Abstract
Although human exposure to low-dose ionizing radiation can occur through a variety of sources, including natural, medical, occupational and accidental, the true risks of low-dose ionizing radiation are still poorly understood in humans. Here, the global transcriptional responses of human skin after ex vivo exposure to low (0.05 Gy) and high (5 Gy) doses of X rays and of time in culture (0 Gy) at 0, 2, 8 and 30 h postirradiation were analyzed and compared. Responses to low and high doses differed quantitatively and qualitatively. Differentially expressed genes fell into three groups: (1) unique genes defined as responsive to either 0.05 or 5 Gy but not both and also responsive to time in culture, (2) specific genes defined as responsive to either 0.05 or 5 Gy but not both and not responsive to time in culture, and (3) dose-independent responsive genes. Major differences observed in ex vivo irradiated skin between transcriptional responses to low or high doses were twofold. First, gene expression modulated by 0.05 Gy was transient, while in response to 5 Gy persistence of modified gene expression was observed for a limited number of genes. Second, neither TP53 nor TGFβ target genes were modulated after exposure to an acute low dose, suggesting that the TP53-dependent DNA damage response either was not triggered or was triggered only briefly.
INTRODUCTION
Human exposure to low-dose ionizing radiation (0.01 to 0.1 Gy) can occur through a variety of sources, including natural (cosmic rays, radionuclides in Earth’s crust), medical (diagnostic imaging and radiotherapy), occupational (nuclear power plant and medical workers), and accidental (nuclear accident, terrorist act). However, the true risks of low-dose radiation are still poorly understood in humans. On the other hand, high-dose acute whole-body irradiation in humans is associated with well-defined spectra of dose-related radiation syndromes. The LD50/60 (lethal dose to 50% of the population within 60 days) for untreated adults after acute X-or γ-radiation exposure varies between 2.6 and 5.5 Gy (1). Cancer is judged to be the main risk from low-dose radiation (2). Currently, risks for ionizing radiation-induced cancer at low doses are extrapolated from the linear no threshold (LNT) dose–response model, assuming a linear relationship between radiation dose and effects (3). However, the validity of the LNT model for low-dose radiation is controversial. Opponents argue that this model exaggerates the risks associated with low-dose radiation and contradicts accumulating findings that demonstrate quantitative and qualitative differences in responses of living cells and tissues to low and high doses of radiation (4, 5). In comparison to low-dose effects, high-dose effects are more easily quantified and experimentally accessible; epidemiological studies to assess risks of low and very low doses are limited by poor dosimetry, the very large sample size required, and the difficulty to tease out radiation exposure from other confounders. Therefore, an understanding of low-dose radiation-associated biological risks in humans will emerge only from the elucidation of the cellular mechanisms involved in the response of humans to low-dose radiation. Scientific and public interests converge in the effort to characterize these mechanisms. The need to enable health risk assessment and provide adequate radioprotection for human exposure to low-dose radiation is the driving force behind such an effort (http://lowdose.energy.gov).
Cellular stress responses induced by radiation are complex. The ability to analyze global gene expression by using microarrays has facilitated the study of these responses (6, 7). Information available on radiation-induced changes in gene expression profiles has been collected primarily in single cell types grown in vitro. Human lymphocytes irradiated in vivo or in vitro represent one of the most studied cell types. Access to occupational workers exposed to radiation and the fact that blood drawing is a relatively easy and noninvasive method to obtain human samples contribute to making human lymphocytes a material of choice for radiation studies. Other cell types used include normal fibroblasts and keratinocytes (8), usually isolated from human skin. The primary focus of such studies has been the identification of a gene signature for radioprotection applications (9–12). Although many studies have analyzed global transcriptional responses to high radiation doses (9, 12–16), ranges of doses, including low doses, have been tested in others (8, 17–24). Consensus findings of large scale transcriptional studies are that responses to radiation are dependent on dose, and gene regulation occurs more rapidly at high doses than at low doses. Although a number of gene signatures have been proposed, no consensus gene signature characteristic of either high or low radiation doses has yet emerged (25). Such signatures, if identified, will likely be cell type- and tissue-specific and dependent on time postexposure. Indeed, studies have shown that transcriptional responses to radiation are cell line- and tissue-specific and are influenced by postexposure time (25). Nevertheless, genes induced in response to genotoxic stress primarily induced by moderate to high radiation doses have been identified (14, 16, 26–28).
Information obtained from in vitro studies is limited, since neither the three-dimensional organization of tissues nor their multiple cell type structure is reproduced in monolayer single cell type cultures. Additionally, the importance of intracellular communication in the cellular response to radiation is overlooked in vitro (29). The use of a mouse model for in vivo studies of low-dose radiation effects in humans is hampered by the lack of a good correlation between mouse and human responses to radiation (12, 30). Thus these effects would be best studied in vivo in humans. However, it is also obvious why the use of human subjects strictly for research purposes is forbidden. To overcome this hurdle, our laboratory has pioneered the use of skin biopsies from patients undergoing radiation therapy for prostate cancer to study in vivo human responses to low-dose radiation at the transcriptional level (31, 32). Furthermore, studying the effects of radiation in human skin is highly relevant since skin is the most abundant organ in humans and represents the interface between us and our environment. Therefore, skin is the first human organ exposed to radiation during occupational, therapeutic and accidental exposures. Human skin is a complex tissue composed of two major distinct layers: epidermis and dermis. The epidermis is the keratin compartment, delimited at the outer surface by the stratum corneum and at the opposite end by the basal layer, which borders the dermis. Keratinocytes are epidermal cells that migrate from the basal layer upward, differentiating along the way. The stratum corneum is composed of dead keratinocytes or corneocytes with high keratin content and lipids. Melanocytes are scattered irregularly along the basal layer and the dermis. The dermis, representing the collagen compartment, contains fibroblasts, resting at the G1 phase of the cell cycle, macrophages and smooth muscle fibers that are part of hair structures. The inner side of the dermis ends with the hypodermis, which contains a varying number of fat cells and carries the major blood vessels and nerves to the skin.
Although skin biopsies are a material of choice for studying in vivo human responses to low-dose radiation, their nature restricts their size and numbers taken from the same patient and thereby limits the experimental design, particularly with respect to various doses and time–response experiments as well as assay choices and number of repetitions. To complement the analysis of in vivo irradiated human skin at the transcriptional level and to enable examination of responses at the post-transcriptional level, we have begun to use postabdominoplasty discarded human skin exposed ex vivo to radiation. This ex vivo irradiated human skin model allows the collection of larger skin samples and confers flexibility to the experimental design. Consequently, assays requiring larger amounts of sample as well experiments with more doses and postirradiation times can be performed. Thus far, responses to low-dose radiation in humans are best documented at the transcriptional level. Therefore, to evaluate the use of ex vivo irradiated human skin for proteomic studies of normal radiobiological responses to low-dose exposure, we began by analyzing such responses at the transcriptional level. Here we present global gene expression analysis data obtained after ex vivo exposure of human skin to low (0.05 Gy) and high (5 Gy) X-ray doses.
MATERIALS AND METHODS
Ex Vivo Human Skin Samples
Skin from two adult female patients (P1 and P2) undergoing abdominoplasty was collected under IRB approval and immediately placed on ice-cold phosphate-buffered saline (PBS), pH 7.2, containing antibiotics (Invitrogen, Carlsbad, CA). Skin from two patients was used to reduce the bias of inherent interindividual variability, which is expected given that humans have unique genotypes. After removal of the fat layer, the skin was cut into ~2 × 2-cm pieces in ice-cold PBS containing antibiotics. Skin samples were placed skin side up into wells of 6-well plates and submerged with DMEM (Invitrogen) supplemented with 10% FBS and antibiotics and incubated at 37°C under a humidified 95% air/5% CO2 atmosphere. Under these culture conditions, ex vivo skin is viable over several weeks (33, 34).
Irradiation and Time-Course Collection of Samples
X-ray doses of 0 (sham), 0.05 and 5 Gy were delivered with an Elekta Synergy clinical irradiator (Stockholm, Sweden) at a dose rate of 4 Gy/min at 15 MeV. Skin samples were irradiated 1 day after collection. Control and irradiated samples were collected in quadruplicate at 0, 2, 8 and 30 h postirradiation.
Total RNA was extracted from skin using the RNeasy purification kit (Qiagen, Valencia, CA), with the following deviation from the manufacturer’s protocol. Prior to application to the RNA column, skin lysates were briefly extracted with 1/5 V of chloroform for lipid removal. Skin samples were homogenized in lysis buffer using a FastPrep® FP120A homogenizer (Qbiogen, Carlsbad, CA). Purified RNA was quantified with a Nanodrop 1000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE) and then monitored for integrity on a 2100 Bioanalyzer (Agilent, Santa Clara, CA).
Microarray Analysis
Microarray analyses were carried out at the University of California Davis Genomic Core Facility. For each of two patients, triplicate or quadruplicates samples, all with RNA Integrity Numbers (RIN) above 7, were analyzed for each treatment and time tested, resulting in 40 samples analyzed for one patient and 36 samples for the other, for a total of 76 samples analyzed. Prior to hybridization on microarrays, 300 ng of total RNA per sample was amplified and biotinylated using an Illumina® Totalprep™ RNA amplification kit (Ambion, Austin, TX). Resulting biotinylated cRNAs were hybridized to Illumina Sentrix® Human Ref-8 Expression BeadChips V3R2 (Illumina Inc., San Diego, CA). Each beadchip contains eight microarrays, and each microarray is represented by 24,526 RefSeq curated gene probes and controls (www.switchtoi.com/resources). After hybridization using Illumina reagents supplied with the beadchips, microarrays were scanned on an Illumina iscan and fluorescence was recorded.
