Abstract
Solution NMR spectroscopy of labeled arrestin-1 was used to explore its interactions with dark-state phosphorylated rhodopsin (P-Rh), phosphorylated opsin (P-opsin), unphosphorylated light-activated rhodopsin (Rh*), and phosphorylated light-activated rhodopsin (P-Rh*). Distinct sets of arrestin-1 elements were seen to be engaged by Rh* and inactive P-Rh, which induced conformational changes that differed from those triggered by binding of P-Rh*. Although arrestin-1 affinity for Rh* was seen to be low (KD > 150 μM), its affinity for P-Rh (KD ∼80 μM) was comparable to the concentration of active monomeric arrestin-1 in the outer segment, suggesting that P-Rh generated by high-gain phosphorylation is occupied by arrestin-1 under physiological conditions and will not signal upon photo-activation. Arrestin-1 was seen to bind P-Rh* and P-opsin with fairly high affinity (KD of ∼50 and 800 nM, respectively), implying that arrestin-1 dissociation is triggered only upon P-opsin regeneration with 11-cis-retinal, precluding noise generated by opsin activity. Based on their observed affinity for arrestin-1, P-opsin and inactive P-Rh very likely affect the physiological monomer-dimer-tetramer equilibrium of arrestin-1, and should therefore be taken into account when modeling photoreceptor function. The data also suggested that complex formation with either P-Rh* or P-opsin results in a global transition in the conformation of arrestin-1, possibly to a dynamic molten globule-like structure. We hypothesize that this transition contributes to the mechanism that triggers preferential interactions of several signaling proteins with receptor-activated arrestins.
Keywords: G protein-coupled receptors, nuclear magnetic resonance, TROSY, bicelles
Arrestin-1, also known as visual or rod arrestin, was the first member of the arrestin family discovered (1). Arrestin-1 preferentially binds light-activated phosphorylated rhodopsin (P-Rh*) and prevents further signaling by direct competition with transducin, the visual G protein (2, 3). Arrestin-1 also binds dark phosphorylated (P-Rh) and light-activated unphosphorylated (Rh*) rhodopsin, albeit with much lower affinity (4, 5). These interactions, as well as arrestin-1 binding to phospho-opsin (P-opsin), remain largely unexplored, despite their biological importance. Rh* is directly produced by light. P-Rh is generated by high-gain phosphorylation of multiple rhodopsin molecules upon activation of a modest subpopulation (6, 7), and P-opsin is generated by the decay of P-Rh*. Arrestin-1 is the second most abundant protein in rod photoreceptors, expressed at a ∼0.8:1 M ratio to rhodopsin (8–10). In dark-adapted rods the bulk of arrestin-1 is localized away from the rhodopsin-containing outer segment. Light induces massive translocation of arrestin-1 into this compartment because of its tight binding to P-Rh* (11, 12). Mammalian arrestin-1 self-associates, and dimers and tetramers predominate at physiological concentrations (13–15). Only the arrestin-1 monomer binds rhodopsin (16). Arrestin-1 monomers, dimers, and tetramers have very different sizes, a fact of importance given that the space between rhodopsin-containing discs in the outer segment is fairly narrow (17). It appears that only monomeric arrestin-1 is small enough to readily diffuse into the interdiscal space, and this has been proposed to be the major factor limiting arrestin concentration in the outer segment under dark conditions (17). The binding of arrestin-1 to any form of rhodopsin affects the arrestin-1 distribution in rod photoreceptors and determines the concentration of the active monomer in the outer segment, which likely contributes to the kinetics of photoresponse and recovery (18). Thus, to fully understand receptor signaling it is important to determine the affinities of these interactions and to identify the arrestin-1 residues involved. This determination is necessary for the elucidation of the molecular mechanisms of arrestin-1 function in the visual system and for building a comprehensive model of its behavior in photoreceptors (19). These data are also critical for the design of arrestin-1 mutants capable of compensating for the effects of rhodopsin mutations that cause congenital human disorders (20).
NMR spectroscopy is well suited for the study of dynamic and relatively low-affinity interactions (21), which constitute the majority of biologically important protein–protein and ligand–protein interactions. Here we took advantage of our recent assignment of >140 amide 1H,15N correlation resonances in the NMR spectrum of arrestin-1 (22) to probe its binding interface and affinity for different functional forms of rhodopsin.
Results
Arrestin-1 Retains Native Structure in the Presence of Anionic Bicelles.
Rhodopsin must be solubilized in an isotropic model membrane system to investigate arrestin-1 binding by solution NMR. Detergent micelles (23) and lipid-detergent bicelles (24) successfully solubilize functional rhodopsin. However, detergents sometimes disrupt the native structure of arrestins. We examined arrestin-1 structure and stability using near-UV CD in the presence of different types of micelles, bicelles, and amphipols (25). Detergents and amphipols destabilized arrestin-1, whereas three different types of bicelles yielded CD spectra very similar to those observed in buffer only (Fig. S1).
Rhodopsin is stable and functional in anionic bicelles (24). Rhodopsin in bicelle-like nanodiscs containing negatively charged lipids binds arrestin-1 with a physiologically relevant affinity (KD∼3–4 nM) (26). We acquired a 2D 1H-15N transverse relaxation optimized spectroscopy (TROSY) spectrum of 50 μM 2H,15N-labeled arrestin-1 in the presence of 4.2% (wt/vol) dimyristoylphosphatidylcholine + diheptanoylphosphatidylcholine (DMPC/D7PC) bicelles containing 20% negatively charged dimyristoylphosphatidylglycerol (DMPG). The NMR spectrum of arrestin-1 in the presence of bicelles was similar to that in buffer (Fig. S2), indicating lack of a bicelle-induced global structural perturbation. Thus, isotropic bicelles are suitable for solution NMR-based studies of arrestin–rhodopsin interactions.
Arrestin-1 Binding to Dark-State Phosphorylated Rhodopsin.
