Abstract
Chloroplasts are the organelles of green plants in which light energy is transduced into chemical energy, forming ATP and reduced carbon compounds upon which all life depends. The expenditure of this energy is one of the central issues of cellular metabolism. Chloroplasts contain ∼3,000 proteins, among which less than 100 are typically encoded in the plastid genome. The rest are encoded in the nuclear genome, synthesized in the cytosol, and posttranslationally imported into the organelle in an energy-dependent process. We report here a measurement of the amount of ATP hydrolyzed to import a protein across the chloroplast envelope membranes—only the second complete accounting of the cost in Gibbs free energy of protein transport to be undertaken. Using two different precursors prepared by three distinct techniques, we show that the import of a precursor protein into chloroplasts is accompanied by the hydrolysis of ∼650 ATP molecules. This translates to a ΔGprotein transport of some 27,300 kJ/mol protein imported. We estimate that protein import across the plastid envelope membranes consumes ∼0.6% of the total light-saturated energy output of the organelle.
Keywords: translocation ATPase, chloroplast translocons at inner and outer envelop membranes, import machineries, protein trafficking, cellular energy budget
The majority of proteins in cells are synthesized in the cytoplasm on free or membrane-bound ribosomes. The fraction of proteins that are translocated across one or more cellular membranes has been estimated to be almost 50% in eukaryotic cells (1) and as high as 30% in bacteria (2, 3). Thus, protein movement out of the cytoplasm across membranes represents a considerable cellular activity involving numerous and varied protein transport systems. Protein translocation across membranes is also generally an energy-requiring process, making it a potentially costly activity for cells (4).
The nature of the energy inputs to the different protein translocation systems has been extensively studied. We know, for example, that the posttranslational transport of proteins into the endoplasmic reticulum requires ATP hydrolysis alone (4–6), and that the transport of proteins into the thylakoid lumen or across the bacterial plasma membrane on the Tat pathway requires only the transmembrane protonmotive force with no contribution of ATP hydrolysis (7, 8). A more complex energy input is required for many other protein transport systems. For instance, the import of proteins into the mitochondrial matrix requires both ATP hydrolysis and the Δψ across the inner mitochondrial membrane (9–11), and bacterial secretion and protein transport to the thylakoid lumen on their respective Sec pathways use ATP hydrolysis and the protonmotive force (3, 7, 12, 13).
In contrast to the abundance of work defining the nature of the energy driving protein transporters, few attempts have been made to quantitate the amount of energy required for these reactions. Without this information, it is not possible to assess the amount of metabolic energy that is spent on the cell’s considerable protein trafficking activity. One such measurement of the energy required for protein transport is the so-called translocation ATPase activity of the bacterial Sec pathway observed with inverted plasma membrane vesicles from Escherichia coli (14, 15). In these experiments it was found that ∼700 ATP molecules were hydrolyzed per prOmpA molecule exported when the membranes were allowed to develop a protonmotive force. This number rose to more than 5,000 ATP per protein translocated when the protonmotive force was dissipated by the addition of ionophores. Because the energy content of the protonmotive force was not quantitated in these studies, it is not possible to know the amount of Gibbs free energy used for transport (ΔGprotein transport) of prOmpA from these experiments.
The sole protein translocation system for which the ΔGprotein transport was experimentally determined (in our laboratory) is the chloroplast Tat (cpTat) pathway responsible for the transport of a subset of proteins from the chloroplast stroma into the thylakoid lumen (16). We chose this system for analysis because it has a simple energy input in the form of the transmembrane protonmotive force; no NTP hydrolysis is required or contributes to this process. Measurements of the drain of the protonmotive force during protein transport revealed that an energetic equivalence of more than 10,000 ATP molecules were spent per protein transported on this pathway. Although this amount of energy seems excessive, we noted that chloroplasts can sustain maximum rates of protein transport on the cpTat pathway and give up less than 3% of their capacity for photosynthetic ATP synthesis.
The high cost of protein transport on the cpTat pathway, as well as that for the uncoupled bacterial Sec pathway, raised the possibility that protein trafficking might impose a large, previously unrecognized drain on a cell’s energy budget. To determine if this is the case, we have been working to expand our studies of the ΔGprotein transport to different membrane transporters. An obvious next choice for our analysis is the translocation of proteins across the chloroplast envelope membranes from the cytoplasm to the chloroplast stroma through the translocons of the outer and inner envelope membranes of chloroplasts, the so-called Toc and Tic machineries. As with the cpTat pathway, this reaction has an experimentally simple energy input, in this case requiring only the hydrolysis of exogenously added ATP with no assistance from the protonmotive force. We report here that this protein import reaction requires the hydrolysis of an average of 650 ATP molecules per precursor imported, which in dark-adapted chloroplasts (17) translate to a ΔGprotein transport of ∼27,300 kJ/mol.
Results
Effect of Inhibitors on Intrinsic Background ATPase Activity in Intact Chloroplasts.
For ease of reference, we define the background ATPase activity as that measured in the absence of protein import substrate, signal ATPase activity as the total ATPase activity measured during protein import minus the background ATPase activity, and the translocation ATPase activity as the signal ATPase activity divided by the amount of protein imported, yielding ATP hydrolyzed per protein imported.
