Abstract
The cyanobacterial phylum encompasses oxygenic photosynthetic prokaryotes of a great breadth of morphologies and ecologies; they play key roles in global carbon and nitrogen cycles. The chloroplasts of all photosynthetic eukaryotes can trace their ancestry to cyanobacteria. Cyanobacteria also attract considerable interest as platforms for “green” biotechnology and biofuels. To explore the molecular basis of their different phenotypes and biochemical capabilities, we sequenced the genomes of 54 phylogenetically and phenotypically diverse cyanobacterial strains. Comparison of cyanobacterial genomes reveals the molecular basis for many aspects of cyanobacterial ecophysiological diversity, as well as the convergence of complex morphologies without the acquisition of novel proteins. This phylum-wide study highlights the benefits of diversity-driven genome sequencing, identifying more than 21,000 cyanobacterial proteins with no detectable similarity to known proteins, and foregrounds the diversity of light-harvesting proteins and gene clusters for secondary metabolite biosynthesis. Additionally, our results provide insight into the distribution of genes of cyanobacterial origin in eukaryotic nuclear genomes. Moreover, this study doubles both the amount and the phylogenetic diversity of cyanobacterial genome sequence data. Given the exponentially growing number of sequenced genomes, this diversity-driven study demonstrates the perspective gained by comparing disparate yet related genomes in a phylum-wide context and the insights that are gained from it.
The Cyanobacteria are one of the most diverse and widely distributed phyla of bacteria. Among photosynthetic prokaryotes, they uniquely have the ability to perform oxygenic photosynthesis; they are considered to be the progenitor of the chloroplast, the photosynthetic organelle found in eukaryotes. Cyanobacteria contribute greatly to global primary production, fixing a substantial amount of biologically available carbon, especially in nutrient-limited environmental niches, from oligotrophic marine surfaces to desert crusts (1, 2). In addition, cyanobacteria are key contributors to global nitrogen fixation (3), and many produce unique secondary metabolites (4). Despite these important traits and substantial interest in developing cyanobacterial strains for biotechnology, there is a paucity and unbalanced distribution of publicly available genomic information from the Cyanobacteria: 40% (29 of 72 species) of the available genomes fall within the closely related marine Prochlorococcus/Synechococcus subclade. Improvements in coverage of sequenced genomes will enable a more accurate and comprehensive understanding of cyanobacterial morphology, niche-adaptation, and evolution.
Taxonomic studies organized the Cyanobacteria into five subsections based on morphological complexity (5). Unicellular forms are split between those that undergo solely binary fission (subsection I, Chroococcales) and those that reproduce through multiple fissions in three planes to create smaller daughter cells, baeocytes (subsection II, Pleurocapsales). Strains in subsection III (Oscillatoriales) divide the vegetative cell solely perpendicular to the growing axis. Organisms in subsections IV (Nostocales) and V (Stigonematales) are able to differentiate specific cells [i.e., heterocysts (for nitrogen fixation)] and may form akinetes (dormant cells) and hormogonia (for dispersal and symbiosis competence). Subsection V is further distinguished by the ability to form branching filaments. Before this study, two subsections (II and V) had no representative genomes, underscoring the dearth in our understanding of these more complex morphological phenotypes.
In this study, 54 strains of cyanobacteria were chosen to improve the distribution of sequenced genomes. The approach is modeled on the phylogenetically driven Genomic Encyclopedia of Bacteria and Archaea (GEBA) (6), and so we refer to our data as the CyanoGEBA dataset (SI Appendix, Table S1 and Dataset S1). The results highlight the value of phylum-wide genome sequencing based on phylogenetic coverage.
Results
Increased Coverage and Diversity of Cyanobacterial Genomes.
Strains were chosen for genome sequencing according to their phylogenetic placement and their physiological relevance to the cyanobacterial research community (e.g., type strains). Beginning with a phylogenetic tree of cyanobacterial small subunit rRNA genes gathered from the greengenes database (7), cultured strains representative of major cyanobacterial branches for which genome sequences were not yet available were chosen for this study. Fifty-four genomes, sequenced using Illumina and 454 technologies, were annotated and assembled, resulting in a collective total of 332 Mb, of which 29 are complete genomes and 25 are assembled to draft genome status (SI Appendix, Table S1).
The cyanobacteria sequenced in this study cover a broad range of morphologies, lifestyles, and metabolisms. The CyanoGEBA dataset includes genomes from six baeocytous (subsection II) and five ramified (subsection V) morphotypes in addition to doubling the number of sequenced genomes from the heterocystous (11 of 18) and filamentous (19 of 29) strains. Diverse types of physiology are also encompassed in our dataset; highly halotolerant cyanobacteria (Halothece sp. PCC 7418 and Dactylococcopsis sp. PCC 8305), a fresh water picocyanobacterium (Cyanobium sp. PCC 6307), and a filamentous chlorophyll a and b containing cyanobacterium (Prochlorothrix hollandica PCC 9006) are represented at the genomic level. The CyanoGEBA data set also includes the largest cyanobacterial genome to date, Calothrix sp. PCC 7103 of 11.6 Mb.
