Abstract
In eukaryotes, the nucleocytoplasmic transport of macromolecules is mainly mediated by soluble nuclear transport receptors of the karyopherin-β superfamily termed importins and exportins. The highly versatile exportin chromosome region maintenance 1 (CRM1) is essential for nuclear depletion of numerous structurally and functionally unrelated protein and ribonucleoprotein cargoes. CRM1 has been shown to adopt a toroidal structure in several functional transport complexes and was thought to maintain this conformation throughout the entire nucleocytoplasmic transport cycle. We solved crystal structures of free CRM1 from the thermophilic eukaryote Chaetomium thermophilum. Surprisingly, unbound CRM1 exhibits an overall extended and pitched superhelical conformation. The two regulatory regions, namely the acidic loop and the C-terminal α-helix, are dramatically repositioned in free CRM1 in comparison with the ternary CRM1–Ran–Snurportin1 export complex. Single-particle EM analysis demonstrates that, in a noncrystalline environment, free CRM1 exists in equilibrium between extended, superhelical and compact, ring-like conformations. Molecular dynamics simulations show that the C-terminal helix plays an important role in regulating the transition from an extended to a compact conformation and reveal how the binding site for nuclear export signals of cargoes is modulated by different CRM1 conformations. Combining these results, we propose a model for the cooperativity of CRM1 export complex assembly involving the long-range allosteric communication between the distant binding sites of GTP-bound Ran and cargo.
Eukaryotic cells are separated into compartments, such as the endoplasmic reticulum, mitochondria, or the nucleus. The nucleus is encompassed by a double-layered membrane, the nuclear envelope, whereas aqueous connections to the cytoplasm are maintained by large macromolecular assemblies, the nuclear pore complexes. However, aside from the regulatory advantages of compartmentalization, the spatial separation also poses a logistic challenge, namely to ensure the efficient exchange of proteins, RNA, and metabolites between these compartments. An elaborate transport system has evolved to achieve the bidirectional transport of proteins and RNAs. Nuclear transport of macromolecules in eukaryotic cells is mainly mediated by soluble transport receptors of the karyopherin-β superfamily termed importins and exportins (1, 2). They share a common structural arrangement of approximately 20 repetitive elements, so-called HEAT repeats. Among them, the prototypical exportin chromosome region maintenance 1 (CRM1) is essential for nuclear depletion of numerous structurally and functionally unrelated protein and ribonucleoprotein cargoes (3–7). Commonly, CRM1–cargo binding depends on a 10- to 15-residue-long, leucine-rich nuclear export signal (NES) within the transport target (8–10), which binds to a hydrophobic cleft (NES binding cleft) on the outer convex surface of CRM1. Efficient cargo binding requires the presence of the small GTPase Ran in its GTP-bound nuclear form (RanGTP). The binding of RanGTP and cargo to CRM1 has been shown to be cooperative, as the affinity for either of the binding partners is increased in the presence of the other protein (11–13). However, the structural basis for this cooperativity has so far been unclear. Subsequent to formation of the stable CRM1–RanGTP–cargo complex, the assembly translocates through the nuclear pore complex into the cytoplasm. GTP hydrolysis by Ran in the cytoplasm is stimulated by the Ran GTPase activating protein (i.e., RanGAP) and further increased by Ran binding proteins (RanBPs), resulting in a release of the cargo. CRM1 in the free form shuttles back into the nucleus for the next round of export.
In recent years, the crystal structures of three different CRM1 export complexes have been solved. They are CRM1 with bound cargo Snurportin1 [CRM1–SPN1; Protein Data Bank (PDB) ID code 3GB8] (10), CRM1 with RanGTP (CRM1–RanGTP; PDB ID code 3NC1) (8)—both representing assembly intermediates—as well as the functional ternary export complex CRM1–RanGTP–SPN1 (PDB ID code 3GJX) (9). Moreover, the structure of one disassembly complex containing CRM1, RanGTP, and RanBP1 (CRM1–RanGTP–RanBP1; PDB ID code 3M1I) (14) has been determined. All these structures have in common that CRM1 adopts a compact ring-like shape of a toroid with the N- and C-terminal regions forming numerous interactions. Alterations between these CRM1 structures are found in the first three HEAT repeats, a highly conserved region involved in RanGTP binding, but the N- and C-terminal HEAT repeats are always in close contact. This led to the proposal that cargo-free CRM1 may retain a ring-like shape (9), which was further supported by single-particle EM and small-angle X-ray scattering analyses (12, 15).
Surprisingly, and in contrast to all other known export complexes, no obvious explanation for the observed cooperative effects were seen; in particular, no direct interactions between cargo and RanGTP in the CRM1–RanGTP–SPN1 complex could be detected. Therefore, two significant differences between these ring-shaped CRM1 structures might be of particular relevance. The first one concerns the so-called acidic loop, a stretch of acidic amino acids forming a more or less extended loop of variable length in many members of the importin-β superfamily, which has been shown to be involved in cargo binding and release (9, 16–18). In the RanGTP-bound CRM1 complex structures, this acidic loop traverses the central opening of the CRM1 toroid and affixes RanGTP to CRM1, in a seatbelt-like fashion, to N- and C-terminal regions that form numerous interactions (8, 9). In the CRM1–RanGTP–RanBP1 complex, this loop is reoriented in a “flipped back” position that brings the loop into proximity of the HEAT repeats, forming the NES binding cleft (14). The binding of the acidic loop to the inward-oriented HEAT repeat helices 11B and 12B may stabilize the NES cleft in a contracted, closed state. The other feature in CRM1 relates to the last HEAT repeat helix 21B, which is located in a parallel orientation to the HEAT repeat helix 21A in the Ran-bound complexes (9, 14). In the cargo-only bound state (i.e., CRM1–SPN1), these two helices are considerably repositioned, and the relative position of the N-terminal HEAT repeats with respect to the C-terminal region and thus the interaction pattern of these two regions is altered (10). The B-helix is now bridging the central opening and interacts with the HEAT repeats that form the NES cleft. The acidic residues located C-terminally of the B-helix (i.e., C-terminal acidic tail) are thought to form close contacts to basic patches at the back side of the NES binding cleft, thereby regulating the NES binding cleft state (19). However, the question of how CRM1 achieves these conformational changes and how binding cooperativity is mediated from the RanGTP binding site to the cargo binding site remains elusive, mainly because no crystal structure of free CRM1 has yet been solved.