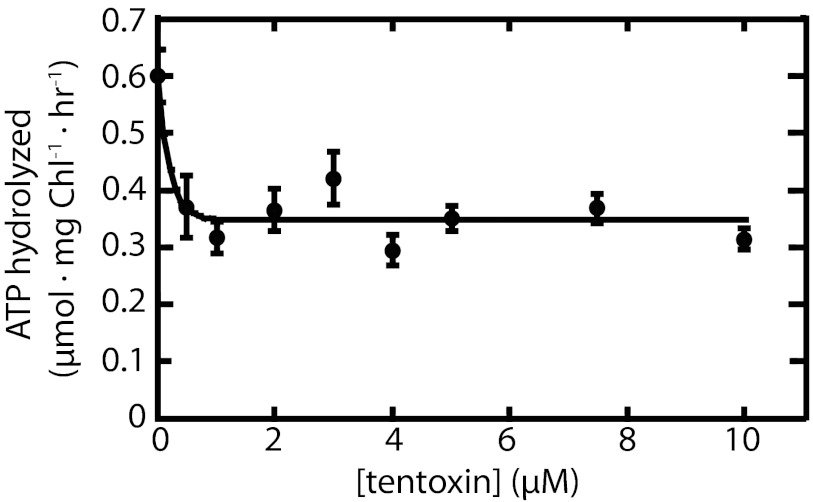
To increase the signal-to-noise ratio of the measurement of the ATP hydrolyzed during protein import, we wanted to minimize the intrinsic background ATPase activity manifested in our isolated chloroplasts. As a first step toward this goal, we examined the effect of tentoxin on the intrinsic rate of ATP hydrolysis in the absence of a protein import substrate. Tentoxin is a well-characterized inhibitor of the reversible chloroplast CF1/CF0 ATPase responsible for photophosphorylation (18–21). Whereas the CF1/CF0 ATPase is relatively inactive in dark-adapted chloroplasts (22), it is nonetheless responsible for a low amount of ATP hydrolysis even in its nonactivated form. This is evidenced by the ability of exogenous ATP to create, through reverse proton pumping, a protonmotive force sufficient to support some protein transport on the Tat pathway (7). Fig. 1 shows that a low-background ATPase activity of 0.6 μmoles ATP hydrolyzed per milligram chlorophyll (Chl) per hour was measured in our samples in the absence of any inhibitors. Compared with the published activity of >100 μmoles, ATP hydrolyzed per milligram Chl per hour seen after coupling factor activation (22, 23), we can conclude that the CF1/CF0 ATPase is indeed in its inactive state. However, this background activity is in the same range as the signal ATPase activity for translocation that might be expected using reasonable assumptions concerning the rates of protein import and coupling stoichiometry. The background activity dropped by 30–50% when the chloroplasts were preincubated for 10 min at room temperature in the dark with 0.5 μM tentoxin and remained constant with higher concentrations of tentoxin up to 10 μM (Fig. 1) or with longer preincubation times (Fig. 2). Because micromolar concentrations of tentoxin are also required to inhibit the ATPase activity of the activated CF1/CF0 coupling factor (18), we can tentatively conclude that approximately half of the background ATPase activity observed in our samples is due to the action of this enzyme, albeit operating in its inactivated form.
Fig. 1.
Inhibition by tentoxin of background ATPase activity in chloroplasts. Chloroplasts were incubated at room temperature in the dark for 10 min in IB containing tentoxin as indicated. ATP mixed with [γ-32P]-ATP was then added to final concentration of 1 mM. Samples were withdrawn at 0 and 30 min after adding ATP. Reactions were stopped by adding an ice-cold acidic charcoal solution. After separating Pi from ATP and other nucleotides using charcoal (Materials and Methods), released [32P] was quantified by scintillation counting. Each point on the graph is the mean of at least three replicates; error bars represent SD.
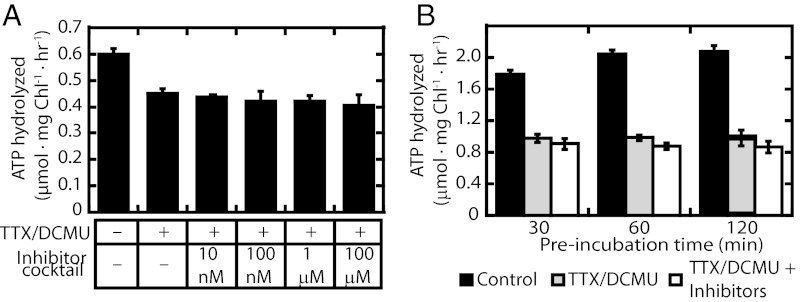
Fig. 2.
Effects of inhibitors of different chloroplast biochemical pathways on background ATPase activity. (A) Chloroplasts were resuspended in IB containing (or not) tentoxin (5 μM), DCMU (10 μM), and where indicated, a mixture of the inhibitors bispyribac-sodium, chloramphenicol, glyphosate, halosulfuron-methyl, penoxsulam, spectinomycin, imazethapyr, clomazone, malathion, and rifampicin, was added, bringing each to the specified final concentration. Chloroplasts were pretreated at room temperature in the dark for 15 min, and then ATP was added to 1 mM (cold ATP mixed with [32P]-ATP). Samples were removed at 0 and 60 min and ATPase activities were determined. (B) Chloroplasts in IB supplemented with 1 mM DTT were incubated at room temperature for 30, 60, and 120 min in the absence or presence of inhibitors. A total of 1 mM ATP ([32P] and cold) was added to start the ATPase reaction at room temperature in the dark. Samples were withdrawn at 0 and 30 min and ATPase activities were determined. Control, no inhibitors; inhibitor cocktail, each inhibitor mentioned at the indicated concentrations; TTX/DCMU, 5 μM tentoxin and 10 μM DCMU; TTX/DCMU + inhibitors, 5 μM tentoxin, 10 μM DCMU, each inhibitor mentioned above at 100 μM. Each point on the graphs is a mean of at least three replicates; error bars represent SD.
In an attempt to further suppress the intrinsic ATPase activity below that observed in the presence of tentoxin, we screened inhibitors of potential ATP-using biosynthetic pathways present in chloroplasts. Table S1 shows a list of the compounds tested, along with the metabolic pathways on which they act, which include RNA (24), protein, amino acid (25, 26) and carotenoid synthesis (27), photophosphorylation (28–34), the Calvin-Benson-Bassham cycle (33, 35), and specific selected enzymes (33, 36, 37). A useful inhibitor for our purposes would be one that lowered the background ATPase activity while having no (or a stimulating) effect on protein import. Accordingly, their effect on protein import was also measured for each potentially promising compound. As can be seen in Table S1, only tentoxin possessed the desirable combination of qualities when compounds were analyzed individually.
Fig. 2 shows experiments in which the background ATPase activity was measured using a combination of multiple inhibitors added together. A 25–50% reduction in ATP hydrolysis was observed with a combination of tentoxin and dichlorophenyl-dimethylurea (DCMU), the latter added to ensure the absence of any potentially confounding effects of the photosynthetic light reactions or recycling of released phosphate by photophosphorylation. The degree of ATPase inhibition with tentoxin and DCMU was relatively independent of the preincubation time. Further addition of a mixture of other inhibitors representing the pathways identified in Table S1 had only a minimal effect on the suppression of background ATPase activity. From these results, we decided to perform our translocation ATPase assays in the presence of tentoxin and DCMU only.
Effects of Time and [ATP] on the Import Reactions.
We next sought to establish the time frame and ATP concentration with which to perform our experiments that would minimize the background ATPase activity and maximize the amount of protein imported into chloroplasts. Recognizing that the intrinsic ATPase activity is likely to be dependent on the ATP concentration used, we determined this rate at 1, 2, and 3 mM ATP, concentrations that would support protein import (38–40). Fig. 3A shows that the background ATPase activity was indeed dependent on ATP concentration and remained fairly constant for up to 60 min. However, the rates measured at 2 and 3 mM ATP were not significantly different.
