Abstract
How diversity evolves and persists in biofilms is essential for understanding much of microbial life, including the uncertain dynamics of chronic infections. We developed a biofilm model enabling long-term selection for daily adherence to and dispersal from a plastic bead in a test tube. Focusing on a pathogen of the cystic fibrosis lung, Burkholderia cenocepacia, we sequenced clones and metagenomes to unravel the mutations and evolutionary forces responsible for adaptation and diversification of a single biofilm community during 1,050 generations of selection. The mutational patterns revealed recurrent evolution of biofilm specialists from generalist types and multiple adaptive alleles at relatively few loci. Fitness assays also demonstrated strong interference competition among contending mutants that preserved genetic diversity. Metagenomes from five other independently evolved biofilm lineages revealed extraordinary mutational parallelism that outlined common routes of adaptation, a subset of which was found, surprisingly, in a planktonic population. These mutations in turn were surprisingly well represented among mutations that evolved in cystic fibrosis isolates of both Burkholderia and Pseudomonas. These convergent pathways included altered metabolism of cyclic diguanosine monophosphate, polysaccharide production, tricarboxylic acid cycle enzymes, global transcription, and iron scavenging. Evolution in chronic infections therefore may be driven by mutations in relatively few pathways also favored during laboratory selection, creating hope that experimental evolution may illuminate the ecology and selective dynamics of chronic infections and improve treatment strategies.
Keywords: population genomics, microbial ecology, ecotype, clonal interference
Bacterial evolution during chronic infections, particularly in pulmonary infections of persons with cystic fibrosis (CF), may involve adaptation to life in biofilms. These aggregations of cells on surfaces are more spatially structured and both generate and experience more diverse environmental conditions than well-mixed planktonic populations, leading to increased biodiversity (1–4). Biofilm diversity often evolves from founding clones and commonly is observed as variation in colony morphology, production of extracellular polymers, motility, and secreted signals or substrates (3, 5, 6). Because isolates recovered from chronic infections of different patients often resemble these biofilm-specific variants (7–9), they may reflect adaptation to similar selective forces in vivo. However, the nature of selection during infections and why multiple types evolve is uncertain; despite several recent surveys of evolution in the lungs of CF patients (10–13), the ecology of chronic infections remains a black box. For this reason, we developed a model enabling long-term selection in biofilms (4) to study how genetic and ecological diversity evolves in a structured environment.
Our simple model involves a daily cycle in which cells must form a biofilm on a plastic bead suspended in minimal medium in a test tube (Fig. 1). Colonized beads then serve to inoculate the next tube containing a fresh bead, so bacteria must remain adherent during transfer and then disperse to colonize a new bead. In this environment, we studied adaptation by a soil isolate of Burkholderia cenocepacia, an opportunistic pathogen responsible for fatal biofilm-associated lung infections in CF patients (14, 15). Although this species has been identified as the most prevalent and pathogenic of the Burkholderia cepacia complex species in CF patients (14), relatively little is known about how it adapts to a biofilm lifestyle or to the CF lung environment (5, 16).
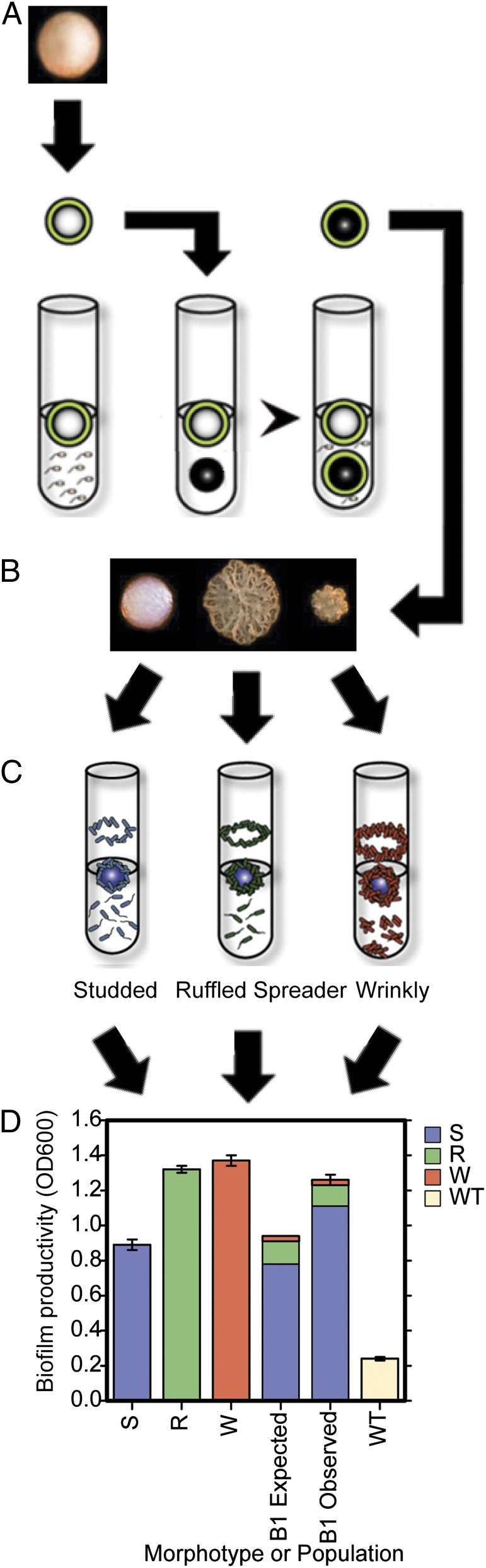
Fig. 1.
Summary of the biofilm experimental evolution and its ecological diversification. (A) B. cenocepacia HI2424 was grown in GMM with a 7-mm polystyrene bead for 24 h. This bead was transferred to a new tube of GMM with an oppositely marked bead. Cells that adhered to the first bead needed to disperse and attach to the oppositely marked bead, which then was transferred to another tube after 24 h of growth. These bead-to-bead transfers were conducted for ∼1,050 generations. (B) After ∼300 generations of biofilm selection, an adaptive radiation of biofilm generalists (S) and specialists (R and W) was observed. Subsequently, each ecotype remained detectable throughout the experiment. (C) Each ecotype has distinct growth patterns when grown in monoculture. S occupies the planktonic phase and displays moderate bead attachment, R exhibits less planktonic growth than S and forms thick biofilms, and W forms clumps and produces copious amounts of biofilm. (D) Ecotypes vary in biofilm production (3); however, when S, R, and W are grown together, total biofilm is greater than expected from their monoculture values and starting frequencies, indicating synergy when all ecotypes are present (Methods). Assays were focused on sequenced clones isolated at 1,050 generations; error bars indicate the 95% confidence interval.
Six B. cenocepacia populations underwent ∼1,050 generations of biofilm selection, all of which diversified into similar sets of three distinct, heritable colony morphotypes from a single ancestral clone (Fig. 1) (4). At least one of these morphotypes—a small, highly rugose “wrinkly” type (17)—is commonly isolated from chronic lung infections of both Burkholderia spp. and Pseudomonas aeruginosa and has been associated with high patient mortality (7, 17). Each morphotype associates with a distinct ecological niche and thus can be considered an ecotype; collectively, ecotypes segregate nutrients and biofilm space in ways that enhance community productivity (Fig. 1D) (4). The persistence of these ecotypes for the duration of the experiment raised several questions about their evolutionary dynamics and mutational causes. Did each type evolve once and persist, or did multiple mutations produce the same ecotypes? In addition, did these types generate further diversity and coevolve, much as Darwin envisioned for the evolution of his famous “tangled bank” (18) of myriad, interdependent plants and animals?
We also sought to test whether mutations favored during prolonged selection in experimental biofilms occurred in targets similar to those identified during genetic surveys of CF lung isolates, including small-colony variants. We applied multiple, complementary genomic approaches to both clones and communities to identify the molecular bases of biofilm adaptation and quantify mutant frequencies over time. These results allowed us to reconstruct a nearly complete evolutionary history of long-term adaptation and diversification in one biofilm population with unprecedented resolution. Comparing these mutations with those found in five other replicate experimental biofilm populations revealed considerable parallelism both in genes known to affect biofilm production and in unexpected pathways. Moreover, many of these mutations occurred in genes that commonly are mutated among isolates of Burkholderia and P. aeruginosa from chronic lung infections, implying that adaptation during chronic infections may be partly driven by selection in biofilms.
