helix-Asp in the T288E background. The double-mutants D311A/T288E, D311L/T288E, and D311N/T288E did not show any detectable substrate phosphorylation, whereas the T288E mutation by itself showed activity similar to, or greater than that of wild-type Aurora A (Fig. 4A). Furthermore, we characterized the in vitro kinetics of the D311L/T288E and D311N/T288E mutants using a fluorescence based assay (see SI Materials and Methods for details). The kinetic data indicate that the two mutants catalyze phosphoryl-transfer at significantly different rates relative to each other, as well as to the wild-type. The rate of D311L/T288E is 3.5-fold higher than D311N/T288E, and D311L/T288E and D311N/T288E are 45- and 160-fold slower than the wild-type, respectively (Fig. S2).
Mutational analysis of HRD-His and E-helix-His.
To determine the role of the HRD-His and E-helix-His in Aurora A kinase activity, we carried out mutational analysis of these EPK-ELK conserved residues in the T288E background. HRD-His to a tyrosine mutation (H254Y) was observed in some AGC kinases. We also mutated the HRD-His to a phenylalanine to disrupt the hydrogen bonds made by the HRD-His. As seen in Fig. 4B, the H254Y mutant does not change Aurora kinase activity, whereas the H254F mutant abolishes kinase activity. In addition to the HRD-His, we mutated the E-helix-His, which is part of the EPK-ELK structural component. The E-helix-His was mutated to a glutamate and alanine because these mutations are naturally observed in some tyrosine kinases. As seen in Fig. 4B, the E-helix-His mutations either abolish Aurora kinase activity, or substantially reduce activity.
Discussion
Conformationally strained polypeptide backbones usually occur near enzyme-active sites (32). In the phospho-carrier protein, HPr, for example, a torsion-angle strain switch in the active site has been suggested to lower the activation barrier (33) during phosphoryl-transfer. Here we perform detailed conformational strain analysis of 2,442 protein kinase crystal structures to show that the catalytically important HRD motif in the active site is under conformational strain. Although the precise role of the strain is unclear at this point, several lines of evidence suggest a regulatory role. First, the ability of certain APKs to catalyze phosphoryl-transfer without the strain, and the associated EPK-ELK structural component, suggests that the strain is not part of the catalytic mechanism. Second, conformational changes associated with regulation, such as formation of the hydrophobic spine, is correlated with the strain-switch. Third, mutations that disrupt hydrogen-bonding interactions with the strained backbone abrogate catalytic activity (Fig. 4). A regulatory role for the strain is also supported by recent NMR studies on PKA, which show that ATP binding induces global conformational changes coupling the ATP and substrate binding lobes (12, 13). This coupling has been suggested to involve the hydrophobic spine connecting the ATP and substrate-binding lobes (16, 17). Our analysis suggests that in addition to the hydrophobic spine, switching of the HRD backbone from relaxed to strained conformation may also contribute to allosteric coupling between ATP and substrate binding lobes (Fig. 5 and Fig. S3).
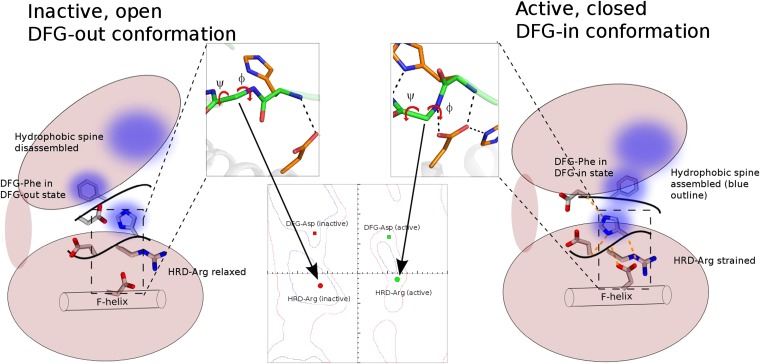
Fig. 5.
