Abstract
The initiation of chromosomal DNA replication is rigidly regulated to ensure that it occurs in a cell cycle-coordinated manner. To ensure this in Escherichia coli, multiple systems regulate the activity of the replication initiator ATP-DnaA. The level of ATP-DnaA increases before initiation after which it drops via DnaA-ATP hydrolysis, yielding initiation-inactive ADP-DnaA. DnaA-ATP hydrolysis is crucial to regulation of initiation and mainly occurs by a replication-coupled feedback mechanism named RIDA (regulatory inactivation of DnaA). Here, we report a second DnaA-ATP hydrolysis system that occurs at the chromosomal site datA. This locus has been annotated as a reservoir for DnaA that binds many DnaA molecules in a manner dependent upon the nucleoid-associated factor IHF (integration host factor), resulting in repression of untimely initiations; however, there is no direct evidence for the binding of many DnaA molecules at this locus. We reveal that a complex consisting of datA and IHF promotes DnaA-ATP hydrolysis in a manner dependent on specific inter-DnaA interactions. Deletion of datA or the ihf gene increased ATP-DnaA levels to the maximal attainable levels in RIDA-defective cells. Cell-cycle analysis suggested that IHF binds to datA just after replication initiation at a time when RIDA is activated. We propose a model in which cell cycle-coordinated ATP-DnaA inactivation is regulated in a concerted manner by RIDA and datA.
Keywords: AAA+, bacteria, initiation regulation, in vitro reconstitution, nucleoprotein complex
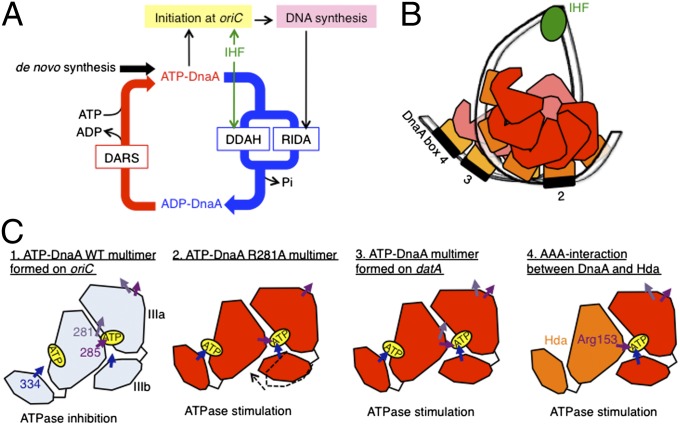
In Escherichia coli, DnaA protein initiates DNA replication by forming a nucleoprotein complex with the chromosomal replication origin oriC (1–4). DnaA consists of four functional domains: (i) Domain I binds to specific proteins, such as DnaB helicase. (ii) Domain II is a flexible linker. (iii) Domain III, the AAA+ domain, has motifs important for ATP-binding/hydrolysis and inter-DnaA interactions (3–6). This domain can be divided to subdomains IIIa and IIIb (5). (iv) Domain IV is the sequence-specific DNA binding domain (5, 7). The minimal oriC (245 bp) contains an AT-rich region that facilitates DNA duplex unwinding, in addition to a DnaA assembly region that contains two high-affinity 9-mer DnaA binding sites (DnaA boxes) called R1 and R4; a moderate-affinity site, R2; and several low-affinity ATP-DnaA binding sites, including τ1 (2–4, 8, 9) (Fig. 1A). Multiple ATP-DnaA molecules bind cooperatively to oriC. Key residues for inter-DnaA interactions include the AAA+ Arg-finger Arg285, which plays a key role in the ATP-dependent interaction between DnaA monomers that activates DnaA complexes during initiation, and Arg281 in AAA+ box VII, which stabilizes inter-DnaA interactions (8, 10). Along with ATP-DnaA, integration host factor (IHF), a nucleoid-associated protein (11), binds to oriC at a specific site (IBS: IHF-binding site) (Fig. 1A) (9, 12), resulting in formation of the initiation complex. IHF bends DNA sharply (∼180°) (11) and enhances DnaA binding and DNA unwinding (9, 12, 13). The binding of IHF to oriC occurs during the preinitiation stage in vivo (14).
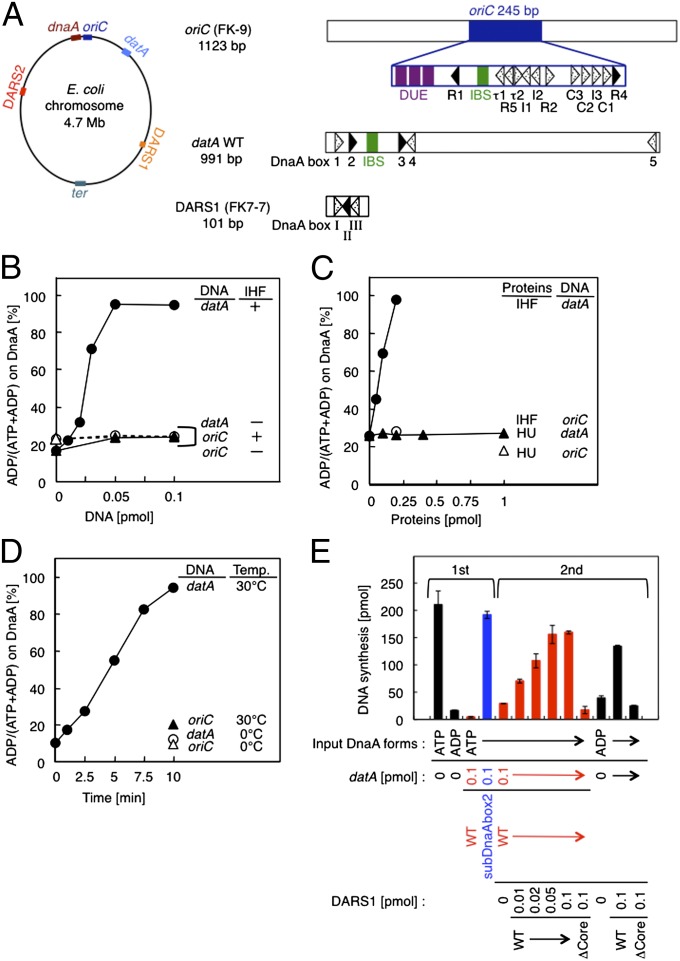
Fig. 1.
