Abstract
Synthetic Escherichia coli consortia engineered for syntrophy demonstrated enhanced biomass productivity relative to monocultures. Binary consortia were designed to mimic a ubiquitous, naturally occurring ecological template of primary productivity supported by secondary consumption. The synthetic consortia replicated this evolution-proven strategy by combining a glucose positive E. coli strain, which served as the system’s primary producer, with a glucose negative E. coli strain which consumed metabolic byproducts from the primary producer. The engineered consortia utilized strategic division of labor to simultaneously optimize multiple tasks enhancing overall culture performance. Consortial interactions resulted in the emergent property of enhanced system biomass productivity which was demonstrated with three distinct culturing systems: batch, chemostat and biofilm growth. Glucose-based biomass productivity increased by ~15, 20 and 50% compared to appropriate monoculture controls for these three culturing systems respectively. Interestingly, the consortial interactions also produced biofilms with predictable, self-assembling, laminated microstructures. This study establishes a metabolic engineering paradigm which can be easily adapted to existing E. coli based bioprocesses to improve productivity based on a robust ecological theme.
Keywords: Synthetic biology, Metabolic engineering, Metabolite exchange, Biofilm, Synthetic consortia
1. Introduction
Naturally occurring microbial populations are optimized by selective pressures and evolution to competitively utilize available resources. Co-occurring species frequently benefit from functional differentiation and metabolite exchange in an ecological strategy termed syntrophy. This consortial strategy is often credited with enabling efficient nutrient cycling and stability against perturbations (Girvan et al., 2005; Kato et al., 2008). Consortial metabolic interactions, generated by eons of selective pressure, can serve as templates for metabolic engineers to build enhanced bioprocess platforms.
The consortia concept is in stark contrast with the traditional bioprocess focus on monocultures and the creation of ‘superbugs’ capable of a wide range of concurrent heterologous processes. Engineering a single microbe to simultaneously optimize multiple tasks which are not consistent with its native functionality represents a major challenge (Brenner et al., 2008). In fact, the ‘superbug’ concept contradicts a common ecological theory regarding stable, competitive ecological functioning; optimization of one trait comes at the price of other traits due to tradeoffs in resource allocation (Carlson and Taffs, 2010; Kneitel and Chase, 2004). When analyzed from an ecological perspective, bioprocesses based on robust consortia should outcompete a single ‘superbug’ at performing multiple, complex tasks (McMahon et al., 2007).
Natural consortia typically host numerous functional guilds including consumers that specialize in catabolizing byproducts of the system’s primary producers (Bateson and Ward, 1988). By consuming byproducts, the consumer guild reduces feedback inhibition and toxicity, improves substrate-product thermodynamic feasibility by modulating concentration dependent free energy ratios and captures reduced carbon and energy that would otherwise be lost. Monoculture-based industrial bioprocesses do not typically exhibit this functionality and often accumulate undesirable levels of byproducts such as acetic acid. Acetic acid is inhibitory, reduces culture yields and can stall Escherichia coli growth at ~5 g/L (De Mey et al., 2007; Lasko et al., 2000; Majewski and Domach, 1990). Byproducts like acetate also represent a waste of substrate carbon which further reduces process productivity, selectivity and yields.
Numerous metabolic engineering studies have explored strategies for reducing acetate accumulation, although each has drawbacks (De Mey et al., 2007). Deletion of the pta-ack pathway, which converts acetyl-CoA into acetate, reduced acetate accumulation but also prevented synthesis of acetyl-phosphate, an important global metabolic regulator (De Mey et al., 2007; Klein et al., 2007; McCleary et al., 1993; Yang et al., 1999). Other strategies have converted acetate into less toxic compounds like isoamyl acetate or acetoin (Aristidou et al., 1994; Dittrich et al., 2005). However, these compounds still represent a loss of substrate and increase host metabolic burden by requiring expression of additional recombinant pathways.
