Significance
Under adverse environmental conditions, bacteria can form specialized antibiotic-resistant colonies called “biofilms.” In Staphylococcus epidermidis biofilms, a protein, Aap, links bacterial cells together but does so only in the presence of zinc ions. We have determined the atomic structure of an adhesive portion of Aap bound to zinc. The protein adopts an elongated, flexible fold with zinc ions bridging two protein chains. The mode of assembly indicates that Aap is likely to form twisted rope-like structures between bacterial cells. These data provide clues about regions of the protein that could be targeted to prevent intercellular adhesion in the developing biofilm.
Keywords: Staphylococci, X-ray crystallography, zinc, self-assembly, G5 domain
Abstract
Staphylococcal bacteria, including Staphylococcus epidermidis and Staphylococcus aureus, cause chronic biofilm-related infections. The homologous proteins Aap and SasG mediate biofilm formation in S. epidermidis and S. aureus, respectively. The self-association of these proteins in the presence of Zn2+ leads to the formation of extensive adhesive contacts between cells. This study reports the crystal structure of a Zn2+-bound construct from the self-associating region of Aap. Several unusual structural features include elongated β-sheets that are solvent-exposed on both faces and the lack of a canonical hydrophobic core. Zn2+-dependent dimers are observed in three distinct crystal forms, formed via pleomorphic coordination of Zn2+ in trans across the dimer interface. These structures illustrate how a long, flexible surface protein is able to form tight intercellular adhesion sites under adverse environmental conditions.
Healthcare-associated infections affect an estimated 1.5–2 million patients annually in the United States, with ∼99,000 annual fatalities (1). Staphylococci represent the most commonly isolated genus in healthcare-associated infections (2) and are the most common cause of infections on implanted devices (3). The key pathogenic mechanism in these infections is formation of a biofilm. A biofilm is a specialized bacterial colony with higher-order organization analogous to that of a tissue in multicellular organisms, in which there is concerted regulation of metabolic activity and gene expression (3). The entire colony is encased in an extensive extracellular matrix that can comprise polysaccharide, protein, nucleic acids, or combinations thereof (4). The extracellular matrix is important for mediating adhesion among neighboring bacteria as well as to diverse surfaces (3). Bacteria within a biofilm are resistant to antibiotics (5) and to host immune defenses (6), reducing the efficacy of available antimicrobials. Understanding the mechanisms of biofilm formation will allow us to combat the significant pathogenic advantages of biofilm-based infectious diseases.
There are species- and strain-specific differences that affect staphylococcal biofilm formation (7), but certain key similarities have been identified. An important group of adhesive proteins includes the Staphylococcus epidermidis protein Aap and its Staphylococcus aureus homologs SasG and Pls. These are multidomain, multifunctional proteins with significant roles in biofilm formation. Exogenous expression of Aap or SasG in non–biofilm-forming cocci is sufficient to mediate adhesion to host cells (8, 9) and to initiate biofilm formation (10, 11). Aap knockout ablates biofilm formation (12).
Aap, SasG, and Pls all have similar domain arrangements. In Aap, the N-terminal portion of the protein is comprised of an A-repeat region, with short (∼16-residue), imperfect sequence repeats, followed by a putative globular (α/β) domain with predicted α-helical and β-sheet content (Fig. 1A). This region of Aap mediates attachment to host cells via interactions with an unidentified receptor (8, 9). The α/β domain is followed by a B-repeat region, which consists of 5–17 nearly identical 128-amino acid sequence repeats that end in a conserved 78-amino acid partial repeat (Fig. 1A and Fig. S1A) (7). A Pro/Gly-rich region extends for 136 amino acids between the B-repeat region and an LPXTG sortase motif, through which the protein is covalently attached to the bacterial cell wall. A proteolytic processing event cleaves Aap between the α/β and B-repeat regions. Proteolytically processed forms of Aap and SasG have been shown to mediate intercellular adhesion even in the absence of polysaccharide, and expression of truncated constructs in non–biofilm-forming staphylococci induced biofilm formation (10, 11). Furthermore, the length of the B-repeat region was shown to be critical for SasG-induced biofilm formation in S. aureus, because SasG constructs with five or more B-repeats supported biofilm formation, whereas constructs with four or fewer B-repeats did not (11).
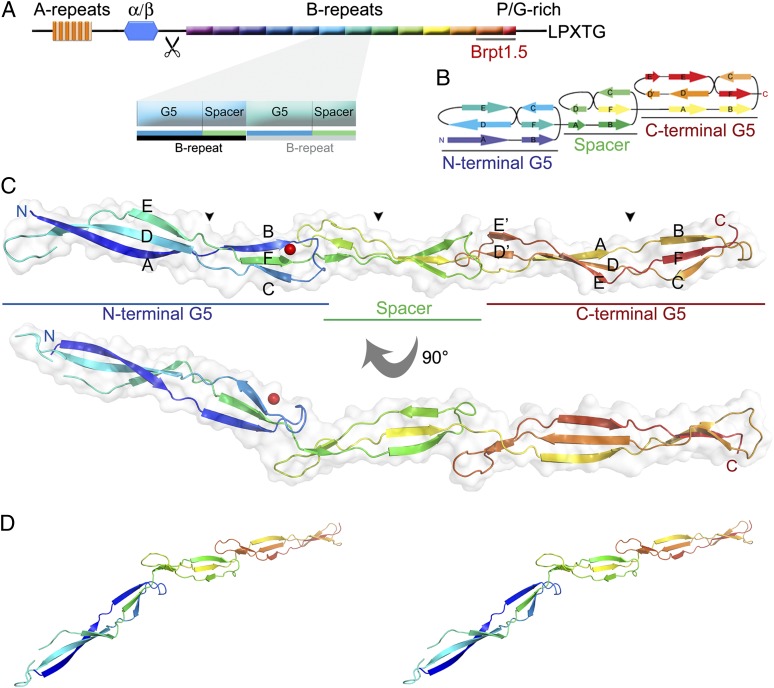
Fig. 1.
Crystal structure of Brpt1.5. The domain arrangements, topology, and overall fold of Brpt1.5. (A) Domain arrangement of Aap, with a zoomed-in view of two successive B-repeats; the Brpt1.5 construct is underlined in gray. The scissors indicate the cleavage site between the α/β domain and B-repeat region. A Pro/Gly-rich collagen-like region lies at the C-terminal end of the B-repeat region. This region terminates in the LPXTG sequence which is required for the covalent cross-linking of Aap to the cell wall by sortase. (B) Topology diagram of the Brpt1.5 fold. Note that strand E is skipped in the spacer domain to highlight the structural similarity of strands B, C, and F. (C) Model of Brpt1.5 shown in cartoon representation colored from blue (N terminus) to red (C terminus) with a transparent surface overlay. Zn2+ is shown as a red sphere. Triple-helix–like regions are indicated by the black arrowheads. (D) Stereo view of Brpt1.5 in cartoon representation, with color scheme used in B and C.
Each B-repeat consists of two domains. The first 78 amino acids correspond to a G5 domain, followed by a 50-residue spacer domain that has regions of high sequence identity with the G5 domain (Fig. S1B). The C-terminal conserved partial repeat at the end of the B-repeat region is a single G5 domain. The G5 domain was first identified in bioinformatics searches and takes its name from the five conserved Gly residues that are considered to be characteristic of the fold. G5 domains tend to occur in two different contexts: single-copy G5 domains attached to soluble enzymatic domains and multiple tandem repeated copies in Gram-positive surface proteins such as Aap and SasG. The structure of a single G5 domain in the Mycobacterium tuberculosis resuscitation factor RpfB revealed an elongated fold consisting of three-stranded β-sheets connected by triple-helix–like twists (13). The spacer domain is found in Aap and SasG B-repeats, alternating with the G5 domains. Not all Gram-positive cell surface proteins with tandem G5 domains contain 50-residue spacer domains; some contain much shorter intervening sequences between G5 domains.