Results
This study focused on a single biofilm population [B1, from a larger experiment (4)] in which three distinct colony morphologies that correspond with unique ecological functions evolved: studded (S), ruffled (R), and wrinkly (W) (Fig. 1C). The S morphotype is the most abundant and retains the capacity to grow planktonically while forming thin, mostly confluent biofilms; therefore it can be considered a generalist. The R morphotype is a specialist and produces more copious biofilm and fewer planktonic cells. The rarest W type primarily grows as a dense, towering biofilm (Fig. 1). To identify the molecular bases of these ecological differences, we sequenced DNA from mixed communities (metagenomes), the complete genomes of representative clones, and alleles of 60 alternative clones from multiple time points. These analyses (Table 1) revealed that early ecotypes (isolated at generation 315) evolved independently by distinct mutations, whereas late ecotypes (isolated at generation 1,050) derived from a common lineage and subsequently evolved by one to four ecotype-specific mutations. Thus, although the composition of the community—a majority S ecotype and minority R and W ecotypes—remained constant, its genetic structure changed and suggested that R and W types had evolved anew from an S haplotype. Within the S lineage alone, five mutations became detectable within ∼315 generations in a population that never dropped below 5 × 107 cells/mL (the minimum number of cells adhering to the bead that was transferred). Given this large population size and the fact that the mutation rate did not evolve (4), genetic drift cannot explain the rapid rise of these mutations. Rather, the following features strongly suggest that nearly all mutations were favored by selection: Mutations in coding sequences were mostly nonsynonymous (nonsynonymous/synonymous ratio = 5.5), intergenic mutations were associated with likely promoters, and deletions affected genes that were plausible targets of selection (Table 1). The rapid rise of these lineages combined with the low per-genome mutation rate (<10−2 per genome per generation) also theoretically should hinder hitchhiking of neutral mutations but does not exclude this possibility.
Table 1.
Mutations detected in one population during 6 mo of selection under a daily cycle of biofilm colonization and dispersal and their predicted functional significance
| Mutation | Ecotype, time (in generations) | Locus | Annotation | Mutational effect | Significance |
| M1 | S, 315 | YP_837186 | yciR | Y355D | Cyclic di-GMP |
| M2 | YP_837213 | Monooxygenase | E481D | ||
| M3 | S, 315 S,R,W, 1050 | YP_835154 | 2-oxoglutarate dehydrogenase | R204S | Metabolism |
| M4 | S, 315 S,R,W, 1050 | YP_834472 | Transcriptional regulator 5′ to lactose dehydrogenase | Del 38, del 39, L40V | Metabolism |
| M5 | S, 315 S,R,W, 1050 | yciR and 94 genes downstream | Del, 95 genes, including M1 & M2 | Cyclic di-GMP | |
| M6 | S, 315 S,R,W, 1050 | YP_834525 | manC | STOP @ 263 | EPS/LPS |
| M7 | S,R,W, 1050 | YP_835839 | 5′ of bacterioferritin | −10 G → A promoter | Iron |
| M8 | S, 1050 | Loss of 49 diverse genes | del, 49 genes | ||
| M9 | S, 1050 | YP_837566 | Succinate dehydrogenase | Synonymous | Metabolism |
| M10 | R, 1050 | YP_834838 | DUF88 | A209P | |
| M11 | R, 1050 | YP_834185 | 5′ of mltA | −10 G → A promoter | Peptidoglycan turnover |
| M12 | R, 1050 | YP_837420 | wspD | L35P | Cyclic di-GMP |
| M13 | W, 1050 | YP_837416 | wspA | A407V | Cyclic di-GMP |
| M14 | W | YP_837421 | wspE | S726L | Cyclic di-GMP |
| M15 | R, 315 | YP_837186 | yciR | A106P | Cyclic di-GMP |
| M16 | W, 315 | YP_837416 | wspA | I196N | Cyclic di-GMP |
| M17 | W, 315 | YP_833988 | rpoC | A1318V | Altered RNAP affinity |
| M18 | W | YP_835839 | 5′ of bacterioferritin | −35 G → A promoter | Iron |
| M19 | S,R,W, 1050 | YP_838893 | McsS | Synonymous | Osmotic shock |
| M20 | All | YP_834392 | Ferrochelatase (in ancestor) | T154A | Iron |
Ecotypes (S, R, and W) are defined as in Fig. 1, and if the mutation is associated with a particular time point, it was identified in a fully sequenced isolate. Most mutations (bold font) were subsequently screened in multiple isolates using Sanger sequencing.
Phylogeny and Ecological Dynamics of Biofilm Diversification.
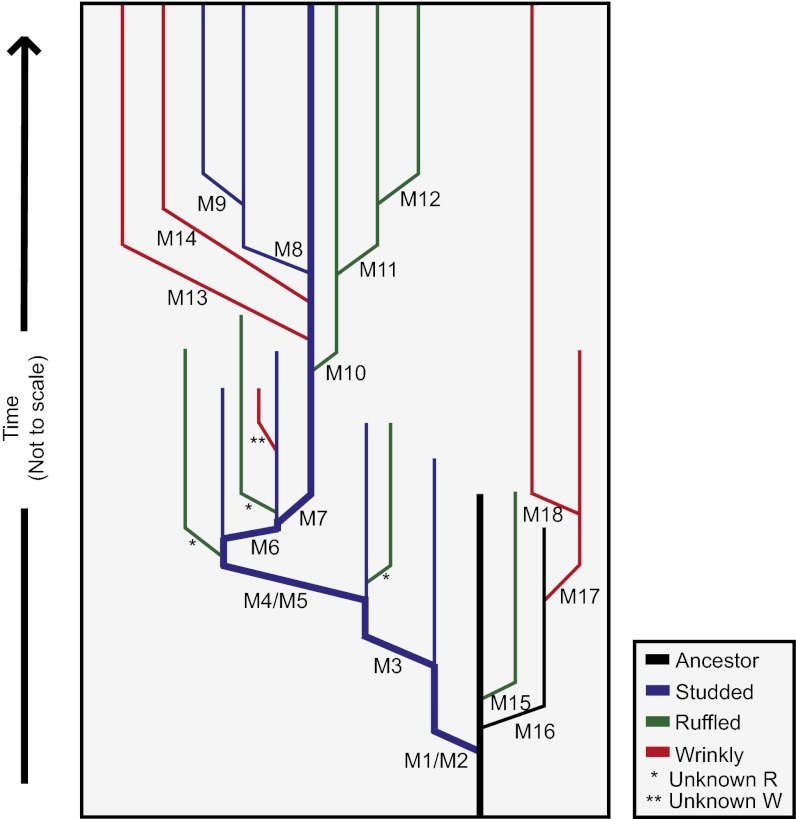
To determine how well the fully sequenced clones represented the ecotype populations, and to evaluate the accuracy of mutation read depth in the metagenomes as estimates of allele frequency, 10 clones of S, R, and W from the early and late communities were screened for key mutations (Table 1). Read depth in the metagenomes correlated well with mutation frequency among screened clones (linear regression: F = 567.5, df =1, P < 0.0001, r2 = 0.99). Between 10 and 20 mutations were reliably detectable at any given time, and perhaps more if lower-quality reads are considered (Table 1, and Table S1). Importantly, sequencing individual clones linked mutations in the metagenome to certain ecotypes and haplotypes (e.g., M14 and M18 defined new W lineages), and these screening data (Tables S2 and S3) enabled a stepwise, most parsimonious phylogeny of this biofilm population (Fig. 2). By combining this phylogeny with estimates of allele frequency, we developed a model of how the genetic and ecological composition of the community evolved throughout the experiment (Fig. 3). The competitive dynamics of genotypes acting both within and between niches became more apparent and suggested a more fluid community structure. In addition, new mutants coexisted with their ancestors for hundreds of generations, in contrast to the sequential replacement that often occurs in unstructured, homogeneous environments. Each ecotype also remained genetically diverse, often by different mutations in genes of similar putative function.
Fig. 2.
Phylogeny of adaptation and diversification in a model biofilm population. Haplotypes were assembled by screening 30 clones from 525 generations and 30 clones from 1,050 generations at variable loci in the metagenome sequences of the community, and their most parsimonious phylogeny was constructed. The ancestor (black line) is rapidly displaced by new mutant ecotypes depicted in different colors; lineages that do not continue were outcompeted by superior lineages within their niche. “Unknown” lineages are ecotypes of uncertain genetic composition, i.e., the mutation defining the R or W morphology is unknown.
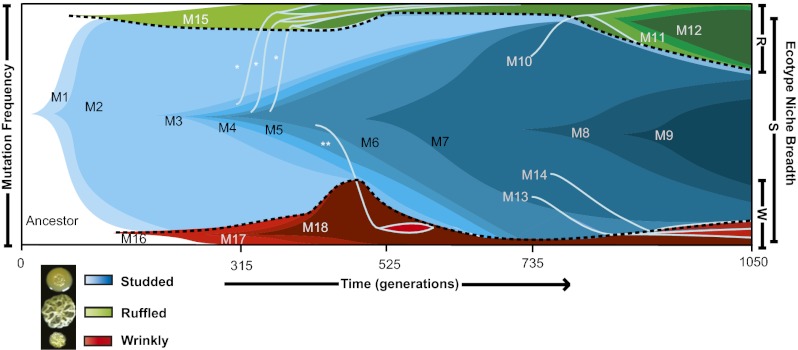
Fig. 3.