Fig. 3.
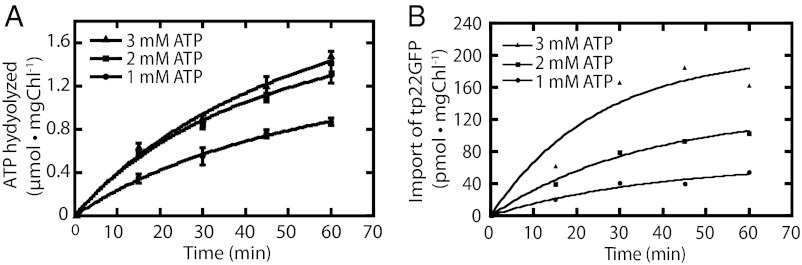
Effects of ATP concentration on background ATPase activity and protein import. Chloroplasts were incubated in the presence of 10 μM DCMU and 5 μM tentoxin at room temperature in the dark for 15 min. Buffer (A) or bacterially expressed tp22-GFP (B), 1 mM DTT and the indicated concentration of ATP were added to start the reactions. Samples were removed at 0, 15, 30, 45, and 60 min. (A) Background ATPase activity. Each point on the graph is the mean of at least three replicates; error bars represent SD. (B) Import assay.
When the rate of protein import into isolated chloroplasts was examined as a function of ATP concentration and time, we observed the expected dependence on the concentration of ATP consistent with previous estimates of a KM of 0.9 mM for this process (38) (Fig. 3B). We also observed that the import reaction continued in these experiments for at least 30 min without slowing, and sometimes even longer. This is longer than that usually reported for in vitro import reactions (38, 41), which may be due to our keeping the chloroplasts in complete darkness during the reaction, thereby avoiding the formation of potentially harmful reactive oxygen species. This phenomenon was not investigated further.
Measurement of the Chloroplast Translocation ATPase Activity.
In addition to minimizing the background ATPase activity operating in our isolated chloroplasts, an increase in the signal-to-noise ratio of the ATPase activity would be achieved by increasing the amount of protein imported. To this end, we incubated chloroplasts with high concentrations of precursors produced using three different techniques. In each instance the translocation ATPase activity was calculated as the amount of ATP hydrolyzed during the protein import reaction minus that hydrolyzed in identical reactions lacking only the import substrate, divided by the number of precursor molecules imported.
Table 1 shows a compilation of seven experiments using radiolabeled precursor of the small subunit of RuBisCO (prSSU) that was produced by overexpression in bacteria from a cDNA possessing a C-terminal His6 tag. After induction in the presence of [3H]-leucine, the precursor was solubilized in 8 M urea and purified by Ni2+ bead chromatography. Use of this precursor in chloroplast import experiments (Fig. S1), along with concurrent measurements of the chloroplast ATPase activity, yielded an average of 847 ± 135 ATP molecules hydrolyzed per protein imported.
Table 1.
Measurement of translocation ATPase activity with bacterially expressed [3H]-prSSU as substrate
| Experiment | Signal ATPase activity (nmol ATP hydrolyzed ⋅mg Chl−1⋅h−1) | Import rate (pmol prSSU imported ⋅mg Chl−1⋅h−1) | Translocation ATPase activity (ATP hydrolyzed per prSSU imported) |
| 1 | 131.0 | 125.0 | 1048 |
| 2 | 66.7 | 79.0 | 844 |
| 3a | 48.7 | 73.1 | 666 |
| 4b | 26.4 | 30.9 | 854 |
| 5 | 56.5 | 76.8 | 736 |
| 6 | 70.2 | 88.6 | 792 |
| 7 | 91.1 | 92.0 | 990 |
| Average | 847 ± 135 |
Chloroplasts were incubated at room temperature in the dark for 30 min in IB containing 5 μM tentoxin and 10 μM DCMU. ATP mixed with [γ-32P]-ATP was then added to the chloroplasts reaching a final concentration of 1 mM, which was then incubated in the dark at room temperature for 15 min. DTT was added to final concentration of 1 mM (except experiment 3). Import reactions were started by adding 42.6 pmol of bacterially expressed [3H]-prSSU (final concentration, 0.71 μM) or the resulting flow-through from concentrating [3H]-prSSU using Ultra-15 Centrifugal Filtration Devices (Amicon, cutoff 10 kDa). Samples were withdrawn at 0 and 30 min. Reactions assayed for ATPase activity were stopped by adding an ice-cold acidic charcoal solution (2% Norit charcoal, 0.25 M HCl, 0.25 M sodium pyrophosphate, and 0.25 M K2HPO4,) After separating Pi from ATP and other nucleotides using charcoal (Materials and Methods), released [32P] was quantified by scintillation counting. Identical samples analyzed for protein import were stopped by adding 600 μL ice-cold IB; chloroplasts were isolated immediately by centrifugation. The amount of imported SSU was analyzed by fluorography and quantified using ImageQuant. a[DTT] was 0.27 mM. b5 μM triphenyltin was included in the chloroplast pretreatment together with tentoxin and DCMU.
We next sought to expand these results by repeating the measurements with a different precursor generated in the RTS 100 continuous-flow in vitro translation system (Materials and Methods). This system yields relatively high amounts of soluble precursor, obviating the need for urea denaturation required of our bacterially expressed precursors before the import experiments. Additionally, precursors synthesized in this manner are added to the import reactions along with chaperones and other elements of the wheat germ translation mix that have been reported to be active in chloroplast import experiments (42). The radiolabeled precursor used in these experiments was GFP preceded by a stromal targeting signal comprising the prSSU transit peptide followed by 22 amino acids of the mature protein (tp22-GFP, Materials and Methods). After translation, both precursor and β-glucuronidase (GUS) translation products were filtered through Zeba Spin Desalting Columns (7K MWCO) to remove small molecules from translation mixtures. Those small molecules include nucleotides, free amino acids, and the substrate for the ATP regeneration system, creatine phosphate. Accordingly, no ATP originating from the wheat germ system contributed to the import reactions. A fluorograph showing the import of this precursor is presented in Fig. S2. Table 2 shows the results of three experiments performed with this precursor, which together yielded a translocation ATPase activity of 210 ± 117 ATP molecules hydrolyzed per protein imported. The translocation ATPase activity observed with this precursor is somewhat lower than that observed with bacterially overexpressed prSSU described previously, but when viewed next to the ∼11,500 ATP equivalents per protein translocated on the cpTat pathway, it is clear that they are both orders of magnitude lower.