The physical and functional properties of the B-repeat region of Aap were characterized previously using constructs consisting of the terminal one or two intact B-repeats along with the C-terminal G5 domain. These constructs were named “Brpt1.5” and “Brpt2.5,” respectively (14). Analytical ultracentrifugation experiments revealed that both constructs self-assembled in the presence of Zn2+, providing a molecular rationale for how the B-repeat region can trigger intercellular adhesion (14). The Zn2+ concentrations required for half-maximal dimerization decreased with increasing repeat length, although the Zn2+ concentrations required in vitro for dimerization of the short construct were well above physiological extracellular Zn2+ levels. However, Zn2+-dependent dimerization at physiologically relevant concentrations was demonstrated in biofilm growth assays. Biofilm formation was inhibited by the metal chelator DTPA and then was restored by supplementation with 5–20 μM ZnCl2, Zn2+ concentrations that are consistent with those reported in human serum (15). Given that SasG-dependent biofilm formation requires at least five tandem B-repeats (11), a cooperative increase in Zn2+ affinity with increasing repeat length may account for Zn2+-dependent self-association at physiological Zn2+ concentrations. A direct role for self-assembly of Aap during biofilm formation is supported by the inhibition of S. epidermidis biofilms by soluble Brpt1.5. Addition of soluble Brpt1.5 with a hexahistidine-tagged maltose-binding protein (His-MBP) fusion tag was able to inhibit biofilm formation only in the presence of sufficient Zn2+ to support self-assembly, as would be expected for a dominant-negative mode of inhibition (14). SasG also was demonstrated to self-assemble in a Zn2+-dependent fashion, and the high sequence identity between these two proteins suggests a shared mechanism for Zn2+ binding and Zn2+-dependent intercellular adhesion (16).
Linked equilibrium analysis of Brpt1.5 revealed that two or three Zn2+ ions bound per dimer, compared with four or five Zn2+ ions bound per Brpt2.5 dimer (14). These experiments were interpreted as support for a mechanism of Aap B-repeat self-association in which Zn2+ is coordinated in trans by two interacting protomers, with each repeat binding Zn2+ in a modular fashion. However, Zn2+-binding sites that bridge a protein–protein interface are notoriously difficult to identify based on sequence characteristics (17). Sequence analysis has yielded few potential motifs for Zn2+ coordination or self-association, and the recently published SasG B-repeat structures did not yield insights into the physiologically relevant Zn2+-binding activity and self-assembly (18). Therefore, we have undertaken the structural characterization of Brpt1.5, the minimal Zn2+-dependent self-association construct from Aap, in the presence of Zn2+. Structures from four crystals with three independent crystal forms show how Zn2+ induces self-association of Brpt1.5 by bridging two protomers. These structural data illustrate the mechanism by which Zn2+-induced intercellular adhesion occurs in staphylococcal biofilms and reveal regions of Aap that could be targeted therapeutically to inhibit early-stage biofilm formation.
Results
Structure of Brpt1.5.
Brpt1.5 was solved to 2.0-Å resolution by molecular replacement (MR) from a Se-Met single-wavelength anomalous dispersion (Se-SAD)-phased model (Table S1). It is markedly nonglobular, with a long axis of 180 Å and a maximum cross-sectional diameter of only 20 Å. Overall, the fold consists mainly of “freestanding” β-sheets that are solvent-exposed on both faces, with overlapping loops at the interdomain intersections (Fig. 1 B–D). The structure may be subdivided into three domains: two G5 domains separated by a domain we refer to as the spacer domain. The G5 domains (amino acids 1–78 and 129–207) each comprise two successive three-stranded β-sheets connected by triple-helix–like regions (black arrowheads in Fig. 1C), consistent with the previously reported single G5 domain structure from RpfB (13) and the recently reported structures of B-repeat constructs from SasG (18).
The spacer domain (amino acids 79–128) assumes an abbreviated form of the G5 domain fold, with similar topology but shorter β-sheets (Fig. 1B). The amino acid sequence of this domain is characterized by clusters with high sequence identity to the canonical G5 domain, separated by sequence gaps (Fig. S1B). Interestingly, the PGXPG motif [considered a defining characteristic of the G5 domain (13)] is absent from the triple-helix–like region of the canonical G5 domains but is present in the triple-helix–like region of the spacer domain. The structural similarity between the G5 domain and spacer domain is striking; it is best observed in the second β-sheet (strands B, C, and F). To highlight the structural similarity, strand E was omitted from the labels in the spacer region, so that the labels for strands B, C, and F are retained between the spacer and G5 domains (Fig. 1B).
Brpt1.5 Is Highly Flexible.
In the course of structure determination, data sets were collected from several different constructs and crystal forms. The construct used for SAD phasing was generated using point mutagenesis to replace the four Leu residues in the native protein with Met (Fig. S1A). (Native Brpt1.5 does not contain any Met residues.) Crystals of this SeMet-labeled “quadruple mutant” diffracted to 2.4 Å. Non–Se-labeled quadruple mutant crystals containing two monomers in the asymmetric unit diffracted to 2.5 Å. Native crystals with two distinct unit cells diffracted to 2.3 and 2.0 Å (Native1 and Native2, respectively). Importantly, the SeMet quadruple mutant and the Native 1 data set were isomorphous, suggesting that the four Met mutations did not affect the overall fold of the protein. However, interesting differences arise between the SeMet/Native1 structures and the two nonisomorphous data sets.
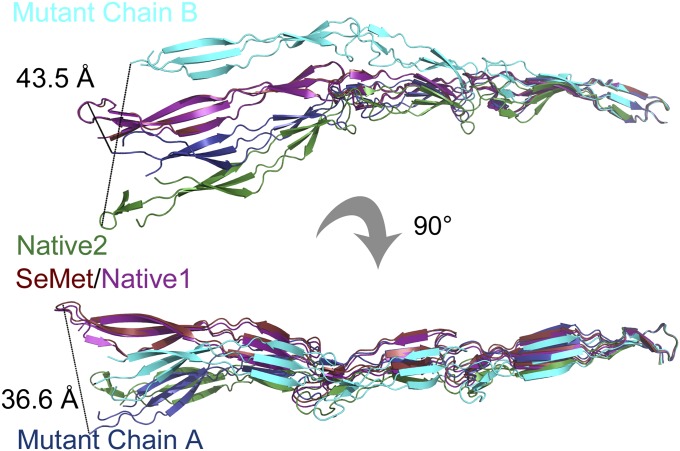
Each crystal form captures the protein in a slightly different conformation, illustrating the intrinsic flexibility of the fold. The summed effect of these differences is shown clearly in Fig. 2; when the C-terminal β-sheet is aligned, the placement of the N-terminal G5 domain varies by up to 40 Å. Structural alignments show that this flexibility is caused by hinge-like behavior at the interdomain interfaces and in the triple-helix–like region, as well as by flexion in the β-sheets (Fig. 2). Although the fold is maintained in an elongated conformation, it is by no means as rigid a structure as previously posited (18).
Fig. 2.
Flexibility of Brpt1.5. The final β-sheet (strands B, C, and F) from the C-terminal G5 domain was aligned for all structures. SeMet (red) and Native1 (magenta) structures overlay almost perfectly. Deviations with other structures arise at the triple-helix–like twist within the G5 domain, at the interdomain interfaces, and within the flexible sheets; Native2 (green) and non-Se labeled mutant chains A (dark blue) and B (cyan) diverge by as much as 44 Å at the tip of the N-terminal G5 domain.
Brpt1.5 Lacks a Canonical Hydrophobic Core.
The extended β-sheets expose the side chains of almost every residue in the protein, most of which are charged or hydrophilic (Fig. S2). Approximately 32% of the residues in Brpt1.5 carry a charge. The only approximation of a well-packed hydrophobic core observed consists of a Phe residue and a Pro residue that form the center of a stack of otherwise polar side chains at each interdomain intersection (Fig. 3A). Interestingly, the residues involved in this hydrophobic packing are conserved in the G5 domains as well as in the spacer domain (Fig. S1), suggesting that this interaction may be an important stabilizing influence on the fold. The only residue in the hydrophobic stack that is not identical in all repeats is N23; it is found as an aspartate in ∼50% of the repeats (Fig. S1).
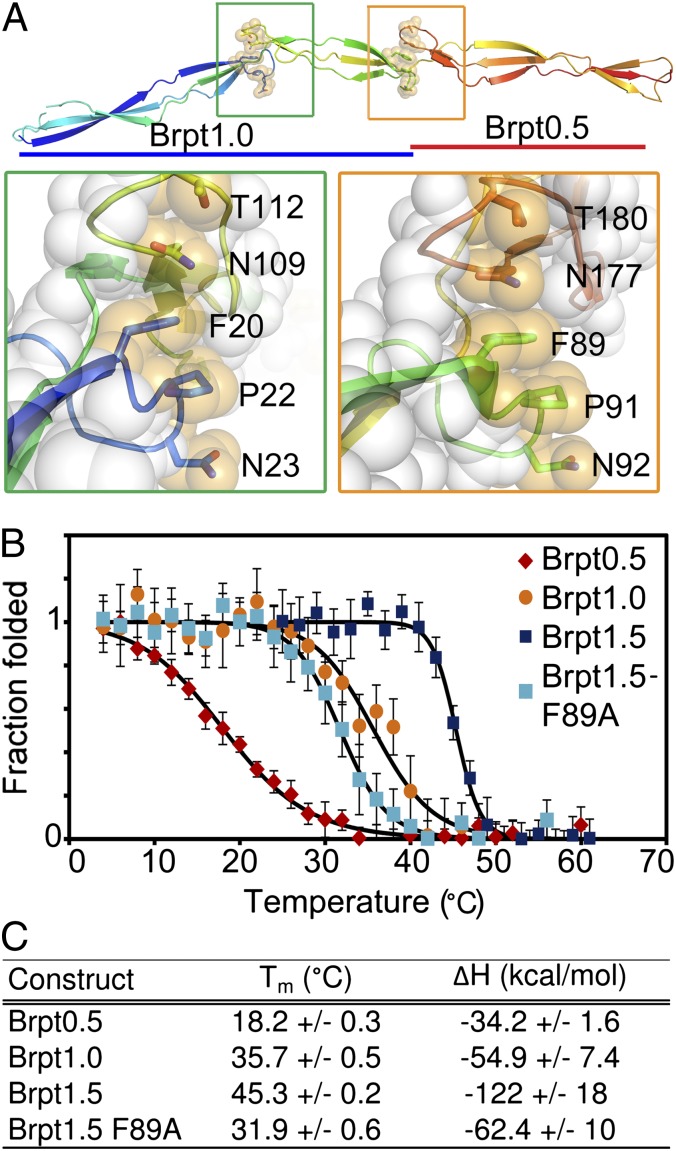
Fig. 3.
