Abstract
Cyanobacteriochromes are cyanobacterial tetrapyrrole-binding photoreceptors that share a bilin-binding GAF domain with photoreceptors of the phytochrome family. Cyanobacteriochromes are divided into many subclasses with distinct spectral properties. Among them, putative phototaxis regulators PixJs of Anabaena sp. PCC 7120 and Thermosynechococcus elongatus BP-1 (denoted as AnPixJ and TePixJ, respectively) are representative of subclasses showing red-green-type and blue/green-type reversible photoconversion, respectively. Here, we determined crystal structures for the AnPixJ GAF domain in its red-absorbing 15Z state (Pr) and the TePixJ GAF domain in its green-absorbing 15E state (Pg). The overall structure of these proteins is similar to each other and also similar to known phytochromes. Critical differences found are as follows: (i) the chromophore of AnPixJ Pr is phycocyanobilin in a C5-Z,syn/C10-Z,syn/C15-Z,anti configuration and that of TePixJ Pg is phycoviolobilin in a C10-Z,syn/C15-E,anti configuration, (ii) a side chain of the key aspartic acid is hydrogen bonded to the tetrapyrrole rings A, B and C in AnPixJ Pr and to the pyrrole ring D in TePixJ Pg, (iii) additional protein-chromophore interactions are provided by subclass-specific residues including tryptophan in AnPixJ and cysteine in TePixJ. Possible structural changes following the photoisomerization of the chromophore between C15-Z and C15-E are discussed based on the X-ray structures at 1.8 and 2.0-Å resolution, respectively, in two distinct configurations.
Keywords: back-to-back dimer, chemotaxis, methyl accepting
Phytochromes are photosensory proteins responsive to red and far-red light that regulate a wide range of physiological processes in many organisms (1, 2). Their N-terminal photosensory modules consist of PAS (Per/ARNT/Sim), GAF (cGMP-phosphodiesterase/adenylate cyclase/FhlA), and PHY (phytochrome-specific) domains. In plant and cyanobacterial phytochromes, a linear tetrapyrrole chromophore, respectively phytochromobilin and phycocyanobilin (PCB), is covalently bound to a conserved Cys residue within the GAF domain. Bacterial and fungal phytochromes covalently bind biliverdin IXα (BV) via a conserved Cys residue N-terminal to the PAS domain. It is generally accepted that light excitation triggers a Z/E isomerization of the C15=C16 double bond between pyrrole rings C and D.
Structural studies of phytochromes have implicated conformational changes that occur during the reversible photoconversion (3–7). The chromophore is buried in a pocket within the GAF domain with C5-Z,syn/C10-Z,syn/C15-Z,anti configuration in the Pr state, and a C5-Z,syn/C10-Z,syn/C15-E,anti configuration in the Pfr state. The so-called “pyrrole water” lies on the α-face of the bilin (8), which is held by the conserved His260, and is thought to play a key role in the stabilization and protonation of bilin rings A, B, and C. Controversially, rotation of ring A, but not ring D, during photoconversion has been reported for the isolated GAF domain of a single thermophilic phytochrome (9) belonging to a subfamily lacking the PAS domain and the knotted architecture.
Cyanobacteriochromes (CBCRs) are a recently identified group of photoreceptors distantly related to the phytochrome family that have been found only in cyanobacteria to date (10). The GAF domain is sufficient for fully reversible photochemistry in CBCRs but not in phytochromes. The CBCR family includes regulators of the chromatic acclimation of phycobiliproteins, RcaE and CcaS (11–13), and regulators of phototaxis, such as PixJ, PixA/UirS, and Cph2 (14–18). There are a huge number of uncharacterized CBCRs, which have been detected in many cyanobacterial genomes (10, 19).
CBCRs are spectrally diverse, covering the visible and near-UV regions of the spectrum. The CBCRs studied to date fall into several subclasses based on primary sequence and spectral properties. The blue/green-type CBCRs represented by TePixJ (PixJ of Thermosynechococcus elongatus BP-1) show photoconversion between violet- or blue-absorbing 15Z (Pv/Pb) and teal- or green-absorbing 15E (Pt/Pg) forms (8, 16, 17, 19–26). The red/green-type CBCRs represented by AnPixJ (PixJ of Anabaena sp. PCC 7120) show photoconversion between red-absorbing 15Z and green-absorbing 15E (Pg) forms (27–32). In this type, it is suggested that the Pr form is almost equivalent to that of the phytochromes and starts a primary photoreaction with Z-to-E isomerization in a mechanism similar to that in the phytochromes, but is finally photoconverted to the unique Pg form (27, 29, 31). Green/red-type CBCRs, represented by CcaS, show photoconversion between green-absorbing 15Z (Pg) and red-absorbing 15E (Pr) forms (11, 12).
The blue/green-type CBCRs are most extensively analyzed so far (8, 16, 17, 19–26). They possess a second conserved Cys residue in addition to the “canonical” Cys residue that binds to C31 of ring A. This second Cys is essential for photoconversion (8, 19, 23, 26). Generally, the blue/green-type CBCRs contain phycoviolobilin (PVB) as chromophore (22). There have been two contradictory models concerning the role of the second Cys during Pb/Pg photoconversion. The reversible-attachment model suggests that Cys is covalently ligated to C10 of PVB in Pb, whereas it is not bound to the chromophore in Pg (8, 19). The stable-double-linkage model suggests that the second Cys is stably ligated to the C5 atom of PCB to produce a PVB-like chromophore in both Pb and Pg (23). Recent studies such as chemical modification, chromophore substitution and mass spectrometry using blue/green(teal)-type cyanobacteriochromes (Tlr1999, Tlr0924, and Cph2) provide significant supports for the reversible attachment model (26, 33, 34). Other types of CBCRs have been found, including the violet/yellow-type (SyCikA) that harnesses the Cys in a modified motif (35) and the violet/orange-type and UV/blue-type CBCRs that have a different conserved second Cys residue in an insert sequence (named “insert-Cys CBCRs,” ref. 36). These CBCRs use PCB as a chromophore (36). These facts indicate that intrinsically red-absorbing linear tetrapyrrole chromophores are much more versatile than expected and that CBCRs harness that versatility in many unexpected ways.
