Abstract
The oncogenic transcription factor c-Myc causes transformation and tumorigenesis, but it can also induce apoptotic cell death. Although tumor suppressors are necessary for c-Myc to induce apoptosis, the pathways and mechanisms are unclear. To further understand how c-Myc switches from an oncogenic protein to an apoptotic protein, we examined the mechanism of p53-independent c-Myc–induced apoptosis. We show that the tumor suppressor protein ARF mediates this switch by inhibiting ubiquitylation of the c-Myc transcriptional domain (TD). Whereas TD ubiquitylation is critical for c-Myc canonical transcriptional activity and transformation, inhibition of ubiquitylation leads to the induction of the noncanonical c-Myc target gene, Egr1, which is essential for efficient c-Myc–induced p53-independent apoptosis. ARF inhibits the interaction of c-Myc with the E3 ubiquitin ligase Skp2. Overexpression of Skp2, which occurs in many human tumors, inhibits the recruitment of ARF to the Egr1 promoter, leading to inhibition of c-Myc–induced apoptosis. Therapeutic strategies could be developed to activate this intrinsic apoptotic activity of c-Myc to inhibit tumorigenesis.
Keywords: oncogene, cancer
The c-Myc protein plays a pivotal role in cell growth, cell cycle progression, and tumorigenesis. Although oncogenic c-Myc protein has been assigned several molecular functions, strong evidence suggests that c-Myc mediates its diverse biological functions through its ability to transcriptionally regulate target genes (1). c-Myc can activate and repress many target genes through several mechanisms, but the best characterized is activation of targets containing the canonical CACGTG Myc E box DNA-binding motif. Both the C-terminal basic region/helix–loop–helix/leucine zipper (b/HLH/LZ) domain and the N-terminal transcriptional domain (TD) are essential for activation and repression of target genes. The TD is not well defined but includes two conserved regions termed Myc box (MB)I and MBII that are necessary for full transcriptional activity (2).
Oncogenic activation of c-Myc occurs when c-Myc is overexpressed or deregulated in tumors. Oncogenic c-Myc causes transformation in cultured cells and tumorigenesis in animal models. In apparent contradiction, oncogenic c-Myc also induces growth inhibition or apoptosis through both p53-dependent and -independent mechanisms that prevent tumorigenicity, but these mechanisms are not well understood (2). The tumor suppressor protein ARF, which is induced by oncogenic c-Myc, plays roles in both mechanisms. In a p53-dependent mechanism, ARF inhibits the E3 ubiquitin ligase Mdm2, leading to stabilization of p53 (3). Independently of p53, on c-Myc activation, ARF becomes nucleoplasmic and directly binds to c-Myc protein. We previously demonstrated that ARF blocks the ability of c-Myc to activate transcription of canonical target genes at promoters and inhibits c-Myc–induced hyperproliferation and transformation (4). However, ARF is essential for p53-independent c-Myc–induced apoptosis (4–7). Recently, we have shown that ARF is also necessary for induction of the direct noncanonical c-Myc target gene Egr1, which is a transcription factor necessary and sufficient for p53-independent c-Myc–induced apoptosis (6). Therefore, ARF binding essentially switches c-Myc from a proliferative to an apoptotic protein.
Here we show that ARF controls this apoptotic switch by inhibiting ubiquitylation of the c-Myc TD. In these studies, we demonstrate that TD ubiquitylation is critical for canonical target gene induction and transformation. Surprisingly, loss of TD ubiquitylation is sufficient for induction of the noncanonical Egr1 target gene and apoptosis. Furthermore, we show that the loss of TD ubiquitylation caused by ARF is due to the inhibition of the interaction of c-Myc with the oncogenic E3 ubiquitin ligase Skp2. Inhibition of endogenous Skp2 expression and ubiquitylation mimics the action of ARF, leading to enhanced Egr1 expression and c-Myc–mediated apoptosis. Conversely, overexpression of Skp2, which commonly occurs in many human tumors (8), inhibits the recruitment of ARF to the Egr1 promoter, leading to inhibition of Egr1 expression and subsequent c-Myc–induced apoptosis. These studies suggest that ubiquitylation controls the intrinsic transcriptional and biological activity of c-Myc.
Results
ARF Inhibits Skp2-Mediated Ubiquitylation of the c-Myc TD Through Competitive Interaction.
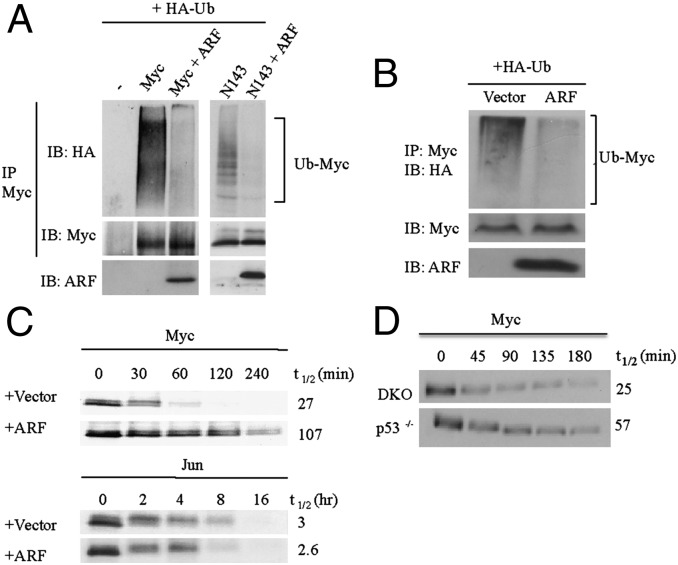
ARF is known to interact with and inhibit the activities of several critical cellular proteins in addition to c-Myc, such as Mdm2 and E2F (9). Because ARF is known to enhance the ubiquitylation and proteolysis of several of its binding partners, including E2F (10), we investigated whether this mechanism was responsible for ARF inhibition of c-Myc activity. In vivo ubiquitylation analyses of both full-length c-Myc protein and an N-terminal c-Myc fragment with or without ARF expression were performed. Surprisingly, the analyses demonstrated that ARF inhibited c-Myc ubiquitylation (Fig. 1A). ARF also inhibited endogenous c-Myc ubiquitylation (Fig. 1B). We then determined the effects of ARF on c-Myc proteolysis by performing cycloheximide chase assays. c-Myc protein was coexpressed with ARF or a vector control, cells were treated with cycloheximide to inhibit protein synthesis, and c-Myc protein degradation was monitored over time. Consistent with the inhibition of ubiquitylation, ARF inhibited the proteolysis of c-Myc (Fig. 1C, Upper). In contrast, there was no stabilization of c-Jun protein when coexpressed with ARF, suggesting that ARF does not cause a generalized inhibition of cellular proteolysis (Fig. 1C, Lower). We previously showed that high levels of endogenous ARF in p53−/− mouse embryo fibroblasts (MEFs) inhibits transformation induced by oncogenic c-Myc using a hydroxytamoxifen (OHT)-activated fusion c-MycER protein (4). Consistent with the results from transient overexpression of ARF and Myc, the proteolysis of c-MycER in these p53−/− MEFs was inhibited compared with c-MycER in ARF−/−p53−/− (DKO) MEFs (Fig. 1D). In addition, ubiquitylation of c-MycER protein was lower relative to total c-Myc protein in p53−/− MEFs compared with DKO MEFs as demonstrated by in vivo ubiquitylation analyses (Fig. S1A). Taken together, these results suggest that ARF inhibits ubiquitylation and proteolysis of c-Myc protein.