Model of strain-switch in kinase activation. The N and C lobes of the kinase are shown as ellipses, and the F-helix is shown as a cylinder in the C lobe. The zoomed in view of the kinase HRD motif region in active and inactive structures is shown (with side-chain removed for clarity) and the peptide flip is shown on the Ramachandran plot. In addition to the HRD-Arg residue (shown on Ramachandran plot as circles), the DFG-Asp ϕ/ψ values are also shown (as rectangles). The hydrophobic spine is shown as blue blurred circles, with a large blue circle in the smaller N lobe representing the spine residues in the N lobe. The hydrogen-bonding network mediated by the EPK-ELK component residues are shown as dotted orange lines.
A functional role for the catalytic loop strain also explains the high degree of conservation of the EPK-ELK component residues (3, 18), which not only hydrogen bond to the strained backbone in the active conformation, but also couple the strain to distal substrate-binding regions. Such coupling, for example, may allow strain-switch mediated conformational changes in the active site to be allosterically communicated to distal substrate binding and regulatory regions, and vice versa. Our model (Fig. 5) suggests that mutations that alter the EPK-ELK–mediated hydrogen-bonding network, such as the Aurora kinase F-helix-Asp to Asn mutation, could lead to loss of catalytic activity by disrupting the allosteric communication network. The substitution of Asn in place of F-helix-Asp could stabilize the relaxed form of the HRD-Arg, as seen in crystal structure of an inactive Rop5 kinase, which naturally conserves an Asn at the F-helix-Asp position (PDB ID 3Q60) (Fig. S4). In the crystal structure of ROP5, the peptide backbone of the HGD motif is flipped and the Asn hydrogen bonds to the carbonyl oxygen of the HGH-Gly backbone (Fig. S4).
The flipping of the HRD-Arg backbone from strained to relaxed conformations could give rise to multiple inactive structures via disruptions in EPK-ELK component-mediated interactions. Crystal structures of Aurora kinase solved in various conformational states (Fig. S5) provide clues to such inactive conformations. In some of the inhibitor-bound Aurora kinase structures (resolution ∼2.0 Å), for example, the HRD backbone exists in a conformational state that is intermediate to the strained and relaxed forms. In these states, the side-chain of HRD-His hydrogen bonds to the F-helix-Asp, breaking the EPK-ELK network seen in active forms (Fig. S5C) (PDB ID 3DJ6). In vivo, it is possible that the strained and relaxed forms exist in equilibrium. The kinetic data for the D311L/T288E and D311N/T288E mutants suggests that the equilibrium between these states can be affected by modifications of the EPK-ELK component-mediated interactions. The D311N/T288E mutant would specifically favor the unstrained conformation as seen in ROP5 (Fig. S4) and, hence, is the slowest among the mutants studied. Because the D311L/T288E mutant favors neither conformation, and the unstrained conformation is more likely to exist in such a scenario, it shows slightly higher activity in comparison with D311N/T288E mutant. The actual rates are still much slower compared to the wild-type (around 45-times and 160-times slower), explaining the lack of activity in Western blots.
Because the catalytic loop strain is observed in both EPKs and ELKs, it is likely that the strain switch represents an ancient allosteric mechanism for mediating phosphoryl-transfer. The strain switch presumably occurs readily in ELKs because ELKs generally conserve small amino acids, such as glycine, at the HRD-Arg position. Rio kinases, and in particular Rio2, are interesting in this regard because they conserve a glycine at the HRD-Arg position across all three kingdoms of life. By selectively conserving a phosphate-coordinating Arg at the HRD-Arg position, EPKs appear to have structurally coupled the catalytic loop strain to the activation loop phosphate, presumably for tighter control of catalytic activity. Such coupling could favor a spring-loaded mechanism of action wherein phosphorylation of the activation loop locks the HRD backbone in a strain conformation.
The proposed strain-switch mechanism also has the potential to inform current drug-discovery efforts on kinases. In the Imatinib-bound structures of Abl kinase, for example, the drug is poised to interact with the backbone atoms of the HRD motif (PDB ID 2HYY). Design of inhibitors that prevent the formation of HRD motif strain could prove to be a unique paradigm in kinase inhibition. Thus, further analysis of the strain-switch in multiple kinase subfamilies, especially using NMR methods, is necessary to fully understand this fundamental feature of kinases.
Materials and Methods
Identification of Strain Conformations.