In our study, we determined the crystal structures of the GAF domains of AnPixJ and TePixJ in Pr and Pg forms at 1.8- and 2.0-Å resolutions, respectively. Comparison of these structures to each other and to available phytochrome structures provides insights into general and diverse aspects of the linear tetrapyrrole-based photoreceptors.
Results and Discussion
Structure Determination.
Crystals of the Pr form of second GAF domain of AnPixJ (AnPixJg2) were obtained using the hanging-drop vapor diffusion method in space group P43212 (a = b = 69.1 Å, and c = 124.1 Å) with one monomer per asymmetric unit (37). The crystal structure of the Pr form of AnPixJg2 was solved at 1.8-Å resolution via iodine single-wavelength anomalous diffraction and refined to an R-factor of 19.7% and a free R-factor of 22.4% (Table S1). The N-terminal 11 residues (including the His-tag used for purification) and the C-terminal nine residues could not be resolved in the final model because of their diffuse electron density, which is probably due to structural flexibility.
Crystals of the Pg form of GAF domain of TePixJ (TePixJg) were obtained using the hanging-drop vapor diffusion method in space group P65 (a = b = 72.9 Å, and c = 166.7 Å) with two monomers per asymmetric unit. The crystal structure of the Pg form of TePixJg was solved at 2.0-Å resolution by the molecular replacement method using the structure of TePixJg apoprotein as a search model and was refined to an R-factor of 18.7% and a free R-factor of 22.3% (Table S1). Two TePixJg molecules in the crystallographic asymmetric unit are quite similar in overall structure with rmsd values of 0.390 Å, except for the flexible N- and C-terminal residues. Here, we describe the structure of TePixJg by referring mainly to that of chain A because the average B factor of chain A (29.636 Å2) is smaller than that of chain B (34.409 Å2).
Overall Fold of the CBCR GAF Domain.
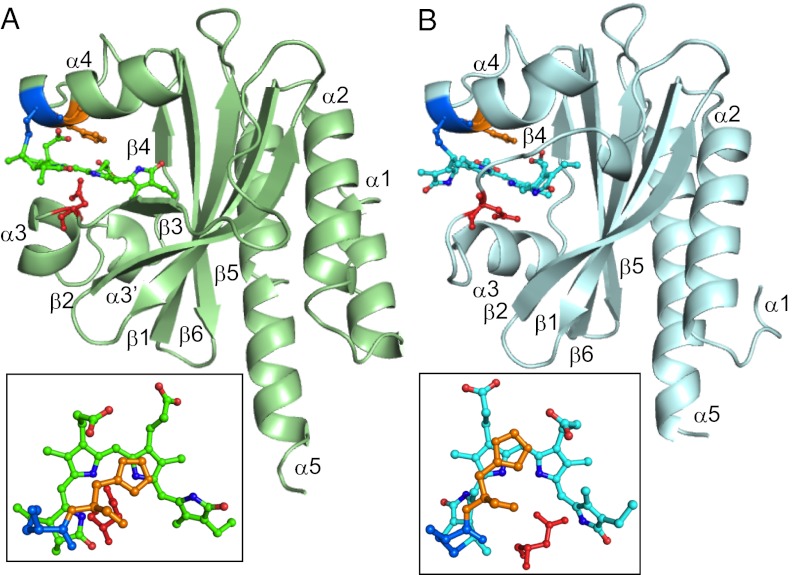
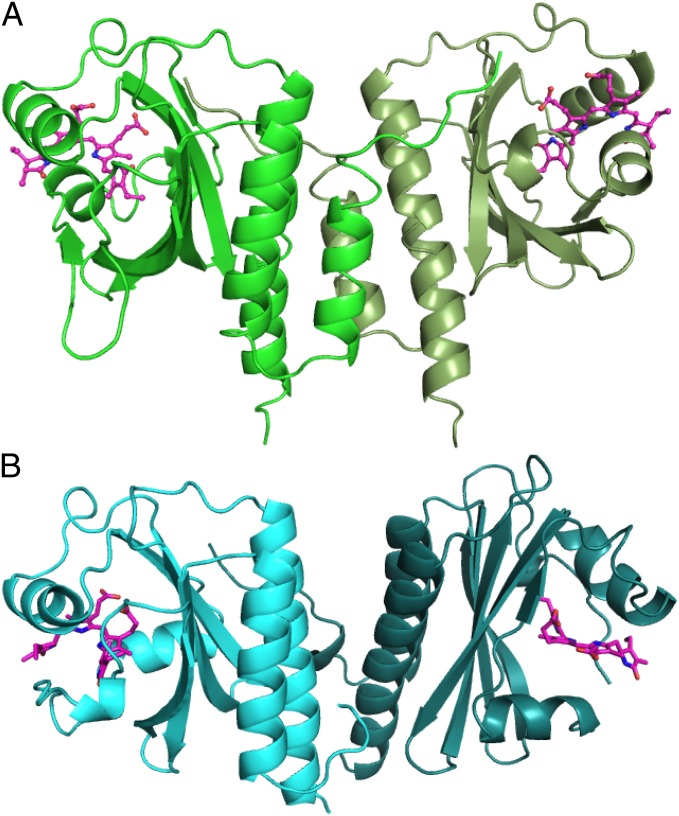
AnPixJg2 and TePixJg adopt typical GAF domain folds (Fig. 1). The GAF domain was divided into three structural units: the central anti-parallel β-sheet consisting of β1–β6, α-helices (α3, α3′, and α4) on the chromophore-binding side, and α helices (α1, α2, and α5) on the back side. These two structures can both be well superimposed with the GAF domain of the cyanobacterial phytochrome Cph1 (Cph1g) (6) in secondary structure level, showing rmsd values of 1.52 Å and 1.58 Å, respectively. The protein structure of the chromophore-binding side of the domain is split into two parts by the cleft in which the crescent-shaped linear tetrapyrrole is embedded. Here, we define the structure on the α-face side of the chromophore as the upper part and the structure on the β-face side the lower part, using the facial definitions of the previous study (8). Fig. 1 and Fig. S1 show side and top views of the whole GAF domain with a chromophore and a few shared key residues (Fig. 1 and Fig. S1 Top and Middle). The linear tetrapyrrole chromophore (PCB for AnPixJg2 or PVB for TePixJg) is inserted into a cleft between the α3 and α4 helices in a similar manner. Three key residues (Cys522, His523, and Asp492 for AnPixJg2; Cys321, His322, and Asp291 for TePixJg; Cys259, His260, and Asp207 for Cph1g), which are critical for chromophore binding, are arranged relative to the chromophore in a manner similar to each other (Fig. 1 and Fig. S1). In all structures, the chromophore is covalently ligated to this Cys and lies between the His and Asp residues on the upper and lower sides, respectively. This seems to be a core structural motif widely conserved among phytochromes (4, 5, 7–9) and CBCRs.