The current study designs, constructs and characterizes synthetic E. coli binary consortia based on the proven ecological template of syntrophy. Syntrophy can be defined broadly as any type of cross feeding between microorganisms (McInerney et al., 2009). Here, the term syntrophy is used to describe nutritional exchanges between individual consortia members which are mutually beneficial (Wintermute and Silver, 2010a). The consortia mimic the common ecological motif of a primary producer/consumer relationship (Taffs et al., 2009). Glucose-consuming E. coli strains serve as the systems’ primary producer while a metabolically engineered glucose-negative strain concurrently consumes primary producer byproducts, improving net biomass production by reducing inhibitory byproduct levels and by utilizing substrate carbon that would otherwise be wasted. Overall consortia functionality differs from wild-type E. coli monoculture functionality by enabling the simultaneous consumption of both glucose and inhibitory byproducts. Wild-type E. coli, due to catabolite repression, will preferentially consume glucose before catabolizing byproducts such as acetate (Nicolaou et al., 2010; Wolfe, 2005). Engineered consortia that simultaneously convert glucose and xylose have been reported but they do not utilize syntrophic metabolite exchange (Eiteman et al., 2008; Eiteman et al., 2009; Minty and Lin, 2010; Trinh et al., 2006). Additional engineered consortia studies have examined interactions between strains that complement auxotrophic deficiencies but with these systems; accumulation of inhibitory, yield lowering byproducts is still a major challenge (Wintermute and Silver, 2010b). The current study focuses on a bioprocess ready platform that demonstrates engineered syntrophy enhancing system performance in batch, continuous, and biofilm based culturing strategies, establishing a new paradigm for metabolic engineering.
2. Materials and methods
2.1. Bacterial strains
E. coli K-12 MG1655 strains were used for all experiments. E. coli MG1655 gene deletion strains were constructed using the KEIO mutant library and successive rounds of P1 viral transductions (Baba et al., 2006; Datsenko and Wanner, 2000). Gene deletions were confirmed via PCR and physiological studies.
2.2. Medium
All reported growth experiments were performed in M9 media (6 g/L NaHPO4, 3 g/L KH2PO4, 1 g/L NH4Cl, 0.5 g/L NaCl, 1 ml/L of 1 M MgSO4•6H2O and 10 ml/L of trace metal stock solution ( 0.55 g/L CaCl2, 0.1 g/L MnCl2*4H20, 0.17 g/L ZnCl2, 0.043 g/L CoCl2•6H2O, 0.06 g/L Na2MoO4•2H2O, 0.06 g/L Fe(NH4)2(SO4)2•6H2O, 0.2 g/L FeCl3•6H2O)). Utilized carbon sources and concentrations are given below. All stock solutions were either autoclaved or filter sterilized separately (Miller, 1992)). Initial medium pH was adjusted to 6.8. Cell enumeration was performed on non-selective Luria-Burtani (LB) media. When necessary, agar was added at 14 g/L.
The experimental glucose concentrations (4 g/L) for batch and chemostat studies were designed to 1) permit comparison with a previously reported study (Trinh et al., 2006) 2) establish glucose-limited chemostat conditions with stabile, predictable steady-state conditions and 3) facilitate comparison of batch and chemostat culture properties.
2.3. Batch culturing
Batch experiments were performed using either shake flasks or batch reactors. Shake flask cultures were grown in 250 ml baffled shake flasks containing 50 ml of M9 minimal media containing an appropriate carbon source (4 g/L glucose or 2 g/L sodium acetate). Shake flasks were incubated at 37°C and agitated at 150 RPM. Samples from the shake flasks were collected aseptically. Total sample volumes collected remained less than ~10% of the total initial culture volume.
Batch reactors (1 L) were operated with 300 ml of M9 with an appropriate reduced carbon source. Batch reactors were incubated at 37°C, agitated with magnetic stir bars at 250 RPM and sparged with air at 1 L/min. Reactors were inoculated with 12 ml of culture, diluted to 0.2 OD600, per strain. Inocula were prepared with fresh overnight cultures grown in M9 minimal media containing either 4 g/L glucose or 2 g/L sodium acetate, depending on strain. Samples were collected aseptically at regular time intervals and analyzed for OD600, pH and extracellular metabolite concentrations.