Statistical Analysis
Arrays were processed with Illumina BeadStudio v3.4.0 with background subtraction but no normalization. We omitted probes from the analysis if the BeadStudio detection P value was greater than 0.05 for all the arrays in the analysis. Of the original 24,526 probes, 16,749 passed this filter. Next, data for all the arrays were imported into the R statistical environment (http://www.R-project.org). The expression values were transformed using the generalized logarithm transformation and normalized using the LMGene R package from our group (35, 36). After preprocessing, an ANOVA model was fitted to data from each probe to screen for potentially differentially expressed genes. A “treatment” factor was derived from each unique combination of dose and time; this factor was then included in a two-way ANOVA model along with a fixed effect for subject. P values from the ANOVA F tests of treatment effect (testing for differences between any levels of the treatment factor) and subject effect were adjusted for multiple testing using the false discovery rate (FDR) method of Benjamini and Hochberg (37). The largest factor in any analysis of gene expression is typically that associated with differences between individuals. In this case, if we perform a three-way ANOVA for each gene and compute the mean squares for each variable (subject, time, dose) and their interactions, and if we then standardize these mean squares to add to 1 for each gene (so that we can compare genes with different levels of expression), the subject effect comprises 80% of the total, followed by time (5%), dose (3%), dose*subject (3%), time*subject (3%), time*dose (2%) and error (1%). It is important to note that the method of analysis identified as differentially expressed genes only those whose change in expression across subjects was large enough to be detected above the background of interindividual variability. This is why studies that use more than one individual (instead of a cell line, for example) are so important. Here the gene expression data represent the average transcriptional response of two individuals.
Probes with an FDR-adjusted P value less than 0.1 for the treatment effect were retained for further investigation. With the reduced set of probes from the above prescreening, a second set of ANOVA models was applied to compare dose effects of 0 and 0.05 Gy, 0 and 5 Gy, and 5 Gy and 0.05 Gy at 2, 8 and 30 h, respectively. The times 0 and 2 h, 0 and 8 h, and 0 and 30 h were also compared at dose 0 Gy to determine the effect of time in culture on gene expression. An unadjusted P value of 0.015 or less was considered statistically significant for the above comparisons (as the reduced set of probes has already been prescreened, further adjustment for false discovery is not required).
Data Analysis
Genes were considered significantly differentially expressed if P values were ≤0.015. The software Metacore by Genego® (version 6.2) (http://www.genego.com/) was used to map significantly differentially expressed genes to functional pathways and networks. The DAVID (version 6.7) functional gene classification tool was used to functionally analyze gene lists (38).
Validation of Microarray Results by RT-qPCR
Reverse transcription quantitative polymerase chain reaction (RT-qPCR) was used to validate some of the changes in gene expression detected by microarray analysis. Total RNA was extracted from additional control and irradiated skin samples from patients P1 and P2 that had been processed at the same time as the samples used in the microarray analysis but stored in RNAlater® (Ambion); additional skin samples for 8 h and 30 h after exposure to 5 Gy were not available for patient P2. The iscript select cDNA synthesis kit (BioRad, Hercules, CA) was used for RT of total RNA primed with oligodT. The expression of four genes, CDKN1A, ING2, CLDN1 and SESN1, was verified by qPCR. Gene specific primers used, designed with the Vector NTI Advance 11 software (Invitrogen), were as follows. CDKN1A: forward 5′TGAAGTGCTTAGTGTACTTGGAGTATTGGG3′ and reverse 5′AGTCTAGGTGGAGAAACGGGAACCA3′; CLDN1: forward 5′GTTGGTAAATCCAACAGCAAGGGAGA3′ and reverse 5′CACAGTGGCTGACTTTCCTTGTGTAGTTTA3′; EGFR: forward 5′TGCCGGTGGCATTTAGGGGT3′ and reverse 5′GTCCGTCCTGTTTTCAGGCCAAG5′; ING2: forward 5′AGGAAAGGGAAGCTTCACCTGTTGA3′ and reverse 5′CCCCTTTGGTTTATAGGTAAGTGAAACACA3′ and SESN1: forward 5′ATCCAAGTCCCATTCTTTGCATGC3′ and reverse 5′ATCCAAGTCCCATTCTTTGCATGC3′. Reference genes were B2M: forward 5′TCACCCCCACTGAAAAAGATGAGTATG3′ and reverse 5′TGCGGCATCTTCAAACCTCCA3′, and GAPDH: forward 5′TGCACCACCAACTGCTTAGCACC3′ and reverse 5′GAGGCAGGGATGATGTTCTGGAGA3′. qPCR with an icycler (BioRad) was performed on 25–50 ng cDNA in the presence of the SsoFast™ EvaGreen® Supermix (BioRad) and gene-specific primers, each at a final concentration of 250 nM, in a 20 μl total volume. For each gene, PCR reactions were performed in triplicate on a given sample. A cycle consisting of 95°C for 10 s and 60°C for 20 s was repeated 45 times. Gene expression ratios were calculated using the comparative Ct method (39).
RESULTS
Genes Significantly Differentially Expressed in Response to an Acute Low or High X-Ray Dose
Significantly differentially expressed genes in response to radiation represent the average response of normal skin from two individuals. In general, for the comparison to be significant and overcome interindividual variability, the average change in gene expression has to be relatively large and consistent in direction (either up- or downregulated), but genes for which the directions differ or in which the effect exists in one patient but not the other are not completely excluded.
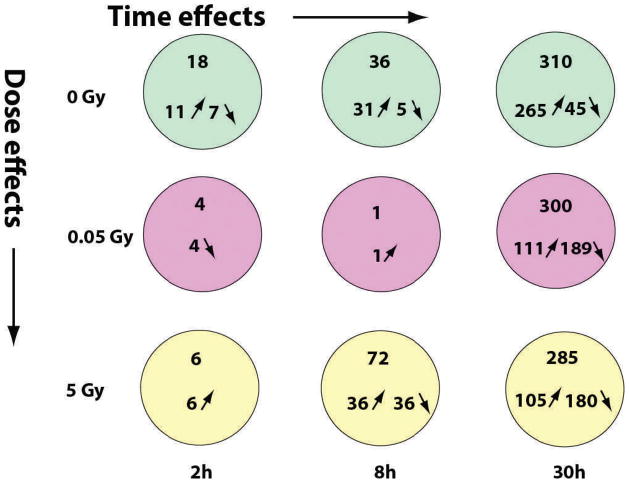
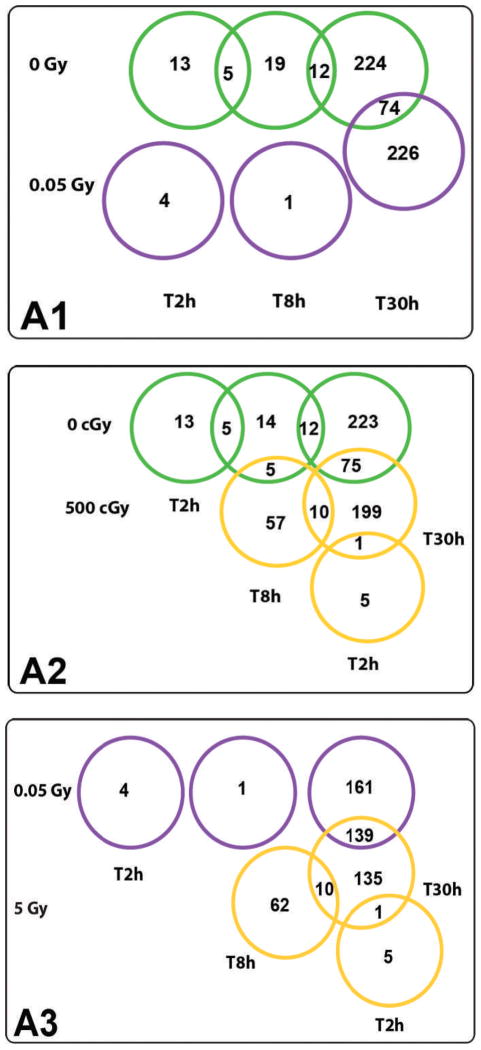
Genes with P values ≤0.015 at 10% FDR were considered significantly differentially expressed. Figure 1 illustrates the numbers of such genes as a function of radiation doses (0, 0.05 and 5 Gy) and postexposure time (2, 8 and 30 h). No skin-specific genes were found among these differentially expressed genes, as determined by a search of the Applied Biosystems human body map database with MetaCore. The number of significantly differentially expressed genes increased with postexposure time but not with radiation dose (Fig. 1). A majority of genes differentially expressed for time in culture were upregulated across all times. In contrast, at 30 h after exposure to 0.05 and 5 Gy, a majority of genes were downregulated (Fig. 1). At 2 h postirradiation, there was little response to either dose. At 8 h postirradiation, there was little response to 0.05 Gy, whereas there were both upregulated and downregulated genes in response to 5 Gy (Fig. 1). At 0 Gy, gene overlap was observed across 2 h and 8 h and across 8 h and 30 h, indicating a persistent change in gene expression that was not related to radiation (Fig. 2A1 and A2). At low dose (0.05 Gy), no gene overlap was observed across the postexposure times tested (Fig. 2A1 and A3), indicating a transient change in expression of the genes identified at a given time, but because of the small number of genes involved at 2 h and 8 h, this mainly indicates that the response to this dose is delayed beyond 8 h. At high dose (5 Gy), some gene overlap was observed across all postexposure times tested (Fig. 2A2 and A3), indicating a persistence of the response for a least a few hours. Gene overlap across doses was observed only at 30 h postexposure (Fig. 2), when the number of significantly differentially expressed genes was highest for all doses tested (Fig. 1).
FIG. 1.
Summary of genes significantly differentially expressed in ex vivo irradiated human skin. Genes significantly differentially expressed (P ≤ 0.015) at indicated X-ray doses and postirradiation times in comparison to the control (time 0 h, dose 0 Gy).
FIG. 2.
Venn diagrams illustrating gene overlap across postirradiation times and doses tested. Panel A1: Comparison between significantly differentially expressed (P ≤ 0.015) genes in response to 0 and 0.05 Gy. Panel A2: Comparison between significantly differentially expressed (P ≤ 0.015) genes in response to 0 and 5 Gy. Panel A3: Comparison between significantly differentially expressed (P ≤ 0.015) genes in response to 0.05 and 5 Gy.
Genes Responsive to 0 Gy or Differentially Expressed Genes Unrelated to Radiation
Inclusion of unirradiated controls for each time tested in our study design enabled the assessment of gene expression affected only by time of the skin samples in culture. As shown in Fig. 1, gene responses to 0 Gy increased with time in culture, and most of them were upregulated at all times. Genes involved in metabolic pathways and represented in protein folding and mRNA processing networks were regulated after 2 h in culture. Pathways affected after 8 h in culture included transport, transcription, DNA damage and immune response, while reproduction, transcription, muscle contraction, cytoskeleton, inflammation and DNA damage repair represent the networks involved. After 30 h in culture, genes modulated in skin cells fell into eight functional groups: membrane proteins, sodium transport, potassium transport, transcription factors, ring finger proteins, proteins with WD (tryptophan and aspartic acid) repeats, kinases and other enzymes. These genes were mainly distributed over transport, development, immune response, DNA damage and cell cycle pathways. Networks involved include muscle contraction, inflammation, reproduction, cell adhesion and translation. Genes overlapping across time in culture (0 Gy) participate in pre-mRNA splicing and ions and protein transport and were almost all upregulated (Table 1).