The 2D 1H-15N correlation NMR [heteronuclear single-quantum coherence (HSQC) or TROSY] has long been used to monitor protein-protein or protein-ligand binding and to map protein residues involved in complex formation. Arrestin-1 binds P-Rh* with high selectivity (27), and arrestin-1 residues engaged by P-Rh* have been extensively mapped (5, 28–34). In vivo, light activation, and subsequent phosphorylation of rhodopsin by GRK1 are essential for high-affinity arrestin-1 binding, which effectively shuts off further signaling (35). Both light-induced Rh* and inactive P-Rh are produced in photoreceptors in vivo (7), but bind arrestin-1 less avidly than P-Rh* (4, 5). However, the dissociation constants (KD) and the elements that interact with Rh* and dark P-Rh remain to be fully elucidated. Here we used NMR titrations to investigate the binding of 2H,15N-arrestin-1 to P-Rh and Rh*.
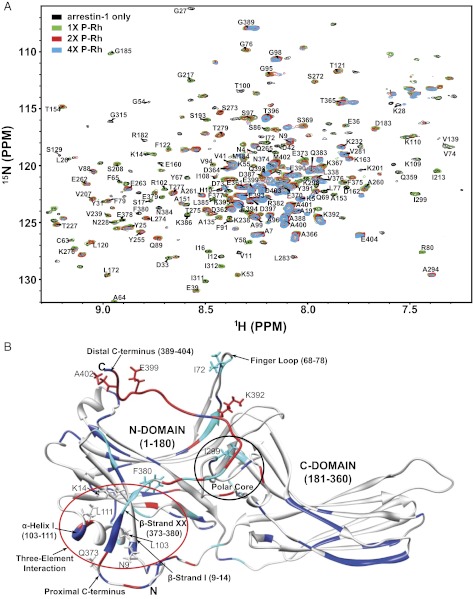
Fig. 1A shows the titration of NMR-labeled arrestin-1 by dark P-Rh. A number of peaks broadened or shifted in response to complex formation, with many peaks broadening beyond detection at the higher levels of P-Rh. We therefore focused our analysis on the sample with a twofold molar excess of P-Rh over arrestin-1, where most peaks were still observable. The peaks shifted rather than split, which indicates that binding was rapid on the NMR time scale. The set of peaks that uniformly shifted the most in response to complex formation were from the distal C terminus of arrestin-1 (Fig. 1B and Fig. S3), indicating that binding to P-Rh changes the conformation of this element. This finding is consistent with the release of the C terminus previously detected in the presence of chemical mimics of the phosphorylated rhodopsin C terminus, heparin, and hexaphosphoinositol (IP6) (22). A few other peaks also shifted significantly or exhibited line broadening (Fig. 1B and Fig. S3). Those peaks were mainly from the “polar core” and “three-element interaction” that were previously shown to be perturbed by IP6 and heparin (22). For three peaks that exhibited relatively large shifts but did not completely disappear, we plotted the variations in chemical shifts as a function of the P-Rh concentration (Fig. S4). Fitting of the data by a 1:1 binding model (9, 26) yielded a KD of 80 ± 30 μM (Fig. S4) for arrestin-1 binding to P-Rh.
Fig. 1.
Arrestin-1 binding to dark-state P-Rh. (A) Superimposed 1H,15N-TROSY spectra of 30 μM 2H,15N-labeled arrestin-1 titrated by P-Rh in 4.2% anionic bicelles, 25 mM Bis●Tris, 100 mM NaCl, 0.1 mM EDTA, 5 mM DTT pH 6.5 at 308 K. The bicelles contained DMPC:DMPG = 4:1 (mol:mol), q = 0.3, where q is the mol:mol ratio of the detergent-like D7PC to DMPC+DMPG. All samples in this work were examined in the same buffer and bicelles and at the same temperature. The molar ratio of arrestin-1 to P-Rh was varied: 1:0 (black), 1:1 (green), 1:2 (red), and 1:4 (cyan). (B) Mapping of P-Rh–induced site-specific NMR chemical-shift changes onto the 3D structure of arrestin-1 (PDB ID 1CF1). Red sites are those for which the backbone amide chemical shifts changed by 0.010 ppm or more. Light blue sites are those for which peaks shifted by less than 0.010 ppm, but underwent significant line-broadening in response to P-Rh. Resonances for dark blue sites neither shifted significantly nor broadened. Because the crystal structure does not resolve residues 1–9, 363–373, and 394–404, these segments were modeled into the depicted structure using X-PLOR-NIH (http://nmr.cit.nih.gov/xplor-nih).
Binding of Arrestin-1 to Unphosphorylated Light-Activated Rhodopsin.
Visual signal transduction is initiated when rhodopsin absorbs a photon, which isomerizes covalently bound 11-cis-retinal, converting rhodopsin into Rh*. Rh* activates transducin and is then rapidly multiphosphorylated at its C terminus (36). The structure of Rh* is significantly different from inactive rhodopsin (37–40). High selectivity of arrestin-1 for P-Rh*, compared with Rh* (5), indicates that arrestin-1 affinity for Rh* must be much lower, although this has not been previously quantitated. Fig. S5A shows overlaid spectra of 30 μM 2H,15N-arrestin-1 with and without 150 μM Rh*. Only a few residues exhibited significant chemical shift changes (Fig. S5B and Fig. S6). Analysis of a full titration (0, 30, 60, and 150 μM Rh*) indicates that binding is relatively weak (KD >150 μM) (Fig. S7). Most resonances exhibiting significant chemical-shift changes corresponded to residues in the 10–70 and 370–385 ranges. These segments include two of three sets of residues known to be involved in the “three-element interaction” between the N-terminal β-strand I (residues 9–14), α-helix I (residues 103–111), and β-strand XX in the proximal C terminus (residues 373–380) (Fig. S5B). The fact that several of the arrestin-1 resonances involved in the three-element interaction were among the handful of resonances that became sharper upon complex formation with Rh* (Fig. S6) is consistent with an increase in local mobility, implying destabilization upon complex formation. These data suggest that the known destabilization of the three-element interaction by P-Rh* is not solely triggered by the phosphorylated rhodopsin C terminus, but that contributions to this destabilization are also made by elements of rhodopsin that change conformation upon light activation. Given that binding of P-Rh (Fig. 1) or P-Rh* (33, 41) results in release of the distal C terminus (residues 385–404), it is notable that binding of Rh* does not do so.
High-Affinity Binding of Arrestin-1 to Phosphorylated Light-Activated Rhodopsin.
Arrestin-1 associates with P-Rh* with high affinity (26, 42, 43), blocking transducin binding sites on rhodopsin (2, 3). To probe structural changes that occur in arrestin-1 upon binding, we titrated 2H,15N-labeled arrestin-1 with P-Rh*.