2.4. Chemostat culturing
Chemostat reactors (1 L) were operated with 300 ml of M9 medium with an appropriate reduced carbon source (4 g/L glucose or 2 g/L sodium acetate). Chemostats were incubated at 37°C and agitated with magnetic stir bars at 250 RPM. High-aeration conditions had an air sparge rate of 1 L min−1 while low-aeration cultures had an air sparge rate of 0.15 L min−1.
Dilution rates (D) were set at 0.1 hr−1 to accommodate the specific growth rate of the acetate specialist strain. Chemostat inocula were prepared in the same manner as reported for batch reactors except 5 ml of inoculum was used for each reactor. Binary systems were inoculated with an initial 1:1 ratio of each strain. Reactors were operated until they achieved steady-state. Steady-state was defined as less than 5% changes in OD600, pH and glucose after a minimum run of 6 residence times. All chemostat experiments were run in triplicate. Biomass concentrations (x) were determined from OD600 vs. cell dry weight (CDW) correlations specific to each reactor condition and strain composition. CDW samples were collected by centrifuging 50 ml of culture broth (1800 g for 20 min). Cell pellets were washed with sterile de-ionized water twice, dried for 24 hours at 99°C in 10 ml glass test tubes, and weighed when the samples reached room temperature.
2.5. Colony biofilm culturing
Colony biofilm cultures were grown according to previously described methods (Anderl et al., 2000; Anderl et al., 2003; Hamilton, 2003; Walters et al., 2003; Zuroff et al., 2010). Colony biofilm culturing systems consisted of M9 agar plates (1% glucose(m/V)) and sterile 0.22 μm pore- 25 mm diameter polycarbonate membranes (GE Water and Process Technologies, K02BP02500). Membranes were aseptically placed on agar plates and inoculated with 100 uL of an exponentially growing culture (diluted to OD600 = 0.1) per strain. Inocula were prepared by collecting exponentially growing cells with centrifugation (1800 g for 10 min) and resuspending in fresh media containing only glucose. Biofilms were incubated at 37°C and aseptically transferred to fresh agar plates every 24 hours. After 72 hours, colony biofilms were aseptically transferred to 5 mL of sterile phosphate buffered saline (PBS) and vortexed vigorously for 30 seconds to separate cells from the membrane. The membrane was discarded and the biofilm suspension was disaggregated for 30 seconds using a tissue homogenizer. The homogenized cell suspension was serially diluted and number of colony forming units (CFU) per biofilm was determined using the drop-plate method (Herigstad et al., 2001).
2.6. Reporter protein based analysis of colony biofilms
Consortia members were transformed with fluorescent reporter protein expressing plasmids to enable visualization of strain position within intact biofilms. Plasmids pRSET-mcitrine and pRSET-td-tomato were used to constitutively express fluorescent yellow-green proteins and red fluorescent proteins respectively (Dr. Tsien, UC, San Diego). Reporter expressing strains were cultivated according to the colony biofilm methods described previously for 7 days. Mature biofilms were covered with TissueTek O.T.C. tissue embedding medium and frozen on dry ice. Embedded biofilms were cut into 5 um thick sections using a Leica CM 1850 cryostat operated at −20°C. Sectioned samples were imaged using epi-fluorescent microscopy (Nikon Eclipse E-800) and standard FITC and TRITC filters. Images were analyzed quantitatively for spatially dependent strain abundance using Imaris (Bitplane) and Metamorph (Molecular Devices) image analysis software.
2.7. Extracellular metabolite analysis
Extracellular metabolite concentrations (glucose, acetate, lactate, fumarate, succinate and formate) were measured using an Agilent 1200 HPLC. Samples were filtered (0.45 um) to remove debris; 20 μl filtrate samples were injected on an HPX-87H column (Bio-Rad) at 40°C, with a 0.005M H2SO4 mobile phase (0.6 ml/min) and analyzed with a variable wave-length detector (VWD) and a refractive index detector (RID). Glucose concentrations were determined using the RID and calibration curves while organic acid concentrations were measured using the VWD (210 nm) and the RID with appropriate calibration curves.