Fig. 1.
ARF inhibits c-Myc TD ubiquitylation and proteolysis. (A) In vivo ubiquitylation assays of c-Myc in Cos7 cells transiently transfected with HA-tagged ubiquitin and empty vector, c-Myc, or c-MycN143, with or without ARF. (B) In vivo ubiquitylation assay of endogenous c-Myc in HeLa cells transiently transfected with HA-Ub and ARF or empty vector. (C) Cycloheximide chase analyses in Cos7 cells cotransfected with c-Myc or Jun and empty vector or ARF. (D) Cycloheximide chase analyses of c-MycER in DKO MycER and p53−/− MycER MEFs treated with 2 μM OHT for 24 h.
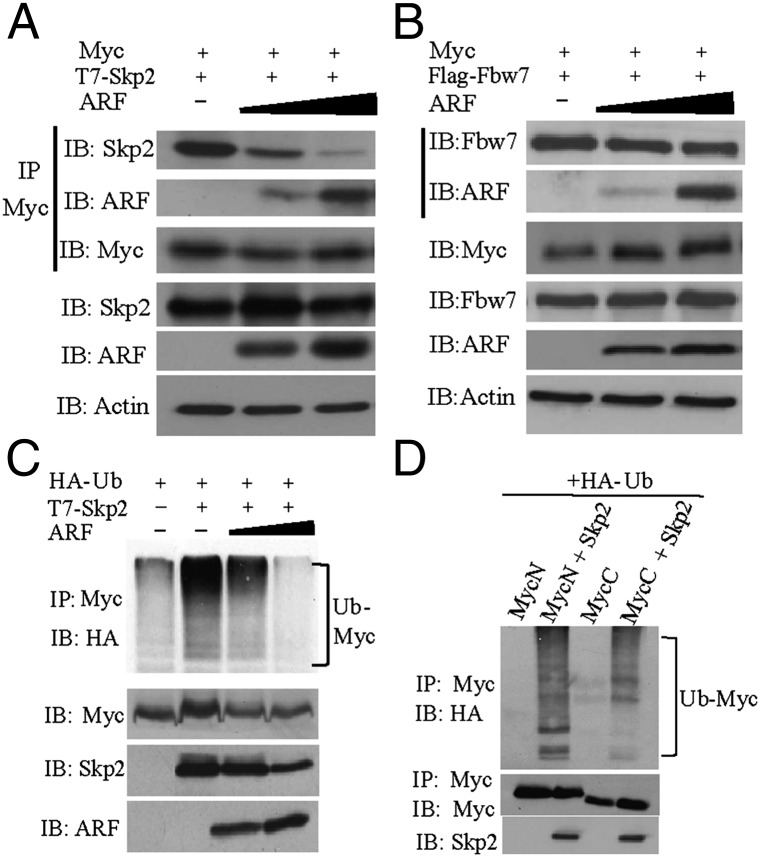
One explanation for the inhibition of ubiquitylation by ARF is that ARF inhibits one or more E3 ubiquitin ligases. Considering that the E3 ubiquitin ligase Skp2 interacts with similar c-Myc domains as ARF and has opposing effects on c-Myc target gene expression and cell cycle progression compared with ARF (4, 11, 12), we investigated whether ARF inhibits the interaction of Skp2 with c-Myc. Skp2 and either full-length c-Myc or the N-terminal 167 amino acids (c-MycN167) containing the TD were coexpressed, and an in vivo coimmunoprecipitation (coIP) assay was performed using c-Myc antibody. To determine whether ARF competes with Skp2 for c-Myc binding, increasing amounts of ARF were expressed in a competitive coIP assay. As ARF levels increased, the amount of Skp2 that coimmunoprecipitated with the c-Myc TD decreased dramatically, whereas the amount of ARF that coimmunoprecipitated with c-Myc increased (Fig. 2A). Similar results were found when the full-length c-Myc protein was used (Fig. S1B). To determine whether ARF inhibits the binding of another c-Myc E3 ubiquitin ligase, we performed the same competitive coIP experiment using Fbw7α, which also interacts with the TD of c-Myc (13–15). In contrast to the inhibition of Skp2 binding, increasing amounts of ARF did not influence the interaction of Fbw7α with c-Myc (Fig. 2B). We next determined whether ARF inhibited the Skp2-mediated ubiquitylation of c-Myc. First we verified that overexpression of Skp2 efficiently ubiquitylates endogenous c-Myc (Fig. S1C), which was previously observed (11). We then expressed Skp2 with increasing amounts of ARF and observed that the ubiquitylation of endogenous c-Myc greatly decreased (Fig. 2C). These results suggest that ARF competitively inhibits the interaction of Skp2 with the TD of c-Myc, resulting in less ubiquitylation of c-Myc.
Fig. 2.
ARF inhibits Skp2 binding to c-Myc and Skp2-mediated c-Myc ubiquitylation. (A) Coimmunoprecipitation performed with anti-Myc followed by immunoblot analyses in Cos7 cells transfected with constant amounts of T7-Skp2 and c-MycN167 and increasing amounts of ARF. (B) Coimmunoprecipitation performed with anti-Myc followed by immunoblot analyses in Cos7 cells transfected with Flag-Fbw7α, c-MycN167, and increasing amounts of ARF. Fbw7α was detected using anti-Flag. (C) In vivo ubiquitylation assay of endogenous c-Myc in HeLa cells transfected with HA-ubiquitin with or without T7-Skp2 and increasing amounts of ARF. (D) In vivo ubiquitylation assay of HA-tagged N- or C-terminal c-Myc in Cos7 cells transfected with HA-ubiquitin and either empty vector or Skp2.