Stability of the freestanding β-sheet fold. (A) Brpt1.5 structure, with the side chains of the hydrophobic stacking residues shown as sticks (colored per backbone) overlaid with transparent spheres (orange). (Left Inset, green box) The hydrophobic stack between the N-terminal G5 domain (blue residues) and spacer domain (yellow residues). (Right Inset, orange box) The interaction between the spacer domain (green residues) and C-terminal G5 domain (orange residues). In both insets the atoms of the remaining residues are shown as transparent white spheres. (B) Circular dichroism thermal denaturation data from Brpt0.5 (red), Brpt1.0 (orange), Brpt1.5 (blue), and the Brpt1.5-F89A mutant (cyan). All melts were fully reversible. The fits to a model of two-state thermal denaturation are superimposed (black). (C) Tm and enthalpy change (ΔH) determined from the thermal denaturation data.
To test the importance of the stacking interaction, we performed circular dichroism thermal denaturation experiments on Brpt0.5 (a single G5 domain), Brpt1.0 (G5 + spacer), and Brpt1.5 (G5 + spacer + G5) (Fig. 3B). The melting temperature (Tm) shows a dramatic stepwise increase with increasing repeat length (Fig. 3C); we attribute this increase to the inclusion of the downstream loop capable of contributing the side chains needed to complete the hydrophobic stack. To test the influence of this interaction further, we mutated F89, the Phe residue in the interface between the spacer and C-terminal G5 domain. Brpt1.5-F89A was destabilized to a degree comparable to the Brpt1.0 construct (Fig. 3 B and C), confirming that the Phe/Pro hydrophobic stacks are the primary source of the enhanced protein stability with increased repeat length.
Zn2+-Induced Brpt1.5 Dimers Are Observed with Variable Coordination Schemes.
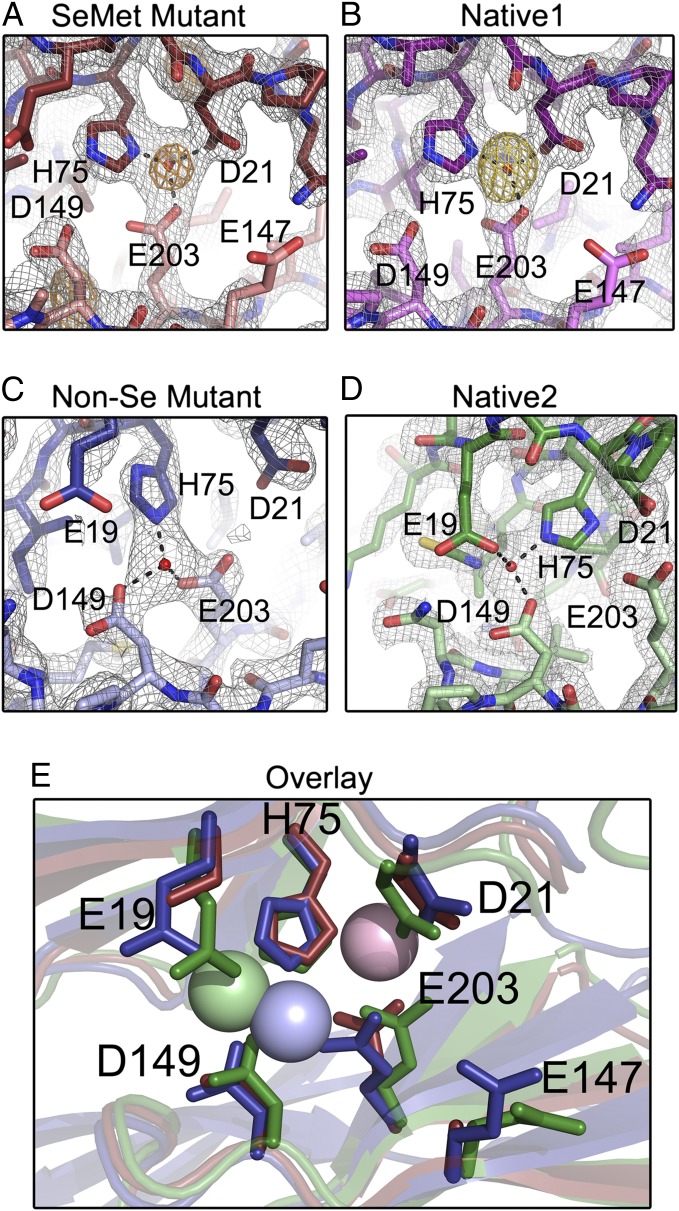
Brpt1.5 was crystallized in the presence of ZnCl2 to capture the Zn2+-dependent self-association complex. Symmetric Zn2+-dependent dimers are observed in each of the four separate data sets (Fig. 4 A–E), including two distinct dimers in the non-Se quadruple mutant data set. An anomalous peak in the SeMet data set (Fig. 5A) was assigned to Zn2+ based on the peak height, position, and X-ray absorption fine-structure (XAFS) data consistent with the presence of Zn2+. An anomalous peak is observed at the same position in the isomorphous Native1 data set, which was collected to high redundancy at the Zn2+ anomalous edge (Fig. 5B). These data unambiguously confirm the presence of Zn2+ in the structures. Furthermore, an overlay of the anomalous map from the Native1 data set with the molecular replacement Fo-Fc map shows that the highest Fo-Fc peak lies at the Zn2+ position (Fig. S3). Zn2+ was omitted from all MR searches, and the highest Fo-Fc peak was used to assign the Zn2+ position in the two remaining data sets (Fig. 5 C and D).
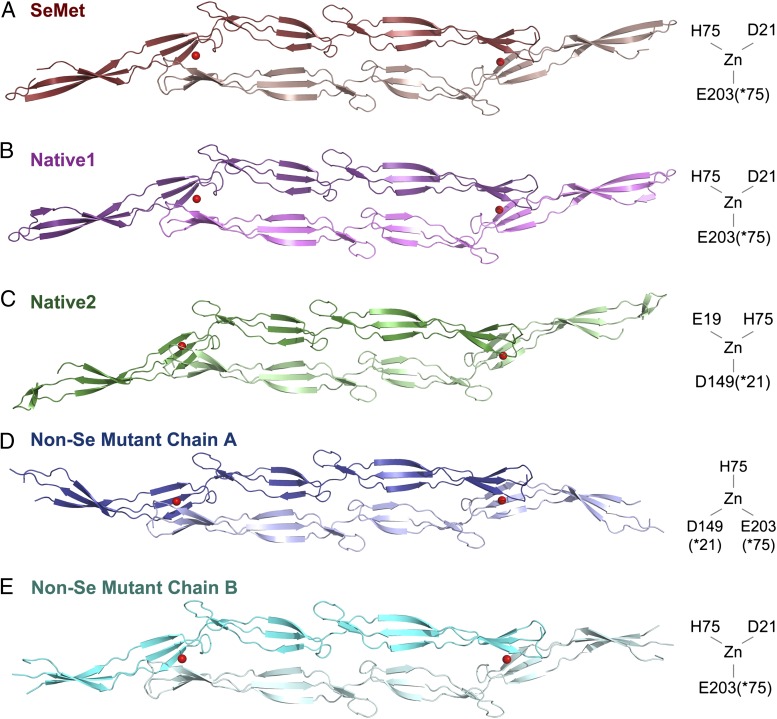
Fig. 4.