datA-dependent DnaA-ATP hydrolysis. (A) Schematic presentation of the chromosomal loci and structures of oriC, dnaA, datA, DARS1/2, and ter. (Left) The location of each site on the E. coli chromosome is indicated. (Right) Open bars indicate the oriC-, datA-, and DARS1-containing fragments used in this study: respectively, FK-9, datA WT, and FK7-7. Arrowheads represent DnaA binding sites that match the 9-mer consensus sequence completely (black) or contain a mismatch(es) (dotted). R5, I1–3, τ1–2, and C1–3 in oriC are low-affinity ATP-DnaA binding sites (1, 4, 9). DnaA boxes 1–5 in datA and DnaA boxes I–III in DARS1 are displayed similarly. IBS (green bars) and AT-rich repeats that facilitate duplex unwinding (AT repeats; purple bars) are also indicated. (B–D) In vitro reconstitution of datA-dependent DnaA-ATP hydrolysis. [α-32P]ATP-DnaA (1 pmol) was incubated under various conditions, and then analyzed by Thin-layer chromatography (TLC). The proportions of ADP-DnaA to total ATP/ADP-DnaA molecules are indicated as percentages (%). (B) ATP-DnaA was incubated at 30 °C for 10 min with the indicated amounts of datA WT (datA) (●, ○) or FK-9 (oriC) (▲, △) in the presence (●, ▲) or absence (○, △) of IHF (0.2 pmol). (C) ATP-DnaA was incubated with the indicated amounts of either IHF (●, ○) or HU (▲, △) in the presence (0.05 pmol) of datA WT (●, ▲) or FK-9 (○, △). (D) Reaction time course was analyzed at 0 °C (○, △) or 30 °C (●, ▲) in the presence of IHF (0.2 pmol) and either datA WT (0.05 pmol) (●, ○) or FK-9 (▲, △). (E) In vitro reconstitution of the DnaA cycle using datA and DARS1. DnaA activity was assessed using an in vitro replication system. In the first stage, IHF (0.4 pmol) and ATP-DnaA or ADP-DnaA (2 pmol) were incubated at 30 °C for 10 min in buffer (25 µL) containing (red bars) or excluding (black bars) datA WT, followed by the addition of DpnII and incubation at 30 °C for 5 min to digest datA DNA. In the second stage, the indicated amounts of DARS1 DNA (FK7-7, 1 µL) were added to samples (27 µL) that had contained ATP-DnaA and datA WT (red bars) or ADP-DnaA (black bars) in the first stage, followed by further incubation at 30 °C for 15 min. After the first- and second-stage reactions, portions (5 µL) were withdrawn, and DnaA initiator activity was analyzed in vitro using a minichromosome replication system in a crude protein extract. In the second stage, subDnaAbox2 (blue bar), a DnaA box 2-substituted derivative of datA that is inactive in DDAH, was used as a negative control (see also Fig. 2B). In the second stage, DARS1 mutant ΔCore (FK7-21), which contains a deletion of DnaA boxes I–III and is inactive in DnaA-nucleotide exchange, was used as a negative control. Error bars represent the SD from two independent experiments.
In E. coli, the cell cycle-coordinated initiation of chromosomal replication is sustained by multiple systems that regulate the activities of DnaA and oriC (1–4, 15). After ATP-DnaA induces replication initiation, DnaA-bound ATP is hydrolyzed by Hda protein in complex with the DNA-loaded β-clamp subunit of DNA polymerase III holoenzyme, yielding initiation-inactive ADP-DnaA (2, 4, 16, 17). This replication-coupled feedback system is called RIDA (regulatory inactivation of DnaA). Hda and DnaA are both AAA+ proteins (2, 4, 5). Two residues play key roles in DnaA-ATP hydrolysis in RIDA: the Hda Arg-finger Arg153 and DnaA Sensor II Arg334 (18). Defective RIDA results in overinitiation and inhibition of cell growth; therefore, DnaA-ATP hydrolysis is critical for preventing extra initiation events (2, 4, 16, 17).
Immediately after initiation, SeqA protein binds to hemimethylated oriC, a product of semiconservative DNA replication of fully methylated oriC, temporarily inhibiting untimely initiation events (2, 4, 15). In addition, dnaA transcription is repressed in a replication-coupled manner by SeqA, and by DnaA itself (2, 4, 19). The dnaA gene promoter contains a DnaA box cluster; ATP-DnaA represses the promoter more tightly than ADP-DnaA (20). Excess DnaA results in replication overinitiation (21).
A specific chromosomal locus known as datA, which spans a ∼1-kb DNA region bearing five DnaA boxes and a single IBS (Fig. 1A), is crucial for repressing untimely initiation events (22–25). datA-deleted cells, like seqA-deleted cells, perform untimely initiations at a level that does not inhibit cell growth (23). Furthermore, deletion of datA or ihf causes rifampicin-resistant initiations at oriC (26, 27). The Bacillus subtilis and Streptomyces coelicolor genomes also have DnaA box clusters analogous to datA that can repress untimely initiations (28, 29).
DnaA binding to datA is thought to reduce the number of DnaA molecules accessible to oriC, thereby inhibiting untimely initiations (22–25). Originally, datA was speculated to bind ∼370 DnaA molecules (22); however, this figure is deduced from indirect measurements not yet supported by direct evidence. It remains unclear how such a large number of DnaA molecules might bind to datA.
Here, we reveal a unique function of datA in DnaA-ATP hydrolysis, termed DDAH (datA-dependent DnaA-ATP hydrolysis). We observed that datA efficiently hydrolyzed ATP bound to DnaA in a manner dependent on both IHF and Arg285 of the DnaA Arg-finger, suggesting that specific DnaA multimer formation is important for DDAH. Deletion of either datA or ihf increased cellular ATP-DnaA levels in a RIDA-independent manner, consistent with the fact that untimely initiations occur in datA and IHF mutant cells (23–25, 27). Because IHF binding to datA in cells was detected specifically at the postinitiation stage, datA function is likely to be temporally regulated.
Results
datA-IHF Complexes Promote DnaA-ATP Hydrolysis.
To assess whether datA-IHF complexes stimulate DnaA-ATP hydrolysis, we incubated ATP-DnaA with a wild-type datA fragment (datA WT) and purified IHF (Fig. 1 B–D). Most of the input ATP-DnaA molecules (1 pmol) were converted into ADP-DnaA after only 10 min at 30 °C in the presence of datA (0.05 pmol) and IHF (0.2 pmol) (Fig. 1B). This activity was specific to datA, because an oriC fragment (FK-9) was inactive in this reaction. These results are consistent with a previous study showing that DnaA has very weak intrinsic DNA-dependent ATPase activity at 37 °C (6, 30).