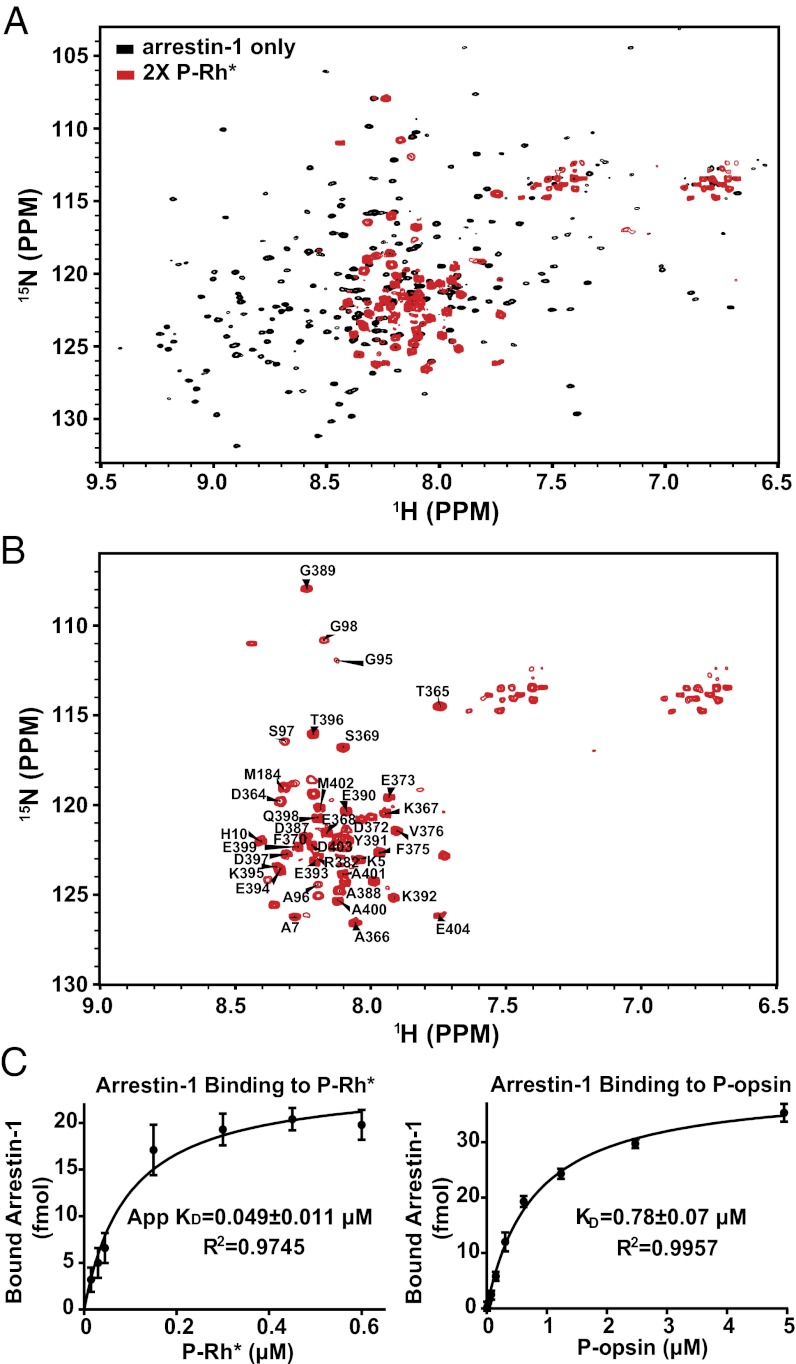
Fig. 2A shows the TROSY spectrum of 30 μM 2H,15N-labeled arrestin-1 in the presence and absence of saturating P-Rh*. Binding of P-Rh* resulted in a dramatic change in the spectrum in which the majority of peaks disappeared (Fig. 2B). The remaining peaks were assigned by tracing the chemical shift changes seen in Fig. 1, which appeared to shift from their positions in free arrestin-1 in the same direction upon complex formation with either P-Rh or P-Rh*. The assignments (Fig. 2B) reveal that the observed arrestin-1 peaks originate from domains shown to be particularly flexible in free arrestin-1 (33, 41), particularly the C-terminal residues 360–404 (see structure in Fig. S8A). Several residues in the body of the N domain (G95, A96, S97, and G98) were also observed, as well as the N-terminal residues K5, A7, and A10. Resonances from most other peaks disappeared upon complex formation because of extensive line broadening. The high intensity of the peaks from the C terminus after complex formation suggests that binding of arrestin-1 to P-Rh* results in freeing of the C terminus of arrestin-1 from tertiary structural interactions, in agreement with distance measurements between the C terminus and the body of the N domain by electron paramagnetic resonance (EPR) and limited proteolysis (33, 41, 44) and also by NMR paramagnetic relaxation experiments (SI Text and Table S1). P-Rh* induced chemical-shift changes for residues G95, S97, and G98, which are located on a short connector between the body of the N domain and the α-helix I (Fig. S8A). Thus, a structural rearrangement of the N domain takes place upon complex formation. This effect was not seen in the titration of arrestin-1 with IP6 or heparin (22), suggesting a possible shift of the α-helix I resulting from disruption of the three-element interaction. Our data confirm that formation of the complex results in a conformational change in the C terminus that enhances its local motional freedom.
Fig. 2.
Binding of arrestin to P-Rh*. (A) (red) TROSY spectrum of 30 μM 2H,15N-labeled arrestin-1 complexed with P-Rh* in bicelles. The spectrum of 30-μM free arrestin-1 (black) shows substantial chemical shift differences for the residues that remain observable. (B) Spectrum of arrestin-1 bound to P-Rh* from A, with labeling of the assigned peaks. (C) Determination of KD for binding of arrestin-1 with P-Rh* (Left) or with P-opsin (Right).