Although Skp2 has been shown to interact with both the TD and HLH/LZ domains of c-Myc (11, 12), the sites of Skp2-mediated ubiquitylation have not been identified. To determine whether Skp2 does ubiquitylate the c-Myc TD and to determine the relative ubiquitylation of c-Myc N and C-terminal domains by Skp2, we expressed the two different halves of c-Myc with and without Skp2 and examined their ubiquitylation status. Although the C-terminal half of mouse c-Myc has the majority of the lysine residues (20 of 27 total), the N-terminal domain was more efficiently ubiquitylated by Skp2 (Fig. 2D). Skp2-induced ubiquitylation was approximately threefold greater on the N-terminal half of c-Myc compared with the C-terminal half. The c-Myc167 N-terminal fragment, which includes the TD, was also efficiently ubiquitylated by Skp2 (Fig. S1D).
Ubiquitylation of the c-Myc TD Is Essential for c-Myc–Induced Transformation.
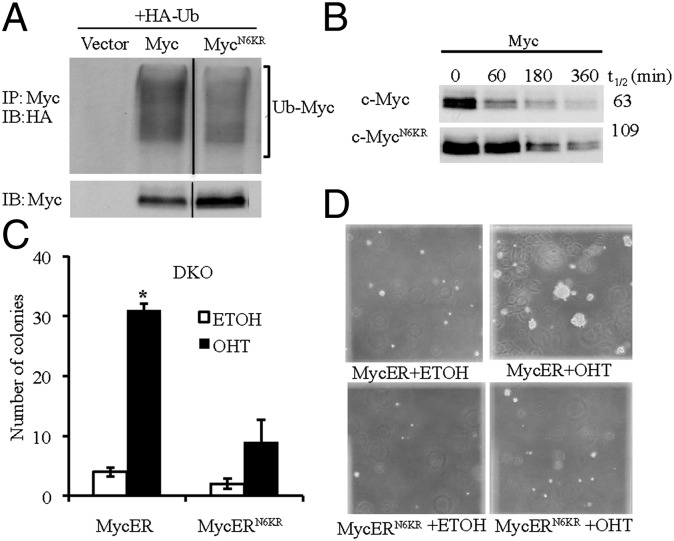
To directly examine the effects of ubiquitylation on c-Myc function, we used site-directed mutagenesis to change lysines to arginines, which retain the positive charge but are unable to be ubiquitylated. Because ARF primarily binds to the TD and inhibits c-Myc target gene induction (4), we targeted the 6 lysines in the TD, which leaves 21 lysines in the remainder of c-Myc protein. All six lysines (51, 52, 127, 144, 149, and 158) are in or near MBI and MBII. We examined the ubiquitylation of this c-Myc protein, termed c-MycN6KR, and found that it was less ubiquitylated compared with the c-Myc protein (Fig. 3A). To examine the effects of loss of c-Myc TD ubiquitylation on c-Myc protein stability, we performed a cycloheximide chase assay. The c-MycN6KR protein was more stable than c-Myc (Fig. 3B). These results suggest that both the N-terminal and C-terminal domain lysines are involved in c-Myc ubiquitylation and proteolysis.
Fig. 3.
Ubiquitylation of the c-Myc TD is essential for c-Myc–induced transformation. (A) In vivo ubiquitylation assays of c-Myc in Cos7 cells transiently transfected with HA-ubiquitin and c-MycN6KR or c-Myc. (B) Cycloheximide chase analyses of c-Myc in Cos7 cells transiently transfected with c-MycN6KR or c-Myc. (C) Anchorage-independent growth analyses of DKO MycER and DKO MycERN6KR MEFs. Asterisk indicates a statistically significant increase in colony formation in OHT-treated DKO MycER cells compared with those treated with ethanol. (D) Representative microscopic images of anchorage-independent growth analyses.
To determine whether loss of TD ubiquitylation also influences c-Myc functions, we generated DKO MEFs expressing either c-MycER or c-MycERN6KR. Immunoblot analysis confirmed that the proteins were expressed at comparable levels and that the exogenous levels were comparable to endogenous c-Myc levels (Fig. S2A). To determine the effect of loss of TD ubiquitylation on the ability of c-Myc to transform DKO MEFs, we performed an anchorage-independent growth assay using soft agar with or without OHT activation. In contrast to the robust transformation induced by activated c-MycER, activated c-MycERN6KR had only weak transforming activity (Fig. 3C). In addition, the size of the foci induced by c-MycERN6KR was relatively small compared with the foci induced by c-MycER (Fig. 3D). These effects on c-Myc–induced transformation in DKO MEFs due to the loss of c-Myc TD ubiquitylation are similar to the effects of ARF interaction with c-Myc observed previously in p53−/− MEFs (4), suggesting that loss of TD ubiquitylation mimics the inhibitory effects of ARF and that TD ubiquitylation is necessary for c-Myc–mediated transformation.
Inhibition of c-Myc TD Ubiquitylation Is Sufficient for c-Myc to Induce Apoptosis.
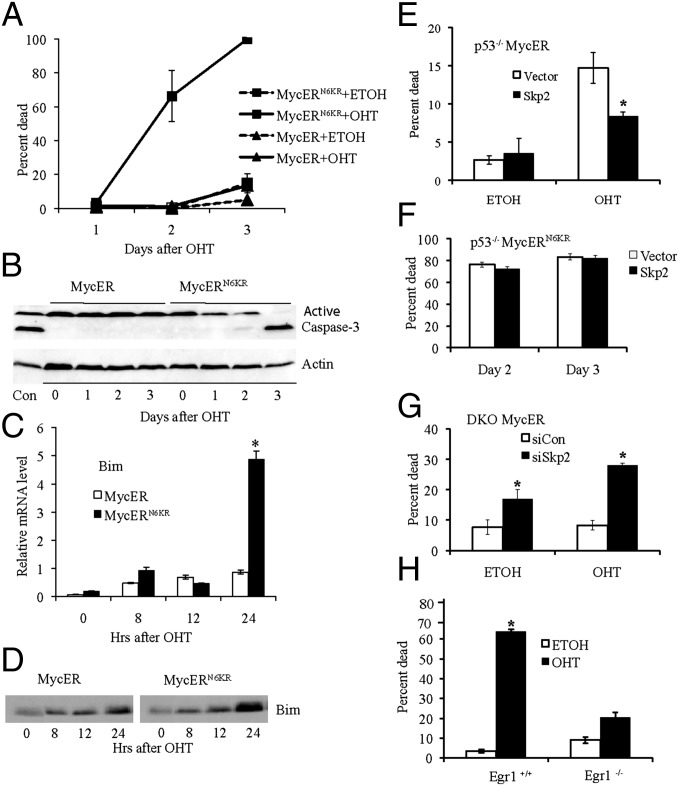
The inhibition of c-Myc–induced transformation caused by loss of TD ubiquitylation may be due to an increase in apoptosis. Therefore, we next examined whether the loss of ubiquitylation is sufficient for c-Myc to induce apoptosis without the need for ARF or p53. We compared the ability of c-MycER and c-MycERN6KR to induce apoptosis in the DKO MEFs shifted to low serum. In contrast to the inability of activated c-MycER to induce cell death without ARF and p53, activated c-MycERN6KR was very efficient in inducing cell death (Fig. 4A). We confirmed that the observed cell death was indeed apoptosis by measuring the cleavage of caspase 3 following activation of c-MycER compared with c-MycERN6KR (Fig. 4B). Because p53-independent apoptosis induced by c-Myc has been linked to an increase in Bim (16), we also compared the ability of c-MycER and c-MycERN6KR to cause an increase in Bim expression in DKO MEFs. Bim mRNA (Fig. 4C) and protein levels (Fig. 4D) were both substantially increased by c-MycERN6KR, but Bim expression was poorly induced by c-MycER. Taken together, these results suggest that inhibition of TD ubiquitylation is essential for c-Myc–mediated apoptosis.
