Abstract
In higher organisms such as vertebrates, it is generally believed that lateral transfer of genetic information does not readily occur, with the exception of retroviral infection. However, horizontal transfer (HT) of protein coding repetitive elements is the simplest way to explain the patchy distribution of BovB, a long interspersed element (LINE) about 3.2 kb long, that has been found in ruminants, marsupials, squamates, monotremes, and African mammals. BovB sequences are a major component of some of these genomes. Here we show that HT of BovB is significantly more widespread than believed, and we demonstrate the existence of two plausible arthropod vectors, specifically reptile ticks. A phylogenetic tree built from BovB sequences from species in all of these groups does not conform to expected evolutionary relationships of the species, and our analysis indicates that at least nine HT events are required to explain the observed topology. Our results provide compelling evidence for HT of genetic material that has transformed vertebrate genomes.
Keywords: transposon, interspersed repeat
Repetitive DNA is abundant in metazoan genomes and is largely composed of transposable elements (TEs). Retrotransposons are a class of TEs that are able to “copy and paste” themselves within the genome via an RNA intermediate (1). Long interspersed element (LINE) retrotransposons encode an endonuclease that nicks the DNA and allows the reverse transcriptase encoded by the element to copy the RNA produced from the TE back into DNA during repair of the nick, integrating the LINE into a new genomic position (2). However, unlike retroviruses, LINEs and other TEs do not encode an envelope protein and are hence unable to disperse horizontally without a vector between species.
Horizontal transfer (HT) of TEs is largely inferred by similarity of DNA sequence; however, where the mechanism of HT has been demonstrated, a vector such as a parasite or virus was involved. For example, both P elements, between species of Drosophila (3), and the Space Invader DNA transposon, between tetrapods (4, 5), are transmitted by arthropod parasites (5, 6). The Sauria short interspersed element (SINE), has been shown to have transferred into a West African rodent poxvirus from the snake, Echis ocellatus, also supporting viruses as mechanisms for retrotransposon HT (7). HT of retrotransposons is significant because conservative estimates of their prevalence indicate that they make up between a third and a half of typical vertebrate genomes. Thus, demonstration of widespread HT for retrotransposons has significant implications for our understanding of genome structure and evolution. In this report we describe a comprehensive analysis of HT of BovB, a LINE about 3.2 kb long, which has previously been described in ruminants, marsupials, squamates, monotremes, and African mammals (8–11).
Results and Discussion
To determine the sequence conservation of BovB across taxa and examine the evidence for HT, we identified BovB sequences in all publicly available genomes and in several low coverage genomic survey (454 shotgun) sequences using RepBase (12) consensus sequences for BovB as BLAST (13) queries (SI Appendix, Tables S2–S4). The BovB sequences available in Repbase (12) include a sequence extracted from the horn-nosed viper (Vipera ammodytes) BovB VA, that contains Chicken Repeat 1 (CR1) elements on both the 3′ and 5′ ends (SI Appendix, Fig. S4). This means that early during its colonization of the squamates, it somehow acquired the CR1 sequences now present at both ends. We used a trimmed version of BovB VA in our sequence similarity searches, noting that use of the untrimmed RepBase BovB VA sequence leads to false discovery in birds and nonsquamate reptiles, as reported in turtles and the tuatara (10) (basal to squamates), which have an abundance of CR1 elements.
Other squamates may have CR1 fragments on their BovB consensus sequences too. However, due to the abundance of CR1 in the squamate genomes and the low coverage reads from which the squamate BovBs were built, all CR1 fragments had to be removed to reliably assemble a BovB consensus from those species. Hence additional sequencing in a greater range of reptiles would be required to determine when CR1 ends were acquired by the squamate BovB lineage. Interestingly the BovB sequences for the python and the copperhead that were extracted from RepBase do not have the CR1-like ends that are present in BovB VA. This could be due to a different repeat building process used by Castoe et al. (14).
Our searches revealed that BovB is highly abundant in cow, sheep, and Afrotheria (basal mammals), with significant portions of these genomes resulting from BovB contribution (Table 1). BovB is thus capable of significantly altering genome structure and therefore function. BovB sequences contribute to more than 1% of anole, opossum, platypus, and wallaby genomes but only exist as relatively few copies in the horse, sea urchin, silkworm, and zebrafish. BovB was not found in the tuatara, turtles, birds, or other mammals. BovB was also not found in mosquitos despite the presence of an RTE element (SI Appendix, Table S3).