3.Results
3.1. Construction and adaptation of glucose negative, acetate specialist strain
A glucose negative E. coli MG1655 strain (ΔptsGΔptsMΔglkΔgcd) was designed using a previously described E. coli stoichiometric model (Carlson, 2009) and built using the KEIO deletion library (materials and methods). The deletion mutant, designated strain 403, was adapted for growth on acetate over 100 generations in a chemostat (D = 0.1/hr, M9 medium with 2 g/L sodium acetate, pH 6.8). Following adaptation, culture samples were streaked for isolation on M9 and acetate agar plates.
Specific growth rates (μ) for five adapted isolates were compared to the un-adapted parent deletion mutant (strain 403). All five isolates exhibited an improved specific growth rate on M9 + 2 g/L sodium acetate as compared to the parent strain (Table 1). Isolate 5 exhibited the largest increase in maximum specific growth rate (μmax = 0.152 hr−1 +/− 0.009) as compared to the parent deletion mutant (μmax = 0.116 hr−1+/− 0.007). This isolate was designated strain 403G100 and was used for all reported experiments.
Table 1.
Growth rate properties of E. coli deletion mutant 403 (ΔptsGΔptsMΔglkΔgcd) following 100 generation acetate-limited chemostat adaptation. Five isolates recovered from the adaptation experiment were analyzed for improved growth on minimal M9 + acetate medium. ‘% increased growth rate’ refers to improvement in isolate specific growth rate relative to nonadapted parent deletion mutant. Specific growth rates are reported ± 1 standard deviation from replicate shake flask cultures. Isolate 5 (strain 403G100) was used for all reported consortia batch, chemostat and biofilm studies.
| Strain | Specific growth rate (hr−1) | Doubling time (hr) | % Increased growth rate |
|---|---|---|---|
| 403 Isolate 1 | 0.140 ± 0.003 | 4.95 | 21% |
| 403 Isolate 2 | 0.138 ± 0.012 | 5.02 | 19% |
| 403 Isolate 3 | 0.132 ± 0.010 | 5.25 | 14% |
| 403 Isolate 4 | 0.128 ± 0.009 | 5.42 | 10% |
| 403G100 Isolate 5 | 0.152 ± 0.009 | 4.56 | 31% |
| 403 Original | 0.116 ± 0.007 | 5.98 | NA |
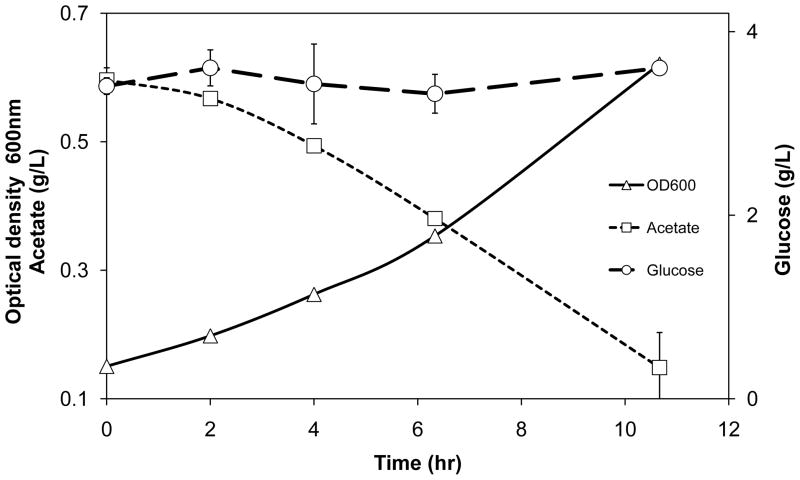
Strain 403G100 preferentially consumed acetate even when cultured in M9 medium containing both sodium acetate and glucose (Fig. 1). Consistent with design, the strain did not grow in M9 medium containing glucose as the sole reduced carbon source.
Figure 1.
Preferential acetate catabolism by glucose negative strain 403G100 cultured in M9 medium containing glucose and sodium acetate. Strain 403G100 did not grow in M9 medium containing glucose as the sole electron donor. Error bars represent ± 1 standard deviation from two independent shake flask experiments.