Fig. 1.
Overall structures of AnPixJg2 (A, green) and TePixJg (B, sky blue) in Pr and Pg forms, respectively. (Insets) Chromophores with key residues of Cys (blue), His (yellow), and Asp (red).
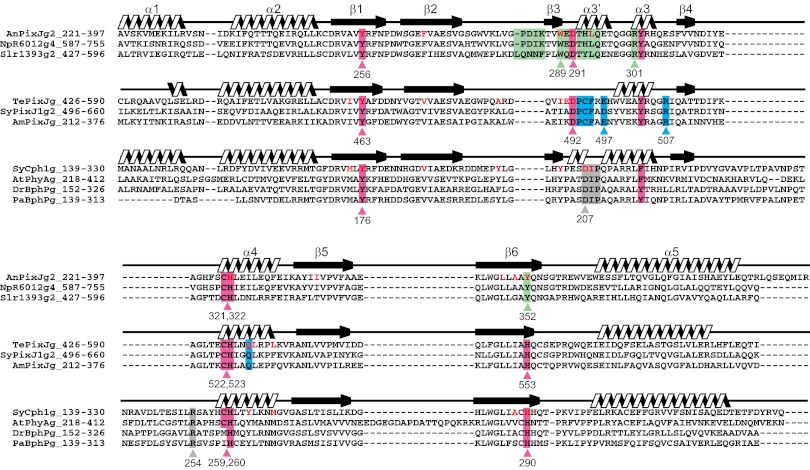
On the other hand, there are some significant differences among these structures in both secondary structure and protein–chromophore interactions (Fig. S2). A long unstructured loop between β4 and α4 of Cph1g, which forms the knot with the neighboring PAS domain (6), is absent in AnPixJg2 and TePixJg. Next to this coil, the α4 helix is similarly positioned between AnPixJg2 and TePixJg, but the α4 helix of Cph1g is significantly closer to the central β sheet. The chromophore-ligating Cys residue is located on the beginning of the α4 helix, so this deviation affects positioning of the chromophores: The chromophore of AnPixJg2 is significantly farther from the central β sheet, compared with that of Cph1g. For example, the distance between the conserved Tyr residue on β1 strand and ring C nitrogen is 9.4 Å in AnPixJg2 and 7.4 Å in Cph2g (Fig. 2; see also Fig. S9). This increased distance and the lack of the knot insert combine to result in shallower insertion of the chromophore within the AnPixJg2 pocket, with the A-ring much more exposed to solvent than in phytochrome crystal structures. Although the highly rotated PVB of TePixJg cannot be compared with the others, the same PCB molecule is initially incorporated into the apoprotein of TePixJg (19). Thus, the assembly process of PCB may be shared between AnPixJg2 and TePixJg. The short α3 and α3′ helices are structurally diverse in these three GAF domains, consistent with the subclass-specific sequences found in these regions. AnPixJg2 possesses a short insertion between β2 and β3, whereas the short β3 strand of AnPixJg2 and Cph1g is an unstructured loop in TePixJg.
Fig. 2.
Multiple alignment of the GAF domains of the phytochromes and CBCRs with the secondary structures. The red/green-type and blue/green-type CBCRs and the phytochromes are structurally aligned. Highly conserved residues among the subgroups are highlighted by pink color. Residues specifically conserved within the subclasses are highlighted by green (red/green-type CBCRs), blue (blue/green-type CBCRs), and gray (phytochromes) colors. Residues that are near the ring D within 5 Å are highlighted by red. Domain names are described such as AnPixJg2. AnPixJg2 means second GAF domain of AnPixJ.
Chromophore and Its Binding Pocket of the Pr Form of AnPixJg2.
AnPixJg2 is covalently bound to PCB, validating the results obtained for AnPixJg2 and other red/green CBCRs by denaturation analysis (27, 32). The PCB chromophore of AnPixJg2 is in a partially extended C5-Z,syn/C10-Z,syn/C15-Z,anti conformation (Fig. 1A and Fig. S3A). The B and C rings of PCB are coplanar, whereas ring A is slightly rotated (12°) and ring D is rotated at an angle of 61° away from this plane (Fig. S4A). The rotated angle of ring D (61°) is much greater than the ∼25–45° reported earlier for the Pr forms of the phytochromes (3, 5, 6) and is in good agreement with the fact that the peak wavelength of the AnPixJg2 Pr (648 nm) is shorter than that of Cph1 (38). Cys321 is covalently connected to the C31 carbon of ring A via a thioether bond, resulting in the S stereochemistry at C31 carbon (Fig. 1A).