TABLE 1.
Genes in our Data Set That Overlap Across Time and Radiation Dose
| Gene symbol | Regulation | Name |
|---|---|---|
| Gene overlap between 2 and 8 h postexposure to 0 Gy | ||
| CSTF3 | 1 | Cleavage stimulation factor, 3′ pre-mRNA, subunit 3 |
| RAB30 | 1 | RAB30, member RAS family |
| SPRR2B | −1 | Small proline rich protein 2B |
| SPRR2C | −1 | Small proline rich protein 2C |
| USP2 | 1 | Ubiquitin specific peptidase 2 |
| Gene overlap between 2 and 30 h postexposure to 0 Gy | ||
| None | ||
| Gene overlap between 8 and 30 h postexposure to 0 Gy | ||
| AGBL4 | 1 | ATP/GTP binding protein like 4 |
| ATP1B4 | 1 | ATP ase Na+/K+ transporting β 4 polypeptide |
| C3orf30 | 1 | Chromosome 3 open reading frame 30 mRNA |
| C21orf66 | 1 | GC rich sequence DNA binding factor 1 |
| CACNA1H | 1 | Calcium channel voltage dependent T type α 1H subunit |
| FXR2 | 1 | Fragile X mental retardation autosomal homolog 2 |
| KCNB2 | 1 | Potassium voltage gated channel, Shab related, member 2 |
| LAMA2 | −1 | Laminin α2 |
| MDGA2 | 1 | MAM domain containing glycosylphosphatidylinositol anchor 2 |
| NXPH2 | 1 | Neurexophilin 2 |
| SF3A2 | 1 | Splicing factor 3a, subunit 2 |
| SLC4A8 | 1 | Solute carrier family 4, sodium carbonate co-transporter member 8 |
| Gene overlap between 2 and 8 h postexposure to 5 Gy | ||
| None | ||
| Gene overlap between 2 and 30 h postexposure to 5 Gy | ||
| LBH | 1 | Limb bud and heart development |
| Gene overlap between 8 and 30 h postexposure to 5 Gy | ||
| BRSK1 | −1 | BR serine/threonine kinase 1 |
| CAMK2B | −1 | Calcium/calmodulin dependent protein kinase 2 β |
| CCDC149 | −1 | Coiled coil domain containing 149 |
| CDKN1A | 1 | Cyclin dependent kinase inhibitor 1A (p21) |
| FAM104B | −1 | Family with sequence similarity 104, member B |
| NUP153* | −1 | Nucleoporin 153 kDa |
| RGS17 | −1 | Regulator of G protein signaling 17 |
| SESN1 | 1 | Sestrin 1 |
| UCRC | −1 | Ubiquinol cytochrome c reductase, complex III subunit X |
| ZNF524 | 1 | Zinc finger protein 524 |
Presence in Our Data Set of Previously Identified Human Radiation-Responsive Genes
Genes significantly differentially expressed in ex vivo irradiated human skin were compared to 24 previously reported human radiation-responsive genes, selected from many for having undergone qRT-PCR validation (Table 2). All 24 genes were upregulated in response to radiation after exposure times as short as 15 min and up to 24 h, and a majority of them have been identified in peripheral blood lymphocytes (PBL) after exposure to doses ranging from 0.5 to 8 Gy (10, 12, 14, 16, 40). Among these 24 previously reported radiation-responsive genes, only three, CDKN1A, CCNG1 and SESN1, were represented in our data set, and all three were upregulated in our ex vivo skin model 8 and/or 30 h after exposure to 5 Gy (Table 2). CDKN1A (cyclin-dependent kinase inhibitor) and CCNG1 (cyclin G1) are both cell cycle regulators that are activated in response to DNA damage. CDKN1A is an inhibitor of cell cycle progression at G1 (41), while cyclin G1 promotes cell cycle progression (42). While only the expression of CDKN1A was significant 8 h after exposure to 5 Gy, expression of both CDKN1A and CCNG1 became significant 30 h postexposure. Furthermore, at this same time, the expression of the CDKN1A gene was repressed in the tissue control (Table 2). Despite the fact that a linear dose response of CDKN1A between 0.02 and 0. 5 Gy has been described in the ML-1 human myeloid cell line 1 to 4 h postirradiation (40), induction of this gene or of the remaining 24 previously reported radiation-responsive genes, was not found in human skin up to 30 h after ex vivo exposure to 0.05 Gy (Table 2). Induction of SESN1, coding for sestrin1, a protein belonging to highly conserved proteins that accumulate in cells exposed to stress (43), was observed in human skin 8 and 30 h postirradiation with 5 Gy (Table 2). If considering gene families, two additional genes out of the 24 previously reported radiation-responsive genes were modulated in ex vivo irradiated human skin (Table 2). Induction of PHF13, coding for a protein known to modulate chromatin structure, was observed in human skin 30 h after irradiation with 0.05 and 5 Gy; exposure of normal keratinocytes to 0.01 Gy led to the upregulation of PHF10, another PHD (plant homeodomain) finger protein required for cell growth (Table 2). In PBL irradiated with 1 or 4 Gy, upregulation of TNFSF9, encoding a cytokine that belongs to the tumor necrosis factor superfamily of ligands, was observed as soon as 15 min postirradiation (16). In human skin, expression of TNFSF 14 and 15 was upregulated in the nonirradiated control tissue at 30 h postexposure, while at that same time, expression of TNFSF15 was downregulated in response to 0.05 and 5 Gy.
TABLE 2.
Presence in Our Data Set of Previously Reported Human Genes Modulated by Radiation
| Radiation doses and times | 0 Gy
|
0.05 Gy
|
5 Gy
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| 2 h | 8 h | 30 h | 2 h | 8 h | 30 h | 2 h | 8 h | 30 h | |
| Gene symbol and category | |||||||||
| Apoptosis | |||||||||
| BAXa | / | / | / | / | / | / | / | / | / |
| BBC3c | / | / | / | / | / | / | / | / | / |
| TNFRSF10Bd | / | / | / | / | / | / | / | / | / |
| TNFSF9b | / | / | TNFSF 14 and 15 up | / | / | TNFSF 15 down | / | / | TNFSF 15 down |
| Cell cycle/Proliferation | |||||||||
| CDKN1Aa,c,f,g,h | / | / | down | / | / | / | / | up | up |
| CCNG1d | / | / | / | / | / | / | / | / | up |
| PCNAa | / | / | / | / | / | / | / | / | / |
| DNA damage/Repair | |||||||||
| DDB2a,g | / | / | / | / | / | / | / | / | / |
| GADD45a,g,h | / | / | / | / | / | / | / | / | / |
| UBE2Be | / | / | / | / | / | / | / | / | / |
| Energy/Translation/Degradation | |||||||||
| FDXRc | / | / | / | / | / | / | / | / | / |
| FHL2d | / | / | / | / | / | / | / | / | / |
| GARSe* | / | / | / | / | / | / | / | / | / |
| SESN1c | / | / | / | / | / | / | / | up | up |
| Immune response | |||||||||
| CXC L10d | / | / | / | / | / | / | / | / | / |
| FCGR1Ag | / | / | / | / | / | / | / | / | / |
| IFNGb | / | / | / | / | / | / | / | / | / |
| Transcription modulators | |||||||||
| EGR1b,f | / | / | / | / | / | / | / | / | / |
| EGR4b | / | / | / | / | / | / | / | / | / |
| JUNb | / | / | / | / | / | / | / | / | / |
| SPOPe | / | / | / | / | / | / | / | / | / |
| TXNLe | / | / | / | / | / | / | / | / | / |
| Zinc finger proteins | |||||||||
| PHF10e | / | / | / | / | / | PHF13 up | / | / | PHF13 up |
| PHPT13c | / | / | / | / | / | / | / | / | / |
Genes induced in CD4+ lymphocytes 24 h postexposure to 0.5 Gy ex vivo (19).
Genes upregulated in PBL after 15 min ex vivo irradiation of human whole blood with 1 and 4 Gy of X rays (16).
Genes induced in PBL in response to ex vivo irradiation with 0.5, 2, 5 and 8 Gy of γ rays, after 6 and 24 h postexposure (12).
Genes upregulated in PBL 12 h after ex vivo exposure to 1 Gy (14).
Genes upregulated in primary normal keratinocytes after irradiation with 0.01 or 2 Gy γ rays (18).
Genes upregulated in G1-arrested primary normal fibroblasts in response to 4 Gy (23).
Genes upregulated in whole blood at 6 and 24 h after total body irradiation with single and multiple 1.5-Gy X-ray doses (10).
Genes induced in ML-1 cells in response to 0.02–0.5 Gy of γ rays after 1 to 4 h postexposure (40).
Confirmation of Differential Expression of Selected Genes by RT-qPCR
The expression of five genes, including two previously known radiation-responsive genes, that were upregulated in ex vivo irradiated skin by microarray analysis was verified by RT-qPCR. Upregulation of the expression of these genes, detected by microarray data analysis, was confirmed by RT-qPCR analysis (Table 3). It should be noted that the validation did not use another aliquot of the same RNA samples as was used for the expression arrays. The RNA for these RT-qPCR runs were from different pieces of skin from the same individuals, and thus more variability would be expected between the PCR and the microarray data. All of the ratios were in the same direction and many of similar size.
TABLE 3.
Validation of Microarray Results by RT-qPCR
| Gene | Postirradiation time and dose | qPCR mRNA ratio (radiation/control)
|
Microarray mRNA ratio (radiation/control) | |
|---|---|---|---|---|
| B2M as reference gene | GAPDH as reference gene | |||
| CDKN1A | 8 h, 5 Gy | 3.2 (P1) | 4.0 (P1) | 1.8 (P1+P2) |
| 30 h, 5 Gy | 3.2 (P1) | 4.0 (P1) | 2.9 (P1+P2) | |
| CLDN1 | 30 h, 0.05 Gy | 2.5 (P1+P2) | 1.5 (P1+P2) | 1.6 (P1+P2) |
| EGFR | 30 h, 0.05 Gy | 20.2 (P1+P2) | 3.9 (P1+P2) | 2.1 (P1+P2) |
| ING2 | 30 h, 0.05 Gy | 1.2 (P1+P2) | 3.1 (P1+P2) | 1.8 (P1+P2) |
| 30 h, 5 Gy | 1.6 (P1) | 1 (P1) | 2.0 (P1+P2) | |
| SESN1 | 8 h, 5 Gy | 1.5 (P1) | 1.9 (P1) | 1.9 (P1+P2) |
| 30 h, 5 Gy | 4.0 (P1) | 2.5 (P1) | 1.9 (P1+P2) | |
Notes. RT-qPCR was performed on total RNA extracted from additional skin samples from each patient (P1 and P2); additional skin samples for 8 h and 30 h after exposure to 5 Gy were not available for patient P2. Expression of five selected genes and two reference genes was analyzed. Calculated gene expression changes based on RT-qPCR analysis represent either a mean between the two patients, or an expression change in one patient (P1). Calculated gene expression changes based on microarray analysis were calculated from the ANOVA coefficients.