Table 1.
Percentage of genome sequence contributed by BovB
| Clade | Species common name | BovB Coverage |
| Monotreme | Platypus | 1.21 |
| Marsupial | Opossum | 1.3 |
| Ruminant | Cow | 18.37 |
| Sheep | 15.21 | |
| Equid | Horse | 0.11 |
| Afrotheria | Elephant | 11.41 |
| Rock Hyrax | 6.86 | |
| Tenrec | 8.12 | |
| Reptile | Anole | 1.36 |
Genome Coverage: Table shows the percentage of the genome that masks as BovB using full-length BovB sequences as the library in RepeatMasker. Note that this is an underestimate of the impact on the genome, as it does not take into account sequences in BovB SINE derived from other sources. In the case of the cow, the total percentage of the genome attributable to BovB and derived SINE would be 25%.
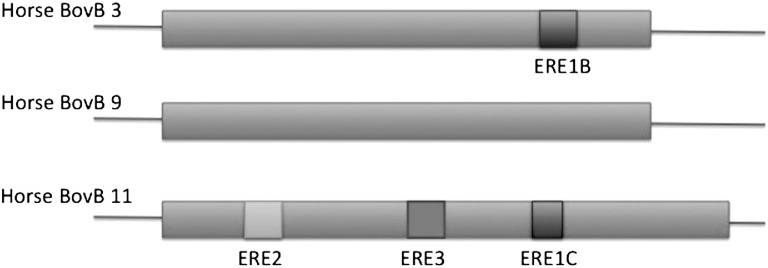
Within the horse genome, just 31 regions were extracted by LASTZ (15) when searched for 80% coverage of the BovB query sequence. We checked to ensure that this was not contamination by searching for 5′-truncated BovB sequences in the genome, which we expected to find if the reverse transcription step of the copy-and-paste movement was truncated due to premature termination. We were able to find >100 5′-truncated BovB per chromosome, indicating that BovB has been undergoing transcription, reverse transcription, and insertion, but at a much more limited scale than in ruminants or afrotheria. Finally, the presence of horse-specific SINEs inserted into some of the full-length horse BovBs indicated that BovB has been present in the horse genome for some time (Fig. 1).
Fig. 1.
SINEs inserted into BovB in the horse genome. This is a visual representation of 3 of 31 nearly full-length horse BovBs according to RepeatMasker (31) using the RepBase (12) horse repeat library. Mid-gray rectangles indicate masking as RTE-1 EC, which is the RepBase equivalent of the horse BovB consensus sequence we constructed. Square gray boxes represent the presence of horse ERE SINE sequences.
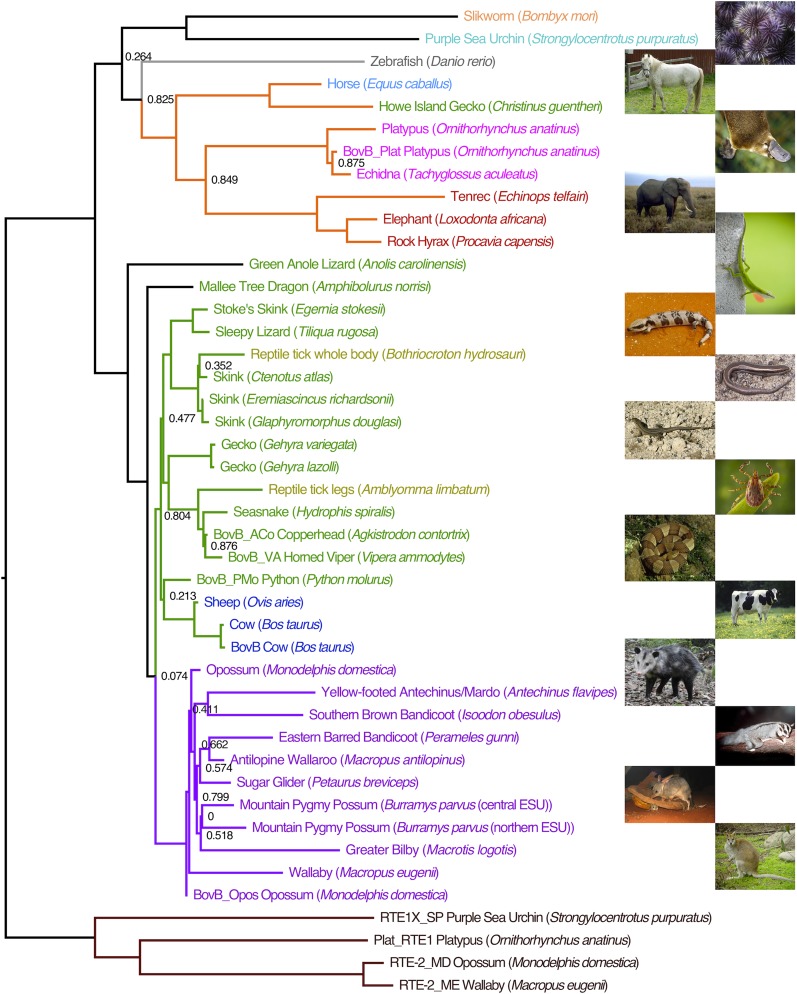
We constructed a consensus from the BovB sequences recovered from each species where possible and conducted phylogenetic analyses using both maximum likelihood (Fig. 2) and Bayesian methods (SI Appendix, Figs. S9 and S10). Both methodologies gave similar tree topologies varying only in the placement of the zebrafish, silkworm, and sea urchin sequences. Excluding these, the consensus sequences were resolved into two major clades of BovB (Fig. 2).