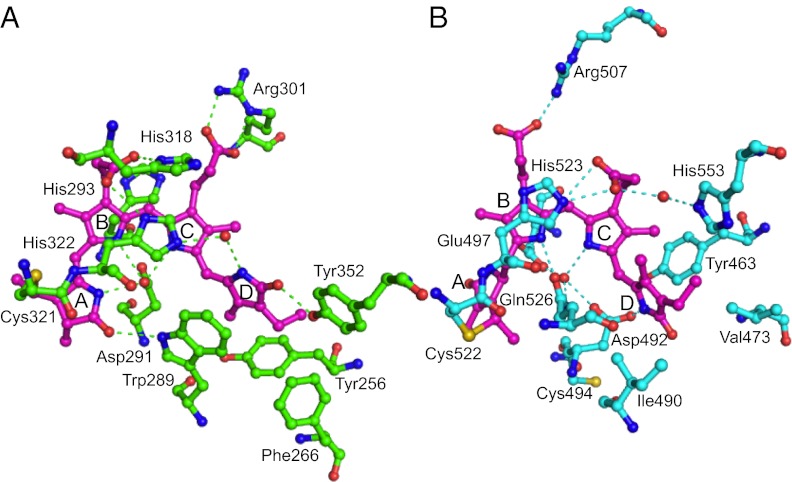
The detailed hydrogen bonding network within the PCB-binding pocket of AnPixJg2 is shown in Fig. 3A and Fig. S4C. The side chain of Cys321 is covalently ligated to the C31 carbon of ring A. One oxygen atom from the side chain of Asp291 is hydrogen bonded to the nitrogen atoms of coplanar rings A, B, and C within 3 Å. Because peak position and molecular extinction coefficient of the Pr form of AnPixJg2 are quite similar to those of phytochromes, the Pr form of AnPixJg2 is proposed to be protonated at the ring C nitrogen atom, as in phytochromes. In the phytochromes, the pyrrole water is stabilized by His260 and could provide its proton to the ring C nitrogen atom. This water molecule is clearly absent in AnPixJg2 Pr (Figs. S5 and S6). Instead, the side chain of Asp291 may be the proton donor for the ring C nitrogen atom in AnPixJg2. The absence of water may be due to the hydrophobic environment under the reverse-oriented imidazole ring of His322, which we inferred from the hydrogen bond network. Thus, the precise position and arrangement of Asp291 of AnPixJg2 is distinctively different from the conserved Asp of DIP motif in phytochromes (4–9) despite of their roughly similar location. Near this Asp291 of AnPixJg2, Trp289 is hydrogen bonded to C1 carbonyl of ring A and is also closely located in parallel with ring D in close proximity (3.4∼3.9 Å), indicative of strong π–π stacking.
Fig. 3.
Chromophore-binding pockets of AnPixJg2 (A) and TePixJg (B).
The ring D carbonyl of C15-Z PCB is hydrogen bonded with the hydroxyl group of Tyr352 in AnPixJg2, whereas it is bonded with the conserved His residue in phytochrome Pr at the same position in the primary sequence (Figs. 2 and 3). Shallower insertion of PCB into AnPixJg2 than Cph1g resulted in the hydrogen bond from opposite side of the ring D carbonyl, and the bond distance is almost identical (Fig. S7).
Chromophore-Binding Pocket of the Pg Form of TePixJ.
The electron density of the chromophore of TePixJg is clearly fitted to PVB (Fig. 1B Inset and Fig. S3B). Notably, the electron density of the methyl carbon C21 is coplanar with ring A, indicative of a C2 = C3 double bond. This clearly shows that the chromophore is not PCB but PVB. Ring A is highly rotated (75°) relative to the slightly rotated B and C rings (21°) (Fig. S4B). Nitrogen atom of ring A faces out from the crescent-shaped tetrapyrrole, which is in contrast with C5-Z,syn of PCB in AnPixJg2. Based on the clear electron density of side chains and C19 carbonyl group (Fig. S3B), ring D can be fitted to a C15-E,anti configuration, which is consistent with acid denaturation analysis (22) and with the only available crystallographic information for the Pfr state of phytochrome (7). Ring D is highly rotated at 73° relative to the coplanar rings B and C. Thus, the overall configuration can be assigned as C10-Z,syn/C15-E,anti of PVB in TePixJg Pg form. It is evident that the chromophore is anchored to Cys522, resulting in the S stereochemistry at C31 carbon like AnPixJg2 (Fig. 1B Inset), but free from the second Cys Cys494, as previously expected based on reversible thiol reactions (26). S stereochemistry of AnPixJg2 and TePixJg is opposite to R stereochemistry of Cph1g (Fig. S1C Lower), although both S and R stereochemistry is detected in the mutant protein (Y263F) structure (39). Isomer selectivity of apoprotein for PCB may be different between AnPixJ/TePixJ and Cph1g.
The detailed hydrogen bonding network within the PVB-binding pocket of TePixJg is shown in Fig. 3B and Fig. S4D. The most notable features of the pocket are hydrogen bonding between the Asp492 side chain and the D ring of PVB and the connection of another oxygen of the same Asp492 side chain to rings B and C via a water molecule. In the Pfr structure of the phytochrome PaBphP (7), the D ring of the C15-E BV chromophore is hydrogen bonded to the Asp side chain of the conserved phytochrome DIP motif, and rings B and C are connected to the Cα carbonyl of the same Asp via the pyrrole water (6). Thus, the hydrogen bond network of the 15E TePixJg Pg state is similar to that of the 15E phytochrome Pfr state, but the precise role of the Asp residue is different. The coplanar rings B and C are covered with His523 imidazole from the upper side, which is similar to AnPixJg2 and to phytochromes. The rotated ring A of PVB is firmly fixed with hydrogen bonds from the side chain of Glu497 to its amide nitrogen at 2.7 Å. Because this Glu497 is somewhat conserved in the blue/green-type subclass of CBCRs (Fig. 2), it may be involved in autoisomerization from PCB to PVB in the assembly or reversible adduct formation between Cys494 and PVB.
Residues for Fixing Propionates and Forming Ring D Cavity.
In all three phytochrome and CBCR structures, one propionate chain of the chromophore is stretched, and another is bent. The ring B propionate is bent in AnPixJg2, whereas the ring C propionate is bent in TePixJg and Cph1g. These propionate chains are fixed by similar but distinctive hydrogen-bonding networks (Fig. S8). The equivalent His residue (His322/523/260 in AnPixJg2/TePixJg/Cph1g), which is highly conserved in most CBCR subfamilies and in phytochromes (Fig. 2), is implicated in fixing the location of the bent propionate. In the Pr-to-Pfr photoconversion process of phytochromes, matrix rearrangement of the ring C propionate is suggested by magic angle spinning NMR spectroscopy (40). Consistently, the hydrogen bond between the ring C propionate and this His residue is broken in the Pfr structure of PaBphP (7). In the case of the CBCRs, similar matrix rearrangement of the bent propionate may also occur during the photoconversion process. On the other hand, an Arg residue (Arg301/507/254 in AnPixJg2/TePixJg/Cph1g), which is conserved within each subclass of GAF domains but specific to the subclasses (Fig. 2), is implicated in fixation of the stretched C8 propionate (Fig. S8).