Fig. 4.
Loss of c-Myc TD ubiquitylation is sufficient for c-Myc–induced p53-independent apoptosis and Skp2 controls c-Myc–mediated p53-independent apoptosis. (A) DKO MycER and DKO MycERN6KR MEFs were assayed for apoptosis in low serum with or without 2 μM OHT. There was a statistically significant increase in cell death in cells expressing MycERN6KR when treated with OHT by day 2. (B) Immunoblot showing active caspase-3 in DKO MycER and MycERN6KR MEFs following activation with OHT in low serum. (C) Real-time RT-PCR analysis of Bim expression in DKO MycER and MycERN6KR MEFs following activation with 2 μM OHT in low serum. (D) Immunoblot analysis of Bim in DKO MycER and MycERN6KR MEFs following activation with 2 μM OHT in low serum. (E) p53−/− MycER MEFs and p53−/− MycER/Skp2 MEFs were assayed for apoptosis 2 d following 2 μM OHT activation in low serum. Asterisk indicates a statistically significant decrease in p53−/− MycER cell death with T7-Skp2 overexpression. (F) p53−/− MycERN6KR MEFs and p53−/− MycERN6KR/Skp2 MEFs were assayed for apoptosis following 2 μM OHT activation in low serum. (G) DKO MycER MEFs treated with control siRNA (siCon) or Skp2 siRNA (siSkp2) for 24 h were assayed for apoptosis in high serum 3 d after activation with 2 μM OHT or ethanol (ETOH). Asterisk indicates a statistically significant increase in DKO MycER cell death with Skp2 siRNA compared with the siCon control. (H) Egr1−/− and parental WT MEFs expressing MycERN6KR were assayed for apoptosis in low serum 2 d after 2 μM OHT activation. Asterisk indicates a statistically significant increase in cell death of WT MEFs treated with OHT compared with the ethanol control.
Considering the dramatic apoptotic effects of inhibiting c-Myc TD ubiquitylation and the ability of ARF to inhibit Skp2-mediated ubiquitylation of c-Myc, we next examined whether Skp2 overexpression could counter ARF-dependent c-Myc–induced apoptosis by performing apoptotic assays with p53−/−MycER MEFs stably overexpressing Skp2. Immunoblot analysis demonstrated that comparable levels of c-MycER were expressed with and without Skp2 overexpression (Fig. S2B). There was little cell death without activation of c-MycER in the presence or absence of Skp2 overexpression (Fig. 4E). In contrast, apoptosis was induced by OHT activation of c-MycER, and this apoptosis was substantially reduced in the OHT-activated cells with Skp2 overexpression (Fig. 4E). In contrast, Skp2 was unable to reduce apoptosis induced by c-MycERN6KR in p53−/− MEFs (Fig. 4F) or DKO MEFs (Fig. S3A). Immunoblot analysis confirmed that comparable levels of c-MycERN6KR were expressed with and without Skp2 overexpression (Fig. S2B). Conversely, to determine whether loss of Skp2-mediated ubiquitylation mimicked the apoptotic effects of ARF, we reduced endogenous Skp2 expression in the DKO MycER MEFs (Fig. S2C). There was little cell death observed on activation of c-MycER with OHT in cells treated with control siRNA, as expected in cells without ARF and p53, but there was an approximately fourfold induction in c-Myc–mediated apoptosis in Skp2 siRNA treated cells (Fig. 4G). Without activation of c-MycER, we detected a smaller increase in the amount of cell death when Skp2 expression was inhibited (Fig. 4G), suggesting Skp2 loss also influences endogenous c-Myc and/or other apoptotic targets.
We also examined the effect of inhibiting Skp2-mediated ubiquitylation on apoptosis in HeLa cells, an epithelial carcinoma line with high endogenous c-Myc and Skp2 expression. Cells were treated with Skp2 siRNA in media with low serum or in high serum, and reduced Skp2 expression was confirmed by immunoblot analysis (Fig. S1E). In addition, ubiquitylation of endogenous c-Myc protein was reduced as demonstrated by immunoblot and ubiquitylation analyses (Fig. S1E). Inhibition of Skp2 expression enhanced apoptosis in low serum by ∼4-fold and by ∼2.5-fold in high serum (Fig. S3B). Taken together, the results in both the DKO MycER MEFs and HeLa cells suggest the inhibition of Skp2 expression reduces c-Myc ubiquitylation, leading to an increase in c-Myc–induced apoptosis. In addition, the results suggest that the opposing actions of ARF and Skp2 that regulate the ubiquitylation status of the TD control c-Myc–driven p53-independent apoptosis.
Ubiquitylation of the c-Myc TD Controls c-Myc Canonical and Noncanonical Transcriptional Activity.
Previous studies have shown that ARF inhibits c-Myc target gene induction (4). Therefore, we examined the effects of TD ubiquitylation on c-Myc target genes and transcriptional activity. We first compared the ability of c-MycER and c-MycERN6KR in DKO MEFs to induce the expression of several well-characterized c-Myc target genes containing the canonical E-box Myc sequence (EMS) site using real-time RT-PCR. The cells were serum starved to decrease the levels of endogenous c-Myc followed by OHT activation of c-MycER or c-MycERN6KR. WT c-MycER induced the expression of all four target genes, nucleolin (Fig. 5A), eIF4E, cdk4, and rcl (Fig. S4 A–C), by two- to fourfold, which is typical for these c-Myc target genes (4). In contrast, c-MycN6KR failed to induce their expression (Fig. 5A; Fig. S4 A–C), suggesting that TD ubiquitylation is essential for induction of c-Myc target genes. To examine the activity of a canonical promoter, we performed luciferase reporter assays using the Myc-responsive 4XEMS promoter with four tandem EMS elements driving luciferase expression. As previously observed with other canonical promoters (6), the 4XEMS promoter was induced by c-MycER in DKO MEFs, but not by c-MycER in p53−/− MEFs or by c-MycERN6KR in DKO or p53−/− MEFs (Fig. S4D). Consistent with these results, transiently expressed c-Myc induced the activity of the 4XEMS promoter by approximately twofold (Fig. S4E), whereas c-MycN6KR failed to induce 4XEMS (Fig. S4E). Taken together, the results suggest that TD ubiquitylation is essential for the canonical transcriptional activity of c-Myc.