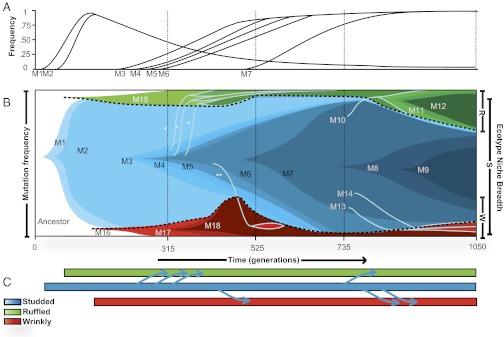
Population genomics and ecological structure of the biofilm community over time. Allele frequencies were determined at four time points (vertical dashed lines), and dynamics were interpolated. (A) Frequency of majority mutations belonging to the dominant haplotype throughout the community. (B) Mutational dynamics within and among niches. Each color transition represents a new haplotype (labeled as in Table 1), and color breadth shows haplotype frequency in the community, to scale. The earliest mutants arose on the ancestral genotype, and subsequent mutations evolved within the ecotypes that subdivided the community. Lines crossing ecotype boundaries (light blue lines) represent invasion of the dominant S haplotype into the R or W niche associated with novel mutations. Horizontal light blue lines highlight the ecological boundaries that evolved within this community. Additional low-frequency mutations were detected in the metagenomes and are reported in Table S1, and other mutants likely evolved before the first samples. *, R isolate with unknown niche-specifying mutation; **, W isolate with unknown niche-specifying mutation. (C) Dynamics of niche invasions by mutants of S over time. Each blue arrow represents the invasion of an S type into an R or W niche.
The S type was the first to achieve high frequency and, as suggested by the genome of an early S clone, evolved by sequential mutations that eventually became globally successful. The dominant S lineage originated with an SNP in yciR (M1), which encodes a protein with three domains: a PAS-sensor, a GGDEF diguanylate cyclase, and an EAL phosphodiesterase (19); this mutation therefore likely affected metabolism of cyclic diguanosine monophosphate (c-di-GMP). Shortly thereafter, a second SNP (M2) occurred 27 genes downstream from this initial mutation on the same haplotype, and this lineage increased to ∼95% of the population (Fig. 3). However, before this lineage could fix, a different yciR SNP occurred and produced the first R ecotype (M15). Similarly, another SNP in a homolog of wspA (M16) occurred in association with the first detected W ecotype. The wsp operon has been characterized extensively in Pseudomonas species (20, 21) for its production of a wrinkly spreader phenotype related to c-di-GMP metabolism. These mutations imply that varied c-di-GMP metabolism was the cause of ecological differentiation and set the stage for prolonged coexistence of S, R, and W (Fig. 3B).
Mutations on the S lineage continued to accumulate but also began to produce new R and W lineages (Fig. 3B). The next S mutation was an SNP in the tricarboxylic acid (TCA) cycle intermediate enzyme, 2-oxoglutarate dehydrogenase (OGDH, M3), which displaced its direct ancestor and also produced a new R variant (Fig. 3B). Then a peculiar set of mutations (M4 and M5) followed in an uncertain order: M4 consisted of a two-codon deletion in a transcriptional regulator, and M5 was a large deletion that removed the mutated yciR locus and the new M1 and M2 alleles. This new M3/M4/M5 haplotype also spawned a successful R type. Because this newer haplotype displaced earlier S haplotypes, the frequencies of M1 and M2 declined but remained detectable (Fig. 3A). A single base-pair deletion (M6) in manC, a gene associated with exopolysaccharide (EPS) and LPS production, occurred next in this dominant lineage and then gave rise to yet another new R and a W variant. These niche invasions involved three new R lineages and one new W lineage (Fig. 3C; the mutations responsible for these invasions are unknown) that drove the original R resident (M15) extinct but did not noticeably affect the original W lineage. The early W lineage also had acquired two new mutations (M17 and M18). The R and W biofilm specialist ecotypes therefore evolved recurrently from a single, dominant S lineage.
The final defining mutation of the dominant lineage (M7) was the most successful mutation in the experiment and altered the promoter of the iron-storage gene bacterioferritin. Remarkably, M18 that evolved on a W haplotype altered the same promoter (Table 1); the dynamics during this period suggest that M7 may have evolved in response to M18. M18 was detected between 315 and 525 generations and was associated with a large gain in W frequency (increase in red fraction in Figs. 3 and 4) to ∼30% of the entire population. Subsequently, the rise of M7 in the main S lineage associated with the decline of W ecotypes to their original ∼10% (decline in red fraction at 600 generations, Figs. 3 and 4). Coincidentally, all R types lacking a bacterioferritin mutation went extinct, and only new R mutants evolving from the M7 haplotype (e.g., M10) preserved this ecotype. However, the frequency of the W lineage with mutation M18 apparently did not change when two new W lineages invaded from the M7 haplotype later in the experiment (Fig. 3), presumably because each of these lineages had a similar bacterioferritin mutation. Once again, the new W types evolved by single mutations in wsp homologs, wspA (M13) and wspE (M14), similar to the original W lineage. These dynamics highlight the global importance of acquiring a mutation in the bacterioferritin promoter sequence and the capacity for small-colony variants to re-evolve.
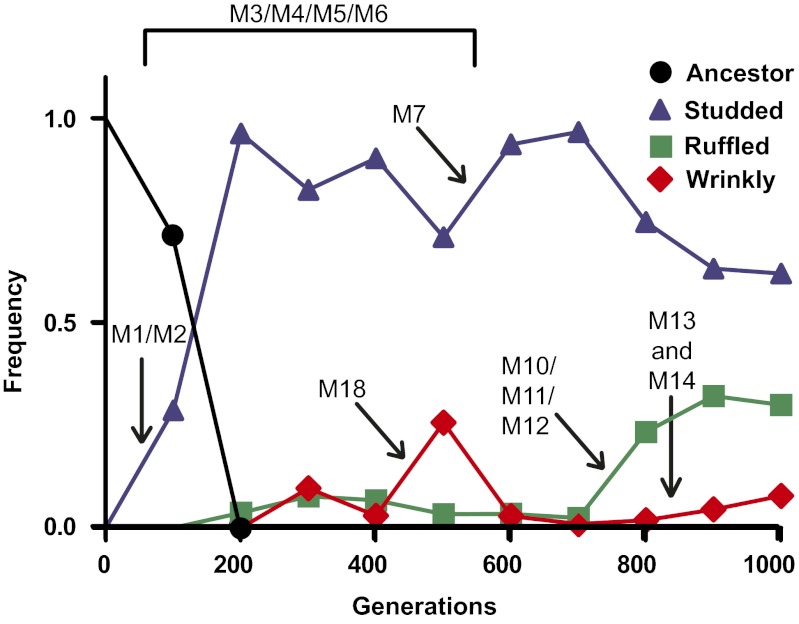
Fig. 4.
Ecotype frequencies in biofilm population B1 over time as evaluated from plate counts. As certain mutations became detectable (arrows and mutation labels), changes in morphotype frequencies were observed.
By retrospective sampling of the population throughout its evolution, we found that the relative frequency of each morphotype, and hence the balance of their respective ecological strategies, indeed fluctuated over time (Fig. 4). Shifting morphotype frequencies were tightly paired with changing allele frequencies, most often when new mutants of the S lineage invaded and displaced existing S, R, or W inhabitants (Fig. 3). This pattern of ecological and genetic invasion from the numerically dominant S niche into the more specialized R and W niches suggests that high-biofilm specialists evolve from generalists, but not vice versa. Therefore the highly productive, synergistic late community (Fig. 1D) (4) mostly comprises ecotypes that are immediate genetic relatives with only one or two mutations defining them.
Clonal Interference Within and Among Niches.
Each ecotype remained genetically diverse throughout the experiment (Fig. 3), indicating that beneficial mutations were relatively common and likely competed with one another (22). This dynamic of clonal interference can effectively reduce the relative benefit of any single mutation or haplotype and may allow multiple lineages to persist. Theory suggests that such competition also can produce a simultaneous sweep of multiple linked mutations (e.g., mutations M3–M6, Fig. 3A), because more than one beneficial mutation may be required to prevail in competition (22, 23).