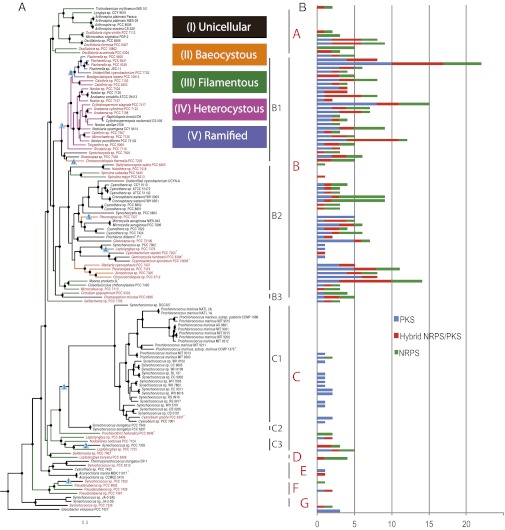
To evaluate the degree to which the 54 genomes improved coverage of the phylum, a species tree was generated using phylogenomic methods by concatenating 31 conserved proteins (8) (Fig. 1A and SI Appendix, Fig. S1). The major subclades of the cyanobacterial tree were highly congruent with the 16S rRNA phylogeny (SI Appendix, Fig. S2) and previous studies that have primarily used this molecular marker (9). A widely used method to measure the diversity in a sample is the phylogenetic diversity metric, which takes branch lengths on a phylogeny as a proxy of diversity. This study’s contribution to phylogenetic diversity was measured by the sum of the length of the 54 branches added by the CyanoGEBA genomes (10.82). To compare this value, randomly sampled subsets of 54 branches across all genomes were averaged (5.28 ± 0.37). Thus our dataset improves the diversity of the phylum approximately twofold (1.92–2.20) (SI Appendix, Table S2). A complementary method to show an improvement in coverage of the phylum is Tree Imbalance, specifically Colless’s Imbalance, which measures how equally distributed branches are on a tree. Again, we observe a decrease in tree imbalance, indicative of a more even distribution of sequenced genomes across the cyanobacterial phylum (SI Appendix, Table S2).
Fig. 1.
Cyanobacterial species tree and the distribution of secondary metabolite biosynthesis. (A) Maximum-likelihood phylogeny of cyanobacteria included in this study (outgroup shown in SI Appendix, Fig. S1). Branches are color coded according to morphological subsection. Taxa names in red are genomes sequenced in this study. Nodes supported with a bootstrap of ≥70% are indicated by a black dot. Morphological transitions that were investigated are denoted by blue triangles, annotated by events 1–8. Phylogenetic subclades are grouped into seven major subclades (A–G), some of which are made up of smaller subgroups. SI Appendix, Table S1 provides reference information for genomes used in this analysis. (B) Distribution of the nonribosomal peptide and polyketide gene clusters.
Surprisingly, of the 292,935 proteins added from this dataset, 21,107 (7.2%) have no detectable similarity to any known protein sequence. Notably, 13% of the proteins from the Leptolyngbya sp. PCC 7375 draft genome are in this sense unique proteins (SI Appendix, Table S3). Likewise, the CyanoGEBA data set contains a large number of clustered regularly interspaced short palindromic repeats (CRISPRs). Of the 54 genomes sequenced in this study, 50 contain CRISPRs (SI Appendix, Table S4); one these genomes, Geitlerinema sp. PCC 7105, contains the highest number of repeat-spacer units observed in cyanobacteria, with 650 units in a total of 15 CRISPR loci.
Morphological Complexity.
Examination of our cyanobacterial tree confirms the multiple and independent acquisition of the filamentous morphology (subsection III), as well as of the ability to form baeocytes (subsection II) (10); three unambiguous reversions and five gains in morphological complexity were revealed (Fig. 1 and SI Appendix, Table S5). Using comparative genomics, we searched for differences in the lineages bracketing these evolutionary transitions, which may represent proteins necessary for these morphological differences. Notably, there is an overlap in the sets of genes that are lost in two of the reversions from filamentous to unicellular morphology; 29 of the 32 proteins (most annotated as hypothetical) that are lost in event 2 correspond to the set of proteins lost in event 3 (SI Appendix, Table S6); this may reflect a similar convergence in the gene loss responsible for these two transitions from filamentous to unicellular phenotypes.
Surprisingly, we find no signature proteins specific to any of the complex morphologies. This also strongly argues for distinct convergences of subsections II and III morphologies. The same holds true when considering the acquisition of the ability to form branching filaments (subsection V within subclade B1). On the contrary, within the monophyletic heterocystous group within subclade B1 (subsections IV and V), the morphological differentiation may be predicated on the concomitant presence of a set of genes, such as the 12 defined for heterocyst formation (SI Appendix, Dataset S2). The ability to undergo this unique cellular differentiation may be due to the presence of regulatory proteins in a common ancestor that lacked the ability to differentiate. This is consistent with previous studies (11) that noted the presence of essential genes for heterocyst development in nonheterocystous cyanobacteria. Similarly, this could explain why several genes previously proposed to underlie other morphological attributes (e.g., hormogonium or akinete formation) (12, 13) are also found spread across the phylum, suggesting they have lineage-specific functions (SI Appendix, Dataset S2). Overall, comparison of the functional categories of Clusters of Orthologous Groups (COG) from the five morphological subsections shows that, in general, more complex morphologies are enriched in genes found in signal transduction and transcription-related functional categories (SI Appendix, Fig. S3), which may be indicative of the importance of regulatory elements in establishing morphological transitions.