Fig. 5.
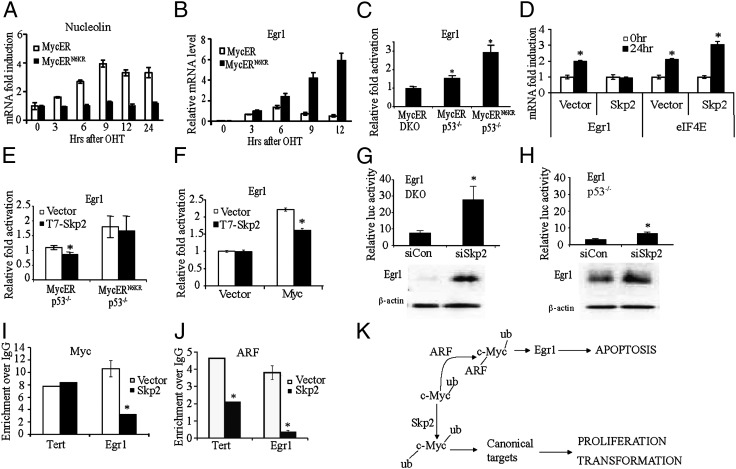
c-Myc TD ubiquitylation controls the induction of canonical target genes and the noncanonical target gene Egr1. (A and B) Real-time RT-PCR analysis of nucleolin (A) and Egr1 (B) in DKO MycER and MycERN6KR MEFs following OHT activation. (C) Luciferase reporter assays of Egr1 promoter activity in DKO MycER, p53−/− MycER, and p53−/− MycERN6KR MEFs. Data are expressed in terms of relative fold activation (OHT/ethanol). Asterisks indicate data in which a statistically significant increase in luciferase activity was observed in OHT-treated cells compared with the ethanol control. (D) Real-time RT-PCR analysis of eIF4E and Egr1 in p53−/− MycER/vector and MycER/Skp2 MEFs following OHT activation. Asterisks indicate data in which a statistically significant increase in mRNA levels was observed after treatment. (E) Luciferase reporter assays of Egr1 promoter activity in p53−/− MycER and p53−/− MycERN6KR MEFs transiently transfected with T7-Skp2 or empty vector. Luciferase data are expressed as in C. Asterisk indicates a statistically significant decrease in induction of Egr1 activity in cells overexpressing Skp2. (F) Luciferase reporter assays of Egr1 promoter activity in Rat1a or Rat1a/Myc cells with and without T7-Skp2 transient overexpression. The asterisk indicates a statistically significant decrease in induction of Egr1 promoter activity in cells overexpressing T7-Skp2. (G and H) Luciferase reporter assays of Egr1 promoter activity in DKO (G, Upper) and p53−/− (H, Upper) MEFS transfected with siSkp2 or control siRNA (siCon). Asterisk indicates a statistically significant increase in Egr1 promoter activity in cells transfected with siSkp2 compared with siCon. Immunoblot analysis of Egr1 expression in DKO (G, Lower) and p53−/− (H, Lower) MEFs treated with siCon or siSkp2. (I and J) ChIP analyses of Myc (I) and ARF (J) were performed using chromatin prepared from p53−/− MycER/Skp2 and p53−/− MycER/vector MEFs after treatment with OHT for 6 h. Asterisk indicates a statistically significant decrease in Myc or ARF binding at the target promoters in cells stably expressing T7-Skp2. (K) Ubiquitylation controls the molecular and biological functions of c-Myc. Ubiquitylation of the c-Myc TD by Skp2 is essential for canonical target gene expression that mediates c-Myc–induced proliferation and transformation. Inhibition of Skp2-mediated ubiquitylation of the c-Myc TD by ARF is sufficient for induction of the noncanonical Egr1 target gene that mediates p53-independent c-Myc–induced apoptosis.
In contrast to the ARF inhibition of c-Myc canonical target gene induction (4), ARF is essential for induction of the noncanonical c-Myc target gene, Egr1 (6). To determine whether loss of TD ubiquitylation of c-Myc mimics the necessity of ARF for c-Myc–induced Egr1 expression, we compared the ability of c-MycER and c-MycERN6KR to induce Egr1 expression in DKO MEFs in low serum using real-time RT-PCR. Unlike c-MycER, activation of c-MycERN6KR caused a significant induction of Egr1 (Fig. 5B). In addition, luciferase reporter assays using an Egr1 promoter construct, which was previously shown to be activated by c-Myc in an ARF-dependent manner (6), demonstrated that the activity of the Egr1 promoter was significantly induced in p53−/− MEFs by both c-MycER and c-MycERN6KR but not by c-MycER in DKO MEFs (Fig. 5C). The Egr1 promoter activity in p53−/− MEFs having both ARF and c-MycERN6KR was enhanced compared with p53−/− MEFs with ARF alone (Fig. 5 C and E). These results suggest, as would be expected, that blocking TD ubiquitylation by mutation is more efficient than inhibiting Skp2-mediated TD ubiquitylation through competitive binding with ARF.
Considering Skp2 overexpression countered the apoptotic effects of endogenous ARF following activation of c-Myc in p53−/−MycER MEFs (Fig. 4E), we next examined whether Skp2 overexpression inhibited the ARF-dependent induction of Egr1 in these same cells. As previously described (6), activation of c-MycER in p53−/−MEFs induced the expression of Egr1 (Fig. 5D), whereas Egr1 induction by activated c-MycER was inhibited when Skp2 was overexpressed (Fig. 5D). In contrast, Skp2 overexpression up-regulated the canonical c-Myc target gene, eIF4E (Fig. 5D), as previously observed (11, 12), countering the inhibitory effects of ARF on canonical c-Myc target gene expression. Furthermore, we determined that overexpression of Skp2 caused a significant decrease in the ability of activated c-MycER (Fig. 5E) and transiently expressed c-Myc (Fig. 5F) to induce the Egr1 promoter in luciferase reporter assays. In contrast, the ability of c-MycERN6KR to induce Egr1 promoter activity was not significantly influenced by Skp2 overexpression (Fig. 5E).