IHF was required for DDAH and stimulated it in a dose-dependent manner (Fig. 1 B and C). In contrast HU, another representative nucleoid-associated protein (11), was completely inactive in this reaction (Fig. 1C). The data in Fig. 1 B and C suggest that the optimal molar ratio of DnaA:IHF:datA for DDAH in vitro is 20:4:1. The DnaA-ATP hydrolysis rate remained constant for 10 min at 30 °C (Fig. 1D); 1.6 molecules of ATP-DnaA were hydrolyzed per min per datA molecule. These data suggest that, like the DNA-clamp-Hda complex in RIDA, datA catalytically promotes DnaA-ATP hydrolysis. RIDA can hydrolyze at least 0.9 molecules of DnaA-ATP per DNA-clamp-Hda complex per minute (18).
In E. coli, reactivation of ADP-DnaA by the DARS- (DnaA reactivating sequence) dependent exchange of ADP for ATP is crucial for timely replication initiation coordinated with the cell cycle (31) (Fig. 1A). The E. coli chromosome contains two such sequences, DARS1 and DARS2, each of which has a DnaA box cluster that binds multiple ADP-DnaA molecules and yields ATP-DnaA by nucleotide exchange. Consistent with this, ADP-DnaA resulting from hydrolysis of ATP-DnaA by RIDA is reactivated in vitro by DARS1 (31). We therefore asked whether ADP-DnaA generated from ATP-DnaA by DDAH could be reactivated for replication in a DARS-dependent manner (Fig. 1E). In the first stage, ATP-DnaA was inactivated in a DDAH-specific manner, and datA DNA was thoroughly digested by DpnII. When the resulting samples were further incubated in the presence of DARS1 and 2 mM ATP, DnaA was fully reactivated in a DARS1-specific and dose-dependent manner (Fig. 1E). These results support the idea that DDAH can regulate the replication initiation cycle in vivo. Exchange of DnaA-bound nucleotide may be stimulated by acidic phospholipids as well as by DARS (1), but we did not examine this pathway in this study.
Sequence Element Requirements of datA for DDAH.
The original characterization of datA suggested that the region containing DnaA boxes 1–5 can bind the maximum number of DnaA molecules in vivo (22). Later studies found that only DnaA boxes 2–3 and IBS are crucial for repressing untimely initiations (24, 25). To identify the specific sequence element prerequisites of datA for DDAH, we first performed deletion analysis using a reconstituted DDAH assay (Fig. 2A). Full DDAH activity was sustained even by datA del5 containing DnaA boxes 2–4 and IBS. Furthermore, the datA del6 fragment, containing only DnaA boxes 2–3 and IBS, also displayed DDAH activity, although the reaction efficiency was moderately reduced. In contrast, the datA WTΔIBS, del5ΔIBS, or del7-9 fragments, which lack DnaA box 2 or 3 or the IBS, were completely inactive (Fig. 2A). Thus, datA del6 is the minimum region required for DDAH, consistent with the results of the previous in vivo studies described above.
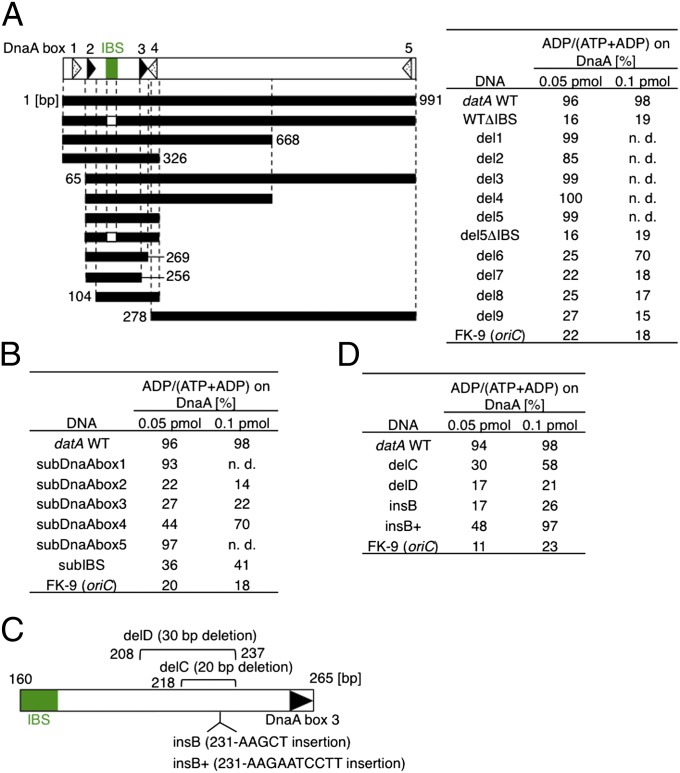
Fig. 2.
DnaA and IHF binding sites for DDAH. (A) The open bar at the top depicts the datA region, as described in Fig. 1A. datA WT DNA and its truncated derivatives (black bars) were incubated with [α-32P]ATP-DnaA (1 pmol) and IHF at 30 °C for 10 min, followed by TLC. When the datA derivatives included were 0.05 pmol, IHF included was 0.4 pmol. When the datA derivatives included were 0.1 pmol, IHF included was 0.8 pmol. Results (%) are shown to the right of the bars. n.d., not determined. (B) Similar experiments were performed using the indicated mutants of datA or oriC FK-9. subDnaAbox1–5 and subIBS contain substitutions of the corresponding DnaA box and IBS, respectively (Fig. S1 and Table S3). (C) Schematic of the relationship between IBS and DnaA box 3. delC, delD, insB, and insB+ mutations are shown with the nucleotide numbers (25). For details in sequences, see Fig. S2. (D) An in vitro reconstituted DDAH system using datA derivatives containing a deletion or an insertion between IBS and DnaA box 3.
Next, we analyzed substitution mutants of DnaA boxes 1–5 and IBS, using a similar assay (Fig. 2B and Tables S1 and S2). These mutants were inactive for DnaA or IHF binding (Fig. S1). The results indicate that DnaA boxes 2–3 and IBS are crucial for DDAH in vitro (Fig. 2B), consistent with the results of the in vivo study described above. Substitution of DnaA box 4 moderately reduced DDAH activity, consistent with the deletion analysis (Fig. 2A). This moderate inhibition might be a consequence of the inhibition of the cooperative binding of DnaA to the datA derivative. DnaA boxes 3 and 4 reside side-by-side, with only 3 bp separating them, suggesting that binding of DnaA to DnaA box 4 might stimulate cooperative binding of DnaA to the DnaA boxes 2–3 region. DnaA box 4 is not required for the repression of untimely initiations in cells growing in M9 medium (24), supporting the idea that the DDAH activity of datA lacking DnaA box 4 is sufficient for repression, at least under such conditions.
Additional deletion/insertion analyses indicate that the distance between IBS and DnaA box 3 is important for DDAH (Fig. 2 C and D), consistent with previously reported in vivo data (25). Specifically, datA delC (20-bp deletion) and insB+ (10-bp insertion) sustained moderate DDAH activity in vitro, whereas datA delD (30-bp deletion) and datA insB (5-bp insertion) were inactive (Fig. 2D). Notably, datA delC and datA insB+ repressed untimely initiation events in vivo as effectively as datA WT, whereas datA delD and datA insB led to untimely initiations (25).