Plastid Evolution.
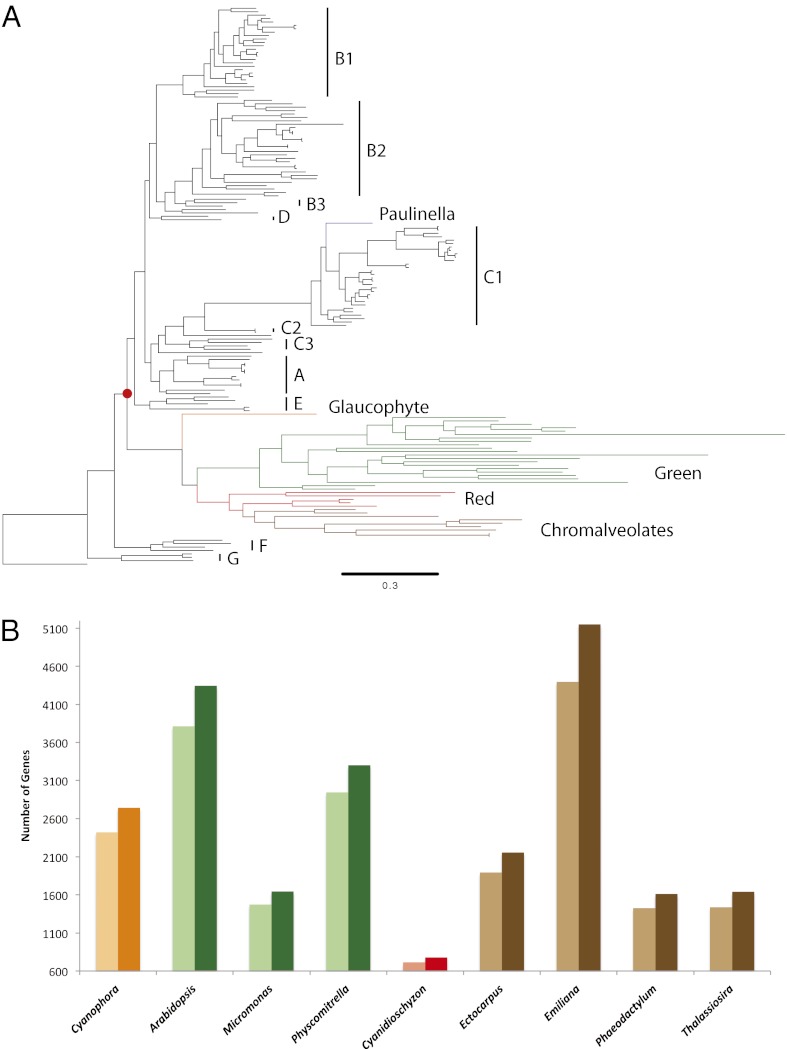
Cyanobacteria have greatly contributed to eukaryotic diversity, most notably in the plant kingdom, by giving rise to photosynthetic organelles via one or more endosymbiotic events. Many studies have attempted to find the closest relative to the original plastid endosymbiont leading to the Archaeplastida lineage. Poor phylogenetic sampling has also yielded conflicting conclusions regarding the identity of the most closely related extant cyanobacteria to the original endosymbiont; some studies claim the absence of a closest relative on the basis of phylogenetic placement, whereas other studies have suggested heterocystous cyanobacteria to be the closest relatives to plastids (14, 15). We investigated the placement of the Archaeplastida lineage within the cyanobacterial phylum by building a “plastidome tree” using a concatenation of 25 conserved plastid proteins. Although most studies support the monophyly of primary plastids (16, 17), others have reported a polyphyletic origin (18, 19). We find strong support for the monophyletic placement of plastids near the base of the cyanobacterial tree (Fig. 2A and SI Appendix, Fig. S4), as previously observed by single loci phylogenetic analysis (9, 20, 21). The short branches near this node imply a possible large radiation event that occurred near the primary endosymbiosis event, as suggested previously (15). Despite the increased coverage through the inclusion of the CyanoGEBA dataset, we cannot identify which lineage was most closely related to the original plastid endosymbiont, finding no support for the claim that heterocystous cyanobacteria are most similar to the original endosymbiont (14). Criscuolo and Gribaldo (15) have previously reported the importance of investigating, at a phylogenomic level, the relation of plastids to the deep-branching Pseudanabaena lineage represented in our clade F (Fig. 1). This clade along with a small subset of unicellular cyanobacteria (clade G) is indeed basal to the plastid branch; however, neither of these two clades represents a distinct sister lineage most closely related to plastids, which makes it difficult to propose them as the original endosymbiont. Our phylogenetic analysis does not definitively reject the hypothesis that plastids emerged from clade F. However, considering that clades A–E are monophyletic, show a sister relationship to plastids, and share a common ancestor, all extant cyanobacteria of these clades are just as closely related to modern day plastids. Given that clades A–E cover representatives of all morphological subsections, with highly diverse physiologies, it is clear that it would be difficult to predict the morphological or metabolic traits of the original endosymbiont with any certainty.