We began to quantify the selective forces acting on each mutational step leading to the dominant haplotype by competition between these mutants and two reference clones: with the founding ancestor and with a highly adapted intermediate S clone (Table 2). Competition versus the original ancestor provides a common reference to judge fitness in the absence of other beneficial variation, whereas competition versus a well-adapted “contemporary” mutant illustrates selective forces among contending adaptive lineages. Each mutant displayed fitness advantages that would displace the ancestor before 70 generations in the absence of clonal interference; in actuality, none of these alleles fixed, and their frequencies increased approximately 10-fold more slowly in the evolving community (Table 2). However, the slight variation in fitness among mutants versus the ancestor did not explain how selection favored the mutants’ rapid succession. In contrast, far greater fitness differences among haplotypes were evident from competitions with the most derived (M3–M7) haplotype, which was considerably fitter versus an early-evolved mutant than versus the original ancestor. The fitness ranks of evolved haplotypes were consistent with the mutational order and demonstrated a large advantage of the yciR deletion (M5). Evidently, selection in this biofilm system is strongly context dependent, favoring mutants that are superior in both the current biotic environment comprising multiple lineages and in the fluctuating abiotic conditions.
Table 2.
Fitness estimates of evolutionary steps within the dominant S haplotype
| Evolved S haplotype | Fitness vs. ancestor | Fitness vs. M3-M7 | Fitness from whole metagenome | Fitness from niche metagenome |
| M1–M2 | 0.45 (0.065) | −0.96 (0.19) | 0.053 | 0.066 |
| M1–M3 | 0.40 (0.061) | −0.84 (0.22) | 0.0091 | 0.029 |
| M3–M5 | 0.51 (0.034) | 0.15 (0.14) | 0.0082 | 0.030 |
| M3–M6 | 0.57 (0.067) | −0.32 (0.17) | 0.015 | 0.035 |
| M3–M7 | 0.52 (0.068) | – | 0.014 | 0.015 |
Haplotypes are defined by mutations listed in Table 1. Empirical fitness (columns 1 and 2) was calculated by direct competitions between clones (column 1, 5–9 replicates, column 2, 11 or 12 replicates) and is reported as selective rate constants with 95% confidence intervals in parentheses. Haplotypes vary modestly in competition versus the ancestor (F = 4.38, P = 0.0058) but vary widely in their fitness versus the most derived genotype (F = 46.0 P < 0.0001), demonstrating very strong selection among co-occurring mutants. Much lower realized fitness differences were inferred from metagenomes (columns 3 and 4) as changes in mutation frequency over time, either throughout the community (column 3) or within niches associated with ecotypes (column 4).
Nonetheless, the extraordinarily high fitness advantages of evolved mutants in pairwise competition far exceed their realized benefits in the community, as judged either from the population dynamics of the entire metagenome or among alleles within a niche (Table 2). The fact that many contending adaptive lineages are found at appreciable frequencies throughout the experiment indeed may have diminished the net advantage of any given haplotype. In addition, more complex frequency- or density-dependent interactions among favored genotypes, such as the cross-feeding interactions described previously (4), may have slowed the rise of adaptive alleles and preserved genetic diversity within the biofilm community.
Genetics and Physiology of Biofilm Adaptation.
The functional significance of the mutations associated with adaptation to prolonged biofilm selection highlights the challenges faced by Burkholderia in this system and the changes that were favored. Four major pathways of adaptation emerged, described below, but some mutations do not fall into these categories, suggesting that selective forces in this biofilm system are manifold and may preserve many alternative alleles.
Altered regulation of c-di-GMP.
Three yciR mutations (M1, M5, and M15) were critical to the early evolution of the biofilm community (Fig. 3). M1 (Y355D) occurred in a highly conserved residue of the GGDEF domain and was associated with S morphology. M15 (A106P) was found in the PAS sensor domain of yciR and in isolation defined R morphology; thus, alternative yciR alleles defined morphological and ecological differences in this system. The third yciR mutation (M5) deleted this locus and 94 other genes and was likely mediated by an IS116-family insertion sequence at the 3′ end of this lesion. This mutation was also associated with S morphology, but its ecological dominance over the two yciR SNPs remains to be explored. Curiously, disrupting this gene in another Burkholderia species had been shown previously to decrease both biofilm production and quorum sensing (24).
In general, wrinkly colony morphologies nearly always are associated with increased c-di-GMP concentrations and in Pseudomonas may be caused by mutations that lead to autoinduction of an associated diguanylate cyclase enzyme, WspR (21). Four nonsynonymous mutations were detected in this operon, each in different clones, including two in wspA (M13 and M16) and one each in wspD (M12) and wspE (M14). All wsp mutations were associated with either an R or W morphotype: wspA and wspE alleles were found in W clones, and wspD was found in the successful R lineage (Fig. 3B). The wspE mutation occurred in an active site for phosphorylation and hence is likely to alter signaling of this pathway. Presumably, each of these four mutations adaptively alters the levels of c-di-GMP in this system, a subject of ongoing study, and also defines particular ecological roles and leads to niche subdivision.
Shifts in central metabolism.
Three beneficial mutations, M2, M3, and M4 (Tables 1 and 2), likely altered central metabolism. M2 is a mutation in a gene annotated as FAD-binding monooxygenase. This type of enzyme may be involved in electron transport, and altering the levels of FAD(H) within the cell may provide a growth advantage in the selective environment. M3 is a nonsynonymous mutation in OGDH (M3), and M4 is a two-codon deletion from a LysR-like transcriptional regulator that is 5′ to a gene encoding lactate dehydrogenase (M4). These mutations could enhance growth in galactose minimal medium, but they also may reflect selection for biofilm production.
Altered mannose metabolism related to EPS and LPS production and adherence.
The globally successful mutation M6 created a premature stop codon in a multidomain gene (manC) that is involved in mannose metabolism and is located within a gene cluster related to LPS biosynthesis. The gene is bifunctional, with an N-terminal mannose-1-phosphate guanylyl transferase domain and a C-terminal mannose-6-phosphate isomerase (PMI) (25). A single-nucleotide deletion occurred in a poly-A tract (a relatively rare sequence in this genome, given its 67% GC content) that preserved the N-terminal domain but truncated the C-terminal PMI domain and likely generated polar effects on the rest of the operon. The effects of this disruption are potentially widespread, because downstream genes are expected to participate in LPS biosynthesis. Late S, R, and W clones produced less biofilm when a functional manC was added on a plasmid (Fig. S1). We hypothesize that this mutation effectively directs mannose toward exopolysaccharide and away from energy production and may increase adherence and coherence as a response to disrupting LPS biosynthesis, as has been reported elsewhere (26, 27).
Enhanced iron competition.
The two different mutations in the bacterioferritin promoter sequence (M18 and M7) occurred in the −35 and −10 σ factor-binding domains, respectively. Quantitative RT-PCR of genotypes with the M7 mutation exhibited increased transcription at this locus (Fig. S2), indicating that these mutants gained capacity for iron storage. Because the experimental environment was not supplemented with iron, this resource likely became limiting. Another iron-associated mutation evolved in the late community in a gene encoding ferredoxin (Table S1), providing further evidence of selection for altered iron metabolism. Thus, competition for iron occurred both within and between niches in the biofilm community, which favored mutants with either M7 or M18, displaced mutants lacking these alleles, and facilitated invasion from the S niche to the R and W niches.
Evolutionary Parallelism Among Six Independently Evolved Biofilm Populations.
Although we have not assembled a detailed evolutionary model of all six biofilm populations (as in Fig. 3), mutations associated with adaptation were identified in the early and late samples of all populations. Exceptional parallelism among adaptive targets occurred: all six populations experienced mutations in the four major functional categories outlined previously (Table 3 and Table S4). At least 26 mutations involved in c-di-GMP metabolism occurred, including seven independent mutations in the yciR gene, 18 independent mutations in the wsp operon, and one in an unclassified diguanylate cyclase. Another commonly mutated area across the populations was a gene cluster involved in LPS biosynthesis, including mutation M6. Twenty independent mutations occurred within this gene cluster among all six populations, thus suggesting that altered LPS may be adaptive in biofilms. Mutations in OGDH also were successful in each population, highlighting this step in the TCA cycle as limiting for the ancestor in this environment. Other classes of mutations occurred in replicate populations as well, including in RNA polymerase subunits rpoC and rpoD (n = 5) and in a galactose metabolism operon (n = 11).
Table 3.