Here, we present crystal structures of free CRM1 exhibiting an extended and pitched superhelical conformation. Single-particle EM studies reveal that, in a noncrystalline environment, free CRM1 exists in equilibrium between extended and more compact conformations. Moreover, molecular dynamics (MD) simulations unravel the contributions of two regulatory regions to the conformational state of CRM1. Combining these results, we propose a model for the cooperativity of CRM1 export complex assembly, which shows how long-range allosteric communication between distant binding sites in proteins orchestrates the tuning of affinities.
Results
Crystal Structure Analysis.
To gain insight into the structure of free CRM1, and to understand the structural requirements for the cooperativity of cargo and Ran binding, we aimed to crystallize it. As all attempts to crystallize full-length mammalian CRM1 have failed so far, we sought to structurally characterize the CRM1 orthologue from the thermophilic fungus Chaetomium thermophilum (ctCRM1) which exhibits a sequence identity of 50% to human CRM1 (Fig. S1). Two different crystal forms of ctCRM1 were obtained belonging to space groups P31 and P212121, respectively. As a result of better crystal quality and higher resolution of the diffraction data, as well as the fact that the trigonal crystals were twinned, the orthorhombic crystals were initially used. The structure was solved by molecular replacement using mouse CRM1 derived from the CRM1–RanGTP−SPN1 complex structure (9) as search model, and refined at 2.94 Å resolution to R and Rfree values of 22.0% and 24.3%, respectively (Table S1).
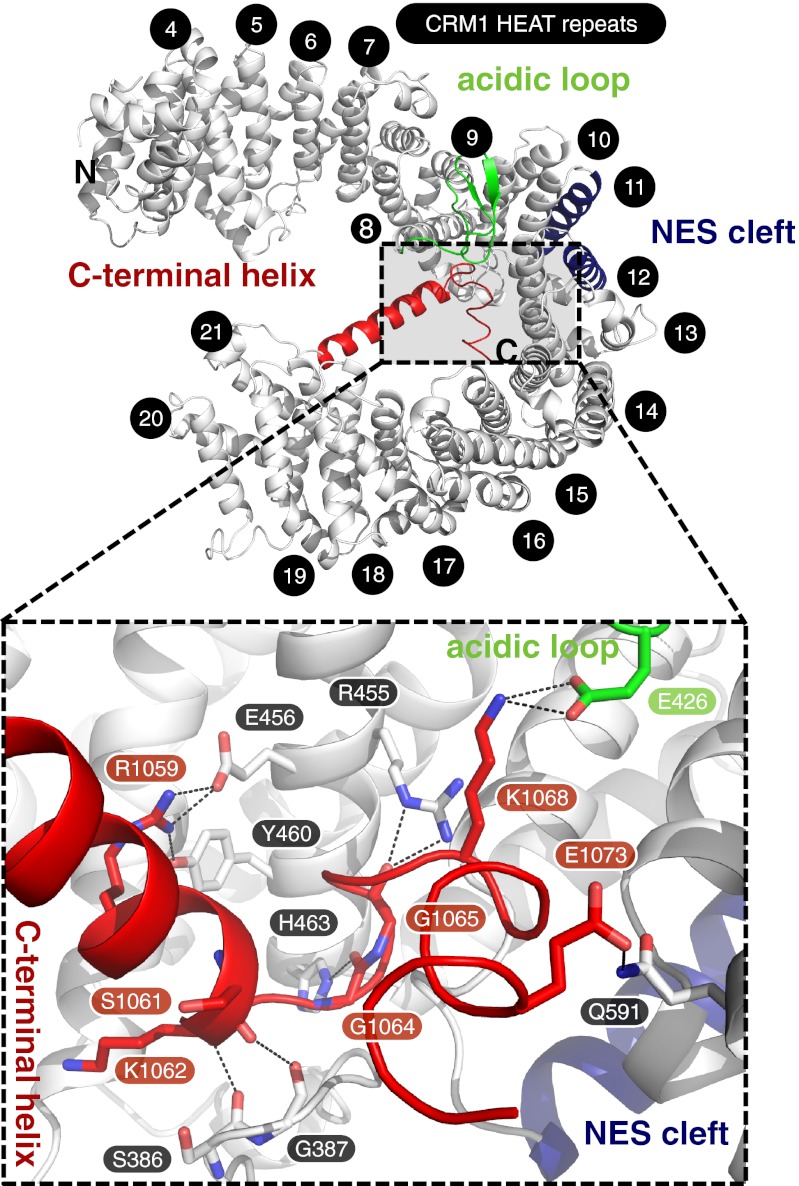
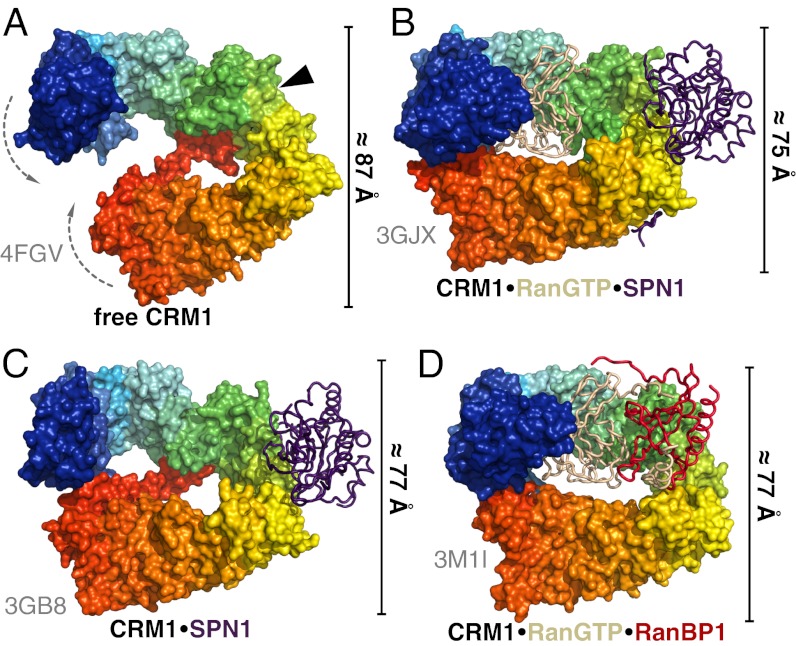
Like mammalian and yeast CRM1, full-length ctCRM1 consists of 21 HEAT repeats (H1–H21; Fig. S1). It adopts an overall extended and pitched superhelical structure (Fig. 1), clearly differing from the known complex structures, in which CRM1 is bent into a distorted toroid and shows less superhelical pitch (9, 10, 14) (Fig. 2 B–D). In all these complexes, N- and C-terminal HEAT repeats of CRM1 interact with each other, involving residues of H2 to H5 on the one side and residues from H21 on the other side. These interactions are completely absent from the cargo-free form of CRM1 (Figs. 1 and 2A). In contrast to the complex structures, the C-terminal half of free CRM1 is shifted from the N-terminal HEAT repeats by approximately 20 Å, increasing the pitch and elongating the superhelix by 12 Å (Fig. 2). As a consequence, the helices 11A and 12A move toward each other, thereby rendering the NES binding cleft between these helices inaccessible for cargo (Fig. 3 and Fig. S2).
Fig. 1.
Crystal structure of free ctCRM1 (gray). The acidic loop (green), the C-terminal helix (red) and the NES cleft (blue) are highlighted. The HEAT repeats are numbered, and termini are labeled. (Lower) Rotated detail view of the interactions of residues from the C-terminal helix with the acidic loop and a patch of CRM1 formed by helices of H8 to H12. Hydrogen bonds and salt bridges are represented by dashed lines, and interacting residues are labeled.
Fig. 2.