The D-ring cavity is mainly hydrophobic, with the nitrogen and carbonyl of the ring D stabilized by hydrophilic interactions in all structures. Ten, ten and eleven residues are positioned near ring D within 5 Å in AnPixJg2, TePixJg and Cph1g, respectively (Fig. 2 and Fig. S9). Most of these residues are superimposable. The equivalent Tyr residue (Tyr256/463/176 in AnPixJ/TePixJ/Cph1), which is highly conserved among the CBCRs and the phytochromes forms the hydrophobic “floor” of the D-ring cavity in all three structures, with only slight differences. Phe266/Val473/Val186 and Trp289/Ile490/Tyr203, which are highly conserved within each subclass of GAF domains, are also adopted for the hydrophobic cavity of ring D. Such loosely common arrangements to fix the propionates and ring D may reflect rather loose structural constraints, which would allow the D ring to move upon excitation.
Insights into the Photoconversion Mechanisms of AnPixJ and TePixJ.
AnPixJg2 and related CBCRs show red/green reversible photoconversion between 15Z Pr and 15E Pg forms. If we assume the side chain of Asp291 moved toward the D-ring nitrogen atom in the C15-E,anti PCB of AnPixJg2 Pg form, as observed in the TePixJg Pg form, the hydrogen bonds between the coplanar rings A, B and C and the side chain of Asp291 must be broken during photoconversion. In this regard, Trp289, which is highly conserved among red/green-type subclass CBCRs, may also play a crucial role in directing chromophore movements during photoconversion. The indole ring of Trp289 binds the pyrrole ring D with a strong π–π interaction in the Pr form of AnPixJg2 (Fig. 3A). The indole NH is also hydrogen bonded to the C1 carbonyl of the ring A. This connection of the rings D and A via Trp289 must be modified when C15-Z PCB is photoconverted to C15-E. To examine the role of Trp289, a Trp residue was replaced with a His residue. The W289H mutant showed the absorption peaks near 550–570 nm but responded reversibly (Fig. S10). Because of very low expression yield, we could not determine the chromophore C15-Z/E configuration. Anyway, the results support the idea that the connection of rings A and D via Trp289 is essential for the red absorption in Pr and the large spectral shift during photoconversion between Pr and Pg. Time-resolved spectroscopy suggested that Pr-to-Pg phototransition consists of at least three steps (31): The first step is red-shift of the absorption, which corresponds to Z-to-E photoisomerization, and the following two steps are progressive blue-shifts of the absorption. These blue-shifting steps might be accounted for by deprotonation of the chromophore ring C nitrogen and/or rotation of the ring A. Similar deprotonation was proposed in phytochromes. We postulated the ring A rotation based on the connection between rings A and D via Trp289. We need more spectroscopic data of wild type and mutagenesis studies for further dissection of the photoconversion and signaling.
The blue/green-type subclass of CBCRs including TePixJg possesses a second conserved Cys residue that covalently binds to the chromophore (8, 19, 23). There have been two contradictory models: The reversible-attachment model presumes that the covalent bond in Pb is released in Pg, resulting in changes in conjugated double bonds of the chromophore (8, 19, 26), and the stable double linkage model presumes that Cys-adduct enables flexible changes in conjugated double bonds during photoconversion. (23). The crystal structure of TePixJg Pg revealed that Cys494 is located far from the chromophore and, instead, forms an intermolecular disulfide bond. Although this covalent bond may be due to an artifact during crystallization, the reversible attachment model seems much more plausible in light of this result and in light of the retention of color during crystallization (Fig. S11), indicating that large chromophore rearrangements have not taken place.
Considering these two CBCR structures together allowed us to evaluate possible ligation sites for Cys494 in the TePixJg Pb state (C15-Z PVB). His293 of AnPixJ, which corresponds to the second Cys residue on the sequence alignment (Fig. 2), is located very close to rings B and C, suggesting that the corresponding Cys494 could be positioned in a distance of covalent bound to C10 of the chromophore.
Upon light excitation of TePixJg Pg, isomerization of C15-E to Z of the chromophore takes place as an initial photoreaction (25), which must disrupt the hydrogen bonding of ring D with the Asp492 side chain. The corresponding Asp291 of AnPixJg2 is released from ring D and binds rings B and C, as mentioned above. Likewise, Asp492 of TePixJg may switch the hydrogen-bonding partner from ring D to rings B and C. Because the second cysteine residue Cys494 is located near Asp492, the switching of Asp492 could attract Cys494 toward C10 of the chromophore to form the covalent bond in the subsequent dark. Time-resolved spectroscopy of a similar Pb/Pg-type CBCR, Tlr0924, revealed at least two intermediates, Lumi-G and Meta-G, which peak at 550∼565 nm (41). Together with chemical modification analysis of another related CBCR, Tlr1999 (26), those intermediates harbor C15-Z PVB, but the adduct is not yet formed between the second Cys and PVB. The transient change from Lumi-G to Meta-G may include the above-mentioned switching of Asp492. On the other hand, the reverse switching of Asp492 from rings B and C to ring D after photoconversion of Pb to Pg would then push Cys494 away from C10, destabilizing the covalent bond between them. It is tempting to speculate that these movements of Cys494 may lead to signaling to the output domain of TePixJg.