Radiation Dose-Responsive Genes
Genes responsive to one radiation dose but not the other fell into two groups: (1) specific genes, corresponding to genes shared with neither time in culture control (0 Gy) nor another dose; and (2) unique genes, which were shared only with the time in culture control but displayed opposite regulation.
1. Genes responsive to 0.05 Gy
Genes with significant altered expression to 0.05 Gy in ex vivo irradiated skin at 2 and 8 h postexposure were all specific genes (Table 4). At 30 h postexposure, 124 specific genes and 37 unique genes were identified. The functional groups represented by these genes are summarized in Table 4. Only a few specific genes were responsive to 0.05 Gy at 2 and 8 h postexposure (Table 4). At 2 h, all four genes were downregulated. NTN4 codes for netrin 4; netrins are related to laminins, which function as structural scaffolds in tissues. The RILPL2 encoded protein, in association with the Rab7 GTPase, controls late endocytic transport. The product of NLRP3, in association with apoptosis associated speck-like protein PYCARD/ASC, functions as an upstream activator of NFκB signaling and regulates inflammation, immune response and apoptosis. At 8 h, upregulation of PRDM1, also known as BLIMP1, was the only significant dose-specific change in gene expression. PRDM1 encodes a repressor of β interferon and TP53 gene expression. At 30 h about half of the specific genes were upregulated and half downregulated. Major pathways identified with these genes were related to development (EGFR and glucocorticoid receptor signaling), proteolysis, transcription (P53 signaling pathway) and translation. Genes such as EGFR (epidermal growth factor receptor), IRS-2 (insulin receptor substrate 2), SUMO1 (SMT3 suppressor of mif two homolog) and CBX4 (chromobox 4) contributed to these pathways and were all upregulated. Almost all unique genes identified at 30 h postexposure were downregulated. Pathways identified included DNA damage (mismatch repair, role of BRCA1 and BRCA2 in DNA repair), estradiol and aldosterone metabolism. Unique genes represented in these pathways were MSH3 (mutS homolog 3), SULTA1 (sulfotransferase family A, member 1), and AKR1D1 (3 oxo 5 beta steroid 4 dehydrogenase), all of which were downregulated.
TABLE 4.
Unique and Specific Genes Modulated in Human Skin at 2 and 8 h after Ex Vivo Exposure to Low or High Radiation Doses
| Gene symbol | Regulation | Name |
|---|---|---|
| Specific genes responsive to 0.05 Gy at 2 and 8 h postirradiation | ||
| At 2 h after exposure | ||
| PSORS1C2 | −1 | Psoriasis susceptibility 1 candidate 2 |
| NTN4 | −1 | Netrin 4 |
| RILPL2 | −1 | Rab interacting lysosomal protein-like 2 |
| NLRP3 | −1 | NLR family, pyrin domain containing 3 |
| At 8 h after exposure | ||
| PRDM1 | 1 | PR domain containing 1 with ZNF domain |
| Unique genes responsive to 0.05 Gy at 2 and 8 h postirradiation | ||
| None | ||
| Specific genes responsive to 5 Gy at 2 and 8 h postirradiation | ||
| At 2 h after exposure | ||
| ACTB | 1 | β actin |
| ATN1 | 1 | Atrophin 1 |
| COX7A2 | 1 | Cytochrome c oxidase subunit VII a polypeptide 2 |
| LBH | 1 | Limb bud and heart development |
| NDUFA1 | 1 | NADH dehydrogenase 1 α complex |
| ZNF581 | 1 | Zinc finger protein 581 |
| At 8 h after exposure | ||
| A1CF | −1 | APOBEC1 complementation factor |
| AGTRAP | 1 | Angiotensin II receptor-associated protein |
| BRSK1 | −1 | BR serine/threonine kinase 1 |
| C17ORF68 | 1 | Chromosome 17 open reading frame 68 |
| C18ORF56 | 1 | Chromosome 18 open reading frame 56 |
| C1ORF110 | −1 | Chromosome 10 open reading frame 110 |
| C8A | −1 | Complement component 8, α polypeptide |
| CABLES1 | 1 | Cdk5 and Abl enzyme substrate 1 |
| CAMK2B | −1 | Calcium/calmodulin dependent protein kinase 2β |
| CBLC | 1 | Cas-Br-M ecotropic retroviral transforming sequence C |
| CCDC149 | −1 | Coiled coil domain containing 149 |
| CDIPT | 1 | CDP diaglycerol inositol 3 phosphatidyltransferase |
| CDKN1A | 1 | Cyclin dependent kinase inhibitor 1A (p21) |
| CSAG3 | −1 | CSA family member 3 |
| CYR61 | −1 | Cysteine rich angiogenic inducer 61 |
| DGKQ | 1 | Diaglycerol kinase theta |
| DNAJB1 | −1 | DNA 5 homolog subfamily B, member 1 |
| FAM104B | −1 | Family with sequence similarity 104, member B |
| FAM82A | −1 | Family with sequence similarity 82, member A1 |
| FLVCR2 | 1 | Feline leukemia virus subgroup C cellular receptor family, member 2 |
| GRM3 | −1 | Glutamate receptor, metabotropic 3 |
| HPS6 | 1 | Hermansky-Pudlak syndrome 6 |
| IL1RAPL1 | −1 | Interleukin 1 receptor accessory protein-like 1 |
| JMJD4 | 1 | Jumonji containing domain 4 |
| KLRB1 | −1 | Killer cell lectin-like receptor subfamily B, member 1 |
| LILRB3 | −1 | Leukocyte immunoglobin-like receptor family B, member 3 |
| NCRNA268 | −1 | Noncoding RNA 268 |
| LOC441426 | −1 | Non-protein coding RNA 268 |
| MGC10997 | −1 | Protein pseudogene MGC10997 |
| MOCOS | 1 | Molybdenum cofactor sulfurase |
| MRPL27 | 1 | Mitochondrial ribosomal protein L27 |
| NECAP1 | −1 | NECAP endocytosis associated 1 |
| NOL10 | −1 | Nucleolar protein 10 |
| NR2E3 | −1 | Nuclear receptor subfamily 2, group E, member 3 |
| ODZ3 | −1 | ODZ, odd Oz/ten-m homolog 3 |
| ORAI1 | −1 | ORAI calcium release-activated calcium modulator 1 |
| PACSIN3 | 1 | Protein kinase C and casein substrate in neurons 3 |
| PARC | 1 | Cullin 9 |
| PEX16 | 1 | Peroxisomal biogenesis factor 16 |
| PIGQ | 1 | Phophatidylinositol glycan anchor biosynthesis class Q |
| PKP3 | 1 | Plakophilin 3 |
| PNPO | 1 | Pyridoxamine 5′ phosphate oxidase |
| POLR2C | 1 | RNA II polymerase DNA directed polypeptide C |
| POMGTN1 | 1 | Protein O-linked mannose β 1,2 acetylglucosaminytransferase |
| PPARGC1A | −1 | Peroxisome proliferator-associated receptor γ coactivator 1 α |
| PTCD1 | 1 | Pentatricopeptide repeat domain 1 |
| RBM45 | 1 | RNA binding motif 45 |
| RCAN2 | −1 | Regulator of calcineurin 2 |
| RGS17 | −1 | Regulator of G protein signaling 17 |
| RNF165 | −1 | Ring finger protein 165 |
| RPL36 | −1 | Ribosomal protein L36 |
| SCAP | 1 | SREBF chaperone |
| SCN2A | −1 | Sodium channel, voltage gated, type II, α subunit |
| SCRIB | 1 | Scribbled homolog |
| SESN1 | 1 | Sestrin 1 |
| SKI | −1 | v-ski sarcoma viral oncogene homolog |
| SLC1A3 | 1 | Solute carrier family 1, member 3 |
| SORBS1 | −1 | Sorbin and SH3 domain containing 1 |
| SRA1 | 1 | Steroid receptor RNA activator 1 |
| STC1 | −1 | Stanniocalcin 1 |
| SUPT6H | 1 | Suppressor of Ty 6 homolg |
| TGIF2 | 1 | TGFB induced factor homebox 2 |
| TLE1 | 1 | Transducing-like enhancer of split 1 |
| TTF2 | 1 | Transcription termination factor RNA polymerase II |
| UCRC | −1 | Ubiquinol cytochrome c reductase, complex III subunit X |
| UGT2B7 | −1 | UDP glucuronosyltransferase 2 family, polypeptide B7 |
| WAC | −1 | WW domain containing adaptor with coiled-coil |
| WDR74 | 1 | WD repeat domain 74 |
| ZBTB9 | 1 | Zinc finger and BTB domain containing 9 |
| ZC3H10 | 1 | Zinc finger CCH-type containing 10 |
| ZFYVE27 | 1 | Zinc finger FYVE domain containing 27 |
| ZNF524 | 1 | Zinc finger protein 524 |
| Unique genes responsive to 5 Gy at 2 and 8h postirradiation | ||
| At 2 h after exposure | ||
| None | ||
| At 8 h after exposure | ||
| AGBL4 | −1 | ATP/GTP binding protein like 4 |
| KCBN2 | −1 | Potassium voltage gated channel, Shab related, member 2 |
| NUP153 | −1 | Nucleoporin 153 KDa |
| SLC29A1 | −1 | Solute carrier family 29, member 1 |
| TXNRD1 | −1 | Thioredoxin reductase 1 |
2. Genes responsive to 5 Gy
Table 4 shows the specific and unique genes whose expression was significantly modulated in human skin 2 and 8 h postirradiation by 5 Gy ex vivo. Downregulation of RPL36, a specific gene encoding a protein whose overexpression has been shown to activate TP53 at 8 h postirradiation, is noteworthy. Among the genes with altered expression at 30 h postirradiation, 108 were specific and 38 unique; their classification into functional groups is shown in Table 5. Major pathways identified with specific genes modulated at 2 h postexposure include development [slit Robo signaling and TGFβ dependent induction of EMT (epithelial mesenchymal transition)], cell adhesion and cytoskeleton remodeling. ACTB, the gene coding for β actin, which was upregulated, is the common denominator in all these pathways. At 8 h postirradiation, specific genes included CDKN1A, which was upregulated. CDKN1A was a player in most of the pathways derived from these genes: transcription, development (TGFβ and Wnt signaling), cytoskeleton remodeling, DNA damage (ATM/ATR regulation of G2/M checkpoint), and cell cycle regulation. The most prominent pathways identified with the 30-h specific genes included cytoskeleton remodeling, development (TGFβ-dependent induction of EMT), cell cycle regulation, cell adhesion and immune response. Modulated genes represented in these pathways included ACTA2 (α actin), PPP3CC (protein phosphatase 3 catalytic subunit γ isoenzyme), and SMAD4 (SMAD family member 4), which were upregulated, and SKP2 (S phase kinase associated protein 2), NEK 8 (never in mitosis gene a related kinase 8), and COL4A5 (collagen type IV), which were downregulated. Although not represented in the pathways mentioned above, TP53INP1 (tumor protein p53 inducible nuclear protein 1), a known stress responsive protein and pro-apoptotic regulator of TP53, was also upregulated. The five unique genes responsive to 5 Gy at 8 h postexposure were all downregulated (Table 4) and were involved in translation, cytoskeleton and response to hypoxia networks. Prominent pathways identified with unique genes modulated 30 h postexposure were DNA damage (ATM/ATR regulation of G2/M and G1/S checkpoints), cell adhesion, development and cytoskeleton remodeling. Unique genes contributing to these pathways included upregulated CDKN1A and CLDN3 (claudin 3) and downregulated FANCD2 (Fanconi anemia complementation group D2), CLDNL14 and FZD7 (frizzled homolog). The upregulation of one gene, LBH, persisted between 2 and 30 h after exposure to 5 Gy but interestingly was not significant at 8 h postirradiation; 10 additional genes showed continuous modified expression between 8 and 30 h postexposure (Table 1 and Fig. 2A2).