Because light-activated rhodopsin decays faster than equilibrium can be reached, a true KD cannot be determined in a simple titration experiment. The apparent KD for arrestin-1 binding to P-Rh* was determined to be 49 ± 11 nM (Fig. 2C), which is similar to the 25- to 50-nM value previously measured using an extra-Meta II assay (42, 43). Some comment on the stoichiometry of the complex is merited. Monomeric rhodopsin was shown to be necessary and sufficient for high-affinity arrestin-1 binding (26, 45). However, recent reports suggest that two types of arrestin-1-rhodopsin complexes are formed upon activation of a high fraction of rhodopsin in native disk membranes, with 1:1 and 1:2 arrestin:rhodopsin stoichiometries (46, 47). In the latter, arrestin-1 apparently binds only one rhodopsin with high affinity, stabilizing its Meta II state (46) and trapping all-transretinal (47), whereas the other molecule is engaged with much lower affinity by distinct arrestin-1 elements localized in the C-domain (47). All forms of rhodopsin used here were solubilized by a large excess of bicelles and most of the perturbations were observed to be in the N domain of arrestin-1 and in the C terminus that is anchored to this part of the molecule (Figs. 1 and 2). These data point to a 1:1 stoichiometry, making it highly unlikely that “auxiliary” interaction with a second rhodopsin contributed to our results.
Binding to P-Rh* Induces a Major Change in the Global Dynamics of Arrestin-1.
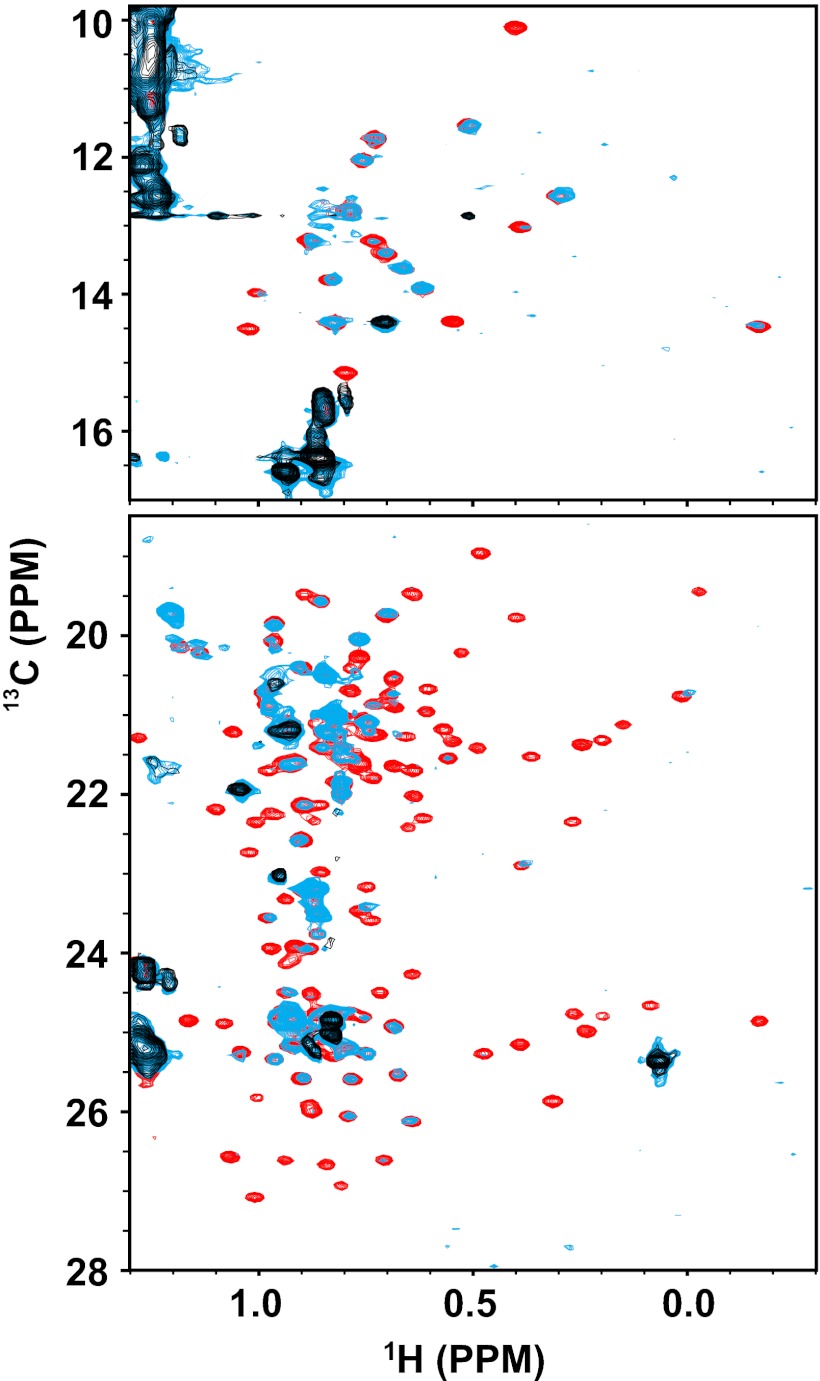
Most arrestin-1 peaks disappear upon binding to P-Rh* (Fig. 2B). A priori, this phenomenon could be a result of the large correlation time associated with the ∼200-kDa size of the arrestin/P-Rh*/bicelle complex. Alternatively, complex formation might result in a global change of conformational dynamics within arrestin-1, with the induced motions occurring in the intermediate rate regime (msec−1–μsec−1) of the NMR time scale. To determine which line-broadening mechanism actually pertains, we collected 1H,13C-methyl-TROSY spectra of ILV-13C-methyl–protonated arrestin-1 in the presence and absence of P-Rh*. It is well established that high-quality methyl-TROSY spectra can be acquired even for complexes with molecular weights well in excess of 200 kDa (48). However, our data (Fig. 3) revealed the same general disappearance of peaks in the methyl-TROSY spectrum that is observed in the 1H,15N-TROSY spectrum of backbone amide peaks. This finding indicates that NMR peak disappearance for arrestin-1 upon binding to P-Rh* is most likely a result of widespread conformational exchange in the intermediate rate regime, leading to extensive exchange-broadening. This is not an unusual phenomenon in protein structure and dynamics: widespread exchange-broadening is a hallmark of proteins in molten globule-like conformational states (49). Binding of P-Rh* apparently induces transition of arrestin-1 to such a state, which evidently encompasses both its N and C domains.
Fig. 3.