The dataset used for the analysis consists of 1,928 PDB files containing a protein kinase-like domain. The 1,928 PDB files were separated into 2,442 individual chains and aligned using MAPGAPS (34), a procedure for rapidly and accurately aligning a large number of sequences. The alignment was used to define structurally equivalent residues in the kinase domain. Torsion-angle values (ϕ and ψ) for the aligned residues were calculated using in-house programs, and classified into three categories based on MolProbity (35) definitions of the Ramachandran plot. Specifically, torsion-angle values (rounded off to the nearest integer) were classified as favored (within 98% contour), disfavored (between 98% contour and 99.8% contour), or disallowed (outside 99.8% contour) based on their location in the Ramachandran plot. Residues with torsion-angle values in the disfavored regions were defined as “strained.” The conformations were further analyzed for unusual N-Cα-C bond-angle values because torsion-angle strain is known to correlate with nonideal N-Cα-C bond-angle values (36, 37). Indeed, in majority of strained conformations, the N-Cα-C bond-angle value for the HRD-Arg deviates from the normal value of around 111° by 2 SDs (Fig. 2B and Table S1). P values reported in the text and Tables S2 and S3 were calculated using the CHITEST module of MS Excel. The list of high-resolution structures with the ϕ/ψ values of the HRD-Arg and N-Cα-C angles is given in Table S1. Table S1 also highlights the active conformations, which were identified based on the following criteria: (i) presence of ATP or ANP in the nucleotide binding pocket, (ii) presence of Lys-Glu salt bridge in the N-lobe, and (iii) closure of the ATP binding N lobe relative to the substrate C lobe (7).
Expression and Purification of Aurora WT and Mutant Proteins.
The wild-type human Aurora A was obtained from Addgene (plasmid 8510) and the gene of interest was excised using BamHI and regenerated XhoI site from the vector (38). The insert was cloned into BamHI/XhoI sites of a modified pet24DBAM vector with an N-terminal 6× His-tag. Mutations were made using the Stratagene QuikChange II kit, and each plasmid was sequenced before expression and some plasmids (T288E, D178L, and D178E) after expression as quality control. Escherichia coli BL21 cells (gift of Z.A.W., University of Georgia) expressing the plasmid were grown at 37 °C until O.D ∼0.6, and induced with isfopropyl-β-D-thiogalactopyranoside at a final concentration of 1 mM at 20 °C and proteins were harvested after induction overnight. Cells were lyzed using lysozyme treatment (0.5 mg/mL) for 30 min and followed by sonication in lysis buffer [50 mM sodium phosphate buffer (pH 7.8), 300 mM NaCl, and 0.1% Nonidet P-40]. Cell debris was discarded and supernatant loaded on Ni-NTA agarose columns (Qiagen) and purified under batch purification protocol.
Kinase Activity Assay Using Western Blots.
Histone H3 was obtained from Roche as lyophilized powder and was reconstituted in kinase buffer (50 mM Tris●HCl pH 7.6, 5 mM MgCl2, 10 mM NaCl) to give final concentration of 1 mg/mL. Reactions were started in 40-μL final volume by adding kinase mixture (kinase buffer + 10 μg Histone H3+ 2 mM ATP) to varying amounts of wild-type and mutant eluate, and the reaction was carried out for 1 h at 37 °C.
Samples were run on 12% (wt/vol) SDS/PAGE and transferred to PVDF membranes. Western blotting was done using primary antibodies, phospho Histone H3 (pSer10) (Cell Signaling #3377), phospho-Aurora (T288) (Cell Signaling #2914), Histone H3 antibody (Cell Signaling #4499), and Aurora antibody (Cell Signaling #4718). Appropriate secondary antibody was used and the bands were visualized using HRP-conjugated secondary antibody and ECL substrate.
Supplementary Material
Acknowledgments
We thank members of the N.K. laboratory for helpful discussions; and Dr. David Blum and Mr. Paul Volny from the Bioexpression and Fermentation facility of the University of Georgia for their help in protein purification. This study was supported in part by National Science Foundation Grant MCB-1149106 (to N.K.), American Cancer Society Grant RSG-10-188-01-TBE (to N.K.), and the Georgia Cancer Coalition (N.K.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1207104110/-/DCSupplemental.