TABLE 5.
Functional Classification of Radiation Dose-Responsive Genes in Human Skin 30 h after Ex Vivo Exposure
| Specific genes responsive to 0.05 Gya | Specific genes responsive to 5 Gyb |
|---|---|
| Phosphatases | Regulation of cell death |
| Protein transport | Mitochondrial and ribosomal proteins |
| Nuclear lumen proteins | Phosphatases |
| Transcription regulation | Cell cycle |
| DNA damage/repair | Catabolism |
| Transmembrane proteins | Transcription regulation |
| Signal transduction | |
| Nucleotide binding proteins | |
| Secreted proteins | |
| Transmembrane proteins | |
| DNA damage response effectors | |
| Immune response activators | |
| Unique genes responsive to 0.05 Gyc | Unique genes responsive to 5 Gyd |
| Transcription factors | Tight junctions |
| Nucleotide binding proteins | Transcription factors |
| Ion binding proteins | |
| Transport proteins |
Notes. Analyses based on DAVID’s functional classification. For each set of genes,
92 out of total of 124,
84 out of a total of 108,
16 out of a total of 37, and
19 out of total of 38 were taken into account.
Dose-Independent Radiation-Responsive Genes
Genes with altered expression in response to both low and high radiation doses, some with opposite responses depending on the time in culture control, belong to this group. In total, 139 of such genes were identified across all times in our data set. About two-thirds of these genes were downregulated, and all were similarly regulated at both radiation doses. The dose-independent radiation-responsive genes were primarily represented in metabolism regulation, transport, DNA damage repair (NHEJ repair), protein folding (membrane trafficking and signal transduction), and immune response pathways, which, with the exception of the protein folding pathway, appeared to be inhibited since the genes involved in these pathways were downregulated. Out of these 139 genes, 85 clustered into eight functional groups: involvement in membrane and cell junctions, regulation of transcription, protein transport, enzymes, catabolism, RNA processing proteins, stress-related proteins, and effectors in transmembrane signaling systems.
Identification of Radiation-Responsive Genes Linked to Carcinogenesis
Genes that have been linked to carcinogenesis and found to be differentially expressed in response to radiation in our data sets included CLDN1, 3 and 14, EGFR and ING2, all of which displayed altered expression at 30 h postirradiation.
Claudins encode a family of small proteins (22 to 27 kDa) that are important in tight junction formation and regulation in epithelial and endothelial cells. Tight junctions play a crucial role in the maintenance of cell polarity and in paracellular transport; expression of claudins is tissue-specific, and the exact combination of claudin proteins within a given tissue is thought to determine the selectivity and strength of tight junction (44). Upregulation of CLDN3 has been reported for several cancers (45), while both the increase and decrease of CLDN1 have been associated with tumorigenesis. Overexpression of CLDN1 has been associated with aggressiveness and increased malignant phenotype in melanoma (46). Decreased expression of CLDN1 has been observed in breast cancer, where a tumor suppressor function for CLDN1 has been suggested (47). Figure 3A illustrates the gene network associated with CLDN1. Upregulation of CLDN1 was observed in response to 0.05 Gy, while CLDN3 was upregulated and CLDN14 downregulated in response to 5 Gy. Genes for claudins 3 and 14 were also modulated in the time in culture control (0 Gy), where their expression was the opposite of that displayed in response to 5 Gy.
FIG. 3.
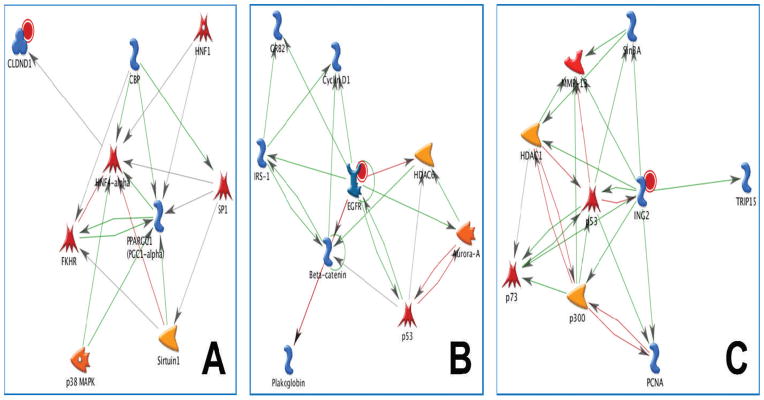
Networks of low-dose radiation responsive genes linked to carcinogenesis. Gene networks built in MetaCore (version 6.2) are shown. Panel A: CLDN1 network. Transcription of CLDN1 is regulated by HNF4α (hepatocyte nuclear factor α); SP1 (transcription factor) and HFN1 (hepatocyte nuclear factor 1) regulate the transcription of HFN4α; FKHR (Fork Head Box O1) physically interacts with HNF4α and decreases its activity; HNF4α is activated through association with CBP (CREB binding protein); FKHR is acetylated by CBP and deacetylated by Sirtuin 1 (NAD dependent deacetylase); CBP regulates the transcription of PPARGC1 (peroxisome proliferator-associated receptor γ coactivator 1α) and activates SP1 via acetylation; SP1 regulates the transcription of Sirtuin 1 and PPARGC1; PPARGC1 is activated through p38MAPK (p38 mitogen-activated protein kinase) phosphorylation and Sirtuin 1 deacetylation; binding of PPARGC1 activates HNF4α. Panel B: EGFR network. EGFR is activated by autophosphorylation upon ligand binding. Interactions of activated EGFR with cellular components lead to cell proliferation. EGFR recruits the GRB2 (growth factor receptor bound protein 2)/Sos (son of sevenless) complex, leading to activation of the Ras pathway; EGFR phosphorylates IRS-1 (insulin receptor substrate 1) and binding of activated IRS-1 activates GRB2; EGFR inhibits β catenin through phosphorylation; β catenin activates the transcription of IRS-1 and cyclin D1 and inhibits plakoglobin by direct binding to it; IRS 1 activates the transcription of cyclin D1; EGFR phosphorylates HDAC6 (histone deacetylase 6) and reduces its activity; HDAC6 activity is increased by Aurora A (serine/threonine protein kinase 6) phosphorylation, and HDAC6 is regulated at the transcriptional level by p53 (cellular tumor antigen p53); nuclear EGFR/STAT5 (signal transducer and activator of transcription 5) regulates the transcription of Aurora A, whose expression is inhibited by p53; phosphorylation of p53 by Aurora A abrogates p53 DNA binding and transactivation activity. Panel C: ING2 network. ING2 activates TRIP15 (COP9 signalosome complex subunit 2), PCNA (proliferating cell tumor antigen), Sin3A (paired amphipathic helix protein A), p73 (tumor protein) HDAC1 (histone deacetylase 1), and p300 (histone acetyltransferase) through direct binding; ING2 and Sin3A activate MMP13 at the transcriptional level; ING2 activates p53, while p53 inhibits ING2 at the transcriptional level; binding of Sin3A activates MMP13, which in turn regulates the transcription of p73 and MMP13; HDAC1 and p300 inhibit each other; HDAC1 activates MMP13; p300 and PCNA inhibit each other; p300 is a coactivator of p53; p53 inhibits MMP13 at the transcriptional level; p53 and p73 activate each other at the transcriptional level.
EGFR is a member of the human epidermal receptor (HER) family of tyrosine kinases expressed at the basal end of epithelial cells. Activation of EGFR in response to ligand-dependent dimerization initiates key signaling pathways leading to cell growth and proliferation, including the mitogen-activated protein kinase (MAPK) and phosphoinositide 3-kinase/AKT pathways. Overexpression of EGFR can cause aberrant cell proliferation and tumorigenesis. EGFR is overexpressed in a variety of human malignancies, including those of the lung and the breast (48). The EGFR-associated gene network is shown in Fig. 3B. In normal cells, EGFR participates in epithelial repair (49). Irradiation of rat hepatocytes with X-ray doses of 1 Gy or higher led to the downregulation of EGFR (50). In our experiments with ex vivo irradiated human skin, EGFR was upregulated in response to 0.05 Gy but not 5 Gy.