Table 2.
Measurement of translocation ATPase activity with RTS in vitro translated [3H] tp22-GFP as substrate
| Experiment | Signal ATPase activity (nmol ATP hydrolyzed ⋅mg Chl−1⋅h−1) | Import rate (pmol tp22-GFP imported ⋅mg Chl−1⋅h−1) | Translocation ATPase activity (ATP hydrolyzed per tp22-GFP imported) |
| 1 | 18.5 | 117.0 | 158 |
| 2 | 11.8 | 92.1 | 128 |
| 3 | 33.6 | 97.7 | 344 |
| Average | 210 ± 117 |
Chloroplasts were incubated at room temperature in the dark for 30 min in IB containing 5 μM tentoxin and 10 μM DCMU. ATP mixed with [γ-32P]-ATP was then added to the chloroplasts forming a final concentration of 1 mM. Reactions were started by adding 36 pmol [3H]-tp22-GFP (final concentration, 0.60 μM) or [3H]-GUS (containing equal amounts of wheat germ as that in [3H]-tp22-GFP), both of which were translated in the RTS wheat germ system (Materials and Methods). Samples were withdrawn at 0 and 60 min. ATPase activity analysis and import assay were as described in Table 1 and in Materials and Methods.
The requirement for minimal media when overexpressing radiolabeled precursors in bacteria reduces the protein yield compared with that obtained with nonradiolabeled overexpression in complete media. Because we wanted to saturate the import machinery for our translocation ATPase assays, we performed a third set of experiments with nonradioactive tp22-GFP overexpressed in bacteria. In this case, we detected the import of the mature GFP protein by immunoblotting (Fig. 4 and Fig. S3). Quantitation of the immunoblots from three such import experiments, coupled with the simultaneous measurements of the import-dependent ATPase activity, is presented in Table 3. The average translocation ATPase activity measured in this way was 637 ± 259 ATP molecules hydrolyzed per protein imported, which is again within the same order of magnitude as that determined in the two previous experiments described previously.
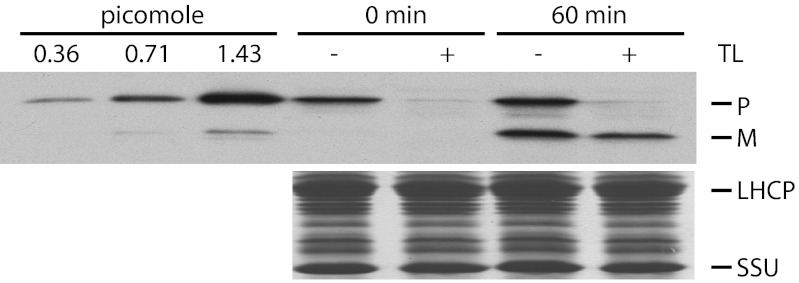
Fig. 4.
Protein import assayed by immunoblotting. Bacterially expressed tp22-GFP was incubated with isolated chloroplasts in the presence of 3 mM ATP in the dark at room temperature for 0 or 60 min, followed by thermolysin (TL, +) or mock protease treatment (−). Samples containing 4 μg Chl were separated by SDS/PAGE followed by immunoblotting against GFP antibody (Upper) or Coomassie staining (Lower). The left three lanes show the signal intensities from the indicated amounts of tp22-GFP loaded onto the gel. LHCP, light harvesting chlorophyll-binding protein; m, mature form of tp22-GFP; p, tp22-GFP; SSU, small subunit of RuBisCO.
Table 3.
Measurement of translocation ATPase activity with bacterially expressed tp22-GFP as substrate
| Experiment | Signal ATPase activity (nmol ATP hydrolyzed ⋅mg Chl−1⋅h−1) | Import rate (pmol tp22-GFP imported ⋅mg Chl−1⋅h−1) | Translocation ATPase activity (ATP hydrolyzed per tp22-GFP imported) |
| 1 | 67.9 | 75.8 | 896 |
| 2 | 161.7 | 254.2 | 636 |
| 3 | 212.9 | 562.0 | 379 |
| Average | 637 ± 259 |
Chloroplasts were incubated at room temperature in the dark for 15 min in IB containing 5 μM tentoxin and 10 μM DCMU. ATP mixed with [γ-32P]-ATP was then added to chloroplasts forming a final concentration of 3 mM, and then 1 mM DTT was added. Import reactions were started by adding 446 pmol bacterially expressed tp22-GFP (final concentration, 7.43 μM) or the resulting flow-through from concentrating tp22-GFP using Ultra-15 Centrifugal Filtration Devices (Amicon, cutoff 10 kDa). Samples were withdrawn at 0 and 30 min. Import reactions were stopped by adding 600 μL of cold IB followed by thermolysin treatment. Proteins were separated by SDS/PAGE followed by immunoblotting against GFP antibody. Analysis of ATPase activity was performed as described in Table 1.
Discussion
The amount of metabolic energy required to transport proteins across biological membranes is a potentially important issue given the large fraction of proteins in both eukaryotes and prokaryotes that function only after being targeting from the cytoplasm across one or more membranes. However, this has received little attention to date.
Although a translocation ATPase activity for the transport of prOmpA on the E. coli Sec pathway has been measured, the energetics of this pathway is complex, requiring obligatory ATP hydrolysis with optional assistance by the protonmotive force. Not surprisingly, dissipation of the protonmotive force caused the number of ATPs hydrolyzed per protein transported to increase from ∼700 to 5,000 or more (14). It was later determined that a single energy-dependent step for the Sec translocon results in the transport of about 50 amino acids of an unfolded linear substrate (43), which is consistent with an independent measurement of 5 ATP hydrolyzed per amino acid transported (44). One might argue that 5,000 ATP hydrolyzed per protein transported is indeed the true cost of this process given that no other energy input beyond ATP hydrolysis is available when the protonmotive force has been removed. However, this might not be correct if the role of the protonmotive force is to prevent retrograde motion of the protein in the translocon, as has been suggested (4, 14). In such a scenario, the translocation simply becomes inefficient in the absence of a protonmotive force because the translocon would be forced to repeat the same energy-consuming steps to achieve an overall forward motion of the protein across the membrane. On the other hand, if the protonmotive force biases the movement of proteins through the translocon, then this energy input must be accounted for in the calculation of the ΔGprotein transport. Accordingly, the actual cost of prOmpA transport through the bacterial Sec pathway must be between the energy content of 700 and 5,000 ATP molecules, some 33,600–240,000 kJ/mol protein transported [assuming a ΔGATP hydrolysis of −48 kJ/mol in E. coli (45)].