References
- 1.Johnson LN, Lewis RJ. Structural basis for control by phosphorylation. Chem Rev. 2001;101(8):2209–2242. doi: 10.1021/cr000225s. [DOI] [PubMed] [Google Scholar]
- 2.Krebs EG. Historical perspectives on protein phosphorylation and a classification system for protein kinases. Philos Trans R Soc Lond B Biol Sci. 1983;302(1108):3–11. doi: 10.1098/rstb.1983.0033. [DOI] [PubMed] [Google Scholar]
- 3.Hanks SK, Hunter T. Protein kinases 6. The eukaryotic protein kinase superfamily: Kinase (catalytic) domain structure and classification. FASEB J. 1995;9(8):576–596. [PubMed] [Google Scholar]
- 4.Jura N, et al. Catalytic control in the EGF receptor and its connection to general kinase regulatory mechanisms. Mol Cell. 2011;42(1):9–22. doi: 10.1016/j.molcel.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taylor SS, Kornev AP. Protein kinases: Evolution of dynamic regulatory proteins. Trends Biochem Sci. 2011;36(2):65–77. doi: 10.1016/j.tibs.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shi Z, Resing KA, Ahn NG. Networks for the allosteric control of protein kinases. Curr Opin Struct Biol. 2006;16(6):686–692. doi: 10.1016/j.sbi.2006.10.011. [DOI] [PubMed] [Google Scholar]
- 7.Nolen B, Taylor S, Ghosh G. Regulation of protein kinases; Controlling activity through activation segment conformation. Mol Cell. 2004;15(5):661–675. doi: 10.1016/j.molcel.2004.08.024. [DOI] [PubMed] [Google Scholar]
- 8.Huse M, Kuriyan J. The conformational plasticity of protein kinases. Cell. 2002;109(3):275–282. doi: 10.1016/s0092-8674(02)00741-9. [DOI] [PubMed] [Google Scholar]
- 9.Johnson LN, Noble ME, Owen DJ. Active and inactive protein kinases: Structural basis for regulation. Cell. 1996;85(2):149–158. doi: 10.1016/s0092-8674(00)81092-2. [DOI] [PubMed] [Google Scholar]
- 10.Johnson LN. Protein kinase inhibitors: Contributions from structure to clinical compounds. Q Rev Biophys. 2009;42(1):1–40. doi: 10.1017/S0033583508004745. [DOI] [PubMed] [Google Scholar]
- 11.Brognard J, Hunter T. Protein kinase signaling networks in cancer. Curr Opin Genet Dev. 2011;21(1):4–11. doi: 10.1016/j.gde.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Masterson LR, et al. Dynamics connect substrate recognition to catalysis in protein kinase A. Nat Chem Biol. 2010;6(11):821–828. doi: 10.1038/nchembio.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Masterson LR, Mascioni A, Traaseth NJ, Taylor SS, Veglia G. Allosteric cooperativity in protein kinase A. Proc Natl Acad Sci USA. 2008;105(2):506–511. doi: 10.1073/pnas.0709214104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levinson NM, et al. A Src-like inactive conformation in the abl tyrosine kinase domain. PLoS Biol. 2006;4(5):e144. doi: 10.1371/journal.pbio.0040144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shan Y, et al. A conserved protonation-dependent switch controls drug binding in the Abl kinase. Proc Natl Acad Sci USA. 2009;106(1):139–144. doi: 10.1073/pnas.0811223106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kornev AP, Taylor SS. Defining the conserved internal architecture of a protein kinase. Biochim Biophys Acta. 2010;1804(3):440–444. doi: 10.1016/j.bbapap.2009.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kornev AP, Taylor SS, Ten Eyck LF. A helix scaffold for the assembly of active protein kinases. Proc Natl Acad Sci USA. 2008;105(38):14377–14382. doi: 10.1073/pnas.0807988105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kannan N, Neuwald AF. Did protein kinase regulatory mechanisms evolve through elaboration of a simple structural component? J Mol Biol. 2005;351(5):956–972. doi: 10.1016/j.jmb.2005.06.057. [DOI] [PubMed] [Google Scholar]
- 19.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298(5600):1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 20.Adams JA. Kinetic and catalytic mechanisms of protein kinases. Chem Rev. 2001;101(8):2271–2290. doi: 10.1021/cr000230w. [DOI] [PubMed] [Google Scholar]