Fig. 2.
Implications on plastid evolution. (A) Maximum-likelihood phylogenetic tree of plastids and cyanobacteria, grouped by subclades (Fig. 1). The red dot (bootstrap support = 97%) represents the primary endosymbiosis event that gave rise to the Archaeplastida lineage, made up of Glaucophytes (orange), Rhodophytes (red), Viridiplantae (green), and Chromaleveolates (brown). The independent primary endosymbiosis in the amoeba Paulinella chromatophora is shown in purple. (B) Number of predicted eukaryotic, nuclear genes transferred from a cyanobacterial endosymbiont. Colors correspond to the lineage organisms as above. Light and dark shades of colors represent before and after adding the CyanoGEBA genomes, respectively.
Plastids have profoundly changed their eukaryotic hosts through endosymbiotic gene transfer (EGT), the relocation of genes from the endosymbiont genome to the host nuclear genome. Because we lack a close relative of the original endosymbiont and because the primary plastid endosymbiosis happened early within the crown cyanobacteria, only an improved coverage of cyanobacterial genomes can increase our ability to predict which genes underwent EGT. We compared predictions of EGT of nuclear genes from plastid-containing eukaryotes before and after the addition of the CyanoGEBA genomes. Nuclear proteins ascribed as the result of plastid EGT were defined as proteins with top BLAST hits from cyanobacterial, in contrast to other bacterial, archaeal, and nonplastid containing eukaryotic genomes. Given these criteria, we can now assign a cyanobacterial origin to many more genes (an average of 13% per genome) in the nuclear genomes of photosynthetic eukaryotes (Fig. 2B and SI Appendix, Tables S7 and S8, Dataset S3).
Distribution of Membrane-Bound Light-Harvesting Complexes.
Because oxygenic photosynthesis is the defining characteristic of cyanobacteria, we investigated the contribution of the CyanoGEBA dataset to surveying the diversity of photosynthetic light-harvesting strategies. The majority of cyanobacteria absorb light mainly with soluble pigment–protein complexes called phycobilisomes, in contrast to eukaryotes, which use membrane-bound light-harvesting complexes (LHCs). However, an increasing number of transmembrane proteins involved in cyanobacterial light harvesting are being identified, such as Pcb and IsiA (22, 23). These proteins are analogous in function to eukaryotic LHCs. Because of the growing number of proteins and names, an overarching nomenclature has been proposed to name this protein family the chlorophyll binding proteins (CBPs), which are characterized by six transmembrane helices and the ability to bind chlorophyll (24).
With the increase in number and diversity of genomes, we find that CBPs are widely distributed across the cyanobacterial phylum: 67% (84 of 126) of cyanobacterial genomes have, in addition to the phycobilisomes, genes that putatively function as membrane-bound light-harvesting proteins. In our phylogenetic analysis, the increase in sequence diversity reveals strong support for various subclades that we have provisionally named CBPIV, -V, and -VI (Fig. 3A and SI Appendix, Fig. S5). Although not yet experimentally demonstrated, members of CBPIV, -V, and -VI are expected to bind chlorophyll because they contain positionally conserved histidine and glutamine residues that ligate chlorophyll in confirmed chlorophyll-binding CBPs (SI Appendix, Fig. S6). Some of these proteins, such as CBPIV, have previously been annotated as PsbC homologs (25), because all CBP proteins are thought to have a common evolutionary origin with the psbC gene (24). Because of the vast enrichment of cyanobacterial protein sequences, the increase from two to six known CBPVI sequences augments phylogenetic resolution (bootstrap support of 85%), allowing us to more confidently assert that there is a separate and distinct CBPVI subfamily. On the basis of our phylogenetic analysis of the CBP family, and consistent with previous studies (26), there seems to be a substantial amount of gene duplication and horizontal gene transfer among CBPIV, -V, and -VI. In some genomes, CBPIV and CBPV are found in a gene cluster with other CBP proteins, including IsiA (Fig. 3C), suggestive of the potential for lateral transfer of gene clusters encoding light-harvesting proteins, as documented in marine cyanobacteria (27). Interestingly, many proteins of the CBPV clade also contain a C-terminal extension (SI Appendix, Fig. S7) with homology to the PsaL subunit of photosystem I (PSI). Notably, two distinct subclades within the CBPV family seem to have independently lost the PsaL domains, reflecting the modularity of this C-terminal extension. Homology modeling and insertion of the PsaL-like domain into the PSI structure (Fig. 3B and SI Appendix, Fig. S8) suggests how the CBPV protein could theoretically be incorporated as an ancillary light-harvesting polypeptide into a monomeric, but not trimeric, PSI. Although scattered observations of members of these CBP protein clades have been made in previously sequenced genomes [predominantly IsiA (CBPIII) and Pcb genes (dCBP, CBPI, and CBPII)] it is clear that the contribution the 54 genomes included in this study substantially increase the number of homologs within the CBP family, allowing for a more thorough understanding of the distribution of distinct subclades within this large membrane-bound light-harvesting protein family.