To our knowledge, the only calculation of the total ΔGprotein transport completed to date was carried out in our laboratory for the transport of OE17 across the thylakoid membrane on the cpTat pathway (16). This pathway uses only the protonmotive force to affect protein transport, with no contribution by ATP hydrolysis. By four independent means, we calculated the energy input for this reaction to be 690,000 kJ/mol protein transported, an energetic equivalent (17) of ∼11,500 ATP per protein. The reason for such a high-energy input for this pathway is unclear, but one may speculate that it is related to the fact that proteins are transported on the cpTat pathway in a folded conformation. It may also be that the transport of a single protein requires multiple attempts, many of which are unsuccessful but costly in terms of energy, much as speculated previously for the Sec pathway operating without the protonmotive force.
A nonparametric bootstrap analysis of all of the experiments described in Tables 1–3 yielded a mean of 647.2 ATP molecules hydrolyzed per protein imported across the two chloroplast envelope membranes through the Toc and Tic translocons, with an associated 95% bias-corrected and accelerated (BCA) bootstrap confidence interval (46) of 460.8–793.1 ATP hydrolyzed per protein transported. This large spread is a reflection of the relatively high background ATPase activity observed in our chloroplasts; the increase in ATP hydrolysis attributable to protein import was sometimes greater than 10%, but averaged 6.2%. It is for this reason that we repeated our measurements 13 times with precursors produced by three different methods.
The precursor concentration used in the experiments of Table 3 are about 10-fold higher than those for the experiments of Tables 1 and 2, resulting in an expected increase in the protein import rate, on average, in the former over the latter. At the same time, the rates of protein import–dependent ATP hydrolysis, the signal ATPase activity, roughly followed the protein import rates. This gave rise to a translocation ATPase activity, expressed as the ratio of the signal ATPase activity divided by the import rate, which is independent of the substrate concentration. This is exactly the behavior expected—more ATP hydrolysis with more protein import—if we were indeed measuring the ATPase activity of the protein import motor(s).
It is also noteworthy that although the same substrate was used in Tables 2 and 3, the translocation ATPase activity in the former appeared to be half of that in the latter. Although we do not know whether these differences are significant, it is perhaps germane that the substrate for the Table 2 experiments was produced in a wheat germ translation medium, which has been reported to contain additional factors such as 14-3-3 proteins and molecular chaperones (42, 47) that can stimulate protein import. Although additional experiments are required to explore this point further, our data are consistent with the view that these factors might serve in some way to increase the efficiency of energy utilization during the import process.
We earlier postulated (16) that the high cost of protein translocation on the cpTat pathway might be explained by one of two possibilities. In the first, we recognize that thylakoids often operate at light levels exceeding that which can be captured for photochemistry and so there may be little evolutionary pressure to maximize the efficiency of protonmotive force-dependent protein transport. In the second, we postulated that this high-energy input is just the inherent cost of protein translocation across biological membranes. Clearly, the figure of 647 ATPs hydrolyzed per protein imported measured here eliminates the second possibility and is closer to the value operating in the bacterial protonmotive force-assisted Sec pathway than to that in the chloroplast Tat pathway. Whether other protein translocation systems throughout the cell will operate with this lower value—for example, an energetic equivalent of less than 1,000 ATP per protein transported—remains to be determined, but we now have two transporters out of three examined in which this appears to be the case. One can calculate that the energy drained from chloroplasts for protein import at saturating rates (48–50) is still only about 0.6% of the total energy-producing potential of the organelle (51). If the assumption is made that the other protein translocation systems operating throughout the cell do so at rates and costs similar to that seen for the chloroplast envelope membranes, then it is possible for the ΔGprotein transport measured here to apply to these other systems and still stay well within the 25–30% of the cell’s total energy budget dedicated to protein synthesis and targeting (52, 53).
Materials and Methods
Plant Materials and Growth Conditions.
Pea (Pisum sativum, var. Little Marvel) seedlings were grown under 10/14 h light/dark periods at 23 °C. Seedlings were harvested 8–10 d after germination.
Isolation of Chloroplasts and Treatment with Inhibitors.
Intact chloroplasts were isolated essentially as described elsewhere (40, 54). Where indicated, chloroplasts were incubated with inhibitors at room temperature in darkness; concentrations of inhibitors and time lengths of treatments are indicated in Tables 1–3 and Figs. 1–3.
Synthesis of Precursor Proteins.
Bacterially expressed radiolabeled prSSU.
A cDNA encoding prSSU from pea was cloned into a pET161/GW/D-TOPO vector (Invitrogen) to produce an in-frame six-histidine tag at the C terminus. [3H]-prSSU was expressed in E. coli strain BL21 CodonPlus as described previously (16) and was purified using Ni-NTA agarose according to the manufacturer’s instructions (Qiagen) and concentrated using Ultra-15 Centrifugal Filtration Devices, again according to the manufacturer’s instructions (Amicon).
In vitro translated tp22-GFP.
tp22-GFP is a chimeric protein created by fusing the coding region of the prSSU transit peptide followed by 22 amino acids from the mature SSU protein to that for GFP. The DNA construct was cloned into pET23a (Invitrogen). [3H]-labeled tp22-GFP was produced by sequential in vitro transcription and translation. The linearized plasmid was transcribed using T7 RNA polymerase. Long-term in vitro translation was conducted using the RTS 100 Wheat Germ CECF Kit (Prime 5, Inc) according to the manual, with modifications. The translation was performed at 24 °C for 24 h with shaking at 900 rpm in a Thermomixer R (Eppendorf). Control vector provided with the kit was used for translation of [3H]-GUS, which served as a mock substrate in the translocation ATPase assays. Both precursor and GUS translation mixtures were filtered through Zeba Desalting Columns (7K MWCO) to remove small molecules from translation mixtures according to the manufacturer’s manual (Thermo Scientific). The efficiency of removal of small molecules by this procedure is high because 99% of unused [3H]-leucine in the translation mixtures was removed.
Bacterially expressed tp22-GFP.
Nonradiolabeled tp22-GFP was also expressed in E. coli strain BL21 CodonPlus and purified with Ni-NTA agarose beads. Protein concentrations were determined by comparison with BSA standards via SDS/PAGE; bands were quantified using ImageQuant.