DDAH Requires DnaA Motifs for Inter-DnaA Interactions and for DNA Binding.
To analyze the mechanism underlying stimulation of DnaA-ATP hydrolysis by the datA-IHF complex, we analyzed the DDAH activity of DnaA mutants in vitro (Fig. 3). In RIDA, Arg334 in DnaA AAA+ Sensor II is crucial for DnaA-ATP hydrolysis (6). This residue is located in close proximity to the phosphate moiety of bound ATP and is proposed to participate directly in catalysis of ATP hydrolysis (6, 32). DnaA R334A was completely inactive in DDAH (Fig. 3A), suggesting that this residue plays a crucial role in DDAH, as it does in RIDA.
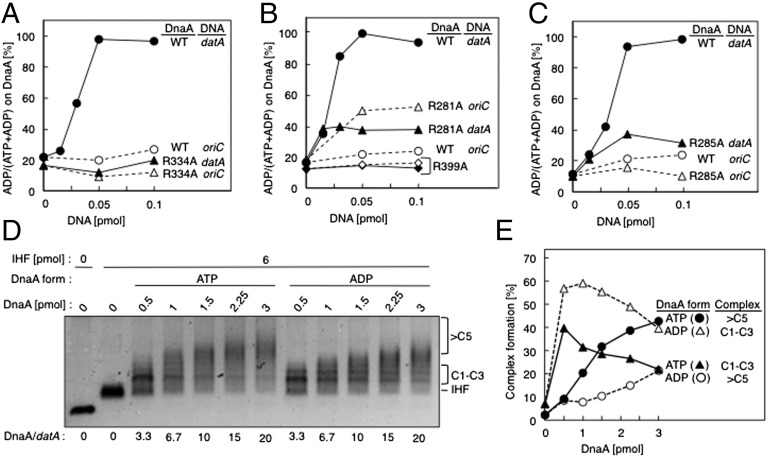
Fig. 3.
DnaA motifs are required for DDAH. (A) The [α-32P]ATP forms (1 pmol) of WT DnaA (●, ○) or DnaA R334A (▲, △) were incubated at 30 °C for 10 min with IHF (0.2 pmol) and the indicated amounts of datA WT (●, ▲) or oriC FK-9 (○,△). (B) The binding of WT DnaA (●, ○), DnaA R281A (▲, △), or DnaA R399A (◆, ◇) to datA (●,▲, ◆) or oriC (○, △, ◇) were similarly analyzed. (C) Similar experiments were performed using WT DnaA (●, ○) and the R285A mutant (▲, △). (D) The indicated amounts of ATP- or ADP-DnaA were incubated with datA del5 (0.15 pmol), IHF (6 pmol), and 150 ng λDNA (as a competitor) at 15 °C for 5 min, followed by EMSA and Gel-star staining. The gel image is shown in black-and-white inverted mode. IHF, IHF-bound datA. C1–C3, datA-IHF carrying 1–3 DnaA molecules. >5C, datA-IHF carrying more than five DnaA molecules. (E) The proportions of higher (>C5; ●, ○) and lower (C1–C3; ▲, △) complexes of ATP- (●, ▲) or ADP-DnaA (○, △) were determined using the data shown in D and are plotted as percentages (%).
The AAA+ Arg-finger Arg285 and box VII Arg281 also play important roles in inter-DnaA interactions within the oriC initiation complex. Both residues are exposed on the surface of the tertiary structure of DnaA. Arg285 promotes the ATP-specific conformation of the initiation complex, as well as replication initiation, by recognizing ATP bound to the flanking DnaA molecule in the DnaA homo-oligomer (8, 32, 33). DnaA Arg281 is required to stabilize DnaA homo-oligomers at oriC (10). Unlike Arg334, these residues are dispensable for DnaA-ATP hydrolysis in RIDA (8). In RIDA, the interaction between DnaA and Hda promotes DnaA-ATP hydrolysis in a manner dependent on the Hda Arg-finger and DnaA Arg334 (18). As in the DnaA-oriC complex, but distinct from RIDA, DDAH required Arg281 and Arg285 (Fig. 3 B and C), supporting the idea that inter-DnaA interactions are crucial for DDAH. However, DnaA R281A and DnaA R285A exhibited slight residual DDAH activity, which may be explained by the lower stability of DnaA complexes formed on datA than those formed on oriC and by the less critical nature of either one of the two residues for DDAH than for initiation at oriC. When DnaA R281A was used, ATP hydrolysis was weakly stimulated even in the presence of oriC (Fig. 3B). One possible explanation for this result is that Arg281 can affect the overall structure of DnaA complexes (Discussion).
DnaA domain IV is the DnaA box-binding region; in the crystal structure, Arg399 in this domain directly binds to the DnaA box (7). Consistent with this finding, DnaA R399A is defective in DNA binding (34), and this mutant protein did not display any DDAH activity (Fig. 3B).
Oligomerization of DnaA on the Minimum Region of datA.
Several low-affinity DnaA boxes have been suggested to be present in the region between DnaA boxes 2–3 (35). To determine whether DnaA oligomers are formed in the region, we performed EMSA using datA del5 incubated at 15 °C for 5 min in the presence of IHF and either ATP- or ADP-DnaA. The ATP-DnaA level was sustained during the incubation (Fig. S3). ATP-DnaA formed complexes carrying at least six DnaA molecules per datA del5 when the input molar ratio was 7–20 DnaA per datA (Fig. 3 D and E). In the presence of ADP-DnaA, such complexes formed significantly less efficiently than with ATP-DnaA; under these conditions, complexes carrying only one to three DnaA molecules were predominant (Fig. 3 D and E). These data suggest that ATP-DnaA forms oligomers on datA more efficiently than ADP-DnaA. Consistent results were obtained by DpnII-digestion protection experiments using datA del5, DnaA, and IHF (Fig. S4). Furthermore, these data are consistent with previous surface plasmon resonance and footprint data for datA (25), as well as with the ATP-DnaA–specific stimulation of assembly on oriC and the dnaA promoter (8, 36).
Taken together, the results suggest that DDAH is a unique mechanism that regulates the hydrolysis of ATP bound to DnaA; DnaA oligomers are formed on the IHF-bound datA DnaA box 2–3 region, stimulating specific inter-DnaA interactions and DnaA conformational changes that promote the ATP-Arg334 interaction and DnaA-ATP hydrolysis.
datA and IHF Required for Lowering ATP-DnaA Levels in Vivo.