- 21.Madhusudan A, Akamine P, Xuong NH, Taylor SS. Crystal structure of a transition state mimic of the catalytic subunit of cAMP-dependent protein kinase. Nat Struct Biol. 2002;9(4):273–277. doi: 10.1038/nsb780. [DOI] [PubMed] [Google Scholar]
- 22.Gibbs CS, Zoller MJ. Rational scanning mutagenesis of a protein kinase identifies functional regions involved in catalysis and substrate interactions. J Biol Chem. 1991;266(14):8923–8931. [PubMed] [Google Scholar]
- 23.Skamnaki VT, et al. Catalytic mechanism of phosphorylase kinase probed by mutational studies. Biochemistry. 1999;38(44):14718–14730. doi: 10.1021/bi991454f. [DOI] [PubMed] [Google Scholar]
- 24.Strong TC, Kaur G, Thomas JH. Mutations in the catalytic loop HRD motif alter the activity and function of Drosophila Src64. PLoS ONE. 2011;6(11):e28100. doi: 10.1371/journal.pone.0028100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Steichen JM, et al. Structural basis for the regulation of protein kinase A by activation loop phosphorylation. J Biol Chem. 2012;287(18):14672–14680. doi: 10.1074/jbc.M111.335091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brown NR, Noble ME, Endicott JA, Johnson LN. The structural basis for specificity of substrate and recruitment peptides for cyclin-dependent kinases. Nat Cell Biol. 1999;1(7):438–443. doi: 10.1038/15674. [DOI] [PubMed] [Google Scholar]
- 27.Leon BC, Tsigelny I, Adams JA. Electrostatic environment surrounding the activation loop phosphotyrosine in the oncoprotein v-Fps. Biochemistry. 2001;40(34):10078–10086. doi: 10.1021/bi010838e. [DOI] [PubMed] [Google Scholar]
- 28.Kannan N, Taylor SS, Zhai Y, Venter JC, Manning G. Structural and functional diversity of the microbial kinome. PLoS Biol. 2007;5(3):e17. doi: 10.1371/journal.pbio.0050017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steinbacher S, et al. The crystal structure of the Physarum polycephalum actin-fragmin kinase: An atypical protein kinase with a specialized substrate-binding domain. EMBO J. 1999;18(11):2923–2929. doi: 10.1093/emboj/18.11.2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Knight JD, Hamelberg D, McCammon JA, Kothary R. The role of conserved water molecules in the catalytic domain of protein kinases. Proteins. 2009;76(3):527–535. doi: 10.1002/prot.22451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crosio C, et al. Mitotic phosphorylation of histone H3: Spatio-temporal regulation by mammalian Aurora kinases. Mol Cell Biol. 2002;22(3):874–885. doi: 10.1128/MCB.22.3.874-885.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Herzberg O, Moult J. Analysis of the steric strain in the polypeptide backbone of protein molecules. Proteins. 1991;11(3):223–229. doi: 10.1002/prot.340110307. [DOI] [PubMed] [Google Scholar]
- 33.Jia Z, Vandonselaar M, Hengstenberg W, Quail JW, Delbaere LT. The 1.6 Å structure of histidine-containing phosphotransfer protein HPr from Streptococcus faecalis. J Mol Biol. 1994;236(5):1341–1355. doi: 10.1016/0022-2836(94)90062-0. [DOI] [PubMed] [Google Scholar]
- 34.Neuwald AF. Rapid detection, classification and accurate alignment of up to a million or more related protein sequences. Bioinformatics. 2009;25(15):1869–1875. doi: 10.1093/bioinformatics/btp342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 1):12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramachandran GN, Sasisekharan V. Conformation of polypeptides and proteins. Adv Protein Chem. 1968;23:283–438. doi: 10.1016/s0065-3233(08)60402-7. [DOI] [PubMed] [Google Scholar]
- 37.Karplus PA. Experimentally observed conformation-dependent geometry and hidden strain in proteins. Protein Sci. 1996;5(7):1406–1420. doi: 10.1002/pro.5560050719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Crane R, Kloepfer A, Ruderman JV. Requirements for the destruction of human Aurora-A. J Cell Sci. 2004;117(Pt 25):5975–5983. doi: 10.1242/jcs.01418. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.