800 MHz 1H,13C-Me-TROSY of arrestin-1 binding to P-Rh*. (Upper) Isoleucine region of arrestin-1 spectrum. (Lower) Leucine and valine region of spectrum. All three samples contained 4% bicelles at 308 K and pH 6.5. Arrestin-1 was perdeuterated except for the protonated methyl groups of isoleucine, leucine, and valine, which were also 13C-labeled. The sample represented by the black spectrum contained 90 μM unlabeled P-Rh*, but no arrestin-1 (only the set of natural abundance 1H,13C-methyl peaks from the bicelles were observed). The sample represented by the red spectrum contained 30 μM arrestin-1 but no P-Rh*. The light-blue spectrum corresponds to a sample containing both 30 μM labeled arrestin-1 and 90 μM unlabeled P-Rh*.
Comparison of the Binding Affinity, Structure, and Dynamics of Arrestin-1 Bound to Phosphorylated Opsin (P-Opsin) vs. P-Rh*.
The regeneration of rhodopsin following photoactivation requires the release of all-transretinal, which occurs while arrestin-1 is still associated with P-Rh*. Previous studies indicated that arrestin-1 binds retinal-free phosphorylated opsin (5); however, it has not been clear whether the conversion of P-Rh* to P-opsin induces additional changes in bound arrestin-1 or triggers arrestin-1 release, as was hypothesized previously (5, 50). Fig. 2C shows determination of KD for arrestin-1 binding to both P-Rh* and P-opsin. Binding to P-opsin was indeed strong [KD = 780 ± 70 nM; which is close to a recently reported KD of 1.5 μM (47)], but is less avid by an order-of-magnitude than binding to P-Rh* (apparent KD = 49 ± 11 nM). The 1H-15N TROSY spectrum of arrestin-1 in the presence of a threefold (saturating) molar excess of P-opsin was nearly identical to that in complex with P-Rh* (Fig. S8B), indicating high similarity in the structure and dynamics of arrestin-1 in the two complexes. Opsin exists in a pH-dependent equilibrium between active-like and inactive conformations (51), with lower pH favoring the former (38, 39). Because both transducin (52) and arrestin-1 (43) stabilize the active Meta II state of rhodopsin, it is likely that bound arrestin-1 shifts this equilibrium toward the active-like state, similar to binding of rhodopsin to the C-terminal peptide of the transducin α-subunit (51). P-opsin and P-Rh* induce the same transition in the global structure of arrestin-1 to what appears to be a partially-disordered state (Fig. 3 and Fig. S8).
Discussion
High-affinity binding of arrestin-1 to P-Rh* is necessary for rapid termination of rhodopsin signaling (35). Arrestin-1 elements engaged by P-Rh* and the conformational changes in the arrestin-1 molecule induced by P-Rh* binding have previously been extensively characterized (19). In contrast, although arrestin-1 binding to other functional forms of rhodopsin—Rh*, dark P-Rh, and P-opsin—has been reported (5), the KD values for most of these interactions have not been determined and the impact of complex formation on the structure of arrestin-1 has not been studied. Here we used NMR to identify arrestin-1 elements engaged by nonpreferred forms of rhodopsin and to probe the nature of conformational changes induced by these interactions. The fact that these solution NMR studies focused on complex formation between a large soluble protein and a large membrane protein, and were carried out using only 30 μM arrestin-1, also exemplifies the advances in NMR sensitivity in recent years, thanks to the availability of very high-field magnets, exploitation of TROSY spin physics (48, 53), and cryoprobe detectors (54).
We found that binding to Rh* affects the N terminus and proximal C terminus (residues 370–385) of arrestin-1 (Figs. S5 and S6). Rh* binding did not result in the freeing of the distal C terminus (residues 385–404), in contrast to binding of the phosphorylated forms of rhodopsin (Figs. 1 and 2 and Fig. S8). This finding provides evidence that a unique conformational change in arrestin is induced by Rh*. Significant chemical-shift changes of residues in the 10–70 and 370–385 ranges, along with sharpening of several peaks upon complex formation (Fig. S6), suggest that active unphosphorylated receptors destabilize the three-element interaction previously believed to be disrupted solely by receptor-attached phosphates (55). The data also indicate that the proximal C terminus can be perturbed without the displacement of the distal C terminus. This possibility has not been previously considered (19). Finally, our estimate of arrestin-1 affinity for Rh* (KD > 150 μM) is consistent with physiological data that wild-type arrestin-1 cannot effectively quench Rh* signaling (20, 35, 56).
We found that both dark P-Rh and P-opsin displace the distal C terminus of arrestin-1 (Fig. 1 and Fig. S8), similar to P-Rh* (33, 41) and the phospho-mimics, heparin and IP6 (22). These data show that receptor-attached phosphates are necessary and sufficient for this conformational rearrangement. The affinity of arrestin-1 for dark P-Rh measured in this work (KD∼80 μM) is comparable to the arrestin-1 homodimerization constant (12, 15). This finding suggests that interaction with P-Rh can significantly affect the arrestin-1 monomer-dimer-tetramer equilibrium in photoreceptors. Only the monomeric form of arrestin-1 binds rhodopsin (15). The concentration of arrestin-1 monomer in the outer segments, calculated based on its distribution in rods and its self-association constants (12), yielded values that are insufficient to explain the unexpectedly short lifetime of active rhodopsin (18). Low-affinity binding of arrestin-1 to additional partners, such as nonpreferred forms of rhodopsin, would increase the supply of arrestin-1 monomer available to P-Rh*, reconciling biochemical and physiological evidence.
Complex formation between arrestin-1 and P-Rh* was seen to result in widespread NMR peak disappearance, with the exception of peaks from highly mobile elements (N-terminal 10 residues, sites 95–98, and the distal C terminus). Because peak loss was seen in both 1H,15N-TROSY and in 1H,13C-methyl-TROSY spectra, it is unlikely that this result is primarily because of the size of the complex. A much more likely explanation is intermediate time-scale conformational exchange peak broadening that arises from a global “loosening” of the arrestin-1 conformation. Although the results do not prove this conclusively, the data are consistent with the notion that the global conformation of arrestin-1 is destabilized by high-affinity binding to P-Rh*, triggering a transition into a molten globule-like structure. High flexibility of both free and rhodopsin-bound arrestin-1 is consistent with rapid H/D exchange observed in both states (28), as well as with the relatively wide distance distributions recently detected in both states by pulsed EPR (57). Based on these observations for arrestin-1, it can be hypothesized that a transition from a flexible but well-ordered structure to a partially disordered ensemble of conformations might be the central mechanism underlying the “activation” of other arrestins (27), which enables association with many signaling proteins. This finding would explain how activation of a given arrestin by a given receptor can result in distinct signaling complexes, leading to different functional outcomes (58, 59).