3.2. Binary consortium batch growth characterization
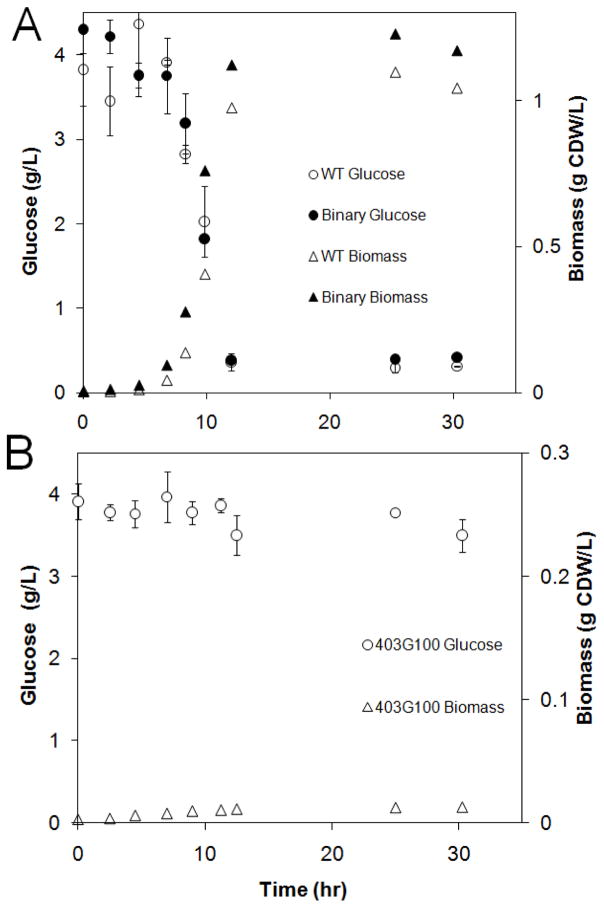
A synthetic binary consortium, comprised of glucose negative 403G100 and wild-type MG1655, was characterized and compared to monoculture properties using batch growth (Fig. 2 and 3). Wild-type monoculture controls reached a final cell density of 1.04 ± 0.02 g CDW/L over 30 hours while the glucose negative strain 403G100 monoculture controls showed negligible growth on glucose (Fig. 2B). The binary consortium produced higher final biomass titers, reaching a final cell density of 1.20 ± 0.04 gCDW/L, a ~15% increase as compared to the monocultures. Consortium and wild-type monoculture volumetric glucose consumption rates were effectively identical; both cultures consumed all glucose within 12 hours (Fig. 2A). The consortium produced ~15% more biomass in the same 12 hour exponential growth phase as compared to the wild-type monoculture resulting in a ~15% higher volumetric biomass productivity (Pbatch = Δx/Δt). Reported values are means from three independent experiments ± one standard deviation. Wild-type monoculture growth rates, glucose yields and acetate yields are consistent with previous publications for this E. coli K-12 laboratory culture (Trinh et al., 2006; Trinh et al., 2008).
Figure 2.
Binary consortium and monoculture batch growth. A) Binary consortium (403G100 + WT) and wild-type monoculture batch growth glucose and biomass concentration time profiles. All experiments were performed with M9 medium supplemented with glucose as the sole reduced carbon source. Error bars represent ± 1 standard deviation from three independent batch experiments. B) Strain 403G100 monocultures did not demonstrate significant growth or glucose consumption. Error bars represent ± 1 standard deviation from three technical replicates of measurements.
Figure 3.