Fig. 3.
Increased sequence coverage reveals distinct and highly supported subclades of putative CBPs. (A) Unrooted maximum-likelihood tree of CBP sequences. Putative CBP clades that have emerged as distinct and phylogenetically well supported are labeled in red, and previously described CBP clades are labeled in black. CP43 protein sequences (encoded by the PsbC gene) are provided as an outgroup. (B) Cartoon representation of unique domain architecture of CBPV from Chroococcidiopsis thermalis PCC 7203 (Chro_2988), based on the two separate homology models of (i) the N-terminal CBP domain (red) and (ii) the C-terminal PsaL-like domain (yellow). Potentially chlorophyll-binding histidine residues are shown in green sticks. (C) Gene cluster containing multiple CBP genes from Anabaena cylindrica PCC 7122 (locus tags labeled above, annotations labeled below).
Secondary Metabolite Analysis.
Much of the natural product chemical diversity observed in nature is attributed to versatile nonribosomal peptide synthase (NRPS) and polyketide synthase (PKS) biosynthetic pathways (4). However, the extent and distribution of the capacity for secondary metabolite synthesis in cyanobacteria has nevertheless been underestimated. We retrieved 384 nonribosomal gene clusters from 126 genomes, 61% from the CyanoGEBA dataset. Our results reveal that 70% of cyanobacterial genomes encode NRPS or PKS gene clusters (Fig. 1B and SI Appendix, Fig. S9); their presence is partly correlated to the genome size (Pearson correlation on total number of NRPS/PKS gene clusters, or on total KS domains, as well as on total C domains: R2 = 0.3, P < 0.0001). Moreover, the distribution is uneven by a skewed frequency of NRPS/PKS in the late branches of our cyanobacterial tree (clades A and B), including all genomes from baeocystous, heterocystous, and ramified morphotypes. Notably, 5.2% of the Fischerella sp. PCC 9339 genome is devoted to NRPS/PKS clusters and contains an unexpected diversity with up to 22 NRPS/PKS clusters (5 NRPS, 10 PKS, and 7 NRPS/PKS hybrids). Although the PCC 9339 genome is in a draft stage, nine of these clusters are located at a contig border and thus partial. Most of the clusters await characterization; however, the potential for the production of microcystins, shinorine, or heterocyst glycolipids can already be predicted (SI Appendix, Fig. S10).
Likewise, gene clusters involved in ribosome-dependent synthesis of diverse peptides through the posttranslational modification of short precursor proteins (28–30) are even more broadly distributed across the phylum (SI Appendix, Fig. S9). The most abundant corresponds to the newly discovered bacteriocin family (30, 31), whereas the terpenes (32) are present in almost all of the 126 genomes. Even the genes encoding cyanobactins (33) were recovered from 10% of the dataset. Strikingly, the Prochlorococcus/Synechococcus subclade seems to lack NRPS gene clusters and harbors only type III PKS; however, they contain an abundance of bacteriocin clusters.
Discussion
With the exponentially growing capacity for sequencing genomes it is becoming increasingly important to focus sequencing efforts so as to obtain a high-value return. Here, we show the benefits of genome sequencing based on a more representative phylogenetic coverage, with the objective of better understanding general characteristics of the phylum, as well as uncovering unique and novel traits of cyanobacterial genomes and subclades.
The addition of the CyanoGEBA genomes lays the foundation for the cyanobacterial phylum to become a model comparative genomic system for understanding the gain and loss of morphological complexity. Given the close relationship between morphology and taxonomy for the Cyanobacteria, the genome sequence data now available from all five morphological subsections have revealed the lack of specific and unique genes that are the genetic determinants underlying these major phenotypes; a similar result emerged from comparative studies of eukaryotic genomes (34).
An increased distribution of sequenced cyanobacterial genomes has also corrected previous biases, such as the limited occurrence and diversity among CBPs. The addition of the CyanoGEBA dataset clearly shows that two-thirds of cyanobacterial genomes actually have membrane-bound CBPs encoded in their genomes, potentially allowing for alternative light-harvesting strategies other than phycobilisomes. Furthermore, the addition of these diverse CBPs has also enabled the placement of phylogenetically well-supported and distinct CBP subclades.
Our results likewise reveal an unexpectedly high frequency and diversity of NRPS/PKS enzyme systems for the production of secondary metabolites. Furthermore, we found that the known ribosomal-dependent pathways for production of small peptides are also frequent and found throughout the lineage. Cyanobacteria have thus adopted multiple parallel strategies for the production of peptides through the modification of short precursors. Ultimately, their chemical diversity may rival or exceed that of the better-known nonribosomal peptides and polyketides. The increased diversity of NRPS/PKS genes now apparent in the cyanobacterial phylum emphasizes one of the many benefits that are gained when using diversity-driven genome sequencing, which the previously biased genome representation of cyanobacteria failed to reveal.