Comparison of CRM1 conformations in different crystal structures. Free ctCRM1 (A), CRM1–RanGTP–SPN1 (B), CRM1–SPN1 (C), and CRM1–RanGTP–RanBP1 (D) are shown. CRM1 is depicted as rainbow colored surface from N (blue) to C terminus (red), whereas the interacting proteins are shown as tube models (SPN1, purple; RanGTP, beige; RanBP1, red). The position of the NES cleft in structures lacking cargo is marked by a black arrowhead. The bars at the side of the individual structures indicate the dimension of the protein in the shown orientation.
Fig. 3.
Comparison of NES cleft conformations between free ctCRM1 (PDB ID code 4FGV; green) and CRM1 bound to SPN1 and RanGTP (PDB ID code 3GJX; red). The NES cleft is shown in cartoon mode (Left) with the centers of mass of the helices 11A and 12A represented by blue spheres and their distances indicated. A surface model (Center) illustrates the differences of the NES clefts between both structures. A superposition of the SPN1 NES from the ternary CRM1–RanGTP–SPN1 complex (Right) highlights the structural changes in the NES cleft, which are incompatible with NES binding in free CRM1.
The extended conformation of free CRM1, and, consequently, the closed state of the NES binding cleft, seems to be stabilized by the rearranged C-terminal helix of CRM1 (HEAT repeat helix 21B), which diagonally crosses the superhelix. The C-terminal helix is found in a similar position as in the CRM1–SPN1 binary complex (10) (Fig. 1 and Fig. S3). Several residues between Arg1059 and Glu1073 belonging to the C-terminal helix and the adjacent C-terminal tail contact the B-helices of H8 to H12 on the opposite side of CRM1 (Fig. 1). Only one salt bridge each has been observed for the C-terminal tail in interaction with the acidic loop (Lys1068 with Glu426) and the back side of the NES cleft (Glu1073 and Gln591; Fig. 1). In contrast, the B-helix of H21 is located at the outer surface of the CRM1 toroid in parallel orientation to helix 21A in the Ran-containing complexes (Fig. S3).
Another important structural difference between free CRM1 and its export complexes concerns the acidic loop, which is inserted between the A- and B-helix of H9 (Figs. S1 and S4). In free ctCRM1, this region contacts the HEAT repeat helices forming the back side of the NES cleft (Fig. S5A). The observed conformation of the acidic loop is clearly different from that in the CRM1–RanGTP and CRM1–RanGTP–SPN1 complexes (in the seatbelt conformation) and instead closely resembles the orientation of the respective part in the CRM1–RanGTP–RanBP1 structure, with the acidic loop in a flipped-back conformation (Fig. S3). Specifically, hydrophobic residues of the acidic loop (Val427, Leu428, Ile429, and Ile437) pack against the surface of helices 10B, 11B, and 12B at the back side of the NES cleft. This results in a major rearrangement of the side chains located in the hydrophobic core of H10, H11, and H12 (Fig. S5 B–D). Met580 is shifted toward the hydrophobic residues of the acidic loop, and the resulting empty space is filled by Met542, the position of which in turn is replaced by Phe569. As a consequence, this rearrangement facilitates a significant rotation and rearrangement of the highly conserved Lys531 and Lys534. The latter enables the helix 11A to move toward helix 12A. This ultimately leads to a closure of the NES cleft, which prevents binding of an NES-bearing cargo (Fig. S2). Mutations of the respective hydrophobic residues within the acidic loop of yeast CRM1 have been shown to cause a reduction of NES-cargo release rate in CRM1–RanGTP–RanBP1 complexes, indicating a direct influence on NES cleft accessibility (14).