Mutations shared among the six evolved biofilm populations, categorized by putative function
| Function | Locus | Annotation | No. of mutations |
| Transcription | YP_833988 | rpoC | 3 |
| YP_837516 | rpoD | 2 | |
| Galactose metabolism | NC | 5′ of galactose metabolism operon | 7 |
| YP_834263 | iciR family (regulator of galactose metabolism) | 3 | |
| YP_834264 | 2-keto-3-deoxy-galactonokinase (galactose metabolism) | 1 | |
| LPS gene cluster | YP_834517 | dTDP-glucose 4,6-dehydratase (3′ of manC) | 1 |
| YP_834518 | Glucose-1-phosphate thymidylyltransferase | 3 | |
| YP_834524 | Glycosyltransferase | 1 | |
| YP_834525 | manC | 4 | |
| YP_834526 | GDP-mannose 4,6-dehydratase | 10* | |
| YP_834528 | fkbM, methyltransferase (5′ of manC) | 2 | |
| YP_834530 | Glycosyl transferase, family 2 | 1 | |
| YP_834532 | Glycosyltransferase | 1 | |
| YP_834533 | capB, polysaccharide synthesis | 1 | |
| OGDH | YP_835153 | OGDH E1 | 7 |
| YP_835154 | OGDH E2 | 2 | |
| c-di-GMP | 95-gene deletion | yciR deletion | 1 |
| YP_837186 | yciR | 7 | |
| YP_839310 | GGDEF, PAS/PAC Sensor | 1 | |
| YP_837415 | wspR | 1 | |
| YP_837416 | wspA | 6 | |
| YP_837420 | wspD | 2 | |
| YP_837421 | wspE | 9 |
A population evolved under planktonic conditions in the same medium, selected because it was the only one of six to evolve an additional colony type, was sequenced also. This population also revealed some parallelism with the biofilm lines, including three mutations in the LPS gene cluster, two mutations in rpoC, and one mutation in yciR (Table S5). These results imply that adaptation to the high-galactose environment lacking the bead (but preserving all other conditions, including constant 37 °C and the potential for biofilm growth on the test tube walls that may have been shed and preserved during transfer) may explain some of these alleles. Alternatively, different alleles in the same gene may be favored by planktonic or biofilm growth. However, none of the wsp mutations associated with R and W types were detected, and three unique mutations in the quorum-sensing regulator cepR (28) evolved in this planktonic population. In general, the high genetic parallelism in adaptive targets suggested that adaptation in this model proceeded along relatively few pathways, despite progressive ecological complexity and the ease of selecting adaptive mutants (which evolve in a matter of days) from the ancestral clone in this system.
Convergent Evolution with CF Lung Isolates.
The same four classes of mutations found in our replicate populations were found in studies of isolates of Burkholderia dolosa and Pseudomonas aeruginosa that evolved during CF infections (Table 4) (10–12). We make this comparison cautiously, because more mutations were reported in these clinical studies than in this system and because database matches may be uncertain. However, the degree of overlap for these functions is remarkable and is statistically improbable to have occurred by chance alone, given the number of observed mutations in each study and the number of genes associated with each function (Table S6 and SI Methods). Moreover, none of the mutations that uniquely evolved in the planktonic population were shared by these studies. Smith et al. (12) reported six independent wspF mutations in their Pseudomonas aeruginosa study, which highlights altered c-di-GMP regulation in certain infectious lineages. Another study of P. aeruginosa reported two more mutations in this operon, wspE and wspC (Table 4) (11), although these mutations could be explained by chance. Eighteen distinct mutations in the wsp operon were selected among our replicate populations. In addition, five OGDH mutations were detected in 112 clinical B. dolosa isolates, suggesting the importance of altered TCA cycle enzyme activity. Likewise, OGDH mutations evolved in all six biofilm populations and were associated with increased fitness in population B1.
Table 4.
Convergence among mutations found in isolates from chronic pulmonary infections of CF patients, as reported for B. dolosa (10) and P. aeruginosa (11, 12) and mutations that evolved in this experimental system (Table 3)
| B. dolosa | Annotation | Mutations | P. aeruginosa | Annotation | Mutations | |
| Transcription | BDAG_04154 | rpoD | 4 | PA14_07520 | rpoD | 1 |
| BDAG_02789 | rpoB | 1 | PA14_08760 | rpoB | 2 | |
| BDAG_02788 | rpoC | 2 | PA14_08780 | rpoC | 1 | |
| PA4462 | rpoN | 5 | ||||
| PA4270 | rpoB | 1 | ||||
| LPS gene cluster | BDAG_02317 | Glycosyltransferase | 10 | PA14_71910 | wbpZ | 2 (ns) |
| BDAG_02321 | Glycosyltransferase (wbpX homolog) | 6 | PA14_71970 | wbpW (manC) | 1 (ns) | |
| BDAG_02323 | manC | 2 | PA3159 | wbpA | 1 (ns) | |
| BDAG_02820 | manC homolog | 1 | ||||
| OGDH | BDAG_00384 | OGDH E3 | 1 | |||
| BDAG_01761 | OGDH E1 | 2 | ||||
| BDAG_01898 | OGDH E3 | 2 | ||||
| Cyclic di-GMP | BDAG_04135 | diguanylate cyclase | 1 (ns) | PA14_16450 | wspC | 1 (ns) |
| PA14_16470 | wspE | 1 (ns) | ||||
| PA14_31330 | EAL | 1 (ns) | ||||
| PA14_56790 | GGDEF/EAL | 1 (ns) | ||||
| PA3703 | wspF | 6 |
Mutations in the same row are not necessarily orthologs. Convergent evolution between systems occurred more often than expected by chance for all functional categories except those labeled ns (not significant) (Table S6 and SI Methods).
One of the more remarkable examples of convergent evolution involved a gene cluster responsible for synthesizing O-antigen LPS, in which manC (mutation M6) is found. Lieberman et al. (10) identified 19 mutations in this locus across 112 clinical isolates and one additional mutation in another manC homolog. Among the six experimental biofilm populations studied here, 20 independent mutations in the homologous and well-conserved gene cluster were found. Mutations in manC evolved in both B. dolosa (n = 2) and B. cenocepacia (n = 4). The remainder of the mutations occurred in similar but not identical sugar transferase genes in both organisms. Similarly, LPS-associated mutations have been detected in clinical P. aeruginosa isolates in the wbpA, wbpW (a manC homolog), and wbpZ genes (11, 29). Given this exceptional mutational parallelism found among relatively few studies, it is likely that some of the same forces that drive biofilm adaptation also contribute to adaptation of these pathogens within the CF lung.
Discussion
Adaptive diversification and niche partitioning have been observed previously in bacterial populations that were evolved experimentally in structured environments (30–33), so the evolved diversity in this biofilm model is unsurprising. However, we lack understanding of the population genetic dynamics of biofilm communities over the longer term and particularly how newly established lineages might coevolve and shape their environments. Further, to our knowledge, experimental selection for the ability of bacteria to both attach and disperse has not been performed. Our previous observation that three distinct ecotypes evolved and persisted in each of six replicate evolved biofilm populations (4) led us to anticipate that each type represented a distinct lineage that had evolved continuously by endemic alleles. We also found that ecotype frequencies varied over time, and biofilm communities became more productive (4), suggesting that the ecotypes had coevolved. However, a detailed population genomic analysis of one biofilm community, made possible by repeated sequencing of clones and the evolving community as a whole, revealed more complex dynamics. Particularly notable was an example of competition over limiting iron between ecotypes that eventually remodeled the community, but contrary to expectations new specialist ecotypes actually had evolved recurrently from the source population of S (Fig. 3). These high-biofilm, small-colony variants evolved by a variety of single mutations in relative few pathways (often in wsp). More broadly, ecotype niche breadth correlated with evolutionary potential, because generalist S genotypes gave rise to new specialist R and W types, but the converse did not occur.
The order of adaptive mutations in each lineage also revealed the prevailing selective forces over time. At the start of the experiment, cells that adhered better to the plastic bead were favored; consequently, the first three successful ecotypes acquired mutations in genes (M1, M15, and M16) that are likely responsible for controlling levels of c-di-GMP, which is known to underlie transition to a sessile lifestyle (20). These alleles also altered colony morphology and excluded the ancestor type, suggesting that variation in c-di-GMP production is sufficient for adaptive differentiation and the subdivision of biofilm labor, perhaps by varying the tendency to adhere or disperse.
Next, selection for enhanced metabolic efficiency likely dominated, because mutations occurred that affected central metabolism and polysaccharide biosynthesis. Each of these mutations appeared to expand the potential of the S ecotype (Fig. 4) and enhanced competition versus earlier mutants (Table 2). These genotypes also subsequently gave rise to new R mutants with these mutations, indicating that they were globally beneficial and not specific to a particular niche. Although these mutations may simply enhance growth on galactose as a sole exogenous carbon source, adaptation to planktonic growth in the same medium occurred by somewhat different mechanisms (Table S5). Thus, these mutations may be specifically adaptive in biofilms by directing resources toward sessile growth.