Previously, receptor binding-induced conformational changes in arrestin have been proposed to involve the movement of the two arrestin domains relative to each other (27), as suggested by progressive reduction of receptor binding by deletions in the interdomain hinge (60). However, the recently reported absence of large changes in the interdomain distances in arrestin-1 upon binding to P-Rh* (57) argues against this model. Our data reveal only localized conformational rearrangements upon binding of arrestin-1 to dark P-Rh (Fig. 1) and Rh* (Fig. S5). For P-Rh* and P-opsin, our data suggest a major change in arrestin-1 dynamics as described above, but does not inform on the nature of the associated conformational changes (Figs. 2 and 3).
Finally, we also found that P-opsin binds to arrestin-1 with relatively high affinity (KD = 780 nM) and induces changes in its structure and dynamics that are similar to those induced by P-Rh*. These data suggest that it is the regeneration of rhodopsin with 11-cis-retinal, rather than the loss of all-trans-retinal by P-Rh*, as previously proposed (50), that promotes arrestin-1 dissociation. Considering that opsin can activate transducin, delayed arrestin-1 release makes perfect biological sense in rods, where high sensitivity requires efficient noise suppression (61).
Methods
Preparation of Arrestin-1 Samples.
Constitutively monomeric arrestin-1-(F85A/F197A) was expressed in isotopically labeled form and purified as previously described (22, 62). Near-UV CD spectroscopy of arrestin-1 and preparation of various forms of rhodopsin are described in SI Text.
NMR Spectroscopy of Arrestin-1.
1H,15N-TROSY NMR titrations of 2H,15N-labeled arrestin-1 with various forms of rhodopsin were carried out at pH 6.5 and 308 K in the presence of 100 mM NaCl. Samples with different ratios of arrestin-1 and rhodopsin were made using a constant arrestin-1 concentration of 30 μM. 1H,13C-Me-TROSY NMR spectra (48) were collected of arrestin-1 that was uniformly perdeuterated, except for the side-chain methyl groups of isoleucine, leucine, and valine residues, which were also 13C-labeled. Additional details are provided in SI Text.
Determination of Dissociation Constants.
A 1:1 binding model (SI Methods) was fit to NMR titration data to estimate KD for complex formation of arrestin-1 with various forms of rhodopsin. To determine the affinity by an alternative method in some cases, we used direct binding assay with radiolabeled arrestin-1 generated in cell-free translation, as previously described (4, 62). Increasing amounts of P-Rh* or P-opsin were added to a fixed concentration of arrestin-1 and the amount of bound arrestin was determined upon incubation at 37 °C for 5 min (P-Rh*) or for 30 min (P-opsin). Because true equilibrium cannot be achieved with P-Rh* as a result of its decay, these data yielded an apparent KD, whereas P-opsin binding was measured at equilibrium, yielding a true KD.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants U54 GM094608 and P01 GM080513 (to C.R.S.); R01 EY011500, GM077561, and GM081756 (to V.V.G.); and 1R01 GM095633 and 1R21EY018435 (to T.M.I.). Q.C. was supported by the Vanderbilt International Scholars Program.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1215176110/-/DCSupplemental.
References
- 1.Kühn H. Light-regulated binding of rhodopsin kinase and other proteins to cattle photoreceptor membranes. Biochemistry. 1978;17(21):4389–4395. doi: 10.1021/bi00614a006. [DOI] [PubMed] [Google Scholar]
- 2.Wilden U. Duration and amplitude of the light-induced cGMP hydrolysis in vertebrate photoreceptors are regulated by multiple phosphorylation of rhodopsin and by arrestin binding. Biochemistry. 1995;34(4):1446–1454. doi: 10.1021/bi00004a040. [DOI] [PubMed] [Google Scholar]
- 3.Krupnick JG, Gurevich VV, Benovic JL. Mechanism of quenching of phototransduction. Binding competition between arrestin and transducin for phosphorhodopsin. J Biol Chem. 1997;272(29):18125–18131. doi: 10.1074/jbc.272.29.18125. [DOI] [PubMed] [Google Scholar]
- 4.Gurevich VV, Benovic JL. Cell-free expression of visual arrestin. Truncation mutagenesis identifies multiple domains involved in rhodopsin interaction. J Biol Chem. 1992;267(30):21919–21923. [PubMed] [Google Scholar]
- 5.Gurevich VV, Benovic JL. Visual arrestin interaction with rhodopsin. Sequential multisite binding ensures strict selectivity toward light-activated phosphorylated rhodopsin. J Biol Chem. 1993;268(16):11628–11638. [PubMed] [Google Scholar]
- 6.Binder BM, Biernbaum MS, Bownds MD. Light activation of one rhodopsin molecule causes the phosphorylation of hundreds of others. A reaction observed in electropermeabilized frog rod outer segments exposed to dim illumination. J Biol Chem. 1990;265(25):15333–15340. [PubMed] [Google Scholar]
- 7.Binder BM, O’Connor TM, Bownds MD, Arshavsky VY. Phosphorylation of non-bleached rhodopsin in intact retinas and living frogs. J Biol Chem. 1996;271(33):19826–19830. doi: 10.1074/jbc.271.33.19826. [DOI] [PubMed] [Google Scholar]
- 8.Strissel KJ, Sokolov M, Trieu LH, Arshavsky VY. Arrestin translocation is induced at a critical threshold of visual signaling and is superstoichiometric to bleached rhodopsin. J Neurosci. 2006;26(4):1146–1153. doi: 10.1523/JNEUROSCI.4289-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanson SM, et al. Each rhodopsin molecule binds its own arrestin. Proc Natl Acad Sci USA. 2007;104(9):3125–3128. doi: 10.1073/pnas.0610886104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Song X, et al. Arrestin-1 expression level in rods: Balancing functional performance and photoreceptor health. Neuroscience. 2011;174:37–49. doi: 10.1016/j.neuroscience.2010.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nair KS, et al. Light-dependent redistribution of arrestin in vertebrate rods is an energy-independent process governed by protein-protein interactions. Neuron. 2005;46(4):555–567. doi: 10.1016/j.neuron.2005.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim M, et al. Robust self-association is a common feature of mammalian visual arrestin-1. Biochemistry. 2011;50(12):2235–2242. doi: 10.1021/bi1018607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schubert C, et al. Visual arrestin activity may be regulated by self-association. J Biol Chem. 1999;274(30):21186–21190. doi: 10.1074/jbc.274.30.21186. [DOI] [PubMed] [Google Scholar]