Zn2+-dependent dimerization of Brpt1.5. Zn2+-dependent dimers with similar architecture are seen in all crystals. Zn2+ ions are shown as red spheres, and protomers are shown in cartoon representation. Zn2+-coordinating residues are schematized to the right of the respective structures. (A) SeMet-labeled quadruple mutant (dark red and pink). (B) Native1 (dark and light violet) (diffraction data were isomorphous with the SeMet data set). (C) Native2 (dark and light green) crystallizes with differing unit cell dimensions, and subtle differences are observed in domain placement. (D and E) Two distinct symmetric dimers are formed by the two chains present in the non–Se-labeled quadruple mutant (chain A, blue; chain B, cyan), with differences in domain placement.
Fig. 5.
Details of Zn2+ coordination in Brpt1.5 dimers. Stick models of Zn2+ coordination sites are shown in the final refined density (2Fo−Fc electron density map, gray mesh, 1.5 σ), with distinct protomers colored in dark or light shades. (A) Se-Met quadruple mutant showing the position of the Zn2+ in anomalous difference density at 0.9792 Å wavelength (Se K edge) (F+ − F− anomalous difference map, orange mesh, 5σ). (B) Native1 structure showing the position of the Zn2+ in anomalous difference density at 1.2821 Å (Zn K edge) wavelength (F+ − F− anomalous difference map, gold mesh, 5 σ). (C) Chain A from the non–Se-labeled quadruple mutant. (D) Native2 structure. (E) Structural alignment of the Zn2+-binding pockets from SeMet (dark red), non-Se mutant chain A (dark blue), and Native2 (green) structures.
The Zn2+-mediated dimer assembles in an antiparallel fashion, with each protomer contributing side chains that coordinate the Zn2+ in trans, as previously predicted (14). The dimer interface primarily comprises the Zn2+-coordination pocket; this intermolecular contact is conserved among all data sets. In each case Zn2+ is coordinated by four ligands in a roughly tetrahedral arrangement. Interestingly, the exact Zn2+-coordination schemes differ in the three crystal forms (Figs. 5E). In the SeMet and Native1 crystals, Zn2+ is coordinated by D21 and H75 (from the N-terminal G5 domain), E203(*75) (from the C-terminal G5 domain of the other protomer; an asterisk indicates the corresponding residue in the N-terminal G5 domain), and a thiocyanate from the crystallization buffer (Fig. 5 A and B). We predict that the involvement of the thiocyanate is an artifact of the crystallization conditions and that this site would be occupied by water in vivo. In the Native2 structure, the ligands are E19, H75, D149 (*21), and a thiocyanate (Fig. 5D). The non-Se crystal has two protomers per asymmetric unit (each forming a crystallographic dimer), with two distinct coordination schemes; in one dimer, the Zn2+ ligands are H75, D149 (*21), E203(*75), and water (Fig. 5C), whereas the other dimer resembles the SeMet/Native1 coordination scheme with a water molecule in place of the thiocyanate (Fig. 4E).
Mutagenesis of Zn2+-Coordinating Residues.
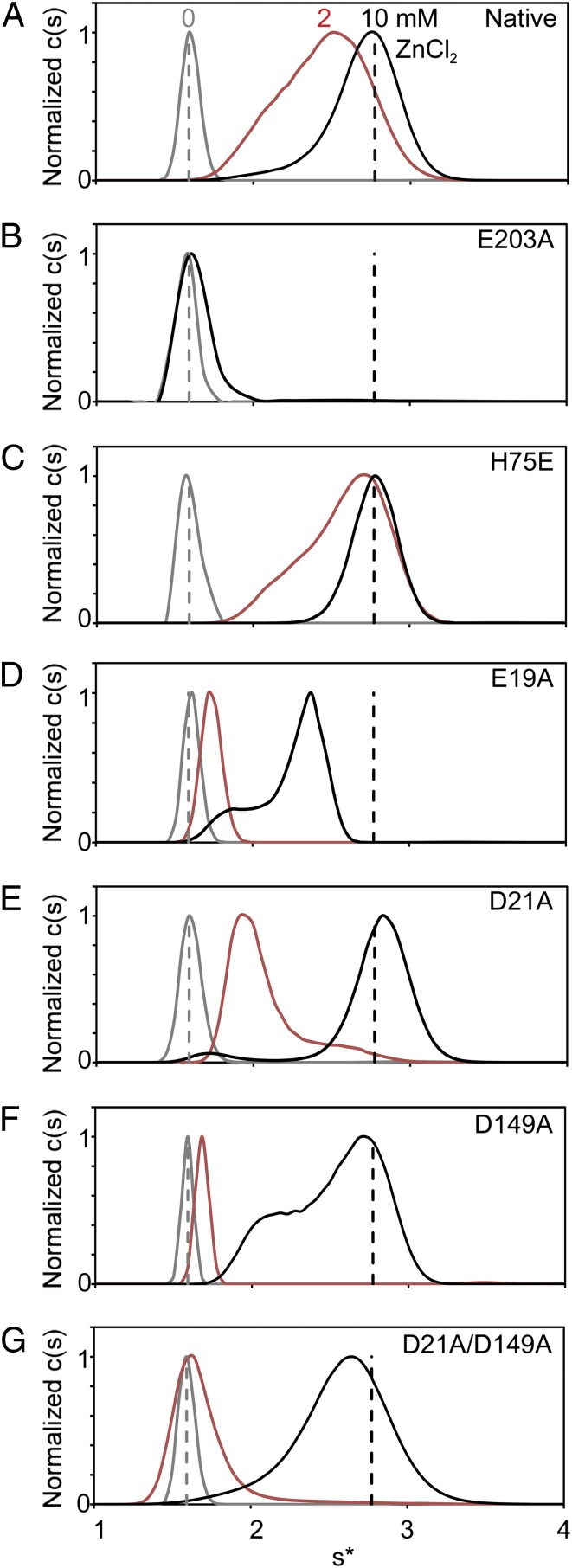
Mutagenesis confirms that each of the observed Zn2+-ligating residues plays a role in dimerization. Dimer formation in the presence of up to 10 mM Zn2+ is ablated by an E203A mutation (Fig. 6 A and B). This result was surprising, because H75 and E203 occupy the same positions in the G5 domain fold (Fig. S1A), and previous research shows a less drastic effect for mutation of H75 (14). In the reported sequences of Aap B-repeats from S. epidermidis strain RP62A, the residues corresponding to position 75/203 are evenly split between His and Glu (Fig. S1A), suggesting the potential for an unusual all-Glu/Asp Zn2+ coordination scheme. An H75E mutant shows no diminution of Zn2+-induced dimerization (Fig. 6C), indicating that Zn2+-induced self-assembly can occur despite B-repeat sequence variations. Dimer formation is reduced by Ala mutations at positions E19, D21, and D149, as well as for a D21/D149 double mutant (Fig. 6 D–G). These results are similar to the previously reported effects of an H75A/H85A double mutant (14). Taken as a whole, these data suggest an interesting mechanism for self-assembly resulting from pleomorphic Zn2+ coordination in trans, in which several amino acids in a symmetric binding pocket play functionally redundant roles. We propose that the multiple coordination schemes observed in the crystal structures represent a compensatory mechanism that allows self-association of a long, highly flexible protein under conditions of torsion and shear stress, despite variability in the residues that mediate Zn2+ coordination.
Fig. 6.
Biophysical characterization of Zn2+ site mutants. The results of sedimentation velocity analytical ultracentrifugation experiments are shown as c(s) distributions. Native Brpt1.5 and Zn2+-coordination mutants were assayed in the presence of 0, 2, and 10 mM ZnCl2 (shown in gray, red, and black, respectively). Dashed vertical lines represent the sedimentation coefficients of native protein at 0 (gray) and 10 (black) mM ZnCl2. (A) Native Brpt1.5. (B) Brpt1.5 E203A in the presence of 0 and 10 mM ZnCl2. (C) Brpt1.5 H75E. (D) Brpt1.5 E19A. (E) Brpt1.5 D21A. (F) Brpt1.5 D149A. (G) Brpt1.5 D21A/D149A.
Discussion
Structural Characteristics of Brpt1.5.
The highly elongated structure of Brpt1.5 is one of the few protein structures containing freestanding β-sheets solved to date. Other reports of proteins with freestanding β-sheets include the recently reported structure of SasG B-repeat constructs, the single G5 domain attached to the catalytic domain of RpfB (Fig. S4A) (13), and a small segment of continuous β-sheet in the Borrelia burgdorferi surface protein OspA (Fig. S4B) (19). In comparison with the latter two, the Brpt1.5 structure almost exclusively comprises freestanding β-sheets, with only minimal stabilizing hydrophobic interactions (Fig. 3A). Furthermore, Brpt1.5, with a predicted isoelectric point of 4.67 and a grand average of hydropathy (GRAVY) hydrophobicity index score of −0.99, is a highly hydrophilic protein compared with most successfully crystallized proteins (20). Brpt1.5 has an unusually low percentage of buried side chains (Table S2), with 68% of its atoms exposed to solvent (21). The hydrophilicity of the sequence (Fig. S2) helps explain the lack of a canonical hydrophobic core.