Iron Competition Engendered Persistence and Evolvability.
As described previously, two independent mutations in the promoter sequence of bacterioferritin led to an ecological revolution that remodeled the community (Figs. 3 and 4). New mutants of S with up-regulated bacterioferritin (Fig. S2) likely succeeded in other niches (Fig. 3C) because of their enhanced competitive ability for limiting iron, and the mutations enabling invasion were novel wsp alleles (Fig. 3). It is surprising that W mutants with the alternative bacterioferritin mutation failed to spread beyond this niche, either by mutations suppressing the wrinkly phenotype or by alternative gain of function. Only the S lineage, which balances fitness between planktonic and biofilm conditions and retains motility (4), was able to spawn new R and W types at least seven times. This S lineage may be more “evolvable” because of its larger population size or because of an absence of negative epistasis between its existing mutations and niche-specializing wsp alleles. Alternatively, the ecological breadth of the S population may have enabled greater access to new mutations that were beneficial in a subset of conditions. These alternative explanations are central to the debate surrounding evolvability (34–36) and may be studied tractably in this model biofilm community.
Evolution During Chronic Infections Is, in Part, a Response to the Biofilm Lifestyle.
Most mutations from published studies of CF lung isolates do not strictly overlap with the mutations found in this study, as is to be expected. Bacteria face many different selective pressures in the CF lung that are absent from our model, such as high levels of antibiotic use, oxygen deprivation when suspended in mucus, and immune system evasion (5, 10, 11, 16, 37, 38). Also, in principle, the fact that Burkholderia and Pseudomonas are in different classes of Proteobacteria should reduce the frequency of convergence. However, most CF isolates produce robust biofilms, and many form small-colony variants (5, 7, 9, 38), and, notably, the colony morphologies described in the literature (38–41) as related to infection outcomes are strikingly similar to those that evolved in this system. Thus, it is remarkable that the causes of phenotypic variation during this experimental biofilm selection and during chronic infections both involved mutations that affected the TCA cycle, iron metabolism, and RNA polymerase. For example, mutations in OGDH occurred frequently in a study of B. dolosa (10) and in this system (Table 4), and this function was up-regulated in two independent studies of clinical P. aeruginosa (13, 42). This form of altered central metabolism may be favored by evolution in biofilms. Iron acquisition also is known to be a major selective pressure in the CF lung (10–12), and it appears to be central to the dynamics of this system, suggesting that perhaps the biofilm environment rather than an iron-limited host may favor these changes. Convergent mutations in subunits of RNA polymerase (RNAP) in clinical B. dolosa and P. aeruginosa (10–12) and in our study are more difficult to explain, given their global effects on transcription. Mutations in major σ factors and polymerase subunits have been widely reported during long-term selection in chemostats (43) and occurred in our planktonic control population as well; these changes may optimize global gene regulation for better growth in minimal medium (44, 45) that requires more anabolism. RNAP mutations in isolates from infections have been associated with selection for antibiotic resistance but alternatively may be responses to selection for to growth in novel nutrient conditions.
Relatively less convergent evolution was found at loci other than wsp that affect c-di-GMP metabolism. Despite several related mutations in infectious P. aeruginosa (11, 12), only one such mutation was reported for B. dolosa (10). These mutations were among the first to evolve in our model, so one possibility is that these mutations became common before clinical sampling. Starkey et al. (40) reported that laboratory-derived rugose small-colony variants with increased intracellular levels of c-di-GMP associated with the ability to persist in CF mouse models. As in our model, the earliest adaptive steps to form a biofilm may require altered c-di-GMP levels, but the way each population accomplishes this task may be variable.
Perhaps the most compelling example of convergent evolution in this system and the mutations identified in CF lung isolates are the many mutations in genes involving LPS (Table 4). Altered LPS production could influence biofilm formation directly because of potentially increased surface adherence and cell-to-cell coherence (26, 27). In this system, complementation of one of the key mutants in this pathway adversely affected biofilm production (Fig. S1). Rather than selection for immune avoidance, altered LPS during chronic infections may underlie biofilm adaptation. A subset of the 19 mutations found in the B. dolosa outbreak restored O-antigen synthesis and may have contributed to virulence in the lung (10), but others did not, suggesting that mutations in the same pathway may reflect different selective pressures. One possibility is that these mutants may be adaptive in different niches within the biofilm, as in our B. cenocepacia system.
Tangled Bank of Biofilms: Perhaps Predictable Dynamics?
What do we make of this coincidence between the adaptive pathways in our system and the mutated loci occurring in chronic infections, and why might prior experimental models (32, 46) have captured fewer of these mutations? We suggest that both the duration of the experiment (6 mo or 1,050 generations) and its requirement to maintain flexible life-history strategies may provide some explanation. Our model required a daily cycle of surface colonization, biofilm growth, dispersal, and then recolonization, i.e., selection for “reversible stickiness.” Under these conditions, pure biofilm-producing specialists might be disadvantaged in the long run because of their inability to disperse and recolonize the next plastic bead. In fact, many early-arising biofilm specialist lineages ultimately were displaced by derivatives of the more flexible S lineage (Fig. 2). Such mutants, successful only in the short term, may resemble those types found in prior studies (31, 32), but the longer-term cyclical requirement for persistence, dispersal, and recolonization may better resemble the dynamic of bacterial populations during chronic infections, because lifestyle flexibility will be needed in a lung environment with diverse conditions (e.g., host immune response, oxygen deprivation, and continuous biofilm establishment and migration).
Phenotypic diversity is known to be common among CF lung isolates of B. cepacia, Burkholderia multivorans, and B. cenocepacia, particularly for biofilm formation and EPS production (5, 13, 47). Seemingly identical genetic relatives isolated from the same lung at the same time may display distinct differences in the ability to form biofilms and may occur within multiple patients (5). Variation in EPS production was associated with different abilities to initiate biofilm formation and form mature biofilms (47). One possible interpretation of these findings is that the Burkholderia isolates within those patients inhabited distinct niches and functioned as a community, even among many other bacterial species in the CF lung (48).
We also explored the high degree of competition both within niches and among ecotypes in these biofilms and suggest that such forces are more intense than in well-mixed environments (49). This interference has the effect of maintaining diversity over longer time scales and could enhance coevolutionary forces. Thus, evolutionary dynamics in biofilms can be considered exceptional, because greater variation is maintained and multiple adaptive mutations are required for any mutant to exclude others (Table 2) (23). However, these dynamics also may be expected and may even become predictable. We offer the hypothesis that the tangled bank of biofilm adaptation (18), in which adaptive radiations fuel a pattern of ecological succession, ultimately drives evolution along proscribed paths. If valid, such a model offers hope for the development of novel therapeutics that target these pathways. More worrisome, however, is the evolution of progressively greater synergy within our model, in which late-stage–related ecotypes are particularly robust (4). Identifying which community members are most central to biofilm resilience therefore becomes paramount, with the caution that the key community member may re-evolve once targeted therapy has ended.
Methods
Strains and Growth Conditions.
The experimental evolution is described in ref. 4. Briefly, a Burkholderia cenocepacia HI2424 isolate was marked with a Tn7::lacZ construct to enable blue/white screening (50). A single lac+ clone was inoculated into M9 salts supplemented with 3% (171 mM) galactose (GMM) containing a polystyrene bead (American Educational Products). Every 24 h, the bead and any adherent bacteria were transferred to a fresh tube of GMM containing an oppositely marked (white or black) polystyrene bead. Cultures were enumerated and characterized by plating on half-concentration agar plates that were incubated at 37 °C for 24 h and at room temperature for 48 h. Cultures for fitness assays were grown under identical conditions except that strains were grown for 24 h in tryptic soy broth (Fisher) from frozen stocks and then were transferred to the selection environment. Biofilm production was quantified using standard methods (3) with sixfold replication of the sequenced S, R, and W clones isolated from the end of the experiment. Mixed-population biofilm production was quantified from an aliquot of the final evolved community. Expected biofilm production (Fig. 1) was calculated by multiplying biofilm production by the fraction of the community associated with each ecotype and then summing these values; contributions of each morph to the observed biofilm produced are estimates based on the frequency of plated morphotypes.
Metagenomic Sequencing and Analysis.