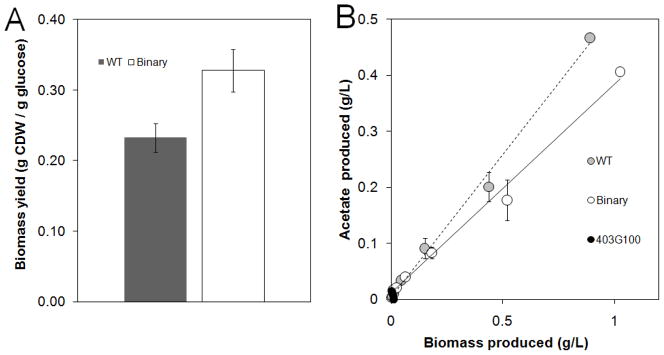
Binary consortium (403G100 + WT) and monoculture batch growth physiological parameters. A) Biomass yield on glucose (g CDW/g glucose) for the engineered binary consortium and wild-type monoculture control. Yields calculated from exponential growth phase data. Glucose negative strain 403G100 did not produce significant growth on glucose. Error bars represent ± 1 standard deviation from three independent batch experiments. B) Exponential growth phase acetate accumulation plotted as a function of biomass from a representative batch growth experiment. Error bars represent ± 1 standard deviation from three technical replicates of measurements.
The binary consortium had an increased biomass yield on glucose and a corresponding reduction in byproduct accumulation as compared to the wild-type monoculture (Fig. 3A). Binary cultures consistently synthesized more biomass per acetate produced during batch growth (Fig. 3B). The reduction in acetate accumulation kept the binary consortium pH higher than the wild-type monoculture likely reducing metabolic stress. In addition, unlike the monoculture, the consortium did not require a metabolically expensive diauxie-based enzyme shift from glucose catabolism to acetate catabolism further improving glucose yields.
3.3. Binary consortium continuous growth characterization
Synthetic consortium properties (wild-type + strain 403G100) were compared to monoculture controls under steady-state chemostat growth (D=0.1/hr). The chemostat dilution rate was selected to accommodate planktonic growth of strain 403G100 (μmax = 0.116 hr−1). The experiments were performed using two distinct aeration regimes, high-aeration and low-aeration (3.3 vpm and 0.5 vpm respectively, vpm = culture volumes per minute). The two aeration regimes were selected because the major metabolite exchanged by the binary culture is thought to be acetate and acetate secretion is negatively correlated to oxygen availability (Alexeeva et al., 2002).
Under both aeration regimes, the binary culture exhibited a ~20% higher steady state biomass concentration which corresponded with a ~20% lower specific glucose uptake rate as compared to the wild-type monocultures (Figs. 4 and 5). Consequently, the binary consortia biomass productivity (Pchemostat = xD) was ~20% higher than the monocultures under both aeration regimes.
Figure 4.

Binary consortium (403G100 + WT) and monoculture biomass concentrations during glucose-limited chemostat cultivation. Cultures were grown continuously in glucose-limited chemostats (D= 0.1/hr) under two different aeration regimes (high-aeration = 1 L/min, low-aeration = 0.15 L/min air sparge). Strain 403G100 monocultures had negligible steady-state biomass concentrations (below detection limit of 0.01 g CDW/L) for both aeration regimes. Error bars represent ± 1 standard deviations from three independent chemostat experiments.
Figure 5.
Binary consortium (403G100 + WT) and monoculture chemostat specific glucose uptake and acetate secretion rates. Cultures were grown continuously in glucose-limited chemostats (D= 0.1/hr) under two different aeration regimes (high-aeration = 1L/min, low-aeration = 0.3 L/min). High-aeration data error bars represent ± 1 standard deviation from three independent chemostat experiments. Low-aeration data error bars represent ± 1 standard deviation from three technical measurements of a representative data set. The low-aeration cultures were highly sensitive to small differences in the rotameter controlled sparge rate. All three low-aeration chemostat experiments exhibited the presented trends.
As expected, high-aeration (3.3 vpm) cultures had higher biomass productivities than low-aeration (0.5 vpm) cultures. Low-aeration growth conditions resulted in ~3-fold higher specific glucose uptake rates and ~3-fold lower biomass productivity, as compared to the high-aeration cultures. Wild-type monoculture acetate and biomass yields and specific glucose uptake rates for the high-aeration chemostats were comparable to previously reported aerobic chemostat values (Trinh et al., 2006; Trinh et al., 2008).
During low-aeration growth, both the binary and wild-type cultures secreted significant amounts of acetate. The binary consortia demonstrated a decreased specific acetate production rate (Fig. 4). During high aeration growth, the binary consortium did not produce measurable amounts of acetate while the wild-type monocultures secreted detectable, albeit small, amounts of acetate.