Despite the global importance of the Cyanobacteria, there has been an unbalanced sequence distribution of the phylum, resulting in a lack of understanding at a genome level of major clades and morphological subsections. The extensive phylogenetically based survey of this single phylum has refined and extended our understanding of plastid evolution, phenotypic differences in morphology, light-harvesting complexes, and secondary metabolisms in cyanobacteria. This study demonstrates the benefits gained from a more balanced representation of sequenced genomes within a phylum.
Materials and Methods
Genome Sequencing and Assembly.
The 54 CyanoGEBA genomes were generated at the US Department of Energy DOE Joint Genome Institute (JGI) using either a combination of Illumina (35) and 454 technologies (36) or only the Illumina technology (SI Appendix, Table S9). Sequence data were assembled using an array of assemblers pending the data generated for a given genome. Assemblers included Newbler, Velvet, and parallel Phrap (High Performance Software). The software Consed was used in the finishing process.
Phylogenetic Analysis.
The species tree was generated by a concatenation of 31 conserved proteins (8). The plastidome tree was generated the same way using 25 conserved plastid proteins (atpH, atpA, atpB, petB, psbA, psbB, psbC, psbD, psbE, psbL, psbH, psaA, psaB, psaC, rpl2, rpl14, rpl16, rps2, rps3, rps4, rps7, rps11, rps19, rpoB, and rpoC2). Maximum-likelihood phylogenetic trees were generated with PhyML 3.0 (37).
Measuring Improved Phylum Sampling.
The phylogenetic diversity metric was measured as described by Wu et al. (6). A maximum-likelihood tree omitting the four outgroup genomes with 51 resamplings of the random set of taxa was used to estimate the contribution of the CyanoGEBA genomes in increasing the phylogenetic diversity of the overall tree. Methods used for the Tree Imbalance analysis are described in SI Appendix.
Comparative Genomic Analysis.
An “all vs. all” BLASTP search was conducted for all cyanobacterial proteins used in this study with an e-value threshold of 1e-10 and a span cutoff of 80%, which was then used to build protein families using the Markov clustering algorithm (MCL), whereby each cluster was considered a protein family (38). Comparative genomics to characterize the protein families lost or gained in specific morphological lineages were based on the MCL protein families.
Prediction of Endosymbiotic Gene Transfer.
Nuclear-encoded proteins from plastid-containing eukaryotes (SI Appendix, Tables S7 and S8) were used as queries to BLASTP against two databases: (i) containing all cyanobacteria, representatives from other bacterial and archaeal phyla, and representatives from nonplastid containing eukaryotes, and (ii) the same as above but using only cyanobacterial genomes available before the CyanoGEBA study. Top-BLAST hits to cyanobacterial proteins were considered genes of cyanobacterial descent, and the total counts for each of the nuclear genomes are presented in SI Appendix, Table S7 and Dataset S3.
Secondary Metabolite Analysis.
Secondary metabolite biosynthesis gene clusters were identified using met2db (39), antiSMASH (40), and NaPDoS (41). Adenylation domain substrate specificity predictions for NRPS enzymes were made using NRPSpreditor2 (42). Annotations were refined manually using CD-search, BLASTP, and InterProScan to identify conserved domains. We estimated the number of gene clusters for each genome using the three methods and containing the minimum of domains needed to perform synthesis. Pearson correlation tests were performed using XLSTAT, v2007-4 (Addinsoft).
Supplementary Material
Acknowledgments
We thank the following people for assistance in aspects of the project including data management, analyses not included here, and discussions: J. Bristow, K. Mavramotis, N. C. Kyrpides, J. Cameron, M. Sutter, D. Stanley, O. Erbilgin, J. Zarzycki, J. Kinney, P. Schwientek, S. Prochnik, and K. K. Niyogi. The work conducted by the US Department of Energy Joint Genome Institute is supported by the Office of Science of the US Department of Energy under Contract DE-AC02-05CH11231. Funding was also provided by the Institut Pasteur, and the Centre National de la Recherche Scientifique Unité de Recherche Associée 2172 are acknowledged for funding. P.M.S. and C.A.K. were also supported by National Science Foundation Grant MCB-0851070. All cyanobacteria represented by genomes sequenced in this study are available from the Institut Pasteur.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the GenBank database (accession nos. CP003610–CP003613, CP003659–CP003665, CP003600, CP003601, CP003602, CP003642–CP003645, CP003646–CP003650, CP003630–CP003638, CP003552, CP003553, CP003554, CP003614–CP003619, CP003590, CP003653–CP003658, CP003594, CP003595, CP003596, ALVM00000000, ALVP00000000, ALVQ00000000, ALWD00000000, ANFQ00000000, ANFJ00000000, ALVJ00000000, CP003495, CP003591, CP003548, CP003592, CP003593, CP003549, CP003550, CP003551, CP003558, CP003559, AJWF00000000, ALVO00000000, ALVI00000000, ALVL00000000, ALVK00000000, ALWB00000000, ALWC00000000, ALWA00000000, ALVY00000000, ALVV00000000, ALVU00000000, ALVZ00000000, ALVN00000000, ALVR00000000, ALVS00000000, ALVW00000000, ANKN00000000, ANKO00000000, ALVJ00000000 and CPOO3940). Genome sequence and annotation data are available at the Joint Genome Institute Integrated Microbial Genomes database, http://img.jgi.doe.gov/cgi-bin/w/main.cgi.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1217107110/-/DCSupplemental.