Based on the data described above, we investigated whether the datA-IHF complexes repress ATP-DnaA levels in vivo. Our previous analyses indicated that RIDA inactivation via the deletion of the hda gene or inactivation of the clamp in asynchronous cells increases the cellular ATP-DnaA level to 70–80% of the total number of ATP/ADP-bound DnaA molecules, but never to 90–100% (16, 17). Earlier experiments also showed that datA deletion slightly (i.e., 5–10%) increased the ATP-DnaA level (37). Therefore, we inferred that DDAH might be a second DnaA-ATP hydrolysis system that plays a prominent role in the absence of functional RIDA. To test this idea, we examined whether DDAH affects cellular ATP-DnaA levels by introducing ΔdatA into RIDA-defective cells (Fig. 4A and Table S3).
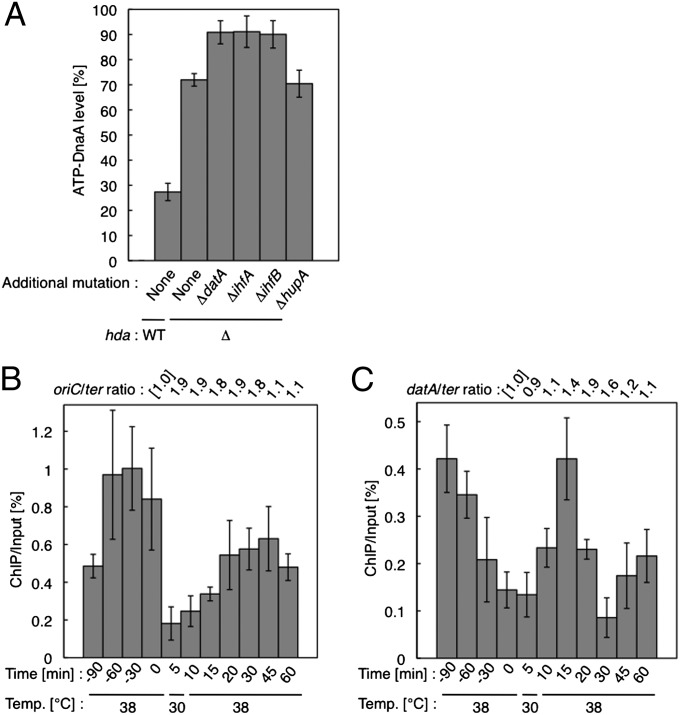
Fig. 4.
Analyses for cellular ATP-DnaA level and IHF binding. (A) KW262-5 (rnhA::Tn3 ΔoriC) (WT), MK86 (Δhda), KX93 (Δhda ∆datA), KX30 (Δhda ∆ihfA), KX31 (Δhda ΔihfB), and KX32 (Δhda ΔhupA) cells were grown at 37 °C in medium containing 32P. DnaA was immunoprecipitated, and recovered DnaA-bound nucleotides were analyzed by TLC. Error bars represent the SD from at least four independent experiments. (B and C) KYA018 (dnaC2) cells growing at 30 °C in supplemented M9 medium were transferred to 38 °C and incubated for 90 min. The cells were then transferred to 30 °C (time 0), incubated for 5 min, and further incubated at 38 °C; samples were withdrawn at the indicated times. The oriC, datA, and ylcC levels before (Input) and after (ChIP) immunoprecipitation using anti-IHF antiserum were determined using real-time quantitative PCR. The ChIP/Input for ylcC (%) was used as a background control for nonspecific IHF binding and was subtracted from the ChIP/Input for oriC and datA. The levels of oriC or datA relative to ter in the Input samples were also quantified using real-time quantitative PCR, and the relative ratios of oriC/ter and datA/ter are expressed relative to the ratio at 0 min (defined as 1). Relative ChIP/Input values for oriC and oriC/ter ratio (B) and the relative ChIP/Input values for datA and datA/ter ratio (C) are shown. Error bars represent the SD from at least three independent experiments.
For this analysis, KW262-5 (ΔoriC ΔrnhA) cells were used as a standard. Deletion of oriC represses lethal overinitiation caused by RIDA inactivation, whereas deletion of the rnhA gene encoding RNaseH I activates alternative origins, allowing the growth of cells lacking oriC (38). DnaA was isolated by immunoprecipitation from the lysates of 32P-labeled exponentially growing cells, and DnaA-bound nucleotides were quantified using TLC, as previously described (16, 17, 31). In KW262-5 cells, ATP-DnaA levels were low (27%) (Fig. 4A). When the hda deletion construct was introduced into KW262-5 cells and the resulting MK86 cells were analyzed, the ATP-DnaA levels increased to 72% (Fig. 4A). Importantly, introducing datA, ihfA, or ihfB deletion constructs further increased the ATP-DnaA levels to 88–97% (Fig. 4A); ihfA and ihfB encode, respectively, the α- and β-subunits of IHF. In contrast, no further increase resulted from deletion of hupA, which encodes the HUα subunit (Fig. 4A). All of these results are consistent with the in vitro data described above, and support the idea that DDAH constitutes a second in vivo DnaA-ATP hydrolysis system.
Coordination Between Initiation of Replication from oriC and Binding of IHF to datA.
IHF binds to oriC in the preinitiation stage and forms an initiation-competent DnaA-oriC complex (14). After initiation, IHF is temporarily released from oriC (14). Therefore, we performed ChIP assays in a temperature-sensitive dnaC2 mutant to investigate whether IHF binding to datA is coordinated with the replication cycle. The dnaC2 mutation inhibits replication initiation, but not the progression of established replisomes at the restrictive temperature. To synchronize the replication cycle, dnaC2 mutant cells that had been growing at 30 °C were incubated for 90 min at 38 °C, the restrictive temperature. To initiate replication, the temperature was rapidly reduced to 30 °C, and then 5-min later the temperature was rapidly shifted to 38 °C to inhibit a second round of replication during further incubations. After isolating DNA-protein complexes using anti-IHF antiserum, we quantified levels of the oriC and datA loci and a control locus ylcC, which has no IHF-specific binding sites (39), by real-time quantitative PCR; the ratio of oriC to ter (the replication terminus) (Fig. 1A) was similarly determined (Fig. 4 B and C).