Fig. 2.
Phylogenetic tree of BovB sequences showing the distribution of BovB across taxa. Maximum likelihood tree built using FastTree (35) from the full-length BovB sequences extracted from full genome sequence and those constructed from low coverage reads. The sequences were aligned with MUSCLE (29) and processed by Gblocks (34) to limit the effect of indels, making an alignment that was 2858 bp long. Local support values are only shown for those nodes with support less than 0.9. Branch colors indicate important BovB clades: marsupials in purple, reptiles/ruminants/ticks in green, monotremes/Afrotheria/horse in orange, and the RTE clade in maroon, used to root the tree. Taxa showing BovB are colored taxonomically, with marsupials in purple, reptiles in green, ruminants in dark blue, arthropods in yellow, Afrotheria in red, monotremes in pink, horse in blue, zebrafish in gray, sea urchin in light blue, and silkworm in orange. The RTEs are in maroon.
The largest clade comprised BovB consensus sequences from the marsupials, ruminants, ticks, and all but one of the squamates examined. Whereas the marsupials robustly grouped together, the resolution within the clade was too low to allow analysis of HT between marsupials. However, as no nonmarsupials are present in this clade, we have concluded that it is likely that BovB was present in the common ancestor of marsupials and potentially no HT has occurred since the divergence of marsupials from other mammals. Analyses with additional taxa will be required to test this hypothesis.
The BovB sequences constructed for the two reptile tick species (Bothriocroton hydrosauri and Amblyomma limbatum) nested within the squamate clade. Although the two tick species were collected from the same host (Tiliqua rugosa), they contributed independent BovB sequences to this analysis, neither of which clustered with the BovB sequences from the host. Both species feed on a diverse range of squamates (16) and the potential exists for contamination from the nucleated red blood cells of the lizard in their gut. For this reason, A. limbatum tick legs were sequenced to remove the potential for contamination. Although contamination is a concern, and can come from various sources, we do not believe it affects our results. The DNA samples for 454 sequencing came from a number of different laboratories and samples were not extracted in one laboratory, or by one person. Pre-PCR and PCR steps were carried out in different laboratories to prevent contamination. Furthermore, the species where we have identified BovB were not amplified/sequenced together but were amplified/sequenced in conjunction with samples where BovB was not detected. If contamination were an issue one would expect the pattern of occurrence to be random, not lineage specific, e.g., all marsupials, reptiles. We describe our controls for false BovB hits in SI Appendix, section 1.7.3. We have also directly tested for contamination by PCR amplifying and sequencing BovB from a subset of critical taxa: horse, both tick species, and the Lord Howe Island Gecko (Christinus guentheri). We were able to validate BovB in these species using freshly extracted DNA from independent specimens that were not sourced from the laboratories where the original samples were obtained, and we describe our methods and report representative results in SI Appendix, section 3.6. Sequences of validation samples have been deposited with GenBank.
It is also important to note that the topology seen in the squamate BovB subtree is not the topology expected from a tree built from gene orthologs or fossil records (SI Appendix, Fig. S8). This indicates that BovB has been moving horizontally among the squamates as well as between them, ruminants, and marsupials.