References
- 1.Partensky F, Hess WR, Vaulot D. Prochlorococcus, a marine photosynthetic prokaryote of global significance. Microbiol Mol Biol Rev. 1999;63(1):106–127. doi: 10.1128/mmbr.63.1.106-127.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garcia-Pichel F, Belnap J, Neuer S, Schanz F. Estimates of global cyanobacterial biomass and its distribution. Algol Stud. 2003;109:213–227. [Google Scholar]
- 3.Zehr JP, et al. Globally distributed uncultivated oceanic N2-fixing cyanobacteria lack oxygenic photosystem II. Science. 2008;322(5904):1110–1112. doi: 10.1126/science.1165340. [DOI] [PubMed] [Google Scholar]
- 4.Welker M, von Döhren H. Cyanobacterial peptides—nature’s own combinatorial biosynthesis. FEMS Microbiol Rev. 2006;30(4):530–563. doi: 10.1111/j.1574-6976.2006.00022.x. [DOI] [PubMed] [Google Scholar]
- 5.Rippka R, Deruelles J, Waterbury JB, Herdman M, Stanier RY. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J Gen Microbiol. 1979;111:1–61. [Google Scholar]
- 6.Wu D, et al. A phylogeny-driven genomic encyclopaedia of Bacteria and Archaea. Nature. 2009;462(7276):1056–1060. doi: 10.1038/nature08656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeSantis TZ, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72(7):5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu M, Eisen JA. A simple, fast, and accurate method of phylogenomic inference. Genome Biol. 2008;9(10):R151. doi: 10.1186/gb-2008-9-10-r151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turner S, Pryer KM, Miao VPW, Palmer JD. Investigating deep phylogenetic relationships among cyanobacteria and plastids by small subunit rRNA sequence analysis. J Eukaryot Microbiol. 1999;46(4):327–338. doi: 10.1111/j.1550-7408.1999.tb04612.x. [DOI] [PubMed] [Google Scholar]
- 10.Schirrmeister BE, Antonelli A, Bagheri HC. The origin of multicellularity in cyanobacteria. BMC Evol Biol. 2011;11:45. doi: 10.1186/1471-2148-11-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang J-Y, Chen W-L, Zhang C-C. hetR and PatS, two genes necessary for heterocyst pattern formation, are widespread in filamentous nonheterocyst-forming cyanobacteria. Microbiology. 2009;155(Pt 5):1418–1426. doi: 10.1099/mic.0.027540-0. [DOI] [PubMed] [Google Scholar]
- 12.Campbell EL, Wong FCY, Meeks JC. DNA binding properties of the HrmR protein of Nostoc punctiforme responsible for transcriptional regulation of genes involved in the differentiation of hormogonia. Mol Microbiol. 2003;47(2):573–582. doi: 10.1046/j.1365-2958.2003.03320.x. [DOI] [PubMed] [Google Scholar]
- 13.Zhou R, Wolk CP. Identification of an akinete marker gene in Anabaena variabilis. J Bacteriol. 2002;184(9):2529–2532. doi: 10.1128/JB.184.9.2529-2532.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deusch O, et al. Genes of cyanobacterial origin in plant nuclear genomes point to a heterocyst-forming plastid ancestor. Mol Biol Evol. 2008;25(4):748–761. doi: 10.1093/molbev/msn022. [DOI] [PubMed] [Google Scholar]
- 15.Criscuolo A, Gribaldo S. Large-scale phylogenomic analyses indicate a deep origin of primary plastids within cyanobacteria. Mol Biol Evol. 2011;28(11):3019–3032. doi: 10.1093/molbev/msr108. [DOI] [PubMed] [Google Scholar]
- 16.Rodríguez-Ezpeleta N, et al. Monophyly of primary photosynthetic eukaryotes: Green plants, red algae, and glaucophytes. Curr Biol. 2005;15(14):1325–1330. doi: 10.1016/j.cub.2005.06.040. [DOI] [PubMed] [Google Scholar]
- 17.Chan CX, Gross J, Yoon HS, Bhattacharya D. Plastid origin and evolution: New models provide insights into old problems. Plant Physiol. 2011;155(4):1552–1560. doi: 10.1104/pp.111.173500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nozaki H, et al. Phylogenetic positions of Glaucophyta, green plants (Archaeplastida) and Haptophyta (Chromalveolata) as deduced from slowly evolving nuclear genes. Mol Phylogenet Evol. 2009;53(3):872–880. doi: 10.1016/j.ympev.2009.08.015. [DOI] [PubMed] [Google Scholar]
- 19.Baurain D, et al. Phylogenomic evidence for separate acquisition of plastids in cryptophytes, haptophytes, and stramenopiles. Mol Biol Evol. 2010;27(7):1698–1709. doi: 10.1093/molbev/msq059. [DOI] [PubMed] [Google Scholar]
- 20.Marin B, Nowack EC, Melkonian M. A plastid in the making: Evidence for a second primary endosymbiosis. Protist. 2005;156(4):425–432. doi: 10.1016/j.protis.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 21.Nelissen B, Van de Peer Y, Wilmotte A, De Wachter R. An early origin of plastids within the cyanobacterial divergence is suggested by evolutionary trees based on complete 16S rRNA sequences. Mol Biol Evol. 1995;12(6):1166–1173. doi: 10.1093/oxfordjournals.molbev.a040289. [DOI] [PubMed] [Google Scholar]