Analysis of the oriC/ter ratio showed that oriC was duplicated within 5 min after the temperature reduction to 30 °C, and that the oriC level was maintained for another 25 min (Fig. 4B). The oriC/ter ratio returned to the original level 45–60 min after the temperature reduction (Fig. 4B), indicating the completion of replication. These data indicate that initiation occurred only once within the initial 5 min at 30 °C. The ChIP data revealed that IHF temporarily dissociated from oriC in an initiation-coupled manner (Fig. 4B), as shown in a previous report (14). In the same culture, the datA region was duplicated 10–20 min after initiation (Fig. 4C). IHF dissociated from datA in the preinitiation stage, bound to this site immediately after initiation, and dissociated again 20–30 min after initiation (Fig. 4C). This dissociation timing may be reasonable given that cellular ATP-DnaA decreases to its basal level about 20 min after initiation (16). Similar results were obtained when the second round of initiation was inhibited by rifampicin (Fig. S5 and SI Results and Discussion). These results indicate that IHF binding to datA is regulated in coordination with the replication cycle (i.e., IHF dissociates from datA during the preinitiation stage and binds to datA after initiation). This behavior of IHF at datA differs from its behavior at oriC, which is appropriate for its role in ensuring timely activation of DDAH in coordination with the replication cycle.
Discussion
In this study, we identified a unique DnaA-regulatory pathway, termed DDAH, which stimulates DnaA-ATP hydrolysis in a datA-IHF-dependent manner. The sequence element requirements for datA in DDAH were the same as those required for repression of untimely initiations in vivo. We also found that deletion of datA or ihf further increased ATP-DnaA levels in RIDA-defective cells. Taken together, these results support the idea that DDAH assists RIDA in regulating ATP-DnaA levels, and restricts initiation of replication in a manner independent of RIDA (Fig. 5A). The chromosomal location of datA is relatively close to those of oriC and dnaA (22) (Fig. 1A), which might enhance interaction of datA with ATP-DnaA molecules expressed from the dnaA locus and localized in the oriC-proximal space (Fig. S6). We hypothesize that DDAH might be a mechanism common to many bacterial species whose genomes contain DnaA box clusters; as noted above, the DnaA box clusters in B. subtilis and S. coelicolor repress untimely initiations (28, 29).
Fig. 5.
A model for the molecular mechanism of DDAH. (A) A revised view of the DnaA activity cycle including DDAH. ATP-DnaA is hydrolyzed by two independent pathways, RIDA and DDAH. IHF stimulates initiation at oriC (9, 12, 13) and DDAH. Thus, IHF plays both positive and negative roles in initiation. DARS reactivates ADP-DnaA by ADP-to-ATP exchange. (B) A model of the structure of the DnaA-IHF-datA complex. ATP-DnaA multimers are formed on regions carrying DnaA boxes 2–4. Sharp DNA bending by IHF stimulates interaction between ATP-DnaA multimers. For simplicity, only DnaA domain III (red or pink polygon) and domain IV (orange square) are shown. (C) A model of DDAH-specific conformational change of DnaA oligomers. For simplicity, only a dimer of DnaA domain III is shown. Domain IIIa (large polygon, IIIa) contains Arg285 of the Arg-finger and Arg281 of Box VII, whereas domain IIIb (small polygon, IIIb) contains Arg334 of Sensor II. In ATP-DnaA-oriC complexes, Arg285 recognizes the neighboring DnaA-bound ATP molecule, and Arg281 supports tight inter-DnaA interactions. These events inhibit the interaction between ATP and Arg334 of Sensor II. In DnaA R281A mutant-oriC complexes, the inter-DnaA interaction is not as tight, resulting in a preponderance of the DnaA Arg334-ATP interaction. In ATP-DnaA-datA complexes, Arg281 modulates the structure of the complex and the inter-DnaA interaction is not as tight, which also allows the DnaA Arg334-ATP interaction. In ATP-DnaA-Hda complexes involved in RIDA, the DnaA-Hda-interaction is not tight, which allows ATP to interact with DnaA Arg334 and Hda Arg153.
Moreover, we found that IHF binds to datA immediately after initiation and that it dissociates from datA 20–30 min after initiation (Fig. 4C). This finding is consistent with observations that ATP-DnaA levels increase during the preinitiation stage and decrease to their basal level during replication (16, 37), and that IHF binds to oriC to promote initiation (14). From these data, we conclude that IHF is crucial for the timely activation of datA after initiation. In other words, given that IHF binds to oriC in the preinitiation stage to stimulate initiation, IHF would play both positive and negative roles in regulating initiation by alternating its binding loci (Fig. 5A and Fig. S6). This feature would be crucial for sustaining coordinated fluctuations in the activities of DnaA and oriC. It remains possible that IHF remains weakly bound to datA throughout the cell cycle, but such binding, if present, might be helpful in fine-tuning the ATP-DnaA level.
The specific requirement of DDAH for IHF, but not HU, is noteworthy: for oriC initiation, the function of IHF can be replaced by that of HU (12). Like IHF, HU induces sharp DNA bending, but its interaction with DNA is not sequence-specific (11). This difference suggests that the exact position of DNA bending is more important in DDAH complexes than in initiation complexes. DnaA binds less stably to datA than to oriC (22), which could increase the requirement for IHF binding to a specific site during the construction of nucleoprotein complexes functional in DDAH. Considering that the AAA+ motifs that support specific inter-DnaA interaction are required for full DDAH activity, we suggest that IHF binding causes DNA looping to promote inter-DnaA interactions between DnaA oligomers formed on DnaA boxes 2 and 3 (Fig. 5B). This process would result in a conformational change of DnaA and stimulation of DnaA-ATP hydrolysis (Fig. 5C). The resultant ADP-DnaA molecules would engage more weakly in cooperative binding, and therefore dissociate from datA, which would accelerate the cyclic binding of ATP-DnaA molecules to datA, leading to an efficient rate of DnaA-ATP hydrolysis.
DnaA complexes actively hydrolyze DnaA-bound ATP on datA, but not on oriC. We propose that ATP-DnaA-oriC complexes in which Arg281 supports tight inter-DnaA interactions inhibit the interaction of Sensor II Arg334 with ATP, thereby inhibiting ATP hydrolysis (Fig. 5C). Tight inter-DnaA interactions are impaired in DnaA R281A mutant-oriC complexes, allowing the interaction of Sensor II Arg334 with ATP and thereby promoting ATP hydrolysis. Similarly, the inter-DnaA interaction in the ATP-DnaA-datA complex is not as tight as that in the ATP-DnaA-oriC complex, thus permitting the interaction of Sensor II Arg334 with ATP and the promotion of ATP hydrolysis (Fig. 5C). Furthermore, Arg281 modulates the structure of the complex, thereby stimulating DDAH. In the ATP-DnaA-Hda complex of RIDA, the DnaA-Hda interaction is weak, facilitating the interaction of ATP with both Sensor II Arg334 and the Hda Arg-finger Arg153, resulting in DnaA-ATP hydrolysis (Fig. 5C).