The increase in apoptosis on inhibition of Skp2 in DKO MEFs without c-MycER activation (Fig. 4G) suggests that loss of ubiquitylation of endogenous c-Myc may also contribute in an increase in Egr1 expression. Indeed, we observed a significant increase in both Egr1 promoter activity (3.6-fold), measured by luciferase reporter assays, and in Egr1 protein levels when cells are treated with Skp2 siRNA (Fig. 5G). As would be expected, the presence of high ARF levels in the p53−/− MEFs results in a higher level of Egr1 protein compared with DKO MEFs, and thus the inhibition of Skp2 expression caused a relatively smaller increase in Egr1 promoter activity (2.1-fold) and Egr1 protein levels (Fig. 5H). In addition, inhibition of Skp2 expression by treating with siRNA in HeLa cells resulted in an increase in Egr1 protein expression (Fig. S4F), which also correlated with the increase in apoptosis with Skp2 siRNA treatment (Fig. S3B). Taken together, we showed by three mechanisms—loss of c-Myc TD ubiquitylation by inhibition of Skp2 expression, ARF competition with Skp2 for c-Myc binding, and expression of a c-Myc with a nonubiquitylatable TD—that inhibition of TD ubiquitylation induces the Egr1 target gene.
To determine whether the inhibition of ARF-dependent Egr1 expression by Skp2 due to competitive interaction with c-Myc occurs at the Egr1 promoter, we used real-time ChIP analysis to examine the recruitment of c-Myc and ARF to the Egr1 promoter in the presence of overexpressed Skp2. We have previously shown that both c-Myc and ARF, but not ARF alone, are recruited to a region ∼1 kb upstream of the transcriptional start site in the Egr1 promoter, which does not contain any canonical Myc binding sites (6). The recruitment of c-Myc to the canonical tert promoter was not influenced by the overexpression of Skp2, whereas c-Myc recruitment to the noncanonical Egr1 promoter was reduced by the overexpression of Skp2 in the p53−/− MycER MEFs (Fig. 5I). These observations are consistent with our previous findings that ARF enhances c-Myc recruitment to the Egr1 promoter (6). The recruitment of ARF to the Egr1 promoter, as well as the tert promoter, was substantially reduced in the p53−/− MycER MEFs cells overexpressing Skp2 (Fig. 5J). Similarly, ARF recruitment to the Egr1 promoter and the tert promoter was also significantly reduced in the p53−/− MycERN6KR MEFs cells overexpressing Skp2 (Fig. S4G), suggesting that the presence of the lysine mutations in the c-Myc TD did not influence the binding of ARF or Skp2 to c-Myc and confirms that the activity of MycERN6KR in inducing Egr1 and apoptosis is not dependent on the binding of ARF. Taken together, the results suggest Skp2 regulates c-Myc ubiquitylation and induction of canonical and noncanonical target gene promoters through competitive interaction with ARF. The results also suggest that ubiquitylation of the c-Myc TD enhances activity and not recruitment of c-Myc at canonical promoters, whereas recruitment and activity of c-Myc to the noncanonical Egr1 promoter are both enhanced by inhibition of TD ubiquitylation.
Egr1 Mediates Apoptosis Induced by Loss of c-Myc TD Ubiquitylation.
Finally, we determined whether Egr1 is necessary for apoptosis induced by loss of TD ubiquitylation of c-Myc, as it is for ARF-dependent c-Myc–induced apoptosis in p53−/− MEFs (6). We generated WT MEFs (Egr1+/+) and Egr1−/− MEFs expressing c-MycERN6KR and confirmed similar expression levels by immunoblot analysis (Fig. S2D). On activation, c-MycERN6KR induced apoptosis in the WT MEFs but was significantly compromised in its ability to induce apoptosis in the Egr1−/− MEFs (Fig. 4H). As another approach, we treated DKO MEFs expressing c-MycN6KR with Egr1 siRNA or control siRNA following activation with OHT and observed a substantial inhibition of c-MycN6KR–induced apoptosis (Fig. S4H). Reduction of Egr1 protein expression was confirmed by immunoblot analysis (Fig. S4H). These results suggest that Egr1 is a key mediator of p53-independent apoptosis induced by loss of TD ubiquitylation of c-Myc.
Discussion
Previous studies have implicated ubiquitylation in influencing c-Myc proliferative and canonical transcriptional activity in addition to proteolysis (15, 17–19). Our results directly show that ubiquitylation of the c-Myc TD is necessary for canonical c-Myc target gene induction and efficient c-Myc–induced transformation. Our results also suggest that the ability of ARF to inhibit canonical transcriptional activity is due to its ability to inhibit Skp2-mediated ubiquitylation of the c-Myc TD. In addition, the loss of ubiquitylation not only inhibits canonical activity but also allows the acquisition of a new noncanonical activity, specifically the induction of the noncanonical c-Myc target gene, Egr1, which then mediates p53-independent c-Myc–induced apoptosis. Therefore, the canonical oncogenic activity of c-Myc, which is enhanced by Skp2, and the noncanonical apoptotic activity induced by ARF are controlling a unique mechanism dependent on ubiquitylation of the c-Myc TD.
Our results suggest that Skp2 preferentially ubiquitylates the c-Myc TD rather than the C-terminal half of c-Myc. In growing cells, it has been shown that half of the lysines in the TD, including 52, 149, and 158, are ubiquitylated (20); however, it is unknown whether the various c-Myc ubiquitin ligases have preferences for specific lysines or domains. Although the c-Myc degron has been mapped to the TD (21, 22), our data suggest that ubiquitylation of both the N- and C-terminal domains contribute to c-Myc proteolysis. Unlike Skp2, which behaves as an oncogene that stimulates c-Myc transcription and proliferation, other c-Myc E3 ubiquitin ligases, such as Fbw7 and TRPC4AP/TRUSS, act as tumor suppressors that inhibit c-Myc transactivation and transformation (14, 23). In contrast to enhanced apoptosis observed upon inhibition of Skp2, loss of Fbw7 binding and ubiquitylation, due to mutation at the Thr58 phosphorylation site of c-Myc, results in less apoptosis (14, 16, 24). Therefore, we conclude that ubiquitylation of the TD regulated by Skp2 and ARF competitive interaction is a key factor controlling c-Myc transcriptional and biological activity, rather than just the levels of c-Myc regulated by proteolysis.