- 14.Imamoto Y, Tamura C, Kamikubo H, Kataoka M. Concentration-dependent tetramerization of bovine visual arrestin. Biophys J. 2003;85(2):1186–1195. doi: 10.1016/S0006-3495(03)74554-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanson SM, et al. Structure and function of the visual arrestin oligomer. EMBO J. 2007;26(6):1726–1736. doi: 10.1038/sj.emboj.7601614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanson SM, et al. A model for the solution structure of the rod arrestin tetramer. Structure. 2008;16(6):924–934. doi: 10.1016/j.str.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Najafi M, Maza NA, Calvert PD. Steric volume exclusion sets soluble protein concentrations in photoreceptor sensory cilia. Proc Natl Acad Sci USA. 2012;109(1):203–208. doi: 10.1073/pnas.1115109109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gross OP, Burns ME. Control of rhodopsin’s active lifetime by arrestin-1 expression in mammalian rods. J Neurosci. 2010;30(9):3450–3457. doi: 10.1523/JNEUROSCI.5391-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gurevich VV, Hanson SM, Song X, Vishnivetskiy SA, Gurevich EV. The functional cycle of visual arrestins in photoreceptor cells. Prog Retin Eye Res. 2011;30(6):405–430. doi: 10.1016/j.preteyeres.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Song X, et al. Enhanced arrestin facilitates recovery and protects rod photoreceptors deficient in rhodopsin phosphorylation. Curr Biol. 2009;19(8):700–705. doi: 10.1016/j.cub.2009.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shi Y, Wu J. Structural basis of protein-protein interaction studied by NMR. J Struct Funct Genomics. 2007;8(2–3):67–72. doi: 10.1007/s10969-007-9021-8. [DOI] [PubMed] [Google Scholar]
- 22.Zhuang TD, Vishnivetskiy SA, Gurevich VV, Sanders CR. Elucidation of inositol hexaphosphate and heparin interaction sites and conformational changes in arrestin-1 by solution nuclear magnetic resonance. Biochemistry. 2010;49(49):10473–10485. doi: 10.1021/bi101596g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ernst OP, Gramse V, Kolbe M, Hofmann KP, Heck M. Monomeric G protein-coupled receptor rhodopsin in solution activates its G protein transducin at the diffusion limit. Proc Natl Acad Sci USA. 2007;104(26):10859–10864. doi: 10.1073/pnas.0701967104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaya AI, Thaker TM, Preininger AM, Iverson TM, Hamm HE. Coupling efficiency of rhodopsin and transducin in bicelles. Biochemistry. 2011;50(15):3193–3203. doi: 10.1021/bi200037j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Popot JL. Amphipols, nanodiscs, and fluorinated surfactants: Three nonconventional approaches to studying membrane proteins in aqueous solutions. Annu Rev Biochem. 2010;79:737–775. doi: 10.1146/annurev.biochem.052208.114057. [DOI] [PubMed] [Google Scholar]
- 26.Bayburt TH, et al. Monomeric rhodopsin is sufficient for normal rhodopsin kinase (GRK1) phosphorylation and arrestin-1 binding. J Biol Chem. 2011;286(2):1420–1428. doi: 10.1074/jbc.M110.151043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gurevich VV, Gurevich EV. The molecular acrobatics of arrestin activation. Trends Pharmacol Sci. 2004;25(2):105–111. doi: 10.1016/j.tips.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 28.Ohguro H, Palczewski K, Walsh KA, Johnson RS. Topographic study of arrestin using differential chemical modifications and hydrogen/deuterium exchange. Protein Sci. 1994;3(12):2428–2434. doi: 10.1002/pro.5560031226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gurevich VV, et al. Arrestin interactions with G protein-coupled receptors. Direct binding studies of wild type and mutant arrestins with rhodopsin, beta 2-adrenergic, and m2 muscarinic cholinergic receptors. J Biol Chem. 1995;270(2):720–731. doi: 10.1074/jbc.270.2.720. [DOI] [PubMed] [Google Scholar]
- 30.Pulvermüller A, Schroder K, Fischer T, Hofmann KP. Interactions of metarhodopsin II. Arrestin peptides compete with arrestin and transducin. J Biol Chem. 2000;275(48):37679–37685. doi: 10.1074/jbc.M006776200. [DOI] [PubMed] [Google Scholar]
- 31.Vishnivetskiy SA, Hosey MM, Benovic JL, Gurevich VV. Mapping the arrestin-receptor interface. Structural elements responsible for receptor specificity of arrestin proteins. J Biol Chem. 2004;279(2):1262–1268. doi: 10.1074/jbc.M308834200. [DOI] [PubMed] [Google Scholar]
- 32.Vishnivetskiy SA, et al. Few residues within an extensive binding interface drive receptor interaction and determine the specificity of arrestin proteins. J Biol Chem. 2011;286(27):24288–24299. doi: 10.1074/jbc.M110.213835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hanson SM, et al. Differential interaction of spin-labeled arrestin with inactive and active phosphorhodopsin. Proc Natl Acad Sci USA. 2006;103(13):4900–4905. doi: 10.1073/pnas.0600733103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hanson SM, Gurevich VV. The differential engagement of arrestin surface charges by the various functional forms of the receptor. J Biol Chem. 2006;281(6):3458–3462. doi: 10.1074/jbc.M512148200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mendez A, et al. Rapid and reproducible deactivation of rhodopsin requires multiple phosphorylation sites. Neuron. 2000;28(1):153–164. doi: 10.1016/s0896-6273(00)00093-3. [DOI] [PubMed] [Google Scholar]