ING2 is a member of the inhibitor of growth (ING) tumor-suppressor family of proteins that integrate histone mark readout and genotoxic stress responses and regulate chromatin homeostasis (51). ING2 is a subunit of the mSIN3A/HDAC1 chromatin modifying complex, which in response to DNA damage relocalizes to the promoter of cell cycle regulatory genes such as cyclin D1 and c-myc, where it associates with histone mark H3K4me3. Binding of ING2 to H3K4me3 stabilizes the mSIN3A/HDAC1 complex at promoters, leading to histone deacetylation and subsequently to transcriptional repression (52). How ING2 orchestrates cellular responses to genotoxic stress is not yet fully understood. Upregulation of ING2 has been associated with colon cancer (53). The ING2 gene network is shown in Fig. 3C. Upregulation of ING2 in response to 0.05 and 5 Gy suggests that the genotoxic stress response governed by this gene is independent of radiation dose.
DISCUSSION
To further our understanding of the effects of low-dose radiation in humans, global transcriptional responses of human skin after ex vivo exposure to low (0.05 Gy) and high (5 Gy) doses of radiation were analyzed and compared. So far, the human transcriptional response to radiation has been analyzed primarily in various cell types grown in vitro (54). However, as initially demonstrated by studies on epithelial cells of the mammary gland, the biological behavior of cells grown in 2D and 3D culture is not identical (55). Therefore, the use of ex vivo irradiated human skin as a model to study radiation-induced cellular changes in humans brings us one step closer to the real, in vivo, skin tissue context.
The quantitative and qualitative differences between transcriptional responses to low and high radiation doses, reported for in vitro grown skin cells (keratinocytes and fibroblasts) (18, 23) were also observed in ex vivo irradiated human skin. In this model, we found the following quantitative differences in gene expression in response to 0.05 and 5 Gy at 2, 8 and 30 h postexposure. First, the overall quantitative response was slightly higher for 5 Gy than for 0.05 Gy. With a 10% FDR and a significance cutoff of P ≤ 0.015, the total numbers of genes with altered expression were 363 at 5 Gy and 305 at 0.05 Gy. Second, downregulation of gene expression was predominant at most times in response to 5 Gy and 0.05 Gy. Third, the major quantitative difference between transcriptional responses to low and high doses was observed at 8 h postexposure; only 1 gene was induced by 0.05 Gy, while 76 genes were modulated by 5 Gy. Initial gene repression and slow onset of the transcriptional response to low-dose radiation have also been reported for other models/systems after in vitro or in vivo exposure. For example, the transcriptional response of primary human keratinocytes irradiated with 0.01 Gy was delayed in comparison to the early gene induction observed at 3 h postirradiation with 2 Gy (18). Exposure of CD4+ lymphocytes to 0.05 Gy (0.45 Gy/min) led to gene downregulation at 3 h postexposure (19). Out of the 78 genes modulated in lymphocytes from occupational workers exposed to very low doses of radiation ranging from 0.696 to 39.088 mSv, 57 were found downregulated compared to 21 upregulated (56). In human skin exposed to 0.01 Gy in vivo, a majority of genes were upregulated at 3 h postexposure (31). For an acute in vivo irradiation, it is likely that 3 h postexposure is past an early event, since gene downregulation up to 1 h was observed in mice after a 0.5-Gy dose of JANUS fission neutrons to the gut (57).
The importance of dose rate on the transcriptional response to radiation has also been reported; in comparison to a low dose rate, more genes were involved in the response to radiation delivered at a high dose rate (58). Because DNA damage decreases at low dose rates, radiotherapy in the treatment of cancer is usually delivered at a high dose rate to maximize DNA damage in malignant cells. Thus, to recapitulate the dose rate used in gene expression studies on in vivo irradiated skin biopsies (31, 32), our study was conducted with a high dose rate (4 Gy/min). Gene expression is a dynamic process influenced by many parameters, including tissue complexity. Indeed, the presence of multiple cell types within tissues and cells at various growth stages adds to the complexity of the response. Therefore, the convergence of the early transcriptional response to low-dose radiation, i.e. gene repression, observed in human skin, keratinocytes, fibroblasts and lymphocytes after in vivo, ex vivo or in vitro exposure at various dose rates, is quite remarkable. In the case of our skin model, one can ask whether only one cell type (most likely keratinocytes) dominated the response. This question may be addressed in future studies.
Qualitative differences in the transcriptional responses induced in human skin irradiated ex vivo with low and high doses were as follows. First, gene expression modulated by 0.05 Gy was transient, as indicated by a total absence of gene overlap up to 30 h postexposure, while during the same period persistence of modified gene expression was observed in response to 5 Gy. Second, these differences enabled the classification of modulated genes into three groups: Group 1 contains specific genes defined as genes responsive exclusively to either the low or high dose but not to both and not to time in culture. Group 2 clusters unique genes defined as responsive to either 0.05 or 5 Gy doses but not both, but also regulated in the opposite direction in the time in culture control, which was included in our experimental design. Group 3 contains dose-independent radiation-responsive genes. These gene groups are reminiscent of the groups defined by Yin et al. when analyzing the transcriptional responses to low and high radiation doses in the brain of mice (22).
Group 1 or dose-specific genes were more abundant in the response to 5 Gy than to 0.05 Gy. Early (2 h postexposure) transcriptional responses to 5 Gy targeted pathways involved in cell adhesion, cytoskeleton remodeling and TGFβ-dependent induction of EMT. At 8 h postirradiation, upregulation of CDKN1A was a determinant in the activation of cell cycle regulation and DNA damage control pathways as well as in sustaining the activity of cytoskeleton remodeling and development signaling through TGFβ and Wnt pathways. The late (30 h postexposure) specific transcriptional response to 5 Gy consisted of activated immune response pathways and maintained activity of cell cycle regulation and cytoskeleton remodeling pathways as well as recapitulation of cell adhesion and TGFβ-dependent induction of EMT pathways. Thus, at 30 h postexposure to 5 Gy, skin cells were still struggling to overcome the insult. By contrast, the early specific transcriptional response to 0.05 Gy was aimed primarily at slowing down cellular processes such as late endocytotic transport and inhibiting cellular responses to stress such as NFκB signaling, inflammation, immune response and apoptosis. This contrasts with the transcriptional response of human skin after an in vivo acute exposure to 0.1 Gy, where upregulation of inflammation and apoptotic gene groups was observed (32). On the other hand, early in the treatment, cell death was infrequent in keratinocytes from human skin exposed in vivo to fractionated radiotherapy with low-dose fractions (59). At 8 h postexposure PRMD1 was the only gene modulated by 0.05 Gy; upregulation of this gene encoding a transcriptional repressor, which regulates cell growth via transcriptional repression of TP53 (60), suggests that skin cells were recovering from the low-dose radiation insult. This recovery effect is further supported by the late transcriptional response to 0.05 Gy, where modulated genes were about half upregulated and half downregulated and were distributed over pathways aimed at cell proliferation, transcription, translation and proteolysis. Pathways/functions overlapping between the response of skin exposed in vivo to 0.01 Gy and ex vivo to 0.05 Gy include transcription and cell proliferation/survival (32).
Genes responsive to 5 Gy and previously identified as radiation-responsive genes (Table 2) included SESN1, CCNG1 and CDKN1A, which belong to Group 1 genes, with the exception of CDKN1A, which is a Group 1 and Group 2 gene. Interestingly, all three genes are TP53 target genes.
Group 2 or unique genes were absent at early times (2 and 8 h) postexposure to 0.05 Gy but were present at 8 h after irradiation with 5 Gy. Unique genes were most abundant at 30 h postirradiation at both low and high doses. Functional overlap between low- and high-dose-responsive unique genes was limited to transcription factors, which is consistent with the fact that transcriptional changes are part of the cellular stress response (40). Unique genes responsive to 0.05 Gy functioned in protein and ion transport and nucleotide binding proteins, while those responsive to 5 Gy were involved in tight junctions. These genes were strongly regulated in response to radiation exposure since their expression was opposite in the time in culture control. The effects of time in culture on global gene expression have also been reported in another study analyzing in vitro transcriptional responses of primary human lymphocytes at 7, 17 and 55 days after exposure to 3 Gy (61).
Group 3, with dose-independent radiation-responsive genes, clustered 139 genes, most of which were downregulated. Functions represented by these genes included involvement in membrane and cell junctions, regulation of transcription and protein transport and catabolism.
Out of 24 previously reported radiation-responsive genes, only three, or five if considering gene families, were found modulated by radiation in human skin (Table 2). The fact that the response to radiation of cells grown in 3D culture is dampened by comparison to their counterparts grown in 2D cultures (62) might account in large part for our observation.
The distinct qualitative transcriptional responses observed in ex vivo irradiated skin exposed to low and high radiation doses do not support the LNT model for low-dose radiation risk extrapolation. Maintenance of genome integrity is essential for cancer prevention (63), and therefore the ability of cells to repair damaged DNA is a determinant for cancer risks. Our study highlights clear differences in the DNA damage response induced by high-dose and low-dose radiation. The response of human skin after ex vivo exposure to an acute high dose/high dose rate involved upregulation of TP53 and TGFβ signaling target genes, whereas neither TP53 nor TGFβ target genes were modulated in response to an acute low-dose/high-dose rate exposure. Thus, in human skin irradiated ex vivo with 0.05 Gy, the TP53-dependent DNA damage response was either not triggered or was triggered only briefly, as further supported by the upregulation of PRDM1, a TP53 transcriptional repressor (60). In response to DNA double-strand breaks (DSBs), ATM (ataxia-telangiectasia mutated), a member of the phosphatidyl inositol 3-kinase-like protein kinases, is activated through autophosphorylation (64). In turn, activated ATM phosphorylates TP53, leading to activation of the TP53 signal transduction pathway. Therefore, failure to activate this pathway in response to an acute low dose may reflect a lack of activation of ATM. Reduced activation of ATM has been reported in normal human cells after exposure to low-dose/low-dose rate radiation (65), and ATM was not activated in fibroblasts irradiated with very low doses (~1 mGy) (66).
In addition, a DNA damage response pathway, novel in its association with radiation and of unknown dependence of ATM, was induced in ex vivo irradiated human skin, as evidenced by upregulation of ING2 in response to low and high radiation doses. ING2 mediates transcriptional repression of cell cycle regulatory genes such as cyclins (52). Direct modulation of cancer-linked genes other than TP53 by radiation may represent additional cancer risks in humans, which will further depend on the genetic makeup of each individual. Ex vivo human skin responded to low-dose radiation by upregulating EGFR, CLDN1 and ING2, and overexpression of these genes can cause tumorigenesis (46, 48, 53).