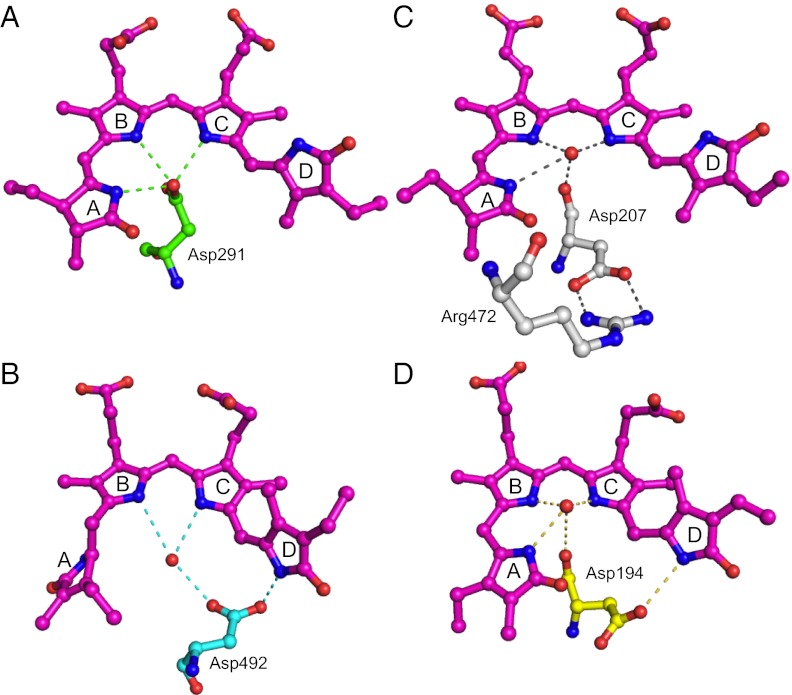
Here we summarize the photoconversion model of the cyanobacteriochromes. The side chain carbonyl of Asp residue forms hydrogen bonds with rings A, B, and C nitrogens in the Z-configured form, whereas it forms hydrogen bond with ring D nitrogen in the E-configured form (Fig. 4 A and B). Switching of these hydrogen bondings may affect the positioning of the Asp residue, possibly resulting in the signaling out to the output domain. Anyway, it must be stressed that the photoconversion-induced switching of the hydrogen bonding seems to be confined within the CBCR GAF domain. On the other hand, phytochrome switches hydrogen bonding of the analogous Asp in Pr (Asp of GAF is bonded to a residue of PHY domain in Cph1) and Pfr (Asp is bonded to ring D nitrogen within GAF in PaBphP) (Fig. 4 C and D) (6, 7). These switches in hydrogen bonding may be the critical difference in the intra- and interdomain signaling between CBCRs and phytochromes. We described insights into photoconversion mechanism of other CBCRs in SI Results and Discussion.
Fig. 4.
Reaction scheme of cyanobacteriochromes and phytochromes with unique Asp residues. (A) Pr form of AnPixJg2. (B) Pg form of TePixJg. (C) Pr form of Cph1. (D) Pfr form of PaBphP.
Quaternary Structure.
In the unit cell of crystals, both AnPixJg2 and TePixJg demonstrate a parallel dimeric arrangement of structure. AnPixJg2 and crystallographic next molecule, related by twofold axis, form a back-to-back configuration, in which C-terminal α helix (α5) in one GAF mainly interacts with α helical bundles in the other GAF (Fig. 5). In the structure of TePixJg, the crystallographic threefold screw operation has generated a back-to-back parallel dimer in a similar manner to AnPixJg2 (Fig. 5).
Fig. 5.
Quaternary structure of AnPixJg2 (A) and TePixJg (B).
These quaternary structures are also homologous to the parallel dimer of bacterial phytochromes, in which the extended helical bundles at the dimer interface are suggested to transmit signals over long distances (7). Similar long distance signaling has also been proposed in the bacterial chemotaxis receptor MCP proteins (42). Even though our structures contain the GAF domains only, secondary structure predictions strongly indicate that α5 of AnPixJg2 and TePixJg is continuously connected to the N-terminal α helix of the neighbor domain (GAF3 in AnPixJ and HAMP in TePixJ). Many other CBCRs are also predicted to harbor similar long helical bundles connected to the neighbor signaling domain. Diverse CBCRs may share a common signaling mechanism.
Materials and Methods
Protein Preparation and Crystallization.
Purification and crystallization of His-tagged AnPixJg2 were described (37). His-tagged TePixJg was first purified by Ni-affinity column from Synechocystis sp. PCC 6803 as reported (20), followed by further purification by using a Superdex 75-gel filtration column (GE Healthcare). The crystallization of TePixJg was carried out at 4 °C under the safety light.
Structure Determination.
All X-ray diffraction data were collected at 100 K on the beamlines of Photon Factory. The structure of AnPixJg2 was determined by the single-wavelength anomalous diffraction (SAD) method using the bound iodide ion. The model was refined against the 1.8-Å native data set with the program Refmac5 (43), with an R-factor of 0.197 and a free-R factor of 0.224 (Table S1). The crystal structure of TePixJg was determined by the molecular replacement method with a program Molrep in CCP4 (44), using the atomic model of the TePixJg apoprotein without the chromophore as the search model. The atomic coordinates, including two molecules of TePixJg in the crystallographic asymmetric unit, were further refined at 2.0 Å. A crystallographic R factor and a free-R factor of the final model are 0.187 and 0.223, respectively (Table S1). Details are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank the staff at beamline BL-5A and BL-17A, KEK, Japan, for their support during the data collection. We also thank Dr. Nathan C. Rockwell for helpful discussion and kind reading of the manuscript. This work was partly supported by Grants-in-Aid for Scientific Research (to R.N., M.I., and G.K.) from the Ministry of Education and Science, Japan, and by PRESTO of Japan Science and Technology Agency (R.N.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3W2Z and 3VV4).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1212098110/-/DCSupplemental.
See Commentary on page 806.