The second major BovB clade includes monotremes, African mammals, the horse, and one species of gecko. The Lord Howe Island gecko appeared to have two subclasses of BovB during the consensus construction process, but only one subclass was deemed of sufficient quality to use in phylogenetic analysis. To get a suitable quality sequence for phylogenetic analysis of the other subclass, significantly more data would be required to build the other BovB subclass in this gecko. There is no suggestion of a vector at present and more widespread sequencing would be required to find a parasite or virus vector that would facilitate the HT of the BovB within this clade. The BovB from the African mammals displayed the relationship expected when building a tree from orthologous sequences, which implies that BovB was present in the common ancestor of Afrotheria and has not moved horizontally between African mammals since its incorporation in the ancestral afrotherian genome.
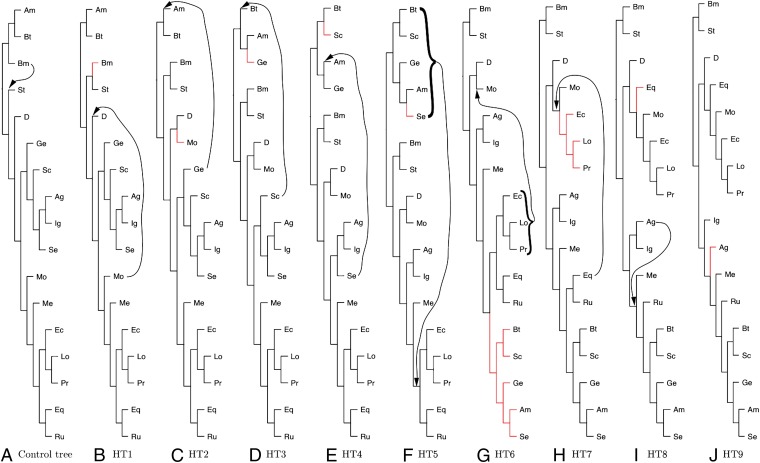
We compared the tree constructed from BovB sequences to the tree constructed from protein orthologs in OrthoDB: Database of Orthologous Groups (17), and TimeTree of Life data (18) using the program SPRIT (19), that estimates the number of required subtree prune and regrafts (SPR) to transform one tree into another. It is apparent from the representation of SPRIT output shown in Fig. 3 that nine SPR are required to explain the observed BovB-based topology. Each SPR corresponds to at least one HT event, therefore we conclude that at least nine interspecies HT events have occurred during the evolutionary history of BovB. This is significantly more than previous estimates of one or two (9, 20) and could increase with the inclusion of new taxa and higher quality data that refines the position of taxa on the BovB and protein ortholog trees.
Fig. 3.
Horizontal transfers. This is a representation of the least number of subtree prune and regrafts (SPR) required to turn the control tree built from protein orthologs (A), into the tree built from the BovB sequences (J) through intermediates B–I. The movement that corresponds to the SPR in the next tree is shown by the arrow and the SPR that made the current tree is shown in red. D, Danio rerio; Eq, Equus caballus; Mo, Monotremata; Ec, Echinops telfairi; Lo, Loxodonta africana; Pr, Procavia capensis; Ig, Iguanidae; Me, Metatheria; Ag, Agamidae; Ru, Ruminantia; Bt, Bothriocroton hydrosauri; Sc, Scincidae; Ge, Gekkonidae; Am, Amblyomma limbatum; Se, Serpentes; Bm, Bombyx mori; and St, Strongylocentrotus purpuratus.
BovB is capable of expanding within a diverse range of species including warm- and cold-blooded animals and shows a large variability in its accumulation of substitutions in different species; showing a low number of substitutions per site in the anole and a very high number in the opossum (Table 2).
Table 2.
Number of substitutions per site
| Species common name | Substitutions per site (∼) |
| Opossum | 0.357 ± 0.006 |
| Cow | 0.110 ± 0.002 |
| Sheep | 0.228 ± 0.004 |
| Horse | 0.229 ± 0.003 |
| Elephant | 0.150 ± 0.003 |
| Anole | 0.076 ± 0.002 |
| Sea Urchin | 0.322 ± 0.014 |
MEGA was used to compute overall mean distances for the nearly full-length BovBs of a selection of species. The Jukes-Cantor model was used due to its lack of inherent assumptions with gamma distribution and 90% partial deletion of missing data.