- 22.La Roche J, et al. Independent evolution of the prochlorophyte and green plant chlorophyll a/b light-harvesting proteins. Proc Natl Acad Sci USA. 1996;93(26):15244–15248. doi: 10.1073/pnas.93.26.15244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laudenbach DE, Straus NA. Characterization of a cyanobacterial iron stress-induced gene similar to psbC. J Bacteriol. 1988;170(11):5018–5026. doi: 10.1128/jb.170.11.5018-5026.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen M, Zhang Y, Blankenship RE. Nomenclature for membrane-bound light-harvesting complexes of cyanobacteria. Photosynth Res. 2008;95(2-3):147–154. doi: 10.1007/s11120-007-9255-0. [DOI] [PubMed] [Google Scholar]
- 25.Kaneko T, et al. Complete genomic sequence of the filamentous nitrogen-fixing cyanobacterium Anabaena sp. strain PCC 7120. DNA Res. 2001;8(5):205–213, 227–253. doi: 10.1093/dnares/8.5.205. [DOI] [PubMed] [Google Scholar]
- 26.Chen M, Hiller RG, Howe CJ, Larkum AWD. Unique origin and lateral transfer of prokaryotic chlorophyll-b and chlorophyll-d light-harvesting systems. Mol Biol Evol. 2005;22(1):21–28. doi: 10.1093/molbev/msh250. [DOI] [PubMed] [Google Scholar]
- 27.Rocap G, et al. Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature. 2003;424(6952):1042–1047. doi: 10.1038/nature01947. [DOI] [PubMed] [Google Scholar]
- 28.Schmidt EW, et al. Patellamide A and C biosynthesis by a microcin-like pathway in Prochloron didemni, the cyanobacterial symbiont of Lissoclinum patella. Proc Natl Acad Sci USA. 2005;102(20):7315–7320. doi: 10.1073/pnas.0501424102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ziemert N, et al. Microcyclamide biosynthesis in two strains of Microcystis aeruginosa: from structure to genes and vice versa. Appl Environ Microbiol. 2008;74(6):1791–1797. doi: 10.1128/AEM.02392-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li B, et al. Catalytic promiscuity in the biosynthesis of cyclic peptide secondary metabolites in planktonic marine cyanobacteria. Proc Natl Acad Sci USA. 2010;107(23):10430–10435. doi: 10.1073/pnas.0913677107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang H, Fewer DP, Sivonen K. Genome mining demonstrates the widespread occurrence of gene clusters encoding bacteriocins in cyanobacteria. PLoS ONE. 2011;6(7):e22384. doi: 10.1371/journal.pone.0022384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Agger SA, Lopez-Gallego F, Hoye TR, Schmidt-Dannert C. Identification of sesquiterpene synthases from Nostoc punctiforme PCC 73102 and Nostoc sp. strain PCC 7120. J Bacteriol. 2008;190(18):6084–6096. doi: 10.1128/JB.00759-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sivonen K, Leikoski N, Fewer DP, Jokela J. Cyanobactins-ribosomal cyclic peptides produced by cyanobacteria. Appl Microbiol Biotechnol. 2010;86(5):1213–1225. doi: 10.1007/s00253-010-2482-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prochnik SE, et al. Genomic analysis of organismal complexity in the multicellular green alga Volvox carteri. Science. 2010;329(5988):223–226. doi: 10.1126/science.1188800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bennett S. Solexa Ltd. Pharmacogenomics. 2004;5(4):433–438. doi: 10.1517/14622416.5.4.433. [DOI] [PubMed] [Google Scholar]
- 36.Margulies M, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437(7057):376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst Biol. 2010;59(3):307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 38.Enright AJ, Van Dongen S, Ouzounis CA. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30(7):1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bachmann B, Ravel J. Chapter 8. Methods for In silico prediction of microbial secondary metabolic pathways from DNA sequence data. Methods Enzymol. 2009;458:181–217. doi: 10.1016/S0076-6879(09)04808-3. [DOI] [PubMed] [Google Scholar]
- 40.Medema MH, et al. antiSMASH: Rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 2011;39(Web Server issue):W339-346. doi: 10.1093/nar/gkr466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ziemert N, et al. The natural product domain seeker NaPDoS: A phylogeny based bioinformatic tool to classify secondary metabolite gene diversity. PLoS ONE. 2012;7(3):e34064. doi: 10.1371/journal.pone.0034064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Röttig M, et al. NRPSpredictor2–a web server for predicting NRPS adenylation domain specificity. Nucleic Acids Res. 2011;39(Web Server issue):W362-367. doi: 10.1093/nar/gkr323. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.