However, analysis of the crystal packing of the orthorhombic crystal form revealed that this conformation of the acidic loop might also be stabilized by the interaction of Glu434, located in the acidic loop, with Gly642, of a symmetry-related molecule. To exclude the possibility that the flipped-back conformation of the acidic loop as well as the overall open conformation of CRM1 are at least in part products of crystal packing, the trigonal ctCRM1 crystal form was investigated as well. The 3.1-Å crystal structure of P31 ctCRM1 was solved by molecular replacement, revealing a different crystal packing of the CRM1 molecules (Table S1). There are two molecules in the asymmetric unit, both showing the extended superhelical structure with the C-terminal helix crossing the CRM1 arch and the acidic loop in the flipped-back position even though the acidic loop is not involved in any crystal contacts (Figs. S4B and S6A). However, there are also two remarkable differences between the structures. First, in the trigonal crystal form, CRM1 adopts a slightly less extended conformation with a reduced superhelical pitch, which also differs between the two molecules in the asymmetric unit, with an rmsd of 1.03 Å (Fig. S6A). Second, H1 to H3, as well as several loops and the C-terminal five residues corresponding to the C-terminal acidic tail, are not clearly defined in the electron density map and thus were not modeled. Overall, these differences reflect the intrinsic high plasticity of free CRM1, which is also known for other transport factors in their free state (20, 21).
EM Structure Analysis.
We next asked whether the extended conformation of free CRM1 could also exist in a noncrystalline environment. For this purpose, ctCRM1 was subjected to the GraFix approach (22) and subsequent single-particle EM analysis (Figs. S7 and S8). Strikingly, free ctCRM1 was detected in at least two different and clearly distinct conformations. Two thirds of the classified particles clearly adopt an extended and pitched superhelical shape similar to that seen in the crystal structures of free ctCRM1 when fitted in the EM model (Fig. 4). Interestingly, the EM model supports a free CRM1 conformation, which adopts an even more extended state than seen in the orthorhombic crystal structure. The other conformer, represented by the remaining third, resembles the shape of a more compact closed ring or distorted toroid, reminiscent of the CRM1 conformation observed in various binary and ternary complexes. These data suggest that free CRM1, also in solution, is able to switch between an extended and a compact conformation.
Fig. 4.
Single-particle EM analysis of free ctCRM1. EM models of the compact (orange) as well as the extended conformation (blue) of free ctCRM1 are shown. The crystal structures of free ctCRM1 and CRM1 in complex with SPN1 and RanGTP are fitted to the envelope models of the EM structures. The position of the NES cleft is marked by a black arrowhead.
MD Simulations.
We next addressed the question whether CRM1 in the extended form represents a strained conformation, and, if so, which structural features prevent closure. We applied MD simulations, which have previously provided insight into the high conformational flexibility of HEAT repeat proteins (23–25). We first compared simulations of WT CRM1 with a deletion mutant lacking the C-terminal helix 21B and the acidic tail. To monitor transitions from the extended toward the compact crystal structure, the progress of the trajectory along the difference vector between those two structures was recorded and used as a reaction coordinate. In five unperturbed simulations of WT, all structures remain in the extended conformation or elongate even further (Fig. 5A). By contrast, 6 of 10 simulations of the helix deletion mutant spontaneously progress toward the compact state within as little as 100 ns (Fig. 5B). Simulations, in which closing and subsequent contact formation between the N- and C-terminal regions of CRM1 are observed, display lower flexibility after closing than those that remain open. These results show that the C-terminal helix is a major determinant restricting the conformational flexibility and thus shifting the population more toward the extended conformation.
Fig. 5.
MD simulations of WT and mutant free ctCRM1. Projections of WT simulations (A, cyan) and simulations with deleted C-terminal helix (B, red) onto the difference vector between the extended and compact structure constitute a measure of how much the protein changes into the compact conformation. (C) Projections onto the plane in the configurational space spanned by the extended, compact, and almost compact crystal structure show that, after deletion of the C-terminal helix, the system adopts the configuration of the almost compact structure (magenta square) rather than the compact conformation (orange square).
We next examined the contribution of the acidic loop. In all but one simulation of the acidic loop deletion mutant, the structure remains in the extended conformation, closely resembling the behavior of WT (Fig. S9A). In contrast, the double deletion mutant lacking both the C-terminal helix and the acidic loop shows conformational changes similar to the deletion mutant lacking the C-terminal helix only (Fig. S9B). This result indicates that the acidic loop plays only a minor role in determining the conformation of CRM1. To characterize the closing motion leading from the extended to compact conformation in more detail, all trajectories were projected onto the subspace defined by the three available crystal structures of free CRM1, the CRM1–SPN1 complex, and the ternary CRM1–RanGTP–SPN1 complex (Fig. 5C). Starting from the extended state, CRM1 does not seem to target the compact conformation seen in the ternary CRM1–RanGTP–SPN1 complex, but rather approaches an almost compact conformation resembling the one seen in the CRM1–SPN1 complex (Fig. 2C). Indeed, the rmsd to the almost compact crystal structure decreases from an initial value of 5.1 Å to a minimum of 2 Å, whereas that to the compact conformation remains at a minimum of 3.9 Å.