In summary, the limited overlap observed between transcriptional responses of ex vivo irradiated human skin to low and high radiation doses adds to the accumulating body of evidence that questions the validity of the LNT model for low-dose radiation risk assessment (67). In addition, the low-dose transcriptional response was transient, while expression of some high-dose-responsive genes persisted over at least 22 h. In this skin model with presumably normal cells where X-ray doses were delivered at a high dose rate, a low dose (0.05 Gy) failed to activate the TP53-dependent DNA damage response, whereas it was activated upon irradiation with a high dose (5 Gy). Upregulation of ING2, which was triggered by both low and high doses, uncovered a novel mechanism in response to radiation-induced DNA damage. For the first time, radiation is linked to the genotoxic stress response induced by ING2 as part of a chromatin-modifying complex known to repress cell cycle regulatory genes (51). Although this ING2-mediated DNA damage response is not yet fully elucidated, it has become clear that chromatin remodeling contributes to cellular DNA damage responses (68). The evolution of the transcriptional response over 30 h in skin exposed ex vivo to 0.05 Gy is consistent with the conclusion from studies conducted by our laboratory on skin exposed in vivo to 0.01 Gy: in response to low-dose radiation, human skin initiates a transcriptional program to enhance survival (32). Thus the ex vivo irradiated human skin model appears suitable for studying radiation effects on a natural tissue. The radiation-induced DNA damage response mediated by ING2 as well as the radiation dose threshold for activation of the TP53-dependent DNA damage response need to be investigated further, particularly at the protein level. The use of ex vivo irradiated skin will afford the amount of tissue needed for such studies.
Acknowledgments
The authors would like to thank the personnel at the Plastic Surgery clinic and the radiation therapists at the University of California Davis Medical Center for assistance with obtaining patient consents and irradiation of ex vivo skin. This work was supported by grants from the U.S. Department of Energy Office of Biological and Environmental Research, DE-FG03-01ER63237, DE-FG02-07ER64341 and DE-SC0001099, Air Force Office of Scientific Research FA9550-06-1-0132 and FA9550-07-1-0146 and the National Institute of Allergy and Infectious Diseases R21/R33 AI080604.
References
- 1.Anno GH, Young RW, Bloom RM, Mercier JR. Dose response relationships for acute ionizing-radiation lethality. Health Phys. 2003;84:565–75. doi: 10.1097/00004032-200305000-00001. [DOI] [PubMed] [Google Scholar]
- 2.Goodhead DT. Understanding and characterisation of the risks to human health from exposure to low levels of radiation. Radiat Prot Dosimetry. 2009;137:109–17. doi: 10.1093/rpd/ncp191. [DOI] [PubMed] [Google Scholar]
- 3.Brenner DJ, Doll R, Goodhead DT, Hall EJ, Land CE, Little JB, et al. Cancer risks attributable to low doses of ionizing radiation: assessing what we really know. Proc Natl Acad Sci U S A. 2003;100:13761–6. doi: 10.1073/pnas.2235592100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tubiana M, Aurengo A, Averbeck D, Masse R. Recent reports on the effect of low doses of ionizing radiation and its dose-effect relationship. Radiat Environ Biophys. 2006;44:245–51. doi: 10.1007/s00411-006-0032-9. [DOI] [PubMed] [Google Scholar]
- 5.Averbeck D. Does scientific evidence support a change from the LNT model for low-dose radiation risk extrapolation? Health Phys. 2009;97:493–504. doi: 10.1097/HP.0b013e3181b08a20. [DOI] [PubMed] [Google Scholar]
- 6.Snyder AR, Morgan WF. Gene expression profiling after irradiation: clues to understanding acute and persistent responses? Cancer Metastasis Rev. 2004;23:259–68. doi: 10.1023/B:CANC.0000031765.17886.fa. [DOI] [PubMed] [Google Scholar]
- 7.Amundson SA, Bittner M, Fornace AJ., Jr Functional genomics as a window on radiation stress signaling. Oncogene. 2003;22:5828–33. doi: 10.1038/sj.onc.1206681. [DOI] [PubMed] [Google Scholar]
- 8.Tachiiri S, Katagiri T, Tsunoda T, Oya N, Hiraoka M, Nakamura Y. Analysis of gene-expression profiles after gamma irradiation of normal human fibroblasts. Int J Radiat Oncol Biol Phys. 2006;64:272–9. doi: 10.1016/j.ijrobp.2005.08.030. [DOI] [PubMed] [Google Scholar]
- 9.Amundson SA, Do KT, Shahab S, Bittner M, Meltzer P, Trent J, et al. Identification of potential mRNA biomarkers in peripheral blood lymphocytes for human exposure to ionizing radiation. Radiat Res. 2000;154:342–6. doi: 10.1667/0033-7587(2000)154[0342:iopmbi]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 10.Amundson SA, Grace MB, McLeland CB, Epperly MW, Yeager A, Zhan Q, et al. Human in vivo radiation-induced biomarkers: gene expression changes in radiotherapy patients. Cancer Res. 2004;64:6368–71. doi: 10.1158/0008-5472.CAN-04-1883. [DOI] [PubMed] [Google Scholar]
- 11.Chaudhry MA. Biomarkers for human radiation exposure. J Biomed Sci. 2008;15:557–63. doi: 10.1007/s11373-008-9253-z. [DOI] [PubMed] [Google Scholar]
- 12.Paul S, Amundson SA. Development of gene expression signatures for practical radiation biodosimetry. Int J Radiat Oncol Biol Phys. 2008;71:1236–44. doi: 10.1016/j.ijrobp.2008.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park WY, Hwang CI, Im CN, Kang MJ, Woo JH, Kim YS, et al. Identification of radiation-specific responses from gene expression profile. Oncogene. 2002;21:8521–8. doi: 10.1038/sj.onc.1205977. [DOI] [PubMed] [Google Scholar]
- 14.Kang CM, Park KP, Song JE, Jeoung DI, Cho CK, Kim TH, et al. Possible biomarkers for ionizing radiation exposure in human peripheral blood lymphocytes. Radiat Res. 2003;159:312–9. doi: 10.1667/0033-7587(2003)159[0312:pbfire]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 15.Mori M, Benotmane MA, Tirone I, Hooghe-Peters EL, Desaintes C. Transcriptional response to ionizing radiation in lymphocyte subsets. Cell Mol Life Sci. 2005;62:1489–501. doi: 10.1007/s00018-005-5086-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Turtoi A, Brown I, Oskamp D, Schneeweiss FH. Early gene expression in human lymphocytes after gamma-irradiation – a genetic pattern with potential for biodosimetry. Int J Radiat Biol. 2008;84:375–87. doi: 10.1080/09553000802029886. [DOI] [PubMed] [Google Scholar]
- 17.Amundson SA, Bittner M, Meltzer P, Trent J, Fornace AJ., Jr Induction of gene expression as a monitor of exposure to ionizing radiation. Radiat Res. 2001;156:657–61. doi: 10.1667/0033-7587(2001)156[0657:iogeaa]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 18.Franco N, Lamartine J, Frouin V, Le Minter P, Petat C, Leplat JJ, et al. Low-dose exposure to gamma rays induces specific gene regulations in normal human keratinocytes. Radiat Res. 2005;163:623–35. doi: 10.1667/rr3391. [DOI] [PubMed] [Google Scholar]
- 19.Gruel G, Voisin P, Vaurijoux A, Grégoire E, Maltere P, Petat C, et al. Broad modulation of gene expression in CD4+ lymphocyte subpopulations in response to low doses of ionizing radiation. Radiat Res. 2008;170:335–44. doi: 10.1667/RR1147.1. [DOI] [PubMed] [Google Scholar]
- 20.Kurpinski K, Jang DJ, Bhattacharya S, Rydberg B, Chu J, So J, et al. Differential effects of x-rays and high-energy 56Fe ions on human mesenchymal stem cells. Int J Radiat Oncol Biol Phys. 2009;73:869–77. doi: 10.1016/j.ijrobp.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 21.Warters RL, Packard AT, Kramer GF, Gaffney DK, Moos PJ. Differential gene expression in primary human skin keratinocytes and fibroblasts in response to ionizing radiation. Radiat Res. 2009;172:82–95. doi: 10.1667/RR1677.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yin E, Nelson DO, Coleman MA, Peterson LE, Wyrobek AJ. Gene expression changes in mouse brain after exposure to low-dose ionizing radiation. Int J Radiat Biol. 2003;79:759–75. doi: 10.1080/09553000310001610961. [DOI] [PubMed] [Google Scholar]
- 23.Ding LH, Shingyoji M, Chen F, Hwang JJ, Burma S, Lee C, et al. Gene expression profiles of normal human fibroblasts after exposure to ionizing radiation: a comparative study of low and high doses. Radiat Res. 2005;164:17–26. doi: 10.1667/rr3354. [DOI] [PubMed] [Google Scholar]
- 24.Amundson SA, Lee RA, Koch-Paiz CA, Mittner ML, Meltzer P, Trent JM, et al. Differential responses of stress genes to low dose-rate gamma irradiation. Mol Cancer Res. 2003;1:445–52. [PubMed] [Google Scholar]
- 25.Amundson SA. Functional genomics in radiation biology: a gateway to cellular systems-level studies. Radiat Environ Biophys. 2008;47:25–31. doi: 10.1007/s00411-007-0140-1. [DOI] [PubMed] [Google Scholar]
- 26.Amundson SA, Bittner M, Chen Y, Trent J, Meltzer P, Fornace AJ., Jr Fluorescent cDNA microarray hybridization reveals complexity and heterogeneity of cellular genotoxic stress responses. Oncogene. 1999;18:3666–72. doi: 10.1038/sj.onc.1202676. [DOI] [PubMed] [Google Scholar]
- 27.Amundson SA, Do KT, Vinikoor LC, Lee RA, Koch-Paiz CA, Ahn J, et al. Integrating global gene expression and radiation survival parameters across the 60 cell lines of the National Cancer Institute Anticancer Drug Screen. Cancer Res. 2008;68:415–24. doi: 10.1158/0008-5472.CAN-07-2120. [DOI] [PubMed] [Google Scholar]