The structural characteristics of the Aap B-repeat would enhance the effectiveness of Aap in both host cell attachment and intercellular adhesion. Studies using atomic force microscopy suggest that Aap exhibits spring-like stretching behavior because of unfolding of B-repeats (22). Our thermal denaturation experiments (Fig. 3) were fully reversible, further supporting the ability of denatured repeats to return readily to the folded state. The ability to oscillate between folded, flexible protein and unfolded polypeptide would facilitate host cell attachment or intercellular adhesion under conditions of shear stress. Several common sites of pathogenic biofilm formation are in regions of bulk fluid flow, such as the surfaces of intravascular catheters, dialysis ports, and heart valves. In these areas, the characteristic flexibility and extensibility that allow the B-repeat to maintain its function would be invaluable.
G5 Domain-Containing Proteins.
Aap, SasG, and Pls are representative of a group of Gram-positive surface proteins containing multiple tandem G5 domains. However, the relative arrangement of G5 domains within a protein varies depending on the organism. Several of the other tandem G5-containing surface proteins also have sequence insertions between the G5 domains, but these insertions have no apparent sequence homology to the spacer domains in the staphylococcal B-repeat–containing proteins. Some other proteins that contain tandem G5 repeats lack sequence insertions altogether. In these cases, the G5 domain still could contribute the upstream and downstream loops necessary to form the interdomain hydrophobic stacks, so we predict that the G5 domains would show cooperative folding similar to that of the Aap B-repeat. It is not clear whether tandem G5 proteins with nonconserved spacer regions will adopt similar stabilizing interactions; furthermore, it is unknown if any of the nonstaphylococcal proteins containing tandem G5 repeats display Zn2+-dependent self-assembly.
A separate group of proteins are those that contain a single G5 domain in tandem with an enzymatic domain (or in some cases, domains of unknown function). The function of the G5 domain in these proteins remains unclear. Based on our results, it is possible that single copies of the G5 domain may act as temperature sensors. When expressed as a single G5 construct, the C-terminal Aap G5 domain is unfolded at room temperature under solution conditions approximating physiological pH and ionic strength (Fig. 3). This result is in contrast to the Tm values of 47 °C and 53 °C for single G5 constructs from SasG (18) and suggests that sequence variations can modulate the thermal stability of the G5 domain. The temperature sensitivity of the G5 domain fold needs to be examined in the context of native proteins that contain a single G5 domain. If similar variations in thermal stability are observed, they may represent a mechanism that allows temperature-specific regulation of catalytic domains when expressed in tandem with single G5 domains.
Zn2+-Dependent Dimerization.
The structures of Brpt1.5 dimers solved in this study reveal interesting insights into Zn2+-dependent self-assembly and its role in biofilm formation. The Zn2+-binding sites are notably lacking in Cys; there is not a single Cys in the entirety of Aap, SasG, or Pls. Although Zn2+ coordination sites lacking Cys are atypical, they are thought to be associated with redox-insensitive Zn2+ binding (17). A nascent staphylococcal biofilm likely would be subjected to oxidative burst from neutrophils, which would make redox-insensitivity an important characteristic for intercellular adhesion. The coordination schemes in Brpt1.5 that use His, Asp, and Glu side chains and a water molecule (or thiocyanate) are similar to interfacial zinc sites found in some other protein complexes, such as those formed by bacterial superantigens (23).
The Zn2+ sites in Brpt1.5 are also notable for their plasticity. The structural and mutagenesis data presented in this and a previous study (14) indicate that the diverse Zn2+ sites are likely to have functional significance. Variations in Zn2+ coordination and protein conformation are observed even within a single crystal form (the non–Se-labeled mutant) or for different crystal forms of the native construct. Although three of the five coordination schemes observed are identical (SeMet, Native1, and non-Se mutant chain B; Fig. 4 A, B, and E), the two variant schemes have very interesting characteristics. For instance, the coordination scheme for the non-Se mutant chain A dimer (Fig. 4D) may be conceptualized as an inversion of the predominant coordination scheme. Considering the position of the residues within the individual G5 domains will help clarify this concept: residues 21 and 149 occupy equivalent positions, as do amino acids 75 and 203. Therefore, in the non-Se mutant chain A structure, the C-terminal G5 domain contributes residues from positions 21 and 75. These residues are in the same positions as those contributed by the N-terminal G5 domain in the predominant coordination scheme. In the same way, the N-terminal G5 domain donates the residue at position 75, which is contributed by the identical residue in the C-terminal G5 domain in the predominant scheme. Overall, this difference in coordination is the result of an asymmetric binding mode at a symmetric interface.
The mutagenesis studies revealed a surprising degree of redundancy in zinc coordination; the E19A, D21A, D149A, and the D21A/D149A mutations each reduced but did not abrogate Zn2+-induced dimerization of Brpt1.5 (Fig. 6). Only the E203A mutation completely prevented dimerization of Brpt1.5. In the case of D21A and D149A single mutants, it is likely that the two residues compensate for one another, based on their identical positions within the G5 domains. The D21A/D149A double mutant shows a further reduction in solution-state dimerization, supporting the functional redundancy of these two residues. The ability of the protein to dimerize even in the absence of both residues is comparable to our previous data on Brpt1.5 H75A/H85A. Although H75 is central to every coordination scheme observed in this study (Fig. 5E), Zn2+-dependent dimerization in the solution state still can occur in the absence of this side chain (14).
In contrast to H75, E203 is indispensable for Zn2+-induced dimerization. E203 and H75 occupy identical positions in the C- and N-terminal G5 domains, respectively, but the E203A mutation eliminates dimer formation under the solution conditions tested. This effect is surprising, because the Native2 structure clearly shows a Zn2+-binding mode without direct participation of E203. However, the well-ordered binding pocket within that structure suggests an additional role for E203A. E203 interacts with H75, one of the residues that does coordinate the Zn2+. Zn2+-binding sites typically require a secondary shell of residues that maintain the proper environment for Zn2+ coordination (24). It is likely that a strong negative charge at either position 75 or 203 is required to maintain the coordination sphere in Brpt1.5, whether or not both residues directly coordinate Zn2+. The negative charge also is likely to play a critical role in maintaining the Zn2+-coordinating residues at the proper pKa. Previous results have shown a strong pH dependence to Zn2+-dependent dimerization, with dimer formation inhibited as the pH decreases below 6.7 (14). The strongly electronegative binding pocket would be expected to elevate the pKa values of the Asp and Glu residues ligating the Zn2+. The E203A mutation therefore may have an indirect but major impact on the chemistry of Zn2+ ligation by altering the electrostatic potential or the local structural arrangement of key residues within the Zn2+-binding pocket. The results with E19A further support the importance of maintaining the proper electrostatic environment. E19 is found at the periphery of the binding site in all structures and participates in Zn2+ binding in only one of the five. However, the E19A mutation decreases Zn2+-dependent dimerization to a greater extent than the D21A/D149A double mutation. We propose that the difference is due to the influence of E19 on the overall electrostatic environment of the Zn2+ binding pocket, in addition to any effects caused by direct binding of Zn2+ by residue E19.
Brpt1.5 is the smallest unit that contains all required elements to support Zn2+-dependent dimerization; however, it should be noted that Brpt1.5 is a short construct from a much larger protein. The S. epidermidis strain used in our studies expresses Aap containing 12.5 repeats. The sequences of each repeat from S. epidermidis RP62A are aligned in Fig. S1A. Interestingly, the Zn2+-coordinating residues identified in this study show a certain amount of variability within the sequence repeats. Position 75 is occupied by a Glu residue in at least 50% of the repeats. The S. aureus homolog SasG contains Glu in this position in 100% of the repeats (18), supporting the importance of a strongly electronegative amino acid in a central position within the binding pocket. The H75E variant demonstrated the ability of repeats with all-acidic Zn2+-binding pockets to maintain dimer formation. Interestingly, E19 is 100% conserved within Aap B-repeats. D21, however, is an Asn residue in ∼50% of the repeats. The level of conservation tracks well with the observed effects of mutations on the solution-state Zn2+-dependent assembly, in that the E19A mutation has a greater effect than D21A.
Selectivity for Zn2+.