Measurement of ATPase Activity.
To avoid ATP synthesis by chloroplasts, all experiments measuring translocation ATPase activity were carried out in very dim room light. Isolated chloroplasts were resuspended in Import Buffer (IB, 50 mM K-Tricine, 0.33 M sorbitol, and 3 mM MgCl2) and kept in the dark on ice at least for 1 h before use to deplete endogenous ATP. Unless indicated, chloroplasts (20 μg Chl) were incubated in presence of 10 μM DCMU and 5 μM tentoxin at room temperature for 15 min in darkness. Concentrations and conditions for other potential inhibitors of chloroplast ATPase activities are indicated in Table S1, Table 1, and Fig. 2. After treatment of chloroplasts with inhibitors, ATP containing ∼0.05 μCi of [γ-32P]-ATP was added to a final concentration of 1–3 mM; samples remained on ice. Protein import reactions were initiated by adding 2–10 μL of precursor to the pretreated chloroplasts and the reactions were transferred to room temperature. Samples were withdrawn for analysis at 0 and 30 or 60 min. Differences in the reaction times of different samples were less than 7 s.
ATPase activity was measured using a charcoal-based method as described (55, 56), with minor modifications. The chloroplast ATPase reactions were stopped by adding 500 μL of charcoal solution [2% (wt/vol) Norit charcoal (Sigma), 0.25 M HCl, 0.25 M sodium pyrophosphate, and 0.25 M K2HPO4,]. The samples were mixed and incubated on ice for 5 min. Phosphate (Pi) released from [γ-32P]-ATP was separated from ATP absorbed on the charcoal by centrifugation at 16,000 × g at 4 °C for 5 min. A total of 140–200 μL of supernatant was added to 4 mL of CytoScint scintillation mixture (Fisher Scientific) and analyzed in a Beckman LS6500 scintillation counter. Samples were counted for 10 min to minimize systemic errors from counting. Measured disintegrations per minute (DPM) numbers (N) were corrected (Nc) based on the elapsed times (t) using the equation: 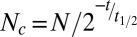 , where t1/2 is the half-life of [32P].
, where t1/2 is the half-life of [32P].
The purity of [3H]-PrSSU and tp22GFP is shown in Fig. S4. When assayed on their own without chloroplasts, the bacterially expressed precursors and flow-through solutions from the concentration devices displayed no more than 4% of the ATPase activities of the corresponding complete (plus chloroplast) reactions.
Protein Import Assays and Thermolysin Treatment.
Protein import reactions were carried out in parallel with ATPase activity measurements, using the same batch of isolated chloroplasts and the same master mixes. As mentioned previously, samples to be assayed for protein import were withdrawn at 0 and 30 or 60 min after addition of precursor protein. Reactions were mixed with 600 μL of ice-cold IB and immediately centrifuged in a microfuge at 4 °C. Chloroplasts were then subjected to SDS/PAGE, followed by either fluorography or immunoblotting. In some experiments, thermolysin digestion of nontranslocated precursor was conducted after the import reaction by addition of 200 μg thermolysin/mL and 5 mM CaCl2 in IB; samples were kept on ice in darkness for 15–30 min. The protease reaction was stopped by the addition of 600 μL of ice-cold IB supplemented with 50 mM EDTA.
The quantities of proteins imported were determined on fluorographs or immunoblots by comparison with known amounts of precursor added to the import reaction. Bands on film were quantitated using ImageQuant, and corrections were applied to account for different numbers of [3H]leucines in the precursor and mature proteins. All quantifications were performed in the linear response range of the films. An extended description of the quantitation procedures is given in SI Text.
Supplementary Material
Acknowledgments
We thank John Perea and John Wu for constructing the tp22-GFP and prSSU-pET161 clones; the members in the S.M.T. laboratory, especially Dr. Shari Lo for valuable discussion, Curtis Tom for assistance in preparing Fig. 1, and Jessica Nguyen and Quynh Tram Phan for their excellent technical assistance. We also thank Drs. Albert J. Fisher and Hagai Yasuor for the gifts of inhibitors: bispyribac sodium, glyphosate, halosulfuron-methyl, penoxsulam, imazethapyr, clomazone, and malathion. We especially thank Dr. Niels G. Waller for performing the nonparametric BCA Bootstrap analysis of our data and for many discussions relating to its meaning. This work was funded by the Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences of the US Department of Energy through Grant DE-FG02-03ER15405.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1115886110/-/DCSupplemental.
References
- 1.Schatz G, Dobberstein B. Common principles of protein translocation across membranes. Science. 1996;271(5255):1519–1526. doi: 10.1126/science.271.5255.1519. [DOI] [PubMed] [Google Scholar]
- 2.Mori T, Ishitani R, Tsukazaki T, Nureki O, Sugita Y. Molecular mechanisms underlying the early stage of protein translocation through the Sec translocon. Biochemistry. 2010;49(5):945–950. doi: 10.1021/bi901594w. [DOI] [PubMed] [Google Scholar]
- 3.Driessen AJ, Nouwen N. Protein translocation across the bacterial cytoplasmic membrane. Annu Rev Biochem. 2008;77:643–667. doi: 10.1146/annurev.biochem.77.061606.160747. [DOI] [PubMed] [Google Scholar]
- 4.Alder NN, Theg SM. Energy use by biological protein transport pathways. Trends Biochem Sci. 2003;28(8):442–451. doi: 10.1016/S0968-0004(03)00167-1. [DOI] [PubMed] [Google Scholar]
- 5.Johnson AE, Haigh NG. The ER translocon and retrotranslocation: Is the shift into reverse manual or automatic? Cell. 2000;102(6):709–712. doi: 10.1016/s0092-8674(00)00059-3. [DOI] [PubMed] [Google Scholar]