- 28.Mori M, Benotmane MA, Vanhove D, van Hummelen P, Hooghe-Peters EL, Desaintes C. Effect of ionizing radiation on gene expression in CD4+ T lymphocytes and in Jurkat cells: unraveling novel pathways in radiation response. Cell Mol Life Sci. 2004;61:1955–64. doi: 10.1007/s00018-004-4147-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feinendegen LE, Muhlensiepen H, Bond VP, Sondhaus CA. Intracellular stimulation of biochemical control mechanisms by low-dose, low-LET irradiation. Health Phys. 1987;52:663–9. doi: 10.1097/00004032-198705000-00020. [DOI] [PubMed] [Google Scholar]
- 30.Dressman HK, Muramoto GG, Chao NJ, Meadows S, Marshall D, Ginsburg GS, et al. Gene expression signatures that predict radiation exposure in mice and humans. PLoS Med. 2007;4:e106. doi: 10.1371/journal.pmed.0040106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goldberg Z, Rocke DM, Schwietert C, Berglund SR, Santana A, Jones A, et al. Human in vivo dose-response to controlled, low-dose low linear energy transfer ionizing radiation exposure. Clin Cancer Res. 2006;12:3723–9. doi: 10.1158/1078-0432.CCR-05-2625. [DOI] [PubMed] [Google Scholar]
- 32.Berglund SR, Rocke DM, Dai J, Schwietert CW, Santana A, Stern RL, et al. Transient genome-wide transcriptional response to low-dose ionizing radiation in vivo in humans. Int J Radiat Oncol Biol Phys. 2008;70:229–34. doi: 10.1016/j.ijrobp.2007.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brill-Almon E, Stern B, Afik D, Kaye J, Langer N, Bellomo S, et al. Ex vivo transduction of human dermal tissue structures for autologous implantation production and delivery of therapeutic proteins. Mol Ther. 2005;12:274–82. doi: 10.1016/j.ymthe.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 34.Sivamani RK, Pullar CE, Manabat-Hidalgo CG, Rocke DM, Carlsen RC, Greenhaigh DG, et al. Stress-mediated increases in systemic and local epinephrine impair skin wound healing: potential new indication for beta blockers. PLoS Med. 2009;6:e12. doi: 10.1371/journal.pmed.1000012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Durbin BP, Hardin JS, Hawkins DM, Rocke DM. A variance-stabilizing transformation for gene-expression microarray data. Bioinformatics. 2002;18 (Suppl 1):S105–10. doi: 10.1093/bioinformatics/18.suppl_1.s105. [DOI] [PubMed] [Google Scholar]
- 36.Huber W, von Heydebreck A, Sultmann H, Poustka A, Vingron M. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics. 2002;18 (Suppl 1):S96–104. doi: 10.1093/bioinformatics/18.suppl_1.s96. [DOI] [PubMed] [Google Scholar]
- 37.Klipper-Aurbach Y, Wasserman M, Braunspiegel-Weintrob N, Borstein D, Peleg S, Assa S, et al. Mathematical formulae for the prediction of the residual beta cell function during the first two years of disease in children and adolescents with insulin-dependent diabetes mellitus. Med Hypotheses. 1995;45:486–90. doi: 10.1016/0306-9877(95)90228-7. [DOI] [PubMed] [Google Scholar]
- 38.Huang da W, Sherman BT, Tan Q, Collins JR, Alvord WG, Roayaei J, et al. The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007;8:R183. doi: 10.1186/gb-2007-8-9-r183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–8. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 40.Amundson SA, Do KT, Fornace AJ., Jr Induction of stress genes by low doses of gamma rays. Radiat Res. 1999;152:225–31. [PubMed] [Google Scholar]
- 41.el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, et al. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–25. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 42.Ohtsuka T, Jensen MR, Kim HG, Kim KT, Lee SW. The negative role of cyclin G in ATM-dependent p53 activation. Oncogene. 2004;23:5405–8. doi: 10.1038/sj.onc.1207693. [DOI] [PubMed] [Google Scholar]
- 43.Budanov AV, Sablina AA, Feinstein E, Koonin EV, Chumakov PM. Regeneration of peroxiredoxins by p53-regulated sestrins, homologs of bacterial AhpD. Science. 2004;304:596–600. doi: 10.1126/science.1095569. [DOI] [PubMed] [Google Scholar]
- 44.Tsukita S, Furuse M, Itoh M. Multifunctional strands in tight junctions. Nat Rev Mol Cell Biol. 2001;2:285–93. doi: 10.1038/35067088. [DOI] [PubMed] [Google Scholar]
- 45.Hewitt KJ, Agarwal R, Morin PJ. The claudin gene family: expression in normal and neoplastic tissues. BMC Cancer. 2006;6:186. doi: 10.1186/1471-2407-6-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leotlela PD, Wade MS, Duray PH, Rhode MJ, Brown HF, Rosenthal DT, et al. Claudin-1 overexpression in melanoma is regulated by PKC and contributes to melanoma cell motility. Oncogene. 2007;26:3846–56. doi: 10.1038/sj.onc.1210155. [DOI] [PubMed] [Google Scholar]
- 47.Myal Y, Leygue E, Blanchard AA. Claudin 1 in breast tumorigenesis: revelation of a possible novel “claudin high” subset of breast cancers. J Biomed Biotechnol. 2010;2010:956897. doi: 10.1155/2010/956897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mitsudomi T, Yatabe Y. Epidermal growth factor receptor in relation to tumor development: EGFR gene and cancer. FEBS J. 2010;277:301–8. doi: 10.1111/j.1742-4658.2009.07448.x. [DOI] [PubMed] [Google Scholar]
- 49.Crosby LM, Waters CM. Epithelial repair mechanisms in the lung. Am J Physiol Lung Cell Mol Physiol. 2010;298:L715–31. doi: 10.1152/ajplung.00361.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maruko A, Ohtake Y, Kawaguchi M, Kobayashi T, Baba T, Kuwahara Y, et al. X-radiation-induced down-regulation of the EGF receptor in primary cultured rat hepatocytes. Radiat Res. 2010;173:620–8. doi: 10.1667/RR1793.1. [DOI] [PubMed] [Google Scholar]
- 51.Bua DJ, Binda O. The return of the INGs, histone mark sensors and phospholipid signaling effectors. Curr Drug Targets. 2009;10:418–31. doi: 10.2174/138945009788185112. [DOI] [PubMed] [Google Scholar]
- 52.Shi X, Hong T, Walter KL, Ewalt M, Michishita E, Hung T, et al. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature. 2006;442:96–9. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kumamoto K, Fujita K, Kurotani R, Saito M, Unoki M, Hagiwara N, et al. ING2 is upregulated in colon cancer and increases invasion by enhanced MMP13 expression. Int J Cancer. 2009;125:1306–15. doi: 10.1002/ijc.24437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roy L, Gruel G, Vaurijoux A. Cell response to ionising radiation analysed by gene expression patterns. Ann Ist Super Sanita. 2009;45:272–7. [PubMed] [Google Scholar]
- 55.Bissell MJ, Barcellos-Hoff MH. The influence of extracellular matrix on gene expression: is structure the message? J Cell Sci Suppl. 1987;8:327–43. doi: 10.1242/jcs.1987.supplement_8.18. [DOI] [PubMed] [Google Scholar]
- 56.Fachin AL, Mello SS, Sandrin-Garcia P, Junta CM, Ghilardi-Netto T, Donadi EA, et al. Gene expression profiles in radiation workers occupationally exposed to ionizing radiation. J Radiat Res (Tokyo) 2009;50:61–71. doi: 10.1269/jrr.08034. [DOI] [PubMed] [Google Scholar]
- 57.Munson GP, Woloschak GE. Differential effect of ionizing radiation on transcription in repair-deficient and repair-proficient mice. Cancer Res. 1990;50:5045–8. [PubMed] [Google Scholar]
- 58.Sokolov MV, Smirnova NA, Camerini-Otero RD, Neumann RD, Panyutin IG. Microarray analysis of differentially expressed genes after exposure of normal human fibroblasts to ionizing radiation from an external source and from DNA-incorporated iodine-125 radionuclide. Gene. 2006;382:47–56. doi: 10.1016/j.gene.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 59.Turesson I, Nyman J, Qvarnström F, Simonsson M, Book M, Hermansson I, et al. A low-dose hypersensitive keratinocyte loss in response to fractionated radiotherapy is associated with growth arrest and apoptosis. Radiother Oncol. 2010;94:90–101. doi: 10.1016/j.radonc.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 60.Yan J, Jiang J, Lim CA, Wu Q, Ng HH, Chin KC. BLIMP1 regulates cell growth through repression of p53 transcription. Proc Natl Acad Sci U S A. 2007;104:1841–6. doi: 10.1073/pnas.0605562104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Falt S, Holmberg K, Lambert B, Wennborg A. Long-term global gene expression patterns in irradiated human lymphocytes. Carcinogenesis. 2003;24:1837–45. doi: 10.1093/carcin/bgg134. [DOI] [PubMed] [Google Scholar]
- 62.Sowa MB, Chrisler WB, Zens KD, Ashjian EJ, Opresko LK. Three-dimensional culture conditions lead to decreased radiation induced cytotoxicity in human mammary epithelial cells. Mutat Res. 2010;687:78–83. doi: 10.1016/j.mrfmmm.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 63.Barcellos-Hoff MH. Cancer as an emergent phenomenon in systems radiation biology. Radiat Environ Biophys. 2008;47:33–8. doi: 10.1007/s00411-007-0141-0. [DOI] [PubMed] [Google Scholar]
- 64.Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 65.Collis SJ, Schwaninger JM, Ntambi AJ, Keller TW, Nelson WG, Dillehay LE, et al. Evasion of early cellular response mechanisms following low level radiation-induced DNA damage. J Biol Chem. 2004;279:49624–32. doi: 10.1074/jbc.M409600200. [DOI] [PubMed] [Google Scholar]
- 66.Rothkamm K, Lobrich M. Evidence for a lack of DNA double-strand break repair in human cells exposed to very low x-ray doses. Proc Natl Acad Sci U S A. 2003;100:5057–62. doi: 10.1073/pnas.0830918100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tubiana M, Feinendegen LE, Yang C, Kaminski JM. The linear no-threshold relationship is inconsistent with radiation biologic and experimental data. Radiology. 2009;251:13–22. doi: 10.1148/radiol.2511080671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pandita TK, Richardson C. Chromatin remodeling finds its place in the DNA double-strand break response. Nucleic Acids Res. 2009;37:1363–77. doi: 10.1093/nar/gkn1071. [DOI] [PMC free article] [PubMed] [Google Scholar]