Previous research showed that Brpt1.5 did not dimerize in the presence of up to 10 mM Mg2+, Ni2+, Sr2+, Mn2+, Co2+, or Ca2+. The selectivity for Zn2+ is interesting, based on the observed plasticity of Zn2+ binding. In each structure reported here, the Zn2+ shows tetrahedral coordination and is ligated by one His residue, two acidic residues, and a solvent molecule (water or thiocyanate). A published survey of metal coordination in high-resolution metalloprotein structures reveals patterns of coordination number (CN), coordination geometry, and preferred ligating residues among the metals (25); these patterns can account for Zn2+ specificity in Brpt1.5. Only Zn2+ and Cu2+ preferentially adopt a CN of 4 or less. In these cases Zn2+ strongly prefers tetrahedral coordination geometry, whereas Cu2+ is found more commonly in a square planar arrangement (25). Fe3+ is found with CN = 4 in several cases, although it more commonly adopts CN = 5 or 6. Ca2+, Mg2+, and Mn2+ typically exhibit CNs of 6, 7, or 8, and there were no examples of Mg2+ or Mn2+ with CNs of 4 or less. Thus, the structural arrangement of ligating residues in Brpt1.5 will favor Zn2+, Cu2+, or Fe3+ binding intrinsically, based on a CN of 3 (CN = 4 with the solvent molecule). Cu2+ is not a likely candidate because of its strong preference for square planar coordination mediated by Cys and His residues (Asp/Glu almost never coordinates Cu2+). Fe3+ occasionally adopts a CN of 4 or less and can be ligated by Asp/Glu residues, although His and Cys are most common. Therefore Zn2+ is the metal ion best suited for ligation in the Brpt1.5 soft metal-binding site(s), given its preference for tetrahedral coordination by His, Asp, or Glu and its bioavailability (particularly in comparison with Fe3+). Indeed, a rigid tetrahedral site is considered highly selective for Zn2+ (26). Even though the Brpt1.5 fold is flexible, and the precise Zn2+-coordinating residues vary, the relative positions of the ligating residues consistently enforce tetrahedral coordination (Fig. 5). These tetrahedral coordination sites are maintained by the second shell of hydrophobic and uncharged hydrophilic residues that orient the Zn2+-coordinating residues and tune their charges and pKa values.
The metal-binding site has additional characteristics that are likely to affect its selectivity. The dimer interface consists of a number of interlocking residues that surround the Zn2+ site, maintaining a close contact between the two Brpt1.5 protomers (Fig. S5). These residues would be unlikely to achieve comparable contact if a metal with a larger atomic radius bound in the same site. Furthermore, Zn2+ has the highest charge-accepting ability among cations that are similar in size and charge (27). This characteristic is of prime importance given the number of acidic residues in the Brpt1.5 metal-binding site.
Comparison with Putative SasG Dimer.
Our structural model of Zn2+-dependent dimerization differs from the one recently proposed by Gruszka et al. (18). They observed dimerization of a construct containing a single G5 domain from SasG. In their model, two protomers interact by coordinating Zn2+ in trans, via a four-residue N-terminal protein extension containing a His residue, in a tip-to-tip dimer assembly. The authors point out that this N-terminal extension is a nonnative cloning artifact. Mutation of the His prevents dimerization of the single G5 construct in the presence of Zn2+, as assayed by size-exclusion chromatography multi-angle laser light scatter (SEC-MALLS). The authors conclude that this result invalidates the data showing Zn2+-induced assembly for SasG constructs in that study and a previous study (16, 18). The authors further argue that the mechanism of Zn2+-dependent intercellular adhesion in biofilms must be distinct from Zn2+-dependent self-assembly of SasG or Aap, based on their experiment with the single-G5 domain construct from SasG. However, we find that Zn2+-induced dimerization of Aap B-repeat constructs has a strong dependence on construct length. A similar lack of assembly is observed in Brpt0.5 (single G5) and Brpt1.0 (G5 + spacer) constructs from Aap. Sedimentation equilibrium analytical ultracentrifugation experiments in the presence of up to 200 or 50 mM ZnCl2 (for Brpt0.5 and Brpt1.0, respectively) show no evidence for higher-order assembly of these short constructs (Fig. S6). In contrast, experiments with Brpt1.5 and Brpt2.5 clearly show the presence of dimer species at concentrations below 10 mM ZnCl2 (Fig. S6). All constructs used in this study contain a single Gly residue at the N terminus after removal of the fusion protein, so the self-assembly is not caused by a nonnative His residue. These results are consistent with the conclusion by Gruszka et al. (18) that the Zn2+-induced dimer observed for their single-G5 construct was an artifact of the construct and crystallization conditions. However, our analytical ultracentrifugation data demonstrate that longer B-repeat constructs are required to support physiologically relevant dimerization in the presence of Zn2+.
Furthermore, the tip-to-tip model of the SasG G5 domain dimer is inconsistent with previous biophysical analyses. Sedimentation velocity experiments showed that the Aap Brpt1.5 dimer was less elongated than the monomer (14). This result is consistent with the side-by-side dimer structure observed in our crystal structures, not a tip-to-tip structure. It is unclear from the reported SEC-MALLS data whether SasG constructs of relevant length show a change in axial ratio in the presence of Zn2+ (18). Examination of the axial ratios would help reveal if the side-by-side dimer observed for Aap B-repeat constructs also occurs for SasG B-repeat constructs. A previous study of SasG B-repeat self-assembly suggests that it does. Geoghegan et al. (16) report surface plasmon resonance (SPR) experiments performed with a different protein construct. This 2.5- repeat SasG construct was coupled to the SPR chip through an N-terminal GST fusion tag. GST is a globular protein, and its presence likely would occlude access to the extraneous N-terminal amino acids that mediate the tip-to-tip dimer. The SPR data support the ability of SasG B-repeats to self-assemble in the presence of Zn2+, even with access to the N terminus blocked, suggesting the formation of a side-by-side antiparallel dimer. Further analysis of SasG constructs lacking the N-terminal extension is necessary to verify fully the ability of longer SasG B-repeat constructs to dimerize in the presence of Zn2+.
Modeling Longer B-Repeat Structures.
Experiments with SasG revealed that the minimum B-repeat length required to mediate biofilm formation is five repeats (11). Models of longer B-repeat Aap constructs and their putative Zn2+-dependent dimers are presented in Fig. S7. These models were generated using sequential structural alignments of N- and C-terminal G5 domains from Brpt1.5. The models show a twisting cable-like assembly in which the antiparallel monomers wrap around one another. Interestingly, it appears that four to five repeats are required for the dimer to make a full 360° twist. This observation suggests there may be a torsional component to dimer assembly in the full-length protein that is absent in the shorter 1.5- and 2.5-repeat constructs, providing a potential molecular explanation for the biological requirement for at least five repeats (11).
Structural Basis of Zinc-Mediated Biofilm Formation.
It is well established that both Aap and SasG play an essential physiological role in the accumulation stage of biofilm formation (10–12, 14, 16). A genetic knockout of Aap inhibited biofilm formation by preventing intercellular adhesion (12). Two independent studies demonstrated that proteolytic cleavage of Aap or SasG upstream of the B-repeat region induced the formation of protein-based biofilms, even in the absence of the biofilm polysaccharide poly-N-acetyl glucosamine (PNAG) (10, 11). Our previous work showed that biofilm formation in S. epidermidis and S. aureus is dependent on Zn2+ and that addition of excess His-MBP-Brpt1.5, in the presence of Zn2+, could inhibit biofilm formation in a dominant-negative fashion (14); similar results were shown by Geoghegan et al. (16) for SasG. The structures we present here reveal the molecular mechanism by which bacterial proteins in this family mediate intercellular adhesion in nascent staphylococcal biofilms, through Zn2+-dependent self-assembly of the B-repeat regions.
When taken together, the known structural, biochemical, and genetic data on Aap and SasG provide a glimpse into early-stage biofilm formation by S. epidermidis and S. aureus. Full-length Aap and SasG have been implicated in binding to host cells via the N-terminal portion of the protein (8, 9); the switch to a biofilm state requires proteolytic processing upstream of the B-repeat region (10, 11). It is assumed that this processing of Aap or SasG must occur to reduce steric hindrance from the N-terminal region and allow the B-repeats from opposing cells to engage. Our crystallographic data reveal that the B-repeats self-assemble in an antiparallel fashion by coordinating Zn2+ in trans, with the potential for pleomorphic coordination that can accommodate the inherent flexibility of the B-repeat fold. Our structure-based model for self-assembly of longer B-repeat constructs (Fig. S7) suggests that the interacting B-repeat regions wrap around one another and that five tandem B-repeats are required for each chain to circumnavigate the other completely, as is consistent with the B-repeat length needed to support biofilm formation in vivo (11). This torsional component of B-repeat self-assembly may be an important feature for increasing the stability of intercellular interactions. For example, in textiles, twisting a bundle of fibers is an effective way to increase the strength of yarns or ropes; tensile stress on the rope induces lateral compression of the constituent fibers, increasing the friction between them (28). This phenomenon, described as early as 1638 by Galileo (29), can increase the overall strength of the yarn or rope. In the case of staphylococcal intercellular adhesion, a twisted cable formed by antiparallel chains of truncated Aap also would undergo lateral compression and potentially would form more extensive intermolecular interactions when under tensile stress. A particularly interesting aspect of this overall mechanism for biofilm formation via Aap or SasG cleavage and self-assembly is that Zn2+ ions and proteases are released by immune cells at sites of inflammation (30, 31). Thus, the host response that is designed to combat colonization is likely to be the very signal that triggers the defensive formation of a biofilm, a colony type that the immune system is unequipped to overcome (6).