The analysis of BovB HT revealed that ticks may have transferred DNA between snakes and lizards and into ruminants and marsupials. Although we cannot identify the exact tick species, it is known that species of Amblyomma and Bothriocroton infest mammals, marsupials, and monotremes, and that Amblyomma sp. are highly important parasites of domestic animals and man in Africa and America (21). Further work is needed to understand why BovB has been so successful at colonizing some genomes, for example the cow and elephant, and so unsuccessful in others, like the horse. In extreme cases such as the cow, almost a quarter of the genome is the result of BovB and derived SINE sequence retrotransposition, with one reported instance of exaptation into a protein-coding gene (22). The timing of HT for BovB is difficult to determine. HT in terrestrial animals could have occurred via a common mechanism/vector before the breakup of Gondwanaland 175–140 Mya (23). Alternatively, it could have occurred much later if migratory birds or insects were transfer partners. In this context it is worth noting that immature stages of Amblyomma sp. are found on wild birds (24). Resolution of these phylogeographic alternatives will have to await the availability of additional genome sequence data.
The frequent horizontal movement of BovB illustrates the significant impact HT has had on animal genomes; expansion of BovB in various lineages has contributed large amounts of sequence (and presumably structural variation) to the genomes of distantly related species. It is tempting to speculate that BovB is not the only retrotransposon to have jumped between species, and further investigation will be required to test this hypothesis. Despite public concern over the transfer of genetic material to create genetically modified organisms, it appears that Mother Nature has been quietly shuffling genomes for some time.
Materials and Methods
A flowchart and detailed description of methods, including perl scripts used are available in SI Appendix.
Presence of BovB in GenBank Data.
A list of genera, families, superfamilies, and orders to be tested for BovB was compiled from information at National Center for Biotechnology Information (NCBI) (25). A BioPerl (26) module, RemoteBlast, was used (script supplied in SI Appendix, section 2) to query the NCBI remote BLAST Nucleotide database using a file of eight BovB/RTE sequences obtained from RepBase (12) and from our own previous analyses (8). Two stringent cut off E values were used to identify significant hits (e = 0 and e ≤ 1e-10) for further analysis.
Identification of BovB Across Taxa with Full Genome Assemblies.
LASTZ (15) was used to identify BovB sequences based on our eight BovB query sequences, with at least 80% length coverage in full genome assemblies. BEDTools (27) were used to merge the LASTZ intervals to get unique BovB sequences based on hits from multiple queries. Sequences were either first clustered, using UCLUST (28), at 70% or 80% identity or directly globally aligned using MUSCLE (29). PILER (30) was then used to get a consensus sequence. If the initial clustering step produced very large clusters, e.g., >2,000 sequences for the elephant and >600 for the cow, the sequences were clustered at 90% and consensus sequences for these clusters were constructed. These 90% cluster consensus sequences were then clustered at 80% to construct consensus sequences that were used to build the BovB for that species. Percent identity used for clustering in various species was: No clustering for platypus, wallaby, sea urchin, zebrafish, silkworm, 70% for opossum and tenrec, 80% for sheep, anole, horse, rock hyrax, and 90% followed by 80% for cow and elephant. For these species, RepeatMasker (31) was used to determine the amount of the genome corresponding to BovB.
Identification of BovB Across Taxa with Genome Survey Sequence Coverage.
There were 65 taxa with low coverage genome survey sequence data containing BovB. A number of these species (shown in the tree in Fig. 2) yielded sufficient hits to build representative BovB sequences for phylogenetic analysis. Sequence reads corresponding to BovB (>60% length or >80% length) were selected for assembly using Phrap (32). Where Phrap built many contigs for single species, the contigs were clustered using UCLUST. Contigs were then aligned and scaffolded to produce full-length BovB sequences using MUSCLE. Alignments/scaffolds were then manually curated and used to build consensus sequences.
Sequencing to Identify Additional Reptile and Monotreme BovB.
Genomic DNA was isolated from Tachyglossus aculeatus, Egernia stokesii, and Tiliqua rugosa and sent to BGI (Hong Kong) for 100-bp paired end sequencing (300-bp library mean insert size). One giga base pair of paired end reads for each species was then used as input for BovB consensus building as described above. These data have been submitted to the EBI Sequence Read Archive (33) under the following project accession ERP001591 and sample accessions T. rugosa (sleepy lizard) ERS195148, E. stokesii (skink) ERS195147, and T. aculeatus (echidna) ERS154930.