Population samples from 43, 71, and 143 d, reflecting ∼315, 735, and 1,050 generations, were grown in the selective environment for 24 h. [These cell-division estimates differ from those reported in ref. 4 and reflect more precise measurements of the growth cycle (SI Methods)]. The bead was vortexed in PBS, and genomic DNA was extracted using standard methods. Libraries for Illumina sequencing were prepared following the manufacturer’s procedures and were sequenced by the Joint Genome Institute (http://jgi.doe.gov) with an Illumina GAII using paired-end 36-bp reads. The average read depth for each metagenome was 189×, 110×, and 141× for the early (315 generations), intermediate (735 generations), and late (1050 generations) time points, respectively. In addition, representative single clones of S, R, and W types were isolated at 315 and 1,050 generations and sequenced at >100× coverage (SI Methods). Subsequently, early and late metagenomes were sequenced from five other replicate biofilm populations (B2–B6) and from a control, planktonically evolved population P2 (Table S7, SI Methods, and ref. 4).
The complete approach for identifying mutations and estimating allele frequencies is described in SI Methods. Mapping of short reads to the B. cenocepacia HI2424 reference sequence (51) was performed using maq-0.7.1 (52) and later was confirmed using BWA (53). Sequences of single clones were analyzed as having two haplotypes to flag regions of poor alignment, and sequences of mixed population metagenomes were analyzed at haplotypes = 20, using clones from these populations as controls for false positives. The high GC content of this genome produced many initial false positives and anomalously low coverage in some genome regions. Thus, all putative mutations were examined manually Table S8 (SI Methods) and most were confirmed by Sanger sequencing.
Phylogeny and Haplotype Identification.
To associate mutations in the metagenome with both clones and ecotypes, single colonies were obtained by growing population samples from 525 and 1,050 generations in the selective environment by plating as described. Ten clones of the three morphotypes at each time point were chosen randomly and frozen for reference. DNA from each clone was isolated, and the loci containing each of the putative mutations were amplified by PCR and sequenced using standard methods (SI Methods).
The estimated frequencies of mutations identified in sequenced clones, in metagenomes, and by manual screening of variant loci from many clones were integrated to produce a phylogeny of how population B1 evolved and diversified. For instance, mutations detected in the metagenome were not yet associated with a clone or ecotype but enabled estimates of their changing frequencies over time, which allowed us to determine their likely order of occurrence. Sequencing multiple isolates at several loci confirmed this order and also allowed us to assemble haplotypes and to associate mutations with ecotypes, i.e., the mutations associated with the colony morphotypes at each stage. This approach also captured multiple putatively adaptive alleles that were undetected in the metagenome. Further details are given in SI Methods.
Fitness Assays.
After overnight growth from clonal freezer stocks and 24 h of solitary growth in the selection environment, competitors were removed from the plastic bead, vortexed in 1 mL of GMM, and standardized by optical density. Competitions of evolved isolates versus the ancestor were started by inoculating 4 mL of GMM with 100 µL of evolved culture and 900 µL of the ancestor; this ratio enabled more accurate assays because of the relatively low fitness of the ancestor. Competitions of earlier S clones versus the M3–M7 haplotype were conducted by using lacZ+ and lacZ− competitors (Table S9). Cultures for competitions were started as described for the ancestral competitions, except that 4 mL of GMM was inoculated with 500 µL of earlier S isolates and 500 µL of M3–M7. Initial ratios were enumerated, and populations were incubated at 37 °C in a New Brunswick TC-7 model roller drum at 50 rpm. After 24 h, the bead was removed, vortexed in 1 mL of PBS, diluted, and plated. Competitions between S clones were enumerated on tryptic soy plates supplemented with X-Gal. Selective values were calculated as:
 |
where t = time (generations), E = frequency of evolved, A = frequency of the ancestor, and s = selective value. Selection rates in the metagenome were calculated using the same formula, substituting mutation frequencies at known times in the numerator. Selection rates of ecotype-specific mutations were calculated by adjusting their frequency to that of the particular ecotype (e.g., a W-specific mutation should never exceed 10% of the total community).
Additional methods are described in SI Methods.
Supplementary Material
Acknowledgments
We thank Laura Benton, Gabrielle Bergeron, Wendy Carlson, Zhang Qian, Marcus Dillon, Crystal Ellis, Kenneth Flynn, Megan McLaughlin, and Christopher Waters for helpful discussions and technical support and thank three anonymous reviewers for their comments. This research was supported by National Institutes of Health Grant 1R15AI082528 and National Science Foundation Career Award DEB-0845851. Collaborative work conducted by the U.S. Department of Energy Joint Genome Institute was supported by the Office of Science of the US Department of Energy under Contract DE-AC02-05CH11231.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the National Center for Biotechnology Information Short-Read Archive, http://www.ncbi.nlm.nih.gov/sra (accession no. SRP004277), in accordance with Joint Genome Institute policy.
See Author Summary on page 822 (volume 110, number 3).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1207025110/-/DCSupplemental.
References
- 1.Stewart PS, Franklin MJ. Physiological heterogeneity in biofilms. Nat Rev Microbiol. 2008;6(3):199–210. doi: 10.1038/nrmicro1838. [DOI] [PubMed] [Google Scholar]
- 2.Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: From the natural environment to infectious diseases. Nat Rev Microbiol. 2004;2(2):95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 3.O’Toole GA, et al. Genetic approaches to study of biofilms. Methods Enzymol. 1999;310:91–109. doi: 10.1016/s0076-6879(99)10008-9. [DOI] [PubMed] [Google Scholar]
- 4.Poltak SR, Cooper VS. Ecological succession in long-term experimentally evolved biofilms produces synergistic communities. ISME J. 2011;5(3):369–378. doi: 10.1038/ismej.2010.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cunha MV, et al. Studies on the involvement of the exopolysaccharide produced by cystic fibrosis-associated isolates of the Burkholderia cepacia complex in biofilm formation and in persistence of respiratory infections. J Clin Microbiol. 2004;42(7):3052–3058. doi: 10.1128/JCM.42.7.3052-3058.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nguyen D, Singh PK. Evolving stealth: Genetic adaptation of Pseudomonas aeruginosa during cystic fibrosis infections. Proc Natl Acad Sci USA. 2006;103(22):8305–8306. doi: 10.1073/pnas.0602526103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Häussler S, et al. Fatal outcome of lung transplantation in cystic fibrosis patients due to small-colony variants of the Burkholderia cepacia complex. Eur J Clin Microbiol Infect Dis. 2003;22(4):249–253. doi: 10.1007/s10096-003-0901-y. [DOI] [PubMed] [Google Scholar]
- 8.Häussler S, et al. Highly adherent small-colony variants of Pseudomonas aeruginosa in cystic fibrosis lung infection. J Med Microbiol. 2003;52(Pt 4):295–301. doi: 10.1099/jmm.0.05069-0. [DOI] [PubMed] [Google Scholar]
- 9.Schneider M, et al. Clinical characteristics associated with isolation of small-colony variants of Staphylococcus aureus and Pseudomonas aeruginosa from respiratory secretions of patients with cystic fibrosis. J Clin Microbiol. 2008;46(5):1832–1834. doi: 10.1128/JCM.00361-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lieberman TD, et al. Parallel bacterial evolution within multiple patients identifies candidate pathogenicity genes. Nat Genet. 2011;43(12):1275–1280. doi: 10.1038/ng.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cramer N, et al. Microevolution of the major common Pseudomonas aeruginosa clones C and PA14 in cystic fibrosis lungs. Environ Microbiol. 2011;13(7):1690–1704. doi: 10.1111/j.1462-2920.2011.02483.x. [DOI] [PubMed] [Google Scholar]
- 12.Smith EE, et al. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc Natl Acad Sci USA. 2006;103(22):8487–8492. doi: 10.1073/pnas.0602138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Huse HK, et al. (2010) Parallel evolution in Pseudomonas aeruginosa over 39,000 generations in vivo. MBio 1(4):e00199-10. [DOI] [PMC free article] [PubMed]
- 14.Coenye T, Spilker T, Van Schoor A, LiPuma JJ, Vandamme P. Recovery of Burkholderia cenocepacia strain PHDC from cystic fibrosis patients in Europe. Thorax. 2004;59(11):952–954. doi: 10.1136/thx.2003.019810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mahenthiralingam E, Urban TA, Goldberg JB. The multifarious, multireplicon Burkholderia cepacia complex. Nat Rev Microbiol. 2005;3(2):144–156. doi: 10.1038/nrmicro1085. [DOI] [PubMed] [Google Scholar]