- 6.Kutay U, Ahnert-Hilger G, Hartmann E, Wiedenmann B, Rapoport TA. Transport route for synaptobrevin via a novel pathway of insertion into the endoplasmic reticulum membrane. EMBO J. 1995;14(2):217–223. doi: 10.1002/j.1460-2075.1995.tb06994.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cline K, Ettinger WF, Theg SM. Protein-specific energy requirements for protein transport across or into thylakoid membranes. Two lumenal proteins are transported in the absence of ATP. J Biol Chem. 1992;267(4):2688–2696. [PubMed] [Google Scholar]
- 8.Braun NA, Davis AW, Theg SM. The chloroplast Tat pathway utilizes the transmembrane electric potential as an energy source. Biophys J. 2007;93(6):1993–1998. doi: 10.1529/biophysj.106.098731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bolender N, Sickmann A, Wagner R, Meisinger C, Pfanner N. Multiple pathways for sorting mitochondrial precursor proteins. EMBO Rep. 2008;9(1):42–49. doi: 10.1038/sj.embor.7401126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mokranjac D, Neupert W. Energetics of protein translocation into mitochondria. Biochim Biophys Acta. 2008;1777(7-8):758–762. doi: 10.1016/j.bbabio.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 11.Neupert W, Brunner M. The protein import motor of mitochondria. Nat Rev Mol Cell Biol. 2002;3(8):555–565. doi: 10.1038/nrm878. [DOI] [PubMed] [Google Scholar]
- 12.Sardis MF, Economou A. SecA: A tale of two protomers. Mol Microbiol. 2010;76(5):1070–1081. doi: 10.1111/j.1365-2958.2010.07176.x. [DOI] [PubMed] [Google Scholar]
- 13.Schiebel E, Driessen AJ, Hartl F-U, Wickner W. Delta mu H+ and ATP function at different steps of the catalytic cycle of preprotein translocase. Cell. 1991;64(5):927–939. doi: 10.1016/0092-8674(91)90317-r. [DOI] [PubMed] [Google Scholar]
- 14.Driessen AJ. Precursor protein translocation by the Escherichia coli translocase is directed by the protonmotive force. EMBO J. 1992;11(3):847–853. doi: 10.1002/j.1460-2075.1992.tb05122.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lill R, et al. SecA protein hydrolyzes ATP and is an essential component of the protein translocation ATPase of Escherichia coli. EMBO J. 1989;8(3):961–966. doi: 10.1002/j.1460-2075.1989.tb03458.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alder NN, Theg SM. Energetics of protein transport across biological membranes. A study of the thylakoid DeltapH-dependent/cpTat pathway. Cell. 2003;112(2):231–242. doi: 10.1016/s0092-8674(03)00032-1. [DOI] [PubMed] [Google Scholar]
- 17.Giersch C, et al. Energy charge, phosphorylation potential and proton motive force in chloroplasts. Biochim Biophys Acta. 1980;590(1):59–73. doi: 10.1016/0005-2728(80)90146-2. [DOI] [PubMed] [Google Scholar]
- 18.Avni A, et al. Tentoxin sensitivity of chloroplasts determined by codon 83 of beta subunit of proton-ATPase. Science. 1992;257(5074):1245–1247. doi: 10.1126/science.1387730. [DOI] [PubMed] [Google Scholar]
- 19.Tucker WC, Du Z, Gromet-Elhanan Z, Richter ML. Formation and properties of hybrid photosynthetic F1-ATPases. Demonstration of different structural requirements for stimulation and inhibition by tentoxin. Eur J Biochem. 2001;268(7):2179–2186. doi: 10.1046/j.1432-1327.2001.02110.x. [DOI] [PubMed] [Google Scholar]
- 20.Pavlova P, et al. Complete inhibition and partial Re-activation of single F1-ATPase molecules by tentoxin: New properties of the re-activated enzyme. J Biol Chem. 2004;279(11):9685–9688. doi: 10.1074/jbc.C400014200. [DOI] [PubMed] [Google Scholar]
- 21.Santolini J, Haraux F, Sigalat C, Moal G, André F. Kinetic analysis of tentoxin binding to chloroplast F1-ATPase. A model for the overactivation process. J Biol Chem. 1999;274(2):849–858. doi: 10.1074/jbc.274.2.849. [DOI] [PubMed] [Google Scholar]
- 22.McCarty RE. The decay of the ATPase activity of light plus thiol-activated thylakoid membranes in the dark. J Bioenerg Biomembr. 2006;38(1):67–74. doi: 10.1007/s10863-006-9007-4. [DOI] [PubMed] [Google Scholar]
- 23.Sherman PA, Wimmer MJ. Kinetic effects of chemical and physical uncoupling on the energy-transducing ATPase from spinach chloroplasts. Eur J Biochem. 1983;136(3):539–543. doi: 10.1111/j.1432-1033.1983.tb07774.x. [DOI] [PubMed] [Google Scholar]
- 24.Surzycki SJ. Genetic functions of the chloroplast of Chlamydomonas reinhardi: Effect of rifampin on chloroplast DNA-dependent RNA polymerase. Proc Natl Acad Sci USA. 1969;63(4):1327–1334. doi: 10.1073/pnas.63.4.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Merotto A, Jr, et al. Cross-resistance to herbicides of five ALS-inhibiting groups and sequencing of the ALS gene in Cyperus difformis L. J Agric Food Chem. 2009;57(4):1389–1398. doi: 10.1021/jf802758c. [DOI] [PubMed] [Google Scholar]
- 26.Yasuor H, et al. Mechanism of resistance to penoxsulam in late watergrass [Echinochloa phyllopogon (Stapf) Koss.] J Agric Food Chem. 2009;57(9):3653–3660. doi: 10.1021/jf8039999. [DOI] [PubMed] [Google Scholar]
- 27.Yasuor H, TenBrook PL, Tjeerdema RS, Fischer AJ. Responses to clomazone and 5-ketoclomazone by Echinochloa phyllopogon resistant to multiple herbicides in Californian rice fields. Pest Manag Sci. 2008;64(10):1031–1039. doi: 10.1002/ps.1604. [DOI] [PubMed] [Google Scholar]
- 28.Knott TG, Robinson C. The secA inhibitor, azide, reversibly blocks the translocation of a subset of proteins across the chloroplast thylakoid membrane. J Biol Chem. 1994;269(11):7843–7846. [PubMed] [Google Scholar]
- 29.Bowler MW, Montgomery MG, Leslie AG, Walker JE. How azide inhibits ATP hydrolysis by the F-ATPases. Proc Natl Acad Sci USA. 2006;103(23):8646–8649. doi: 10.1073/pnas.0602915103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McCarty DR, Keegstra K, Selman BR. Characterization and localization of the ATPase associated with pea chloroplast envelope membranes. Plant Physiol. 1984;76(3):584–588. doi: 10.1104/pp.76.3.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bowman BJ, Slayman CW. The effects of vanadate on the plasma membrane ATPase of Neurospora crassa. J Biol Chem. 1979;254(8):2928–2934. [PubMed] [Google Scholar]