Acetate specialist monocultures produced negligible biomass (below detection limit of 0.01 g CDW/L) and consumed negligible glucose during glucose only chemostat experiments (Fig. 4).
3.4. Binary consortium colony biofilm characterization
Binary consortium (403G100 + wild-type) and appropriate monocultures were also grown as biofilms to compare culture productivity under non-planktonic conditions. Biofilms represent a common, naturally occurring physiological growth state for microbes and a promising bioprocess platform with high cell densities, high volumetric productivities and increased stress tolerance (Gross et al., 2007; Rosche et al., 2009).
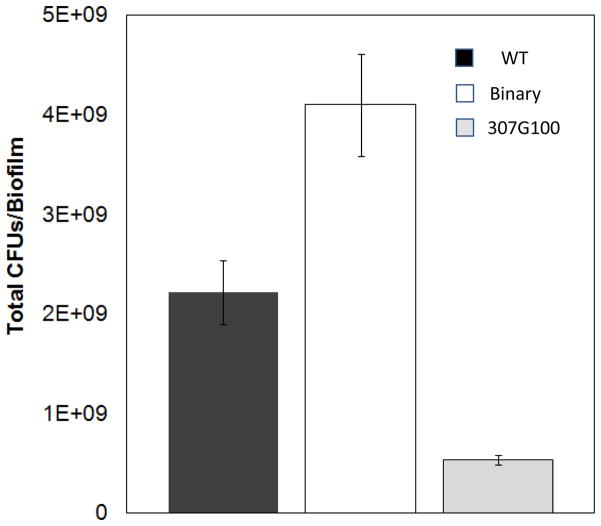
When grown for 72 hours, the binary consortium exhibited higher culture productivities (Pbiofilm = ΔCFU/Δt) than either monoculture individually. The binary system produced approximately 50% more CFUs per biofilm than the sum of the wild-type and 403G100 monocultures (Fig. 6). The large improvement in productivity is likely due to the close spatial proximity of the two strains which facilitates and rewards syntrophic exchanges.
Figure 6.
Colony forming units per biofilm (CFUs/biofilm) for the binary consortium (403G100 + WT) and monoculture controls. Biofilms were cultured on M9 agar containing 1% (w/v) glucose as the sole reduced carbon source. CFUs were enumerated on non-selective (LB) agar. The binary consortium produced more CFUs/biofilm than the sum of monocultures. Error bars represent ± 1 standard deviation from at least three independently cultured biofilms.
3.5 Dual engineered binary consortium
The primary producer/secondary consumer template was further developed with a dual engineered binary consortium. An engineered primary producer strain (E. coli MG1655ΔaceAΔldhAΔfrdA) was designed and constructed to emphasize metabolite exchange thought to be important to the observed syntrophic division of labor. The strain was engineered to prevent acetate from serving as the sole reduced carbon source via an aceA deletion (glyoxylate shunt) and to increase flux toward acetate via removal of lactate and fumarate fermentative pathways (ΔldhA and ΔfrdA respectively). The original gene deletion mutant was adapted to growth on glucose minimal medium over 100 generations of carbon-limited chemostat growth (M9 + 4g/L glucose, D=0.3 hr−1). Recovered isolates were screened for increases in specific growth rate relative to original deletion mutant. The selected isolate designated strain 307G100 demonstrated a μmax of 0.49 ± 0.02 hr−1 compared to 0.43 ± 0.02 hr−1 for the original deletion mutant. Specific growth rates were averaged from three individual experiments, ± one standard deviation. As designed, strain 307G100 did not grow on acetate as the sole reduced carbon source (data not shown).