Phylogenetic Analyses.
Consensus sequences were aligned with MUSCLE, and multiple alignments were processed with Gblocks (34) to select conserved blocks for use in phylogenetic analysis. We used three independent tree building tools to construct phylogenies from the refined multiple alignments; FastTree (35), RAxML (36), and BEAST (37). All three methods used general time reversible (GTR) model and gamma approximation on substitution rates. Sprit (19) was used to calculate the minimum subtree prune and regraft (SPR) distance between the BovB phylogeny (FastTree) and the control phylogeny based on gene orthologs (17).
Supplementary Material
Acknowledgments
We thank Evgeny Zdobnov for the ortholog tree, David Chapple for providing access to Lord Howe Island Gecko sequence data, and Michael Lee for his impeccable advice.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The survey sequence data have been deposited in the Dryad database, http://datadryad.org [doi nos.: 10.5061/dryad.f1cb2/23 (Amphibolurus norrisi), 10.5061/dryad.f1cb2/46 (Eremiascincus richardsonii), 10.5061/dryad.f1cb2/47 (Glaphyromorphus douglasi), 10.5061/dryad.f1cb2/26 (Gehyra variegate), 10.5061/dryad.f1cb2/27 (Gehyra lazelli), 10.5061/dryad.f1cb2/6 (Bothriocroton hydrosauri), 10.5061/dryad.f1cb2/28 (Hydrophis spiralis), 10.5061/dryad.f1cb2/15 (Isoodon obesulus), 10.5061/dryad.f1cb2/16 (Macrotis lagotis), and 10.5061/dryad.f1cb2/19 (Petaurus breviceps)]; and European Bioinformatics Institute Sequence Read Archive (EBI SRA), http://www.ebi.ac.uk/ena/ (accession nos. ERS195148 [Tiliqua rugosa (Sleepy Lizard) paired end reads], ERS195147 [Egernia stokesii (Skink) paired end reads], and ERS154930 [Tachyglossus aculeatus (Echidna) paired end reads], validation sequences deposited in GenBank with accession nos. KC352670, KC352671, KC352672, KC352673, and KC352674).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1205856110/-/DCSupplemental.
References
- 1.Jurka J, Kapitonov VV, Kohany O, Jurka MV. Repetitive sequences in complex genomes: Structure and evolution. Annu Rev Genomics Hum Genet. 2007;8:241–259. doi: 10.1146/annurev.genom.8.080706.092416. [DOI] [PubMed] [Google Scholar]
- 2.Garcia-Perez JL, Doucet AJ, Bucheton A, Moran JV, Gilbert N. Distinct mechanisms for trans-mediated mobilization of cellular RNAs by the LINE-1 reverse transcriptase. Genome Res. 2007;17(5):602–611. doi: 10.1101/gr.5870107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartolomé C, Bello X, Maside X. Widespread evidence for horizontal transfer of transposable elements across Drosophila genomes. Genome Biol. 2009;10(2):R22. doi: 10.1186/gb-2009-10-2-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pace JK, 2nd, Gilbert C, Clark MS, Feschotte C. Repeated horizontal transfer of a DNA transposon in mammals and other tetrapods. Proc Natl Acad Sci USA. 2008;105(44):17023–17028. doi: 10.1073/pnas.0806548105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gilbert C, Schaack S, Pace JK, 2nd, Brindley PJ, Feschotte C. A role for host-parasite interactions in the horizontal transfer of transposons across phyla. Nature. 2010;464(7293):1347–1350. doi: 10.1038/nature08939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Silva JC, Loreto EL, Clark JB. Factors that affect the horizontal transfer of transposable elements. Curr Issues Mol Biol. 2004;6(1):57–71. [PubMed] [Google Scholar]
- 7.Piskurek O, Okada N. Poxviruses as possible vectors for horizontal transfer of retroposons from reptiles to mammals. Proc Natl Acad Sci USA. 2007;104(29):12046–12051. doi: 10.1073/pnas.0700531104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adelson DL, Raison JM, Edgar RC. Characterization and distribution of retrotransposons and simple sequence repeats in the bovine genome. Proc Natl Acad Sci USA. 2009;106(31):12855–12860. doi: 10.1073/pnas.0901282106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gentles AJ, et al. Evolutionary dynamics of transposable elements in the short-tailed opossum Monodelphis domestica. Genome Res. 2007;17(7):992–1004. doi: 10.1101/gr.6070707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kordis D. Transposable elements in reptilian and avian (sauropsida) genomes. Cytogenet Genome Res. 2009;127(2-4):94–111. doi: 10.1159/000294999. [DOI] [PubMed] [Google Scholar]