It has been proposed that datA binds 60–370 DnaA molecules (22, 34), thereby restricting the association of DnaA molecules with oriC for replication initiation (22–25, 35). The results of this study do not completely exclude this possibility; however, the mechanism by which datA absorbs so many DnaA molecules, if indeed it does so, remains unclear. The number of DnaA molecules proposed to bind to datA in vivo (i.e., 60–370 molecules) has been deduced indirectly from experiments that analyzed the de-repression of dnaA or mioC transcription upon introduction of low- or multicopy plasmids bearing datA (22, 35). The binding of DnaA molecules to the promoter regions of these genes, which carry a DnaA box cluster, represses transcription (35). Given that ADP-DnaA oligomers formed on DNA are less stable and functional than the ATP-DnaA oligomers involved in transcriptional repression (20, 36), an increase in cellular ADP-DnaA level induced by extracopies of datA should stimulate de-repression of transcription, potentially resulting in overestimation of the number of datA-binding DnaA molecules. Moreover, the observation that an IBS-substituted datA mutant failed to repress untimely initiations, even though it sustained a basal level of multiple DnaA binding (25), is consistent with the mechanism and the in vivo significance of DDAH.
Materials and Methods
Protein, DNA, E. coli Strains, DpnII Inhibition by ATP-DnaA, and EMSA.
Protein, DNA, E. coli strains, DpnII inhibition by ATP-DnaA, and EMSA are described in the SI Materials and Methods.
In Vitro Reconstitution of DnaA-ATP Hydrolysis by datA.
[α-32P]ATP-DnaA was prepared by incubation of apo-DnaA at 0 °C for 15 min in buffer containing 3 μM [α-32P]ATP, as previously described (6). [α-32P]ATP-DnaA (1 pmol) was then incubated with various amounts of DNA or proteins, as indicated in the figure legends, in 15 µL of buffer H [20 mM Tris●HCl (pH 7.5), 100 mM potassium glutamate, 10 mM magnesium acetate, 2 mM ATP, 8 mM DTT, and 100 µg/mL BSA]. DnaA-bound nucleotides were recovered on nitrocellulose filters, extracted with HCOOH, and analyzed by TLC, as previously described (18).
In Vitro Reactivation of ADP-DnaA by DARS1.
In vitro reconstitution of DARS1 reaction was performed as previously described (31). Minichromosome replication was performed using M13KEW101 and crude extracts, as previously described (31).
ChIP and Synchronization of Replication in Vivo.
The procedures for ChIP and replication synchronization were performed as previously described (40). For details, see SI Material and Methods.
Supplementary Material
Acknowledgments
This study was supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology and the Japan Society for the Promotion of Science (Grants 22370064 and 11J03114); K.K. was supported by a predoctoral fellowship from the Japan Society for the Promotion of Science.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1212070110/-/DCSupplemental.
References
- 1.Messer W. The bacterial replication initiator DnaA. DnaA and oriC, the bacterial mode to initiate DNA replication. FEMS Microbiol Rev. 2002;26(4):355–374. doi: 10.1111/j.1574-6976.2002.tb00620.x. [DOI] [PubMed] [Google Scholar]
- 2.Katayama T, Ozaki S, Keyamura K, Fujimitsu K. Regulation of the replication cycle: Conserved and diverse regulatory systems for DnaA and oriC. Nat Rev Microbiol. 2010;8(3):163–170. doi: 10.1038/nrmicro2314. [DOI] [PubMed] [Google Scholar]
- 3.Kaguni JM. Replication initiation at the Escherichia coli chromosomal origin. Curr Opin Chem Biol. 2011;15(5):606–613. doi: 10.1016/j.cbpa.2011.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leonard AC, Grimwade JE. Regulation of DnaA assembly and activity: Taking directions from the genome. Annu Rev Microbiol. 2011;65:19–35. doi: 10.1146/annurev-micro-090110-102934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Erzberger JP, Pirruccello MM, Berger JM. The structure of bacterial DnaA: Implications for general mechanisms underlying DNA replication initiation. EMBO J. 2002;21(18):4763–4773. doi: 10.1093/emboj/cdf496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nishida S, et al. A nucleotide switch in the Escherichia coli DnaA protein initiates chromosomal replication: evidnece from a mutant DnaA protein defective in regulatory ATP hydrolysis in vitro and in vivo. J Biol Chem. 2002;277(17):14986–14995. doi: 10.1074/jbc.M108303200. [DOI] [PubMed] [Google Scholar]
- 7.Fujikawa N, et al. Structural basis of replication origin recognition by the DnaA protein. Nucleic Acids Res. 2003;31(8):2077–2086. doi: 10.1093/nar/gkg309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kawakami H, Keyamura K, Katayama T. Formation of an ATP-DnaA-specific initiation complex requires DnaA Arginine 285, a conserved motif in the AAA+ protein family. J Biol Chem. 2005;280(29):27420–27430. doi: 10.1074/jbc.M502764200. [DOI] [PubMed] [Google Scholar]
- 9.Ozaki S, Katayama T. Highly organized DnaA-oriC complexes recruit the single-stranded DNA for replication initiation. Nucleic Acids Res. 2012;40(4):1648–1665. doi: 10.1093/nar/gkr832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Felczak MM, Kaguni JM. The box VII motif of Escherichia coli DnaA protein is required for DnaA oligomerization at the E. coli replication origin. J Biol Chem. 2004;279(49):51156–51162. doi: 10.1074/jbc.M409695200. [DOI] [PubMed] [Google Scholar]
- 11.Swinger KK, Rice PA. IHF and HU: Flexible architects of bent DNA. Curr Opin Struct Biol. 2004;14(1):28–35. doi: 10.1016/j.sbi.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 12.Hwang DS, Kornberg A. Opening of the replication origin of Escherichia coli by DnaA protein with protein HU or IHF. J Biol Chem. 1992;267(32):23083–23086. [PubMed] [Google Scholar]
- 13.Grimwade JE, Ryan VT, Leonard AC. IHF redistributes bound initiator protein, DnaA, on supercoiled oriC of Escherichia coli. Mol Microbiol. 2000;35(4):835–844. doi: 10.1046/j.1365-2958.2000.01755.x. [DOI] [PubMed] [Google Scholar]