- 36.Kühn H, Hall SW, Wilden U. Light-induced binding of 48-kDa protein to photoreceptor membranes is highly enhanced by phosphorylation of rhodopsin. FEBS Lett. 1984;176(2):473–478. doi: 10.1016/0014-5793(84)81221-1. [DOI] [PubMed] [Google Scholar]
- 37.Li J, Edwards PC, Burghammer M, Villa C, Schertler GF. Structure of bovine rhodopsin in a trigonal crystal form. J Mol Biol. 2004;343(5):1409–1438. doi: 10.1016/j.jmb.2004.08.090. [DOI] [PubMed] [Google Scholar]
- 38.Scheerer P, et al. Crystal structure of opsin in its G-protein-interacting conformation. Nature. 2008;455(7212):497–502. doi: 10.1038/nature07330. [DOI] [PubMed] [Google Scholar]
- 39.Park JH, Scheerer P, Hofmann KP, Choe HW, Ernst OP. Crystal structure of the ligand-free G-protein-coupled receptor opsin. Nature. 2008;454(7201):183–187. doi: 10.1038/nature07063. [DOI] [PubMed] [Google Scholar]
- 40.Choe HW, et al. Crystal structure of metarhodopsin II. Nature. 2011;471(7340):651–655. doi: 10.1038/nature09789. [DOI] [PubMed] [Google Scholar]
- 41.Vishnivetskiy SA, et al. The role of arrestin alpha-helix I in receptor binding. J Mol Biol. 2010;395(1):42–54. doi: 10.1016/j.jmb.2009.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pulvermüller A, et al. Functional differences in the interaction of arrestin and its splice variant, p44, with rhodopsin. Biochemistry. 1997;36(30):9253–9260. doi: 10.1021/bi970772g. [DOI] [PubMed] [Google Scholar]
- 43.Schleicher A, Kühn H, Hofmann KP. Kinetics, binding constant, and activation energy of the 48-kDa protein-rhodopsin complex by extra-metarhodopsin II. Biochemistry. 1989;28(4):1770–1775. doi: 10.1021/bi00430a052. [DOI] [PubMed] [Google Scholar]
- 44.Palczewski K, Pulvermüller A, Buczyłko J, Hofmann KP. Phosphorylated rhodopsin and heparin induce similar conformational changes in arrestin. J Biol Chem. 1991;266(28):18649–18654. [PubMed] [Google Scholar]
- 45.Tsukamoto H, Sinha A, DeWitt M, Farrens DL. Monomeric rhodopsin is the minimal functional unit required for arrestin binding. J Mol Biol. 2010;399(3):501–511. doi: 10.1016/j.jmb.2010.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sommer ME, Hofmann KP, Heck M. Arrestin-rhodopsin binding stoichiometry in isolated rod outer segment membranes depends on the percentage of activated receptors. J Biol Chem. 2011;286(9):7359–7369. doi: 10.1074/jbc.M110.204941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sommer ME, Hofmann KP, Heck M. Distinct loops in arrestin differentially regulate ligand binding within the GPCR opsin. Nat Commun. 2012;3:995. doi: 10.1038/ncomms2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tugarinov V, Kay LE. Methyl groups as probes of structure and dynamics in NMR studies of high-molecular-weight proteins. ChemBioChem. 2005;6(9):1567–1577. doi: 10.1002/cbic.200500110. [DOI] [PubMed] [Google Scholar]
- 49.Redfield C. Using nuclear magnetic resonance spectroscopy to study molten globule states of proteins. Methods. 2004;34(1):121–132. doi: 10.1016/j.ymeth.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 50.Hofmann KP, Pulvermüller A, Buczyłko J, Van Hooser P, Palczewski K. The role of arrestin and retinoids in the regeneration pathway of rhodopsin. J Biol Chem. 1992;267(22):15701–15706. [PubMed] [Google Scholar]
- 51.Vogel R, Siebert F. Conformations of the active and inactive states of opsin. J Biol Chem. 2001;276(42):38487–38493. doi: 10.1074/jbc.M105423200. [DOI] [PubMed] [Google Scholar]
- 52.Emeis D, Kühn H, Reichert J, Hofmann KP. Complex formation between metarhodopsin II and GTP-binding protein in bovine photoreceptor membranes leads to a shift of the photoproduct equilibrium. FEBS Lett. 1982;143(1):29–34. doi: 10.1016/0014-5793(82)80266-4. [DOI] [PubMed] [Google Scholar]
- 53.Pervushin K, Riek R, Wider G, Wüthrich K. Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc Natl Acad Sci USA. 1997;94(23):12366–12371. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kovacs H, Moskau D, Spraul M. Cryogenically cooled probes—A leap in NMR technology. Prog NMR Spectr. 2005;46:131–155. [Google Scholar]
- 55.Vishnivetskiy SA, et al. An additional phosphate-binding element in arrestin molecule. Implications for the mechanism of arrestin activation. J Biol Chem. 2000;275(52):41049–41057. doi: 10.1074/jbc.M007159200. [DOI] [PubMed] [Google Scholar]
- 56.Burns ME, et al. Deactivation of phosphorylated and nonphosphorylated rhodopsin by arrestin splice variants. J Neurosci. 2006;26(3):1036–1044. doi: 10.1523/JNEUROSCI.3301-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim M, et al. Conformation of receptor-bound visual arrestin. Proc Natl Acad Sci USA. 2012;109(45):18407–18412. doi: 10.1073/pnas.1216304109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim J, et al. Functional antagonism of different G protein-coupled receptor kinases for beta-arrestin-mediated angiotensin II receptor signaling. Proc Natl Acad Sci USA. 2005;102(5):1442–1447. doi: 10.1073/pnas.0409532102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ren XR, et al. Different G protein-coupled receptor kinases govern G protein and beta-arrestin-mediated signaling of V2 vasopressin receptor. Proc Natl Acad Sci USA. 2005;102(5):1448–1453. doi: 10.1073/pnas.0409534102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vishnivetskiy SA, Hirsch JA, Velez MG, Gurevich YV, Gurevich VV. Transition of arrestin into the active receptor-binding state requires an extended interdomain hinge. J Biol Chem. 2002;277(46):43961–43967. doi: 10.1074/jbc.M206951200. [DOI] [PubMed] [Google Scholar]
- 61.Luo DG, Xue T, Yau KW. How vision begins: An odyssey. Proc Natl Acad Sci USA. 2008;105(29):9855–9862. doi: 10.1073/pnas.0708405105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gurevich VV, Benovic JL. Arrestin: Mutagenesis, expression, purification, and functional characterization. Methods Enzymol. 2000;315:422–437. doi: 10.1016/s0076-6879(00)15859-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.