- 16.Coutinho CP, de Carvalho CC, Madeira A, Pinto-de-Oliveira A, Sá-Correia I. Burkholderia cenocepacia phenotypic clonal variation during a 3.5-year colonization in the lungs of a cystic fibrosis patient. Infect Immun. 2011;79(7):2950–2960. doi: 10.1128/IAI.01366-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mann EE, Wozniak DJ. Pseudomonas biofilm matrix composition and niche biology. FEMS Microbiol Rev. 2012;36(4):893–916. doi: 10.1111/j.1574-6976.2011.00322.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Darwin C. On the Origin of Species by Means of Natural Selection. 1st Ed. London: Murray; 1859. [Google Scholar]
- 19.Cairrão F, Chora A, Zilhão R, Carpousis AJ, Arraiano CM. RNase II levels change according to the growth conditions: Characterization of gmr, a new Escherichia coli gene involved in the modulation of RNase II. Mol Microbiol. 2001;39(6):1550–1561. doi: 10.1046/j.1365-2958.2001.02342.x. [DOI] [PubMed] [Google Scholar]
- 20.Hickman JW, Tifrea DF, Harwood CS. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc Natl Acad Sci USA. 2005;102(40):14422–14427. doi: 10.1073/pnas.0507170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Güvener ZT, Harwood CS. Subcellular location characteristics of the Pseudomonas aeruginosa GGDEF protein, WspR, indicate that it produces cyclic-di-GMP in response to growth on surfaces. Mol Microbiol. 2007;66(6):1459–1473. doi: 10.1111/j.1365-2958.2007.06008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sniegowski PD, Gerrish PJ. Beneficial mutations and the dynamics of adaptation in asexual populations. Philos Trans R Soc Lond B Biol Sci. 2010;365(1544):1255–1263. doi: 10.1098/rstb.2009.0290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park S-C, Krug J. Clonal interference in large populations. Proc Natl Acad Sci USA. 2007;104(46):18135–18140. doi: 10.1073/pnas.0705778104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huber B, et al. Genetic analysis of functions involved in the late stages of biofilm development in Burkholderia cepacia H111. Mol Microbiol. 2002;46(2):411–426. doi: 10.1046/j.1365-2958.2002.03182.x. [DOI] [PubMed] [Google Scholar]
- 25.Sousa SA, et al. The Burkholderia cepacia bceA gene encodes a protein with phosphomannose isomerase and GDP-D-mannose pyrophosphorylase activities. Biochem Biophys Res Commun. 2007;353(1):200–206. doi: 10.1016/j.bbrc.2006.12.016. [DOI] [PubMed] [Google Scholar]
- 26.Saldías MS, Ortega X, Valvano MA. Burkholderia cenocepacia O antigen lipopolysaccharide prevents phagocytosis by macrophages and adhesion to epithelial cells. J Med Microbiol. 2009;58(Pt 12):1542–1548. doi: 10.1099/jmm.0.013235-0. [DOI] [PubMed] [Google Scholar]
- 27.Lau PC, Lindhout T, Beveridge TJ, Dutcher JR, Lam JS. Differential lipopolysaccharide core capping leads to quantitative and correlated modifications of mechanical and structural properties in Pseudomonas aeruginosa biofilms. J Bacteriol. 2009;191(21):6618–6631. doi: 10.1128/JB.00698-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tomlin KL, et al. Quorum-sensing mutations affect attachment and stability of Burkholderia cenocepacia biofilms. Appl Environ Microbiol. 2005;71(9):5208–5218. doi: 10.1128/AEM.71.9.5208-5218.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yedid G, Ofria CA, Lenski RE. Historical and contingent factors affect re-evolution of a complex feature lost during mass extinction in communities of digital organisms. J Evol Biol. 2008;21(5):1335–1357. doi: 10.1111/j.1420-9101.2008.01564.x. [DOI] [PubMed] [Google Scholar]
- 30.Boles BR, Thoendel M, Singh PK. Self-generated diversity produces “insurance effects” in biofilm communities. Proc Natl Acad Sci USA. 2004;101(47):16630–16635. doi: 10.1073/pnas.0407460101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rainey PB, Travisano M. Adaptive radiation in a heterogeneous environment. Nature. 1998;394(6688):69–72. doi: 10.1038/27900. [DOI] [PubMed] [Google Scholar]
- 32.Kirisits MJ, Prost L, Starkey M, Parsek MR. Characterization of colony morphology variants isolated from Pseudomonas aeruginosa biofilms. Appl Environ Microbiol. 2005;71(8):4809–4821. doi: 10.1128/AEM.71.8.4809-4821.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Déziel E, Comeau Y, Villemur R. Initiation of biofilm formation by Pseudomonas aeruginosa 57RP correlates with emergence of hyperpiliated and highly adherent phenotypic variants deficient in swimming, swarming, and twitching motilities. J Bacteriol. 2001;183(4):1195–1204. doi: 10.1128/JB.183.4.1195-1204.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Masel J, Trotter MV. Robustness and evolvability. Trends Genet. 2010;26(9):406–414. doi: 10.1016/j.tig.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Elena SF, Sanjuán R. The effect of genetic robustness on evolvability in digital organisms. BMC Evol Biol. 2008;8:284. doi: 10.1186/1471-2148-8-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanjuán R, Elena SF. Epistasis correlates to genomic complexity. Proc Natl Acad Sci USA. 2006;103(39):14402–14405. doi: 10.1073/pnas.0604543103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee B, et al. Mucoid Pseudomonas aeruginosa isolates maintain the biofilm formation capacity and the gene expression profiles during the chronic lung infection of CF patients. APMIS. 2011;119(4-5):263–274. doi: 10.1111/j.1600-0463.2011.02726.x. [DOI] [PubMed] [Google Scholar]
- 38.Häussler S. Biofilm formation by the small colony variant phenotype of Pseudomonas aeruginosa. Environ Microbiol. 2004;6(6):546–551. doi: 10.1111/j.1462-2920.2004.00618.x. [DOI] [PubMed] [Google Scholar]
- 39.Chantratita N, et al. Biological relevance of colony morphology and phenotypic switching by Burkholderia pseudomallei. J Bacteriol. 2007;189(3):807–817. doi: 10.1128/JB.01258-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Starkey M, et al. Pseudomonas aeruginosa rugose small-colony variants have adaptations that likely promote persistence in the cystic fibrosis lung. J Bacteriol. 2009;191(11):3492–3503. doi: 10.1128/JB.00119-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Byrd MS, et al. Direct evaluation of Pseudomonas aeruginosa biofilm mediators in a chronic infection model. Infect Immun. 2011;79(8):3087–3095. doi: 10.1128/IAI.00057-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hoboth C, et al. Dynamics of adaptive microevolution of hypermutable Pseudomonas aeruginosa during chronic pulmonary infection in patients with cystic fibrosis. J Infect Dis. 2009;200(1):118–130. doi: 10.1086/599360. [DOI] [PubMed] [Google Scholar]
- 43.King T, Seeto S, Ferenci T. Genotype-by-environment interactions influencing the emergence of rpoS mutations in Escherichia coli populations. Genetics. 2006;172(4):2071–2079. doi: 10.1534/genetics.105.053892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Conrad TM, et al. RNA polymerase mutants found through adaptive evolution reprogram Escherichia coli for optimal growth in minimal media. Proc Natl Acad Sci USA. 2010;107(47):20500–20505. doi: 10.1073/pnas.0911253107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Philippe N, Crozat E, Lenski RE, Schneider D. Evolution of global regulatory networks during a long-term experiment with Escherichia coli. Bioessays. 2007;29(9):846–860. doi: 10.1002/bies.20629. [DOI] [PubMed] [Google Scholar]
- 46.Bantinaki E, et al. Adaptive divergence in experimental populations of Pseudomonas fluorescens. III. Mutational origins of wrinkly spreader diversity. Genetics. 2007;176(1):441–453. doi: 10.1534/genetics.106.069906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zlosnik JE, et al. Differential mucoid exopolysaccharide production by members of the Burkholderia cepacia complex. J Clin Microbiol. 2008;46(4):1470–1473. doi: 10.1128/JCM.02273-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zemanick ET, Sagel SD, Harris JK. The airway microbiome in cystic fibrosis and implications for treatment. Curr Opin Pediatr. 2011;23(3):319–324. doi: 10.1097/MOP.0b013e32834604f2. [DOI] [PubMed] [Google Scholar]
- 49.Barrick JE, Lenski RE. Genome-wide mutational diversity in an evolving population of Escherichia coli. Cold Spring Harb Symp Quant Biol. 2009;74:119–129. doi: 10.1101/sqb.2009.74.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ellis CN, Cooper VS. Experimental adaptation of Burkholderia cenocepacia to onion medium reduces host range. Appl Environ Microbiol. 2010;76(8):2387–2396. doi: 10.1128/AEM.01930-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.LiPuma JJ Spilker T, Coenye T, Gonzalez CF (2002) An epidemic Burkholderia cepacia complex strain identified in soil. Lancet. 359:2002–2003. doi: 10.1016/S0140-6736(02)08836-0. [DOI] [PubMed] [Google Scholar]
- 52.Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18(11):1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]