- 14.Cassler MR, Grimwade JE, Leonard AC. Cell cycle-specific changes in nucleoprotein complexes at a chromosomal replication origin. EMBO J. 1995;14(23):5833–5841. doi: 10.1002/j.1460-2075.1995.tb00271.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Waldminghaus T, Skarstad K. The Escherichia coli SeqA protein. Plasmid. 2009;61(3):141–150. doi: 10.1016/j.plasmid.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 16.Kurokawa K, Nishida S, Emoto A, Sekimizu K, Katayama T. Replication cycle-coordinated change of the adenine nucleotide-bound forms of DnaA protein in Escherichia coli. EMBO J. 1999;18(23):6642–6652. doi: 10.1093/emboj/18.23.6642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kato J, Katayama T. Hda, a novel DnaA-related protein, regulates the replication cycle in Escherichia coli. EMBO J. 2001;20(15):4253–4262. doi: 10.1093/emboj/20.15.4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakamura K, Katayama T. Novel essential residues of Hda for interaction with DnaA in the regulatory inactivation of DnaA: unique roles for Hda AAA Box VI and VII motifs. Mol Microbiol. 2010;76(2):302–317. doi: 10.1111/j.1365-2958.2010.07074.x. [DOI] [PubMed] [Google Scholar]
- 19.Riber L, Løbner-Olesen A. Coordinated replication and sequestration of oriC and dnaA are required for maintaining controlled once-per-cell-cycle initiation in Escherichia coli. J Bacteriol. 2005;187(16):5605–5613. doi: 10.1128/JB.187.16.5605-5613.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Speck C, Weigel C, Messer W. ATP- and ADP-dnaA protein, a molecular switch in gene regulation. EMBO J. 1999;18(21):6169–6176. doi: 10.1093/emboj/18.21.6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simmons LA, Breier AM, Cozzarelli NR, Kaguni JM. Hyperinitiation of DNA replication in Escherichia coli leads to replication fork collapse and inviability. Mol Microbiol. 2004;51(2):349–358. doi: 10.1046/j.1365-2958.2003.03842.x. [DOI] [PubMed] [Google Scholar]
- 22.Kitagawa R, Mitsuki H, Okazaki T, Ogawa T. A novel DnaA protein-binding site at 94.7 min on the Escherichia coli chromosome. Mol Microbiol. 1996;19(5):1137–1147. doi: 10.1046/j.1365-2958.1996.453983.x. [DOI] [PubMed] [Google Scholar]
- 23.Kitagawa R, Ozaki T, Moriya S, Ogawa T. Negative control of replication initiation by a novel chromosomal locus exhibiting exceptional affinity for Escherichia coli DnaA protein. Genes Dev. 1998;12(19):3032–3043. doi: 10.1101/gad.12.19.3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ogawa T, Yamada Y, Kuroda T, Kishi T, Moriya S. The datA locus predominantly contributes to the initiator titration mechanism in the control of replication initiation in Escherichia coli. Mol Microbiol. 2002;44(5):1367–1375. doi: 10.1046/j.1365-2958.2002.02969.x. [DOI] [PubMed] [Google Scholar]
- 25.Nozaki S, Yamada Y, Ogawa T. Initiator titration complex formed at datA with the aid of IHF regulates replication timing in Escherichia coli. Genes Cells. 2009;14(3):329–341. doi: 10.1111/j.1365-2443.2008.01269.x. [DOI] [PubMed] [Google Scholar]
- 26.Morigen , Molina F, Skarstad K. Deletion of the datA site does not affect once-per-cell-cycle timing but induces rifampin-resistant replication. J Bacteriol. 2005;187(12):3913–3920. doi: 10.1128/JB.187.12.3913-3920.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Von Freiesleben U, Rasmussen KV, Atlung T, Hansen FG. Rifampicin-resistant initiation of chromosome replication from oriC in ihf mutants. Mol Microbiol. 2000;37(5):1087–1093. doi: 10.1046/j.1365-2958.2000.02060.x. [DOI] [PubMed] [Google Scholar]
- 28.Smulczyk-Krawczyszyn A, et al. Cluster of DnaA boxes involved in regulation of Streptomyces chromosome replication: From in silico to in vivo studies. J Bacteriol. 2006;188(17):6184–6194. doi: 10.1128/JB.00528-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okumura H, et al. Regulation of chromosomal replication initiation by oriC-proximal DnaA-box clusters in Bacillus subtilis. Nucleic Acids Res. 2012;40(1):220–234. doi: 10.1093/nar/gkr716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sekimizu K, Bramhill D, Kornberg A. ATP activates dnaA protein in initiating replication of plasmids bearing the origin of the E. coli chromosome. Cell. 1987;50(2):259–265. doi: 10.1016/0092-8674(87)90221-2. [DOI] [PubMed] [Google Scholar]
- 31.Fujimitsu K, Senriuchi T, Katayama T. Specific genomic sequences of E. coli promote replicational initiation by directly reactivating ADP-DnaA. Genes Dev. 2009;23(10):1221–1233. doi: 10.1101/gad.1775809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Erzberger JP, Mott ML, Berger JM. Structural basis for ATP-dependent DnaA assembly and replication-origin remodeling. Nat Struct Mol Biol. 2006;13(8):676–683. doi: 10.1038/nsmb1115. [DOI] [PubMed] [Google Scholar]
- 33.Ozaki S, et al. A common mechanism for the ATP-DnaA-dependent formation of open complexes at the replication origin. J Biol Chem. 2008;283(13):8351–8362. doi: 10.1074/jbc.M708684200. [DOI] [PubMed] [Google Scholar]
- 34.Blaesing F, Weigel C, Welzeck M, Messer W. Analysis of the DNA-binding domain of Escherichia coli DnaA protein. Mol Microbiol. 2000;36(3):557–569. doi: 10.1046/j.1365-2958.2000.01881.x. [DOI] [PubMed] [Google Scholar]
- 35.Hansen FG, Christensen BB, Atlung T. Sequence characteristics required for cooperative binding and efficient in vivo titration of the replication initiator protein DnaA in E. coli. J Mol Biol. 2007;367(4):942–952. doi: 10.1016/j.jmb.2007.01.056. [DOI] [PubMed] [Google Scholar]
- 36.Gon S, et al. A novel regulatory mechanism couples deoxyribonucleotide synthesis and DNA replication in Escherichia coli. EMBO J. 2006;25(5):1137–1147. doi: 10.1038/sj.emboj.7600990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Katayama T, Fujimitsu K, Ogawa T. Multiple pathways regulating DnaA function in Escherichia coli: Distinct roles for DnaA titration by the datA locus and the regulatory inactivation of DnaA. Biochimie. 2001;83(1):13–17. doi: 10.1016/s0300-9084(00)01206-2. [DOI] [PubMed] [Google Scholar]
- 38.Kogoma T. Stable DNA replication: Interplay between DNA replication, homologous recombination, and transcription. Microbiol Mol Biol Rev. 1997;61(2):212–238. doi: 10.1128/mmbr.61.2.212-238.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grainger DC, Hurd D, Goldberg MD, Busby SJ. Association of nucleoid proteins with coding and non-coding segments of the Escherichia coli genome. Nucleic Acids Res. 2006;34(16):4642–4652. doi: 10.1093/nar/gkl542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cho BK, Knight EM, Barrett CL, Palsson BO. Genome-wide analysis of Fis binding in Escherichia coli indicates a causative role for A-/AT-tracts. Genome Res. 2008;18(6):900–910. doi: 10.1101/gr.070276.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.