Because the plane defined by the three aforementioned crystal structures may not contain all relevant dynamics, and to identify the dominant structural changes, we performed a principal component analysis of the combined ensemble of all trajectories of the C-terminal deletion mutant. Fig. S9C shows projections onto the first two eigenvectors of all trajectories, including all variants. As indicated by the inserted ribbon structures, motions along the first principal component analysis eigenvector predominantly describe a change in the diameter of the ring, whereas dynamics along the second eigenvector represent a change in the pitch of the superhelical structure (Movies S1 and S2). Whereas the first eigenvector is similar to the vector connecting the extended and compact crystal structures, significant dynamics are also seen along the second eigenvector, i.e., changes in the pitch without changes in the ring diameter. This motion is not contained in the differences among the three crystal structures.
Next we asked whether the conformational transition from the almost compact to the extended state of CRM1 may affect the cargo affinity of the CRM1 NES binding cleft. To this end, we examined structural changes of the binding cleft and investigated if these depend on the CRM1 conformation. Accordingly, each of the trajectories was subdivided into four states—extended/open, extended/closed, compact/open, and compact/closed—corresponding to the respective state of the overall CRM1 conformation and that of the NES binding cleft, respectively. We then determined from the simulations the conditional probabilities that the binding cleft is open (i.e., high-affinity state), given that the overall conformation is extended or, respectively, compact. Indeed, the probability of observing an NES binding configuration of the NES cleft is larger by a factor of approximately three for the compact CRM1 conformation than for the extended, low-affinity one, with significances lower than 1% and 3% for the double deletion and helix deletion mutant, respectively (Table S2). This population shift corresponds to a marked increase of the free energy difference between the closed and open NES cleft conformations of ∆∆G of 2.7 ± 1.2 kJ/mol for the C-terminal helix deletion mutant and of ∆∆G of 3.5 ± 1.3 kJ/mol for the double deletion mutant.
Discussion
The structural basis for the cooperative binding of cargo and RanGTP to the exportin CRM1 has so far remained an open question. All crystal structures of CRM1 in various complexes with RanGTP and/or SPN1 or RanGTP and RanBP1 showed a toroid, a distorted ring-shaped molecule, in which N- and C-terminal HEAT repeats interact (Fig. 2 B–D and Fig. S3).
Here we demonstrate that CRM1 in the free state can adopt a compact, ring-like structure, and an extended, superhelical structure. The structures of free CRM1 obtained from different crystal forms, as well as single-particle EM and MD simulations, indicate a high intrinsic flexibility. The MD simulations show that the C-terminal helix contributes to stabilizing the extended conformation like a ratchet, whereas the acidic loop is of less importance for the overall structure. Moreover, the extended conformation exhibits a high degree of flexibility that is lost in the closing process as a result of interactions between the terminal HEAT repeats that increase the rigidity of CRM1.
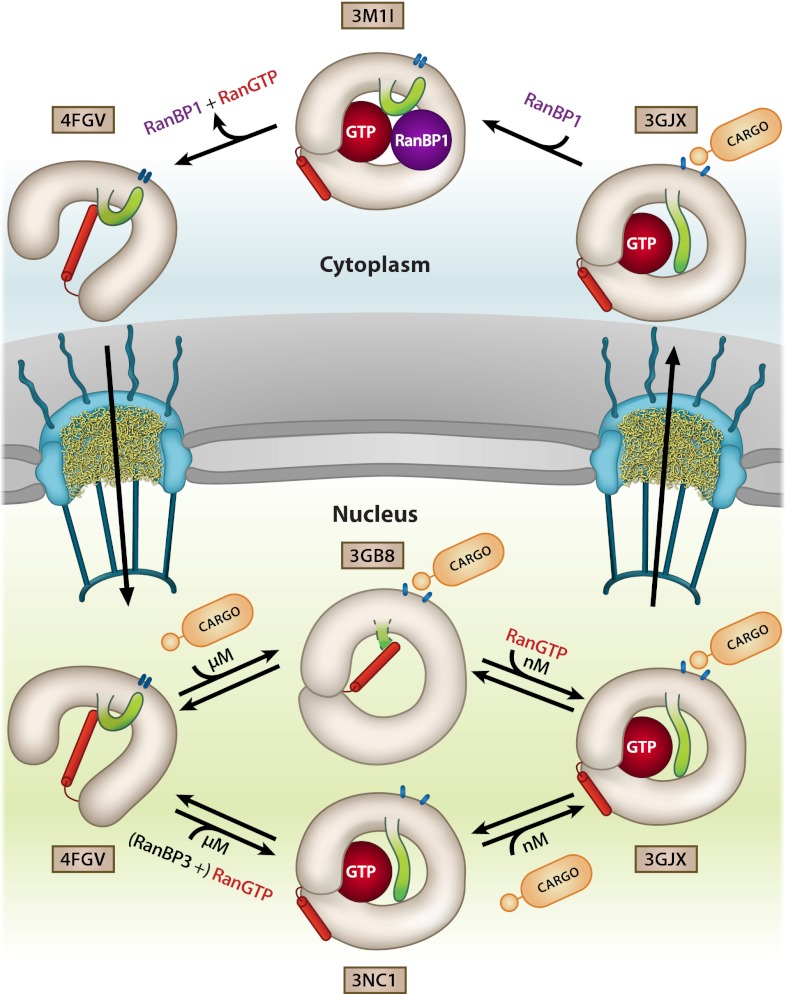
The cooperative binding of RanGTP and cargo by CRM1 was previously suggested to be achieved mainly by local subtle structural changes in the CRM1 toroid (9, 19). Binding of either of the two interaction partners leads to rearrangements in CRM1 altering the binding properties of the second binding site. However, in light of the results presented here, the cooperativity of cargo and RanGTP binding can occur in both pathways for export complex assembly that can be envisioned. As suggested by our EM data, free CRM1 is predominantly in an extended conformation, most likely with the acidic loop flipped back, intricately interacting with the HEAT repeats involved in NES binding, and stabilized by the C-terminal helix 21B (Fig. 6). For binding of RanGTP first, helix 21B has to be released and move into a parallel orientation to helix 21A, as the crossing C-terminal helix is partially occupying the volume required for RanGTP binding (Fig. S3). The release and reorientation of helix 21B seems to be an unfavorable process, which could be causative for the low micromolar affinity of RanGTP to CRM1 (11, 12). During RanGTP binding, the extended structure condenses by interaction of N- and C-terminal regions, both contacting RanGTP. Additionally, the acidic loop is released from the flipped-back position and moves toward the seatbelt position. As a consequence, the NES cleft retains its flexibility and becomes more accessible to cargo by changing the conformation, thereby enhancing the affinities for cargo by a factor of approximately 500 from a low micromolar to a low nanomolar range (11). These large structural rearrangements of the acidic loop and the C-terminal helix of H21 lead to a compact structure with only little superhelical pitch.