Materials and Methods
Molecular Cloning.
Brpt1.5 expression plasmids were generated using the Invitrogen Gateway System as described (14). Point mutants were generated by Quik-change site-directed mutagenesis (Stratagene) using either Phusion polymerase (New England BioLabs) or pfu (Stratagene). Met mutagenesis sites were selected based on sequence alignments of proteins containing a G5 domain; primer sequences are shown in Table S3. L152M, L179M, and D149A mutations were generated from degenerate priming using L24M, L51M, and D21A primers, respectively.
Protein Expression and Purification.
Brpt1.5 and non-Se mutants were expressed as described (14), except that plasmid amplification was performed in NovaBlue cells (Novagen), and protein expression was performed in BLR cells (Novagen). Briefly, 1 L LB medium was inoculated with stationary-phase overnight culture, grown at ∼37 °C to an OD600 between 1.0 and 1.2, then cold-shocked in an ice-water bath until reaching ∼8 °C. Ethanol was added to a final concentration of 2% (vol/vol), and protein expression was induced overnight at 20 °C with 0.2 mM isopropylthio-β-galactoside. SeMet-labeled quadruple mutant Brpt1.5-L24M/L51M/L152M/L179M was expressed in BLR cells (Novagen) using a nonauxotrophic SeMet-labeling protocol (32) and the same induction protocol. Protein-expressing cells were harvested by centrifugation, resuspended in 20 mM Tris (pH 7.4), 150 mM NaCl, 1× Complete protease inhibitor (Roche), and frozen at −80 °C. Cells were thawed, lysed by sonication, and then centrifuged to pellet cell debris. Brpt1.5 was purified from the supernatant using a Hi-Trap Chelating HP Ni column (GE Healthcare). The fusion protein was cleaved using tobacco etch virus (TEV) protease, and then Brpt1.5 was purified using gravity-flow Ni-NTA and Superdex 75 gel filtration columns, followed by MonoQ ion-exchange chromatography as needed. All chromatography materials were supplied by GE Healthcare and all chemicals by Fisher Scientific, except where noted.
Protein Crystallization and Data Collection.
All crystals were grown in batch format from protein stocks at 8–15 mg/mL in 20 mM Tris (pH 7.4), 150 mM NaCl, 2 mM ZnCl2. For Se-Met L24M/L51M/L152M/L179M crystals, 3 μL protein was added to 4 μL of a precipitant solution of 0.12 M potassium thiocyanate, 0.1 M Tris (pH 7.9), 28% (wt/vol) polyethylene glycol monomethyl ether (PEG-MME) 2000, and 3% (wt/vol) glycerol. For Native1 and Native2 crystals, 3 μL protein was added to 4 μL of 0.06 M potassium thiocyanate, 0.1 M Tris (pH 7.9), 28% (wt/vol) PEG-MME 2000, and 2% (wt/vol) PEG 3350 (Sigma). For non-Se quadruple mutant crystals, 4 μL protein was added to 6 μL 0.12 M potassium thiocyanate, 0.1 M Tris (pH 7.9), 28% (wt/vol) PEG-MME 2000, and 0.5% (wt/vol) β-octylglucopyranoside. Crystals were cryoprotected in 0.06–0.12 M potassium thiocyanate, 0.1 M Tris (pH 7.9), and 45–50% (wt/vol) PEG-MME 2000. Data were collected at APS beamlines 24-ID-C and 24-ID-E as 0.8–1° oscillations. Inverse beam collection was used on the SeMet data set. The data collection strategy for the non-Se mutant data set was formulated based on high-mosaicity preliminary images, leading to the suboptimal data truncation caused by detector placement.
Phasing, Refinement, and Validation.
Data sets were integrated and scaled using HKL2000 (HKL Research, Inc.) (33). Experimental phases and initial model for the SeMet data set were generated in Phenix (34). Initial MR phases for all other data sets were generated in PhaserMR (35), using model fragments consisting of single G5 domains and spacer domains or spacer + C-terminal G5. The Native2 data set was anisotropy corrected before phasing (36). All models were rigid-body refined in Phenix or Buster (Global Phasing Limited) after MR (34, 37). For Native 2 and non–Se-labeled quadruple mutant models the highly flexible N-terminal region (residues 1–9 and 46–62) was removed and iteratively rebuilt as refinement progressed. The final models were developed after further cycles of building and refinement, carried out with Coot (38) and Buster (37), respectively. Initial TLS (translation/libration/screw) groups were determined using the TLS motion determination server (39) and implemented during Buster refinement for SeMet, Native1, and Native2 structures. Noncrystallographic symmetry was taken into account during refinement of the non–Se-labeled mutant structure.
All structures were validated using Molprobity (40) and submitted to the RCSB databank (www.rcsb.org/pdb) with structure codes 4FUM, 4FUN, 4FUO, and 4FUP. When density was too weak to place model side chains accurately (even contouring below 1σ), residues were left as pseudoalanine stubs in the final structure. Missing side chains were modeled in Coot before the surface-charge distribution was calculated in Chimera for Fig. S2 (41). The SeMet structure was used to generate all figures, except where otherwise noted.
Models of Brpt5.5 dimers shown in Fig. S7 were developed using sequential alignments of N- and C- terminal G5 domains. When the G5 domains from one protomer were aligned, the G5 domains from the interacting partner showed good agreement in strands B, C, and F. The deviations of strands A, D, and E within the unaligned protomer were consistent with the flexibility of the domain. These strands are omitted from the model for clarity.
Analytical Ultracentrifugation and Circular Dichroism.
Analytical ultracentrifugation experiments were carried out in a Beckman XL-I analytical ultracentrifuge using an An-60 Ti rotor, as previously described (14). For sedimentation velocity 8 μM of wild-type or mutant protein was centrifuged at 48,000 rpm in the presence of 0, 2, or 10 mM ZnCl2. Data were analyzed with Sedfit and are reported as normalized continuous c(s) distributions (42). Single-speed sedimentation equilibrium experiments were performed at 36,000 on Brpt0.5 and Brpt1.0. Brpt1.5 was analyzed at 24,000 rpm, and Brpt2.5 at 18,000 rpm. These speeds were chosen to generate similar monomer values of σ, the reduced buoyant molecular weight, for all constructs. Scans were performed in the presence of varying concentrations of ZnCl2 noted on the graphs. Data were analyzed with WinNonlin (www.rasmb.org) to determine the average molecular weight. Circular dichroism experiments were carried out in an Aviv 215 circular dichroism spectrophotometer using 0.5-mm quartz cuvettetes. Wavelength scans from 260 to 190 nm were used to confirm the secondary structure content of mutants and truncations. Wavelength scans performed at 4 °C on Brpt0.5, Brpt1.0, and Brpt1.5-F89A overlaid perfectly with wild-type protein. Thermal denaturation experiments on Brpt0.5, Brpt1.0, Brpt1.5, and Brpt1.5-F89A were performed at 54, 24, 42, and 50 μM concentrations, respectively, in 10 mM Tris (pH 7.5), 150 mM NaF. All melts were perfectly reversible. The data were normalized to pre- and posttransition baselines; then the fraction folded was fitted in SigmaPlot as described (43).
Supplementary Material
Acknowledgments
We thank C. C. Brescia for initial crystallization trials and R. A. Kovall, T. B. Thompson, A. A. Weiss, A. G. Menon, and W. E. Miller for advice and comments on the manuscript. This project was supported by Grant R01 GM094363 from the National Institutes of Health, a pilot grant from the Midwest Center for Emerging Infectious Diseases at the University of Cincinnati, and funds from the State of Ohio Eminent Scholar program (to A.B.H.). This work is based upon research conducted at the Northeastern Collaborative Access Team beam lines of the Advanced Photon Source, supported by Award RR-15301 from the National Center for Research Resources at the National Institutes of Health. Use of the Advanced Photon Source is supported by the US Department of Energy, Office of Basic Energy Sciences, under Contract W-31-109-ENG-38.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Crystallography, atomic coordinates, and structure factors reported in this paper have been deposited in the Protein Data Bank (www.pdb.org) (structure codes 4FUM, 4FUN, 4FUO, and 4FUP).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1208134110/-/DCSupplemental.