Our model is that the ubiquitylation of the TD is critical for the induction of canonical c-Myc target genes, such as cdk4, cyclin D2 and eIF4E, which mediate c-Myc-induced cell cycle progression under normal conditions (Fig. 5K). Oncogenic c-Myc induces ARF expression, which then blocks the binding of Skp2 and perhaps other ligases, preventing TD ubiquitylation necessary for c-Myc-mediated proliferation and transformation. Simultaneously, the loss of ubiquitylation triggers a transcriptional switch that allows c-Myc to induce the noncanonical Egr1 target gene, which then mediates c-Myc-induced p53-independent apoptosis (Fig. 5K). Tumorigenesis could occur if either the expression of ARF or Egr1 is suppressed or if Skp2 expression or activity is increased, all of which have been found in many different types of human tumors (8, 25–28). Since Skp2 is a c-Myc target gene, positive feedback would enhance c-Myc activity and compete with ARF (29). Conversely, loss of Skp2 would cause growth inhibition (30) or apoptosis. This idea is supported by studies in glioblastoma and laryngeal cancer that demonstrates an inhibition of Skp2 induces apoptosis (31, 32). Therefore, the ubiquitylation of the c-Myc TD, which controls the intrinsic transcriptional and apoptotic activities of c-Myc, can be targeted to develop novel therapeutic strategies to eradicate tumor cells and prevent c-Myc-driven tumorigenesis. Since the Myc-ARF-Egr1 pathway is p53-independent, unlike other c-Myc-induced apoptotic pathways, the ubiquitylation-dependent apoptotic activity of c-Myc can be activated in tumors that have lost p53, which represent half of all human tumors.
Materials and Methods
coIP and Immunoblot Analysis.
coIP analyses were performed as described previously (33), except the immunoprecipitation (IP) was performed with anti-Myc (C-33; Santa Cruz) followed by immunoblotting with the indicated antibodies. Myc IBs were performed with anti-MycN100 (34), unless otherwise described. The other antibodies used for immunoblotting were anti-Skp2 (H-435; Santa Cruz), anti-mouse ARF (54-75; Millipore), anti-human p14 ARF (Bethyl Laboratories), anti-HA (12CA5), anti-T7 (69522; Novagen), anti-Flag (M5; Sigma), anti-Egr1(C-19; Santa Cruz), anti-Jun (Millipore), anti-Bim (BD Pharmingen), anti–β-actin (Sigma-Aldrich), or caspase-3 (BD Pharmingen).
In Vivo Ubiquitylation Analysis.
In vivo ubiquitylation assays were performed as described previously (22) with some modifications. Briefly, cells were transiently transfected with CMV-ubiquitin-HA and the indicated vectors. The cells were then treated with proteasome inhibitor MG132 (10 μM) for 4 h posttransfection, lysed, and boiled for 10 min in lysis buffer with 1% SDS. Lysates were diluted 1:2 with lysis buffer without SDS and incubated with 4 μg of anti-Mycfl (Millipore) overnight at 4 °C. Precipitates were analyzed by immunoblotting with anti-HA.
Cycloheximide Chase Analysis.
Cycloheximide chase analysis was performed as described previously (33). Detailed methods are in SI Materials and Methods.
Anchorage-Independent Growth Assays.
MEFs were plated at a density of 1.5 × 104 cells/60-mm dish in soft agar containing DMEM plus 10% FCS. The cells were fed with media containing 10% FCS with 2 μm OHT or ethanol every other day. Four weeks after seeding, cells were stained with 0.005% crystal violet, and colonies >190 μm were counted from triplicate plates.
Apoptosis Assays.
Apoptosis assays were performed as described previously (4, 6) with some modifications described in SI Materials and Methods.
ChIP Analyses.
MEFs were plated at 6 × 106 cells/150-mm dish and the next day were treated with 2 μM OHT or ethanol for 6 h. Cross-linking and ChIP analyses were performed as described previously (6) with anti-Myc (C-33), anti-mouse ARF, and normal rabbit IgG (Upstate). Purified DNA was subjected to real-time RT-PCR using the primers listed in SI Materials and Methods. Results are reported as the mean of enrichment over IgG calculated by dividing the percent of input from the IP by the percent of input from the IgG control ± SD.
Quantitative Real-Time RT-PCR.
MEFs were plated at 3.5 × 106 cells/10-mm dish, and 24 h later the cells were shifted to media containing 0.1% (vol/vol) calf serum (CS) for 72 h. cDNA was prepared, real-time RT-PCR was performed, and results were calculated as previously described (33). All real-time RT-PCR analyses were performed in triplicate with the primers listed in SI Materials and Methods. Results are reported as the mean of target gene mRNA levels relative to β-actin mRNA levels. Each time point is normalized to time 0 to give the relative fold induction ± SD.
Luciferase Assays.
Luciferase assays for analysis of Egr1 promoter activation were performed as described previously (6). Detailed methods can be found in SI Materials and Methods.
Data Analysis and Statistics.
All data generated are the product of no less than three independent experiments. Statistical significance was assessed through pairwise comparisons using Student t test. Comparisons of data sets yielding P < 0.05 were considered to have statistically significant differences.
Supplementary Material
Acknowledgments
We thank E. Lee for the anti-HA (12CA5) antibodies, G. Kato for the CMV-HA epitope-tagged ubiquitin expression plasmid (pMT123), M. Cole for the pCBS-FLAG-c-Myc vector, and W. Tansey for pCGT-T7-Skp2, pCGN-HA-Myc1-220, and pCGN-HA-Myc 221-439 vectors. We also thank R. Eisenman, M. Eilers, and W. Tansey for critical discussions and reading of the manuscript. This work was supported by National Institutes of Health Grants R01 CA125760 (to S.R.H.) and T32 CA119925 (to Q.Z. and E.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. K.B. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1208334110/-/DCSupplemental.