Fig. 6.
Model for cooperative CRM1 export complex assembly and disassembly showing its conformational variability and the important structural features in different states of the transport cycle. CRM1 is shown in the respective conformations and colored in gray with the acidic loop highlighted in green. The C-terminal helix of CRM1 is shown in red, and the NES binding cleft is represented by blue ovals. The PDB ID codes of the individual crystal structures used are indicated.
In the vice-versa assembly procedure, SPN1 binds first to CRM1 (Fig. 6). In general, NES binding is weak (i.e., micromolar Kd) in the absence of RanGTP, but might be increased as other domains of the cargo bind to CRM1 in addition (10, 11). The association of CRM1 and cargo requires CRM1 in a more toroid-like shape to open the NES binding cleft. The MD simulations suggest a pronounced population shift of the NES cleft toward its open, high-affinity conformation upon CRM1 compaction, which points to a conformational selection mechanism rather than a clear-cut induced fit. In this toroid conformation, the helix 21B might still span the CRM1 interior as observed in the CRM1–SPN1 binary complex structure (10), but is incompatible with the arrangement of the molecules as present in the ternary export complex of CRM1–RanGTP–SPN1 (Fig. S6B). The structure of CRM1 in this binary CRM1–SPN1 complex reveals a high flexibility of the terminal regions as well as the acidic loop, suggesting its release from the flipped-back position upon cargo binding. This conformation with the helix 21B in a kinked and presumably more strained conformation would increase the affinity for RanGTP ∼500-fold (11, 12) by facilitating the release and rearrangement of helix 21B (Fig. S6B). Upon RanGTP binding, CRM1 has to arrest properly in the ring-like shape for a stable ternary complex formation.
Taken together, in both scenarios, upon binding of one protein, CRM1 is transformed from a more or less extended conformation to an almost compact one. This intermediate state with reoriented helix 21B and acidic loop changes the structural properties of the binding site for the missing partner and leads to an increase of affinity. In any case, the C-terminal helix plays a critical role as its deletion causes a dramatic decrease of cooperativity in complex formation and thus leads to high-affinity cargo binding even in the absence of RanGTP (26). Detailed biochemical analysis revealed that deletion of the last 9 aa of CRM1 (C-terminal acidic tail) or mutation of the acidic residues therein to alanine (1064EIPEEMCD1071 to 1064AIPAAMCA1071) leads to an approximately 10-fold increase of CRM1-cargo affinity (15.6 µM to 1.7 µM), independent of the presence of RanGTP (19). Furthermore, this effect is dramatically increased when deletion of the C-terminal acidic tail is combined with mutations in the acidic loop (430VLV432 to 430AAA432). In this double mutant, cargo affinity of free CRM1 is more than 600-fold higher with respect to WT (15.6 µM to 0.0025 µM), and thus comparable to the affinity of CRM1 for cargo in the presence of RanGTP (0.0015 µM for SPN1). However, of the acidic tail residues analyzed in this study, only Glu1073 of free ctCRM1 is in direct contact with the side chain of Gln591, which is the counterpart of Lys594 in human CRM1 (Fig. 1). This suggests a more global electrostatic attraction effect of positive and negative charged patches in the two regions of CRM1 rather than direct ionic interactions.
In summary, CRM1 exhibits large overall structural dynamics, in line with other transport receptors such as importin-β, exportin-t, and Cse1 (27). The cooperativity of cargo and RanGTP binding is achieved by the conformational switch of two distant regions, the acidic loop and the extensive motion of the C-terminal helix. This helix is a unique feature of CRM1 and represents an additional regulatory mechanism not observed in any other nuclear transport receptor. CRM1 dynamics reveal how the interplay between the distant and not-overlapping binding sites of RanGTP and cargo on CRM1 can be mediated and fine-tuned. It remains an open question whether the observed structural flexibility of CRM1 is also important for its function in cell cycle control during mitotic progression.
Materials and Methods
Detailed experimental procedures are described in SI Materials and Methods.
Protein Expression and Purification.
GST-ctCRM1 was expressed from pET24d in Escherichia coli BL21(DE3) (Merck) at 20 °C. The protein was purified on a GSTrap column (GE Healthcare) followed by tobacco etch virus protease-mediated GST cleavage and a final gel filtration with a Superdex 200 column (GE Healthcare). Pure ctCRM1 was concentrated to 20 mg/mL and stored at −80 °C.