- 11.Zhao FQ, Qi J, Schuster SC. Tracking the past: Interspersed repeats in an extinct Afrotherian mammal, Mammuthus primigenius. Genome Res. 2009;19(8):1384–1392. doi: 10.1101/gr.091363.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jurka J, et al. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 2005;110(1-4):462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- 13.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 14.Castoe TA, et al. Discovery of highly divergent repeat landscapes in snake genomes using high-throughput sequencing. Genome Biol Evol. 2011;3:641–653. doi: 10.1093/gbe/evr043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harris RS. Improved Pairwise Alignment of Genomic DNA. University Park, PA: Pennsylvania State Univ; 2007. [Google Scholar]
- 16.Smyth M. Distribution of three species of reptile ticks, Aponomma Hydrosauri (Denny), Amblyomma Albolimbatum Neumann, and Amb. Limbatum Neumann, I. Distribution and hosts. Aust J Zool. 1973;21(1):91–101. [Google Scholar]
- 17.Waterhouse RM, Zdobnov EM, Tegenfeldt F, Li J, Kriventseva EV. OrthoDB: The hierarchical catalog of eukaryotic orthologs in 2011. Nucleic Acids Res. 2011;39(Database issue, suppl 1):D283–D288. doi: 10.1093/nar/gkq930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hedges SB, Dudley J, Kumar S. TimeTree: A public knowledge-base of divergence times among organisms. Bioinformatics. 2006;22(23):2971–2972. doi: 10.1093/bioinformatics/btl505. [DOI] [PubMed] [Google Scholar]
- 19.Hill T, et al. SPRIT: Identifying horizontal gene transfer in rooted phylogenetic trees. BMC Evol Biol. 2010;10:42. doi: 10.1186/1471-2148-10-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kordis D, Gubensek F. Unusual horizontal transfer of a long interspersed nuclear element between distant vertebrate classes. Proc Natl Acad Sci USA. 1998;95(18):10704–10709. doi: 10.1073/pnas.95.18.10704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roberts FHS. The Australian Species of Aponomma and Amblyomma (Ixodoidea) Aust J Zool. 1953;1(1):111. [Google Scholar]
- 22.Iwashita S, et al. A tandem gene duplication followed by recruitment of a retrotransposon created the paralogous bucentaur gene (bcntp97) in the ancestral ruminant. Mol Biol Evol. 2006;23(4):798–806. doi: 10.1093/molbev/msj088. [DOI] [PubMed] [Google Scholar]
- 23.Upchurch P. Gondwanan break-up: Legacies of a lost world? Trends Ecol Evol. 2008;23(4):229–236. doi: 10.1016/j.tree.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 24.Gonzalez-Acuña D, et al. First record of immature stages of Amblyomma tigrinum (Acari: Ixodidae) on wild birds in Chile. Exp Appl Acarol. 2004;33(1-2):153–156. doi: 10.1023/b:appa.0000030015.18088.e6. [DOI] [PubMed] [Google Scholar]
- 25.Sayers EW, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2012;40(Database issue):D13–D25. doi: 10.1093/nar/gkr1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stajich JE, et al. The Bioperl toolkit: Perl modules for the life sciences. Genome Res. 2002;12(10):1611–1618. doi: 10.1101/gr.361602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quinlan AR, Hall IM. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26(6):841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 29.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Edgar RC, Myers EW. PILER: Identification and classification of genomic repeats. Bioinformatics. 2005;21(Suppl 1):i152–i158. doi: 10.1093/bioinformatics/bti1003. [DOI] [PubMed] [Google Scholar]
- 31. Smit AFA, Hubley R, Green P (1996–2004) RepeatMasker Open-3.0.)
- 32.Gordon D, Abajian C, Green P. Consed: A graphical tool for sequence finishing. Genome Res. 1998;8(3):195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- 33. European Nucleotide Archive (2012) Available at www.ebi.ac.uk/ena/home. Accessed November 20, 2012.
- 34.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17(4):540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 35.Price MN, Dehal PS, Arkin AP. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS ONE. 2010;5(3):e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stamatakis A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22(21):2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 37.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7(1):214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.