References
- 1.Lüscher B, Vervoorts J. Regulation of gene transcription by the oncoprotein MYC. Gene. 2012;494(2):145–160. doi: 10.1016/j.gene.2011.12.027. [DOI] [PubMed] [Google Scholar]
- 2.Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nat Rev Cancer. 2008;8(12):976–990. doi: 10.1038/nrc2231. [DOI] [PubMed] [Google Scholar]
- 3.Sherr CJ, et al. p53-Dependent and -independent functions of the Arf tumor suppressor. Cold Spring Harb Symp Quant Biol. 2005;70:129–137. doi: 10.1101/sqb.2005.70.004. [DOI] [PubMed] [Google Scholar]
- 4.Qi Y, et al. p19ARF directly and differentially controls the functions of c-Myc independently of p53. Nature. 2004;431(7009):712–717. doi: 10.1038/nature02958. [DOI] [PubMed] [Google Scholar]
- 5.Gregory MA, Qi Y, Hann SR. The ARF tumor suppressor: Keeping Myc on a leash. Cell Cycle. 2005;4(2):249–252. [PubMed] [Google Scholar]
- 6.Boone DN, Qi Y, Li Z, Hann SR. Egr1 mediates p53-independent c-Myc-induced apoptosis via a noncanonical ARF-dependent transcriptional mechanism. Proc Natl Acad Sci USA. 2011;108(2):632–637. doi: 10.1073/pnas.1008848108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Z, Hann SR. The Myc-nucleophosmin-ARF network: A complex web unveiled. Cell Cycle. 2009;8(17):2703–2707. doi: 10.4161/cc.8.17.9418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan CH, Lee SW, Wang J, Lin HK. Regulation of Skp2 expression and activity and its role in cancer progression. Scientific World Journal. 2010;10:1001–1015. doi: 10.1100/tsw.2010.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dominguez-Brauer C, Brauer PM, Chen YJ, Pimkina J, Raychaudhuri P. Tumor suppression by ARF: Gatekeeper and caretaker. Cell Cycle. 2010;9(1):86–89. doi: 10.4161/cc.9.1.10350. [DOI] [PubMed] [Google Scholar]
- 10.Rizos H, Scurr LL, Irvine M, Alling NJ, Kefford RF. p14ARF regulates E2F-1 ubiquitination and degradation via a p53-dependent mechanism. Cell Cycle. 2007;6(14):1741–1747. doi: 10.4161/cc.6.14.4428. [DOI] [PubMed] [Google Scholar]
- 11.Kim SY, Herbst A, Tworkowski KA, Salghetti SE, Tansey WP. Skp2 regulates Myc protein stability and activity. Mol Cell. 2003;11(5):1177–1188. doi: 10.1016/s1097-2765(03)00173-4. [DOI] [PubMed] [Google Scholar]
- 12.von der Lehr N, et al. The F-box protein Skp2 participates in c-Myc proteosomal degradation and acts as a cofactor for c-Myc-regulated transcription. Mol Cell. 2003;11(5):1189–1200. doi: 10.1016/s1097-2765(03)00193-x. [DOI] [PubMed] [Google Scholar]
- 13.Welcker M, et al. The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc Natl Acad Sci USA. 2004;101(24):9085–9090. doi: 10.1073/pnas.0402770101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yada M, et al. Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbw7. EMBO J. 2004;23(10):2116–2125. doi: 10.1038/sj.emboj.7600217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Popov N, Schülein C, Jaenicke LA, Eilers M. Ubiquitylation of the amino terminus of Myc by SCF(β-TrCP) antagonizes SCF(Fbw7)-mediated turnover. Nat Cell Biol. 2010;12(10):973–981. doi: 10.1038/ncb2104. [DOI] [PubMed] [Google Scholar]
- 16.Hemann MT, et al. Evasion of the p53 tumour surveillance network by tumour-derived MYC mutants. Nature. 2005;436(7052):807–811. doi: 10.1038/nature03845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adhikary S, et al. The ubiquitin ligase HectH9 regulates transcriptional activation by Myc and is essential for tumor cell proliferation. Cell. 2005;123(3):409–421. doi: 10.1016/j.cell.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 18.Muller J, Eilers M. Ubiquitination of Myc: Proteasomal degradation and beyond. Ernst Schering Found Symp Proc. 2008;1:99–113. doi: 10.1007/2789_2008_103. [DOI] [PubMed] [Google Scholar]
- 19.Thomas LR, Tansey WP. Proteolytic control of the oncoprotein transcription factor Myc. Adv Cancer Res. 2011;110:77–106. doi: 10.1016/B978-0-12-386469-7.00004-9. [DOI] [PubMed] [Google Scholar]
- 20.Kim W, et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol Cell. 2011;44(2):325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salghetti SE, Kim SY, Tansey WP. Destruction of Myc by ubiquitin-mediated proteolysis: Cancer-associated and transforming mutations stabilize Myc. EMBO J. 1999;18(3):717–726. doi: 10.1093/emboj/18.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gregory MA, Hann SR. c-Myc proteolysis by the ubiquitin-proteasome pathway: Stabilization of c-Myc in Burkitt’s lymphoma cells. Mol Cell Biol. 2000;20(7):2423–2435. doi: 10.1128/mcb.20.7.2423-2435.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choi SH, Wright JB, Gerber SA, Cole MD. Myc protein is stabilized by suppression of a novel E3 ligase complex in cancer cells. Genes Dev. 2010;24(12):1236–1241. doi: 10.1101/gad.1920310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rottmann S, Wang Y, Nasoff M, Deveraux QL, Quon KC. A TRAIL receptor-dependent synthetic lethal relationship between MYC activation and GSK3beta/FBW7 loss of function. Proc Natl Acad Sci USA. 2005;102(42):15195–15200. doi: 10.1073/pnas.0505114102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reed SI. Deathproof: new insights on the role of skp2 in tumorigenesis. Cancer Cell. 2008;13(2):88–89. doi: 10.1016/j.ccr.2008.01.023. [DOI] [PubMed] [Google Scholar]
- 26.Joslin JM, et al. Haploinsufficiency of EGR1, a candidate gene in the del(5q), leads to the development of myeloid disorders. Blood. 2007;110(2):719–726. doi: 10.1182/blood-2007-01-068809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu J, et al. Concurrent down-regulation of Egr-1 and gelsolin in the majority of human breast cancer cells. Cancer Genomics Proteomics. 2007;4(6):377–385. [PubMed] [Google Scholar]
- 28.Weber JD, et al. p53-independent functions of the p19(ARF) tumor suppressor. Genes Dev. 2000;14(18):2358–2365. doi: 10.1101/gad.827300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bretones G, et al. SKP2 oncogene is a direct MYC target gene and MYC down-regulates p27(KIP1) through SKP2 in human leukemia cells. J Biol Chem. 2011;286(11):9815–9825. doi: 10.1074/jbc.M110.165977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu L. Skp2 knockout reduces cell proliferation and mouse body size: And prevents cancer? Cell Res. 2010;20(6):605–607. doi: 10.1038/cr.2010.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu J, Tan L, Li H, Wang Q, Ji W. Suppression of S-phase kinase-associated protein 2 induces apoptosis and inhibits tumor growth in human laryngeal cancer. ORL J Otorhinolaryngol Relat Spec. 2010;72(4):205–214. doi: 10.1159/000314689. [DOI] [PubMed] [Google Scholar]
- 32.Lee SH, McCormick F. Downregulation of Skp2 and p27/Kip1 synergistically induces apoptosis in T98G glioblastoma cells. J Mol Med (Berl) 2005;83(4):296–307. doi: 10.1007/s00109-004-0611-7. [DOI] [PubMed] [Google Scholar]
- 33.Li Z, Boone D, Hann SR. Nucleophosmin interacts directly with c-Myc and controls c-Myc-induced hyperproliferation and transformation. Proc Natl Acad Sci USA. 2008;105(48):18794–18799. doi: 10.1073/pnas.0806879105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spotts GD, Patel SV, Xiao Q, Hann SR. Identification of downstream-initiated c-Myc proteins which are dominant-negative inhibitors of transactivation by full-length c-Myc proteins. Mol Cell Biol. 1997;17(3):1459–1468. doi: 10.1128/mcb.17.3.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.