Crystallization and Crystal Structure Determination.
ctCRM1 was crystallized by vapor diffusion. Orthorhombic crystals were flash-cooled in liquid nitrogen after soaking in reservoir solution containing, additionally, 14% (vol/vol) glycerol. A data set was collected at BL14.1 operated by the Helmholtz-Zentrum Berlin at the BESSY II electron storage ring (Berlin-Adlershof, Germany) (28). Data were processed with iMOSFLM (29) and SCALA (30). The structure was solved with PHASER (31) by using the crystal structure of mouse CRM1 as search model (PDB ID code 3GJX) (9). The model was refined with PHENIX (32) at 2.94 Å resolution to R and Rfree values of 22.0% and 24.2%, respectively (Table S1). Data of the trigonal crystals were collected at beamline P14 (PETRA III, European Molecular Biology Laboratory, Hamburg, Germany) and processed by using XDS (33) and XSCALE. The structure was solved by means of molecular replacement with PHASER by using the previously solved orthorhombic ctCRM1 structure.
EM Preparation and Image Processing.
ctCRM1 was subjected to the GraFix protocol (22). Negatively stained particles were imaged by using a CM200FEG (Philips) at a magnification of 155,000× (1.85 Å/pixel). Particles were picked, contrast transfer function correction was performed per Sander et al. (34), and further image processing was done in Imagic (35). A starting model was generated by using angular reconstitution facilitated by a voting algorithm (36). Two distinct conformations were detected in the dataset and it was split based on cross correlations. Final 3D models were obtained at a resolution of ∼20 Å.
MD Simulations and Analysis.
Simulations were performed by using Gromacs 4.5 (37) together with the Amber99sb force field (38) and the extended simple point charge water model (39) in a constant particle number/pressure/temperature ensemble. Electrostatic interactions were calculated by using particle-mesh Ewald summation (40). We used four simulation systems: the WT CRM1 extended state (PDB ID code 4FGV), a C-terminal deletion mutant, an acidic loop deletion mutant, and a system containing both deletions. To compare our trajectories with the crystal structures available for CRM1, i.e., the CRM1–RanGTP–SPN1 complex (compact structure, PDB ID code 3GJX, from mouse), the CRM1–SPN1 complex (almost compact structure, PDB ID code 3GB8, from human), and the cargo-free system (extended structure, PDB ID code 4FGV, presented here), a common subset of Cα atoms was selected from residues that are identical among the three orthologues, which was determined by sequence alignment and restricted to α-helical regions of the extended conformation.
Supplementary Material
Acknowledgments
We thank Jil Schrader, Maren Müller, and Stephanie Schell for excellent technical assistance; Ralph Kehlenbach for critical reading of the manuscript; and Lin-Ta Hsu for creating the schematic of the transport cycle. This work was supported by Deutsche Forschungsgemeinschaft Sonderforschungsbereich 860 (to R.F.), a scholarship from Boehringer Ingelheim Fonds (to D.H.), the Scottish Universities Physics Alliance (U.Z.), and the UK National Physical Laboratory (U.Z.). Synchrotron data collection was supported by the Helmholtz-Zentrum.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4FGV and 4HZK); and the EMDataBank (ID codes EMD-2110 and EMD-2111).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1215214110/-/DCSupplemental.
References
- 1.Cook A, Bono F, Jinek M, Conti E. Structural biology of nucleocytoplasmic transport. Annu Rev Biochem. 2007;76:647–671. doi: 10.1146/annurev.biochem.76.052705.161529. [DOI] [PubMed] [Google Scholar]
- 2.Görlich D, Kutay U. Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Biol. 1999;15:607–660. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- 3.Fornerod M, Ohno M, Yoshida M, Mattaj IW. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90(6):1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 4.Stade K, Ford CS, Guthrie C, Weis K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 1997;90(6):1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- 5.Gadal O, et al. Nuclear export of 60s ribosomal subunits depends on Xpo1p and requires a nuclear export sequence-containing factor, Nmd3p, that associates with the large subunit protein Rpl10p. Mol Cell Biol. 2001;21(10):3405–3415. doi: 10.1128/MCB.21.10.3405-3415.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moy TI, Silver PA. Nuclear export of the small ribosomal subunit requires the ran-GTPase cycle and certain nucleoporins. Genes Dev. 1999;13(16):2118–2133. doi: 10.1101/gad.13.16.2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kehlenbach RH, Dickmanns A, Gerace L. Nucleocytoplasmic shuttling factors including Ran and CRM1 mediate nuclear export of NFAT In vitro. J Cell Biol. 1998;141(4):863–874. doi: 10.1083/jcb.141.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Güttler T, et al. NES consensus redefined by structures of PKI-type and Rev-type nuclear export signals bound to CRM1. Nat Struct Mol Biol. 2010;17(11):1367–1376. doi: 10.1038/nsmb.1931. [DOI] [PubMed] [Google Scholar]
- 9.Monecke T, et al. Crystal structure of the nuclear export receptor CRM1 in complex with Snurportin1 and RanGTP. Science. 2009;324(5930):1087–1091. doi: 10.1126/science.1173388. [DOI] [PubMed] [Google Scholar]
- 10.Dong X, et al. Structural basis for leucine-rich nuclear export signal recognition by CRM1. Nature. 2009;458(7242):1136–1141. doi: 10.1038/nature07975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paraskeva E, et al. CRM1-mediated recycling of snurportin 1 to the cytoplasm. J Cell Biol. 1999;145(2):255–264. doi: 10.1083/jcb.145.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Petosa C, et al. Architecture of CRM1/Exportin1 suggests how cooperativity is achieved during formation of a nuclear export complex. Mol Cell. 2004;16(5):761–775. doi: 10.1016/j.molcel.2004.11.018. [DOI] [PubMed] [Google Scholar]