- 32.Van der Bend RL, Duetz W, Colen AM, Van Dam K, Berden JA. Differential effects of triphenyltin and 8-azido-ATP on the ATP synthesis, ATP-Pi exchange, and ATP hydrolysis in liposomes containing ATP synthase and bacteriorhodopsin. Arch Biochem Biophys. 1985;241(2):461–471. doi: 10.1016/0003-9861(85)90571-5. [DOI] [PubMed] [Google Scholar]
- 33.Bamberger ES, Avron M. Site of action of inhibitors of carbon dioxide assimilation by whole lettuce chloroplasts. Plant Physiol. 1975;56(4):481–485. doi: 10.1104/pp.56.4.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heldt HW, Chon CJ, Maronde D. Role of orthophosphate and other factors in the regulation of starch formation in leaves and isolated chloroplasts. Plant Physiol. 1977;59(6):1146–1155. doi: 10.1104/pp.59.6.1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Flügge UI, Freisl M, Heldt HW. Balance between metabolite accumulation and transport in relation to photosynthesis by isolated spinach chloroplasts. Plant Physiol. 1980;65(4):574–577. doi: 10.1104/pp.65.4.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lienhard GE, Secemski II. P 1, P 5-Di(adenosine-5′)pentaphosphate, a potent multisubstrate inhibitor of adenylate kinase. J Biol Chem. 1973;248(3):1121–1123. [PubMed] [Google Scholar]
- 37.Robinson SP, Portis AR., Jr Adenosine triphosphate hydrolysis by purified rubisco activase. Arch Biochem Biophys. 1989;268(1):93–99. doi: 10.1016/0003-9861(89)90568-7. [DOI] [PubMed] [Google Scholar]
- 38.Flügge UI, Hinz G. Energy dependence of protein translocation into chloroplasts. Eur J Biochem. 1986;160(3):563–570. doi: 10.1111/j.1432-1033.1986.tb10075.x. [DOI] [PubMed] [Google Scholar]
- 39.Scott SV, Theg SM. A new chloroplast protein import intermediate reveals distinct translocation machineries in the two envelope membranes: Energetics and mechanistic implications. J Cell Biol. 1996;132(1-2):63–75. doi: 10.1083/jcb.132.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Theg SM, Bauerle C, Olsen LJ, Selman BR, Keegstra K. Internal ATP is the only energy requirement for the translocation of precursor proteins across chloroplastic membranes. J Biol Chem. 1989;264(12):6730–6736. [PubMed] [Google Scholar]
- 41.Clark SA, Theg SM. A folded protein can be transported across the chloroplast envelope and thylakoid membranes. Mol Biol Cell. 1997;8(5):923–934. doi: 10.1091/mbc.8.5.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qbadou S, et al. The molecular chaperone Hsp90 delivers precursor proteins to the chloroplast import receptor Toc64. EMBO J. 2006;25(9):1836–1847. doi: 10.1038/sj.emboj.7601091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van der Wolk JP, de Wit JG, Driessen AJ. The catalytic cycle of the Escherichia coli SecA ATPase comprises two distinct preprotein translocation events. EMBO J. 1997;16(24):7297–7304. doi: 10.1093/emboj/16.24.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tomkiewicz D, Nouwen N, van Leeuwen R, Tans S, Driessen AJ. SecA supports a constant rate of preprotein translocation. J Biol Chem. 2006;281(23):15709–15713. doi: 10.1074/jbc.M600205200. [DOI] [PubMed] [Google Scholar]
- 45.Tran QH, Unden G. Changes in the proton potential and the cellular energetics of Escherichia coli during growth by aerobic and anaerobic respiration or by fermentation. Eur J Biochem. 1998;251(1-2):538–543. doi: 10.1046/j.1432-1327.1998.2510538.x. [DOI] [PubMed] [Google Scholar]
- 46.Efron B, Tibshirani R. An Introduction to the Bootstrap. Boca Raton, FL: Chapman & Hall/CRC; 1994. p. p 456. [Google Scholar]
- 47.May T, Soll J. 14-3-3 proteins form a guidance complex with chloroplast precursor proteins in plants. Plant Cell. 2000;12(1):53–64. doi: 10.1105/tpc.12.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cline K, Henry R, Li C, Yuan J. Multiple pathways for protein transport into or across the thylakoid membrane. EMBO J. 1993;12(11):4105–4114. doi: 10.1002/j.1460-2075.1993.tb06094.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dabney-Smith C, van Den Wijngaard PW, Treece Y, Vredenberg WJ, Bruce BD. The C terminus of a chloroplast precursor modulates its interaction with the translocation apparatus and PIRAC. J Biol Chem. 1999;274(45):32351–32359. doi: 10.1074/jbc.274.45.32351. [DOI] [PubMed] [Google Scholar]
- 50.Pilon M, Weisbeek PJ, de Kruijff B. Kinetic analysis of translocation into isolated chloroplasts of the purified ferredoxin precursor. FEBS Lett. 1992;302(1):65–68. doi: 10.1016/0014-5793(92)80286-p. [DOI] [PubMed] [Google Scholar]
- 51. Hall DO (1976) The coupling of photophosphorylation to electron transport in isolated chloroplasts. Topics in Photosynthesis: Vol 1 the Intact Chloroplast, ed Barber J (Elsevier Scientific Publishing Company, Amsterdam), Vol 1, pp 135–170.
- 52.Rolfe DF, Brown GC. Cellular energy utilization and molecular origin of standard metabolic rate in mammals. Physiol Rev. 1997;77(3):731–758. doi: 10.1152/physrev.1997.77.3.731. [DOI] [PubMed] [Google Scholar]
- 53.Harper M-E, Antoniou A, Bevilacqua L, Bezaire V, Monemdjou S. Cellular energy expenditure and the importance of uncoupling. J Anim Sci. 2002;80(E-Suppl 2):E90–E97. [Google Scholar]
- 54.Shi LX, Theg SM. A stromal heat shock protein 70 system functions in protein import into chloroplasts in the moss Physcomitrella patens. Plant Cell. 2010;22(1):205–220. doi: 10.1105/tpc.109.071464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bais R. A rapid and sensitive radiometric assay for adenosine triphosphatase activity using Cerenkov radiation. Anal Biochem. 1975;63(1):271–273. doi: 10.1016/0003-2697(75)90215-8. [DOI] [PubMed] [Google Scholar]
- 56.Zhang XP, Lee KI, Solinger JA, Kiianitsa K, Heyer WD. Gly-103 in the N-terminal domain of Saccharomyces cerevisiae Rad51 protein is critical for DNA binding. J Biol Chem. 2005;280(28):26303–26311. doi: 10.1074/jbc.M503244200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.