References
- 1.Klevens RM, et al. Estimating health care-associated infections and deaths in U.S. hospitals, 2002. Public Health Rep. 2007;122(2):160–166. doi: 10.1177/003335490712200205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hidron AI, et al. National Healthcare Safety Network Team Participating National Healthcare Safety Network Facilities NHSN annual update: Antimicrobial-resistant pathogens associated with healthcare-associated infections: Annual summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2006-2007. Infect Control Hosp Epidemiol. 2008;29(11):996–1011. doi: 10.1086/591861. [DOI] [PubMed] [Google Scholar]
- 3.Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: A common cause of persistent infections. Science. 1999;284(5418):1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 4.Branda SS, Vik S, Friedman L, Kolter R. Biofilms: The matrix revisited. Trends Microbiol. 2005;13(1):20–26. doi: 10.1016/j.tim.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 5.Patel R. Biofilms and antimicrobial resistance. Clin Orthop Relat Res. 2005;437(437):41–47. doi: 10.1097/01.blo.0000175714.68624.74. [DOI] [PubMed] [Google Scholar]
- 6.Vuong C, et al. Polysaccharide intercellular adhesin (PIA) protects Staphylococcus epidermidis against major components of the human innate immune system. Cell Microbiol. 2004;6(3):269–275. doi: 10.1046/j.1462-5822.2004.00367.x. [DOI] [PubMed] [Google Scholar]
- 7.Rohde H, et al. Polysaccharide intercellular adhesin or protein factors in biofilm accumulation of Staphylococcus epidermidis and Staphylococcus aureus isolated from prosthetic hip and knee joint infections. Biomaterials. 2007;28(9):1711–1720. doi: 10.1016/j.biomaterials.2006.11.046. [DOI] [PubMed] [Google Scholar]
- 8.Macintosh RL, et al. The terminal A domain of the fibrillar accumulation-associated protein (Aap) of Staphylococcus epidermidis mediates adhesion to human corneocytes. J Bacteriol. 2009;191(22):7007–7016. doi: 10.1128/JB.00764-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roche FM, Meehan M, Foster TJ. The Staphylococcus aureus surface protein SasG and its homologues promote bacterial adherence to human desquamated nasal epithelial cells. Microbiology. 2003;149(Pt 10):2759–2767. doi: 10.1099/mic.0.26412-0. [DOI] [PubMed] [Google Scholar]
- 10.Rohde H, et al. Induction of Staphylococcus epidermidis biofilm formation via proteolytic processing of the accumulation-associated protein by staphylococcal and host proteases. Mol Microbiol. 2005;55(6):1883–1895. doi: 10.1111/j.1365-2958.2005.04515.x. [DOI] [PubMed] [Google Scholar]
- 11.Corrigan RM, Rigby D, Handley P, Foster TJ. The role of Staphylococcus aureus surface protein SasG in adherence and biofilm formation. Microbiology. 2007;153(Pt 8):2435–2446. doi: 10.1099/mic.0.2007/006676-0. [DOI] [PubMed] [Google Scholar]
- 12.Hussain M, Herrmann M, von Eiff C, Perdreau-Remington F, Peters G. A 140-kilodalton extracellular protein is essential for the accumulation of Staphylococcus epidermidis strains on surfaces. Infect Immun. 1997;65(2):519–524. doi: 10.1128/iai.65.2.519-524.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ruggiero A, et al. Crystal structure of the resuscitation-promoting factor (DeltaDUF)RpfB from M. tuberculosis. J Mol Biol. 2009;385(1):153–162. doi: 10.1016/j.jmb.2008.10.042. [DOI] [PubMed] [Google Scholar]
- 14.Conrady DG, et al. A zinc-dependent adhesion module is responsible for intercellular adhesion in staphylococcal biofilms. Proc Natl Acad Sci USA. 2008;105(49):19456–19461. doi: 10.1073/pnas.0807717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wellinghausen N, Rink L. The significance of zinc for leukocyte biology. J Leukoc Biol. 1998;64(5):571–577. doi: 10.1002/jlb.64.5.571. [DOI] [PubMed] [Google Scholar]
- 16.Geoghegan JA, et al. Role of surface protein SasG in biofilm formation by Staphylococcus aureus. J Bacteriol. 2010;192(21):5663–5673. doi: 10.1128/JB.00628-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maret W. Zinc coordination environments in proteins determine zinc functions. J Trace Elem Med Biol. 2005;19(1):7–12. doi: 10.1016/j.jtemb.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 18.Gruszka DT, et al. Staphylococcal biofilm-forming protein has a contiguous rod-like structure. Proc Natl Acad Sci USA. 2012;109(17):E1011–E1018. doi: 10.1073/pnas.1119456109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li H, Dunn JJ, Luft BJ, Lawson CL. Crystal structure of Lyme disease antigen outer surface protein A complexed with an Fab. Proc Natl Acad Sci USA. 1997;94(8):3584–3589. doi: 10.1073/pnas.94.8.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Slabinski L, et al. XtalPred: A web server for prediction of protein crystallizability. Bioinformatics. 2007;23(24):3403–3405. doi: 10.1093/bioinformatics/btm477. [DOI] [PubMed] [Google Scholar]
- 21.Fraczkiewicz R, Braun W. Exact and efficient analytical calculation of the accessible surface areas and their gradients for macromolecules. J Comput Chem. 1998;19(3):319–333. [Google Scholar]
- 22.Hu Y, Ulstrup J, Zhang J, Molin S, Dupres V. Adhesive properties of Staphylococcus epidermidis probed by atomic force microscopy. Phys Chem Chem Phys. 2011;13(21):9995–10003. doi: 10.1039/c0cp02800b. [DOI] [PubMed] [Google Scholar]
- 23.Auld DS. Zinc coordination sphere in biochemical zinc sites. Biometals. 2001;14(3-4):271–313. doi: 10.1023/a:1012976615056. [DOI] [PubMed] [Google Scholar]
- 24.Maret W, Li Y. Coordination dynamics of zinc in proteins. Chem Rev. 2009;109(10):4682–4707. doi: 10.1021/cr800556u. [DOI] [PubMed] [Google Scholar]
- 25.Harding MM. Geometry of metal-ligand interactions in proteins. Acta Crystallogr D Biol Crystallogr. 2001;57(Pt 3):401–411. doi: 10.1107/s0907444900019168. [DOI] [PubMed] [Google Scholar]
- 26.Dudev T, Lim C. Principles governing Mg, Ca, and Zn binding and selectivity in proteins. Chem Rev. 2003;103(3):773–788. doi: 10.1021/cr020467n. [DOI] [PubMed] [Google Scholar]
- 27.Dudev T, Lim C. Metal binding affinity and selectivity in metalloproteins: Insights from computational studies. Annu Rev Biophys. 2008;37:97–116. doi: 10.1146/annurev.biophys.37.032807.125811. [DOI] [PubMed] [Google Scholar]
- 28.Pan N, Brookstein D. Physical properties of twisted structures II. Industrial yarns, cords, and ropes. J Appl Polym Sci. 2002;83(3):610–630. [Google Scholar]
- 29. Galilei G (1954) Dialogues Concerning Two New Sciences, trans. Crew H, De Salvio A (Dover Publications, New York)
- 30.Sobotka AK, Malveaux FJ, Marone G, Thomas LL, Lichtenstein LM. IgE-mediated basophil phenomena: Quantitation, control, inflammatory interactions. Immunol Rev. 1978;41:171–185. doi: 10.1111/j.1600-065x.1978.tb01464.x. [DOI] [PubMed] [Google Scholar]
- 31.Metz M, Maurer M. Mast cells—key effector cells in immune responses. Trends Immunol. 2007;28(5):234–241. doi: 10.1016/j.it.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 32.Doublié S. Preparation of selenomethionyl proteins for phase determination. Methods Enzymol. 1997;276:523–530. [PubMed] [Google Scholar]
- 33.Otwinowski Z, Minor W. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 34.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Strong M, et al. Toward the structural genomics of complexes: Crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2006;103(21):8060–8065. doi: 10.1073/pnas.0602606103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bricogne G, et al. (2011) Buster (Global Phasing Ltd., Cambridge, UK)
- 38.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Painter J, Merritt EA. TLSMD web server for the generation of multi-group TLS models. J Appl Cryst. 2006;39(1):109–111. [Google Scholar]
- 40.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 1):12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pettersen EF, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 42.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys J. 2000;78(3):1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Greenfield NJ. Using circular dichroism collected as a function of temperature to determine the thermodynamics of protein unfolding and binding interactions. Nat Protoc. 2006;1(6):2527–2535. doi: 10.1038/nprot.2006.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.