References
- 1.Chen M, Chory J. Phytochrome signaling mechanisms and the control of plant development. Trends Cell Biol. 2011;21(11):664–671. doi: 10.1016/j.tcb.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rockwell NC, Lagarias JC. A brief history of phytochromes. ChemPhysChem. 2010;11(6):1172–1180. doi: 10.1002/cphc.200900894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wagner JR, Zhang J, Brunzelle JS, Vierstra RD, Forest KT. High resolution structure of Deinococcus bacteriophytochrome yields new insights into phytochrome architecture and evolution. J Biol Chem. 2007;282(16):12298–12309. doi: 10.1074/jbc.M611824200. [DOI] [PubMed] [Google Scholar]
- 4.Wagner JR, Brunzelle JS, Forest KT, Vierstra RD. A light-sensing knot revealed by the structure of the chromophore-binding domain of phytochrome. Nature. 2005;438(7066):325–331. doi: 10.1038/nature04118. [DOI] [PubMed] [Google Scholar]
- 5.Yang X, Stojkovic EA, Kuk J, Moffat K. Crystal structure of the chromophore binding domain of an unusual bacteriophytochrome, RpBphP3, reveals residues that modulate photoconversion. Proc Natl Acad Sci USA. 2007;104(30):12571–12576. doi: 10.1073/pnas.0701737104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Essen LO, Mailliet J, Hughes J. The structure of a complete phytochrome sensory module in the Pr ground state. Proc Natl Acad Sci USA. 2008;105(38):14709–14714. doi: 10.1073/pnas.0806477105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang X, Kuk J, Moffat K. Crystal structure of Pseudomonas aeruginosa bacteriophytochrome: photoconversion and signal transduction. Proc Natl Acad Sci USA. 2008;105(38):14715–14720. doi: 10.1073/pnas.0806718105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rockwell NC, et al. A second conserved GAF domain cysteine is required for the blue/green photoreversibility of cyanobacteriochrome Tlr0924 from Thermosynechococcus elongatus. Biochemistry. 2008;47(27):7304–7316. doi: 10.1021/bi800088t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ulijasz AT, et al. Structural basis for the photoconversion of a phytochrome to the activated Pfr form. Nature. 2010;463(7278):250–254. doi: 10.1038/nature08671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ikeuchi M, Ishizuka T. Cyanobacteriochromes: A new superfamily of tetrapyrrole-binding photoreceptors in cyanobacteria. Photochem Photobiol Sci. 2008;7(10):1159–1167. doi: 10.1039/b802660m. [DOI] [PubMed] [Google Scholar]
- 11.Hirose Y, Shimada T, Narikawa R, Katayama M, Ikeuchi M. Cyanobacteriochrome CcaS is the green light receptor that induces the expression of phycobilisome linker protein. Proc Natl Acad Sci USA. 2008;105(28):9528–9533. doi: 10.1073/pnas.0801826105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirose Y, Narikawa R, Katayama M, Ikeuchi M. Cyanobacteriochrome CcaS regulates phycoerythrin accumulation in Nostoc punctiforme, a group II chromatic adapter. Proc Natl Acad Sci USA. 2010;107(19):8854–8859. doi: 10.1073/pnas.1000177107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kehoe DM, Grossman AR. Similarity of a chromatic adaptation sensor to phytochrome and ethylene receptors. Science. 1996;273(5280):1409–1412. doi: 10.1126/science.273.5280.1409. [DOI] [PubMed] [Google Scholar]
- 14.Yoshihara S, Suzuki F, Fujita H, Geng XX, Ikeuchi M. Novel putative photoreceptor and regulatory genes Required for the positive phototactic movement of the unicellular motile cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 2000;41(12):1299–1304. doi: 10.1093/pcp/pce010. [DOI] [PubMed] [Google Scholar]
- 15.Wilde A, Fiedler B, Börner T. The cyanobacterial phytochrome Cph2 inhibits phototaxis towards blue light. Mol Microbiol. 2002;44(4):981–988. doi: 10.1046/j.1365-2958.2002.02923.x. [DOI] [PubMed] [Google Scholar]
- 16.Song JY, et al. Near-UV cyanobacteriochrome signaling system elicits negative phototaxis in the cyanobacterium Synechocystis sp. PCC 6803. Proc Natl Acad Sci USA. 2011;108(26):10780–10785. doi: 10.1073/pnas.1104242108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Narikawa R, et al. Novel photosensory two-component system (PixA-NixB-NixC) involved in the regulation of positive and negative phototaxis of cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 2011;52(12):2214–2224. doi: 10.1093/pcp/pcr155. [DOI] [PubMed] [Google Scholar]
- 18.Moon YJ, et al. Cyanobacterial phytochrome Cph2 is a negative regulator in phototaxis toward UV-A. FEBS Lett. 2011;585(2):335–340. doi: 10.1016/j.febslet.2010.12.007. [DOI] [PubMed] [Google Scholar]
- 19.Ishizuka T, et al. The cyanobacteriochrome, TePixJ, isomerizes its own chromophore by converting phycocyanobilin to phycoviolobilin. Biochemistry. 2011;50(6):953–961. doi: 10.1021/bi101626t. [DOI] [PubMed] [Google Scholar]
- 20.Yoshihara S, Katayama M, Geng X, Ikeuchi M. Cyanobacterial phytochrome-like PixJ1 holoprotein shows novel reversible photoconversion between blue- and green-absorbing forms. Plant Cell Physiol. 2004;45(12):1729–1737. doi: 10.1093/pcp/pch214. [DOI] [PubMed] [Google Scholar]
- 21.Ishizuka T, et al. Characterization of cyanobacteriochrome TePixJ from a thermophilic cyanobacterium Thermosynechococcus elongatus strain BP-1. Plant Cell Physiol. 2006;47(9):1251–1261. doi: 10.1093/pcp/pcj095. [DOI] [PubMed] [Google Scholar]
- 22.Yoshihara S, et al. Reconstitution of blue-green reversible photoconversion of a cyanobacterial photoreceptor, PixJ1, in phycocyanobilin-producing Escherichia coli. Biochemistry. 2006;45(11):3775–3784. doi: 10.1021/bi051983l. [DOI] [PubMed] [Google Scholar]