- 13.Askjaer P, Jensen TH, Nilsson J, Englmeier L, Kjems J. The specificity of the CRM1-Rev nuclear export signal interaction is mediated by RanGTP. J Biol Chem. 1998;273(50):33414–33422. doi: 10.1074/jbc.273.50.33414. [DOI] [PubMed] [Google Scholar]
- 14.Koyama M, Matsuura Y. An allosteric mechanism to displace nuclear export cargo from CRM1 and RanGTP by RanBP1. EMBO J. 2010;29(12):2002–2013. doi: 10.1038/emboj.2010.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fukuhara N, Fernandez E, Ebert J, Conti E, Svergun D. Conformational variability of nucleo-cytoplasmic transport factors. J Biol Chem. 2004;279(3):2176–2181. doi: 10.1074/jbc.M309112200. [DOI] [PubMed] [Google Scholar]
- 16.Vetter IR, Arndt A, Kutay U, Görlich D, Wittinghofer A. Structural view of the Ran-Importin beta interaction at 2.3 A resolution. Cell. 1999;97(5):635–646. doi: 10.1016/s0092-8674(00)80774-6. [DOI] [PubMed] [Google Scholar]
- 17.Chook YM, Blobel G. Structure of the nuclear transport complex karyopherin-beta2-Ran x GppNHp. Nature. 1999;399(6733):230–237. doi: 10.1038/20375. [DOI] [PubMed] [Google Scholar]
- 18.Cook AG, Fukuhara N, Jinek M, Conti E. Structures of the tRNA export factor in the nuclear and cytosolic states. Nature. 2009;461(7260):60–65. doi: 10.1038/nature08394. [DOI] [PubMed] [Google Scholar]
- 19.Fox AM, Ciziene D, McLaughlin SH, Stewart M. Electrostatic interactions involving the extreme C terminus of nuclear export factor CRM1 modulate its affinity for cargo. J Biol Chem. 2011;286(33):29325–29335. doi: 10.1074/jbc.M111.245092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Conti E, Müller CW, Stewart M. Karyopherin flexibility in nucleocytoplasmic transport. Curr Opin Struct Biol. 2006;16(2):237–244. doi: 10.1016/j.sbi.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 21.Zachariae U, Grubmüller H. A highly strained nuclear conformation of the exportin Cse1p revealed by molecular dynamics simulations. Structure. 2006;14(9):1469–1478. doi: 10.1016/j.str.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 22.Kastner B, et al. GraFix: Sample preparation for single-particle electron cryomicroscopy. Nat Methods. 2008;5(1):53–55. doi: 10.1038/nmeth1139. [DOI] [PubMed] [Google Scholar]
- 23.Kappel C, Zachariae U, Dölker N, Grubmüller H. An unusual hydrophobic core confers extreme flexibility to HEAT repeat proteins. Biophys J. 2010;99(5):1596–1603. doi: 10.1016/j.bpj.2010.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zachariae U, Grubmüller H. Importin-beta: Structural and dynamic determinants of a molecular spring. Structure. 2008;16(6):906–915. doi: 10.1016/j.str.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 25.Grinthal A, Adamovic I, Weiner B, Karplus M, Kleckner N. PR65, the HEAT-repeat scaffold of phosphatase PP2A, is an elastic connector that links force and catalysis. Proc Natl Acad Sci USA. 2010;107(6):2467–2472. doi: 10.1073/pnas.0914073107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dong X, Biswas A, Chook YM. Structural basis for assembly and disassembly of the CRM1 nuclear export complex. Nat Struct Mol Biol. 2009;16(5):558–560. doi: 10.1038/nsmb.1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cook AG, Conti E. Nuclear export complexes in the frame. Curr Opin Struct Biol. 2010;20(2):247–252. doi: 10.1016/j.sbi.2010.01.012. [DOI] [PubMed] [Google Scholar]
- 28.Mueller U, et al. Facilities for macromolecular crystallography at the Helmholtz-Zentrum Berlin. J Synchrotron Radiat. 2012;19(pt 3):442–449. doi: 10.1107/S0909049512006395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Battye TGG, Kontogiannis L, Johnson O, Powell HR, Leslie AGW. iMOSFLM: A new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):271–281. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Collaborative Computational Project, Number 4 The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50(pt 5):760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 31.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40(pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66(pt 2):125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sander B, Golas MM, Stark H. Automatic CTF correction for single particles based upon multivariate statistical analysis of individual power spectra. J Struct Biol. 2003;142(3):392–401. doi: 10.1016/s1047-8477(03)00072-8. [DOI] [PubMed] [Google Scholar]
- 35.van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M. A new generation of the IMAGIC image processing system. J Struct Biol. 1996;116(1):17–24. doi: 10.1006/jsbi.1996.0004. [DOI] [PubMed] [Google Scholar]
- 36.Singer A, Coifman RR, Sigworth FJ, Chester DW, Shkolnisky Y. Detecting consistent common lines in cryo-EM by voting. J Struct Biol. 2010;169(3):312–322. doi: 10.1016/j.jsb.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hess B, Kutzner C, van der Spoel D, Lindahl E. GROMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 38.Hornak V, et al. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins. 2006;65(3):712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Berendsen HJC, Grigera JR, Straatsma TP. The missing term in effective pair potentials. J Phys Chem. 1987;91:6269–6271. [Google Scholar]
- 40.Darden T, York D, Pedersen L. Particle mesh Ewald - an N log(N) method for Ewald sums in large systems. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.