- 23.Ishizuka T, Narikawa R, Kohchi T, Katayama M, Ikeuchi M. Cyanobacteriochrome TePixJ of Thermosynechococcus elongatus harbors phycoviolobilin as a chromophore. Plant Cell Physiol. 2007;48(9):1385–1390. doi: 10.1093/pcp/pcm106. [DOI] [PubMed] [Google Scholar]
- 24.Ulijasz AT, et al. Cyanochromes are blue/green light photoreversible photoreceptors defined by a stable double cysteine linkage to a phycoviolobilin-type chromophore. J Biol Chem. 2009;284(43):29757–29772. doi: 10.1074/jbc.M109.038513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rockwell NC, Martin SS, Gulevich AG, Lagarias JC. Phycoviolobilin formation and spectral tuning in the DXCF cyanobacteriochrome subfamily. Biochemistry. 2012;51(7):1449–1463. doi: 10.1021/bi201783j. [DOI] [PubMed] [Google Scholar]
- 26.Enomoto G, Hirose Y, Narikawa R, Ikeuchi M. Thiol-based photocycle of the blue and teal light-sensing cyanobacteriochrome Tlr1999. Biochemistry. 2012;51(14):3050–3058. doi: 10.1021/bi300020u. [DOI] [PubMed] [Google Scholar]
- 27.Narikawa R, Fukushima Y, Ishizuka T, Itoh S, Ikeuchi M. A novel photoactive GAF domain of cyanobacteriochrome AnPixJ that shows reversible green/red photoconversion. J Mol Biol. 2008;380(5):844–855. doi: 10.1016/j.jmb.2008.05.035. [DOI] [PubMed] [Google Scholar]
- 28.Kim PW, et al. Second-chance forward isomerization dynamics of the red/green cyanobacteriochrome NpR6012g4 from Nostoc punctiforme. J Am Chem Soc. 2012;134(1):130–133. doi: 10.1021/ja209533x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim PW, et al. Femtosecond photodynamics of the red/green cyanobacteriochrome NpR6012g4 from Nostoc punctiforme. 1. Forward dynamics. Biochemistry. 2012;51(2):608–618. doi: 10.1021/bi201507k. [DOI] [PubMed] [Google Scholar]
- 30.Kim PW, et al. Femtosecond photodynamics of the red/green cyanobacteriochrome NpR6012g4 from Nostoc punctiforme. 2. Reverse dynamics. Biochemistry. 2012;51(2):619–630. doi: 10.1021/bi2017365. [DOI] [PubMed] [Google Scholar]
- 31.Fukushima Y, et al. Photoconversion mechanism of a green/red photosensory cyanobacteriochrome AnPixJ: Time-resolved optical spectroscopy and FTIR analysis of the AnPixJ-GAF2 domain. Biochemistry. 2011;50(29):6328–6339. doi: 10.1021/bi101799w. [DOI] [PubMed] [Google Scholar]
- 32.Chen Y, et al. Photophysical diversity of two novel cyanobacteriochromes with phycocyanobilin chromophores: Photochemistry and dark reversion kinetics. FEBS J. 2012;279(1):40–54. doi: 10.1111/j.1742-4658.2011.08397.x. [DOI] [PubMed] [Google Scholar]
- 33.Savakis P, et al. Light-induced alteration of c-di-GMP level controls motility of Synechocystis sp. PCC 6803. Mol Microbiol. 2012;85(2):239–251. doi: 10.1111/j.1365-2958.2012.08106.x. [DOI] [PubMed] [Google Scholar]
- 34.Rockwell NC, Martin SS, Lagarias JC. Mechanistic insight into the photosensory versatility of DXCF cyanobacteriochromes. Biochemistry. 2012;51(17):3576–3585. doi: 10.1021/bi300171s. [DOI] [PubMed] [Google Scholar]
- 35.Narikawa R, Kohchi T, Ikeuchi M. Characterization of the photoactive GAF domain of the CikA homolog (SyCikA, Slr1969) of the cyanobacterium Synechocystis sp. PCC 6803. Photochem Photobiol Sci. 2008;7(10):1253–1259. doi: 10.1039/b811214b. [DOI] [PubMed] [Google Scholar]
- 36.Rockwell NC, Martin SS, Feoktistova K, Lagarias JC. Diverse two-cysteine photocycles in phytochromes and cyanobacteriochromes. Proc Natl Acad Sci USA. 2011;108(29):11854–11859. doi: 10.1073/pnas.1107844108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Narikawa R, Muraki N, Shiba T, Ikeuchi M, Kurisu G. Crystallization and preliminary X-ray studies of the chromophore-binding domain of cyanobacteriochrome AnPixJ from Anabaena sp. PCC 7120. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2009;65(Pt 2):159–162. doi: 10.1107/S1744309108044151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yeh KC, Wu SH, Murphy JT, Lagarias JC. A cyanobacterial phytochrome two-component light sensory system. Science. 1997;277(5331):1505–1508. doi: 10.1126/science.277.5331.1505. [DOI] [PubMed] [Google Scholar]
- 39.Mailliet J, et al. Spectroscopy and a high-resolution crystal structure of Tyr263 mutants of cyanobacterial phytochrome Cph1. J Mol Biol. 2011;413(1):115–127. doi: 10.1016/j.jmb.2011.08.023. [DOI] [PubMed] [Google Scholar]
- 40.Rohmer T, et al. Light-induced chromophore activity and signal transduction in phytochromes observed by 13C and 15N magic-angle spinning NMR. Proc Natl Acad Sci USA. 2008;105(40):15229–15234. doi: 10.1073/pnas.0805696105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Freer LH, et al. Chemical inhomogeneity in the ultrafast dynamics of the DXCF cyanobacteriochrome Tlr0924. J Phys Chem B. 2012;116(35):10571–10581. doi: 10.1021/jp302637u. [DOI] [PubMed] [Google Scholar]
- 42.Hazelbauer GL, Falke JJ, Parkinson JS. Bacterial chemoreceptors: High-performance signaling in networked arrays. Trends Biochem Sci. 2008;33(1):9–19. doi: 10.1016/j.tibs.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murshudov GN, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Potterton E, Briggs P, Turkenburg M, Dodson E. A graphical user interface to the CCP4 program suite. Acta Crystallogr D Biol Crystallogr. 2003;59(Pt 7):1131–1137. doi: 10.1107/s0907444903008126. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.