Functionality of the dual engineered consortium was tested using batch shake flask and colony biofilm cultures. During batch growth, the dual engineered consortium (strains 307G100 + 403G100) and the control monoculture (strain 307G100) exhibited nearly identical volumetric glucose consumption rates (Fig. 7A). The consortium demonstrated a ~10% increase in biomass productivity after the 24 hour exponential growth phase relative to the monoculture control (Fig. 7A). As expected, the consortium accumulated less acetate than the monoculture. Acetate concentrations, measured during stationary phase, were 2.83 ± 0.18 and 2.41 ± 0.01 for the 307G100 monoculture and dual engineered consortium respectively. The dual engineered consortium exhibited a ~50% increase in biomass productivity relative to the monoculture controls when cultured as a biofilm (Fig. 7B).
Figure 7.
Culturing properties of a dual engineered binary consortium comprised of glucose positive strain 307G100 (ΔaceAΔldhAΔfrdA ) and glucose negative strain 403G100. A) Biomass and glucose concentration time profiles for dual engineered binary consortium and monoculture control during batch shake flask cultivation. Data collected from four independent shake flask experiments with different sampling intervals. B) Colony biofilm cultivation data for the dual engineered binary consortium and appropriate monoculture controls grown on M9 medium with glucose as the sole reduced carbon source. Error bars represent ± 1 standard deviation from three independently cultured biofilms.
The dual engineered biofilm cultures demonstrated an additional emergent property; the strains self-organized into a laminated biofilm (Fig. 8A). The two strains were tagged with different fluorescent proteins (pRSET-mcitrine and pRSET-td-tomato, Tsien, San Diego) to permit visual identification. Control biofilms, comprised of two 307G100 strains expressing different fluorescent proteins, did not exhibit the same lamination nor did they grow to the same thickness as the engineered consortium (Fig. 8C). Quantitative image analysis of the average fluorescent intensity supported the visual observation of consortium based lamination (Fig. 8B and 8D). The spatial assembly of the acetate consumer strain (403G100 pRSET-td-tomato) at the biofilm air interface is expected since oxygen concentrations are highest at this location. Hence, the spatial pattern formation observed in the current study is the result of engineered reaction, spatial substrate availability and diffusion phenomena (Kondo and Miura, 2010). Self-assembly of strain specific biofilm spatial structures have previously been observed in synthetic microbial consortia built upon very different culturing techniques and genetic platforms (Brenner and Arnold, 2011).
Figure 8.
Epi-fluorescence micrographs and quantitative image analysis of dual engineered binary consortium (403G100 + 307G100) and strain 307G100 monoculture colony biofilms. A) Dual engineered binary consortium biofilm micrograph with glucose negative strain 403G100 expressing reporter protein td-tomato (red) and glucose positive strain 307G100 expressing reporter protein m-citrine (green). Biofilm image oriented with air-interface on left. Glucose negative strain 403G100 localized primarily at the air interface; red cells at the membrane interface are a result of daily aerobic biofilm plate transfers. Dark regions within biofilm are an artifact of cryosectioning thick biofilms. B) Micrograph of control biofilm comprised of two 307G100 strains each expressing a different fluorescent protein (td-tomato, m-citrine). C) Average fluorescence intensity of red and green reporter proteins as a function of position within biofilm for the dual engineered consortium. Depth is measured from the air interface into the biofilm. D) Average fluorescence intensity as a function of position within the biofilm for strain 307G100 expressing either a red or green fluorescence reporter protein. Fluorescence intensity versus position data is the mean of four line segments chosen at random biofilm positions. Error bars represent ± 1 standard deviation.
4. Discussion
Synthetic binary consortia engineered for syntrophy demonstrated increased biomass productivity under three distinct culturing conditions. This emergent property, based on consortial interactions, improved batch and chemostat culture biomass productivities by approximately 15% and 30% respectively and improved biofilm productivities by approximately 50%. The presented strategy is in strong contrast with traditional metabolic engineering optimization approaches. These approaches frequently aim to optimize glucose-based productivity by deleting genes associated with inefficient metabolic routes or futile cycles (Calhoun et al., 1993; Hua et al., 2003; Trinh et al., 2006). The current work optimizes glucose yields by removing one consortium member’s ability to utilize glucose. When the glucose negative strain is cultured with a glucose consuming strain, the resulting binary consortia can simultaneously optimize both glucose and byproduct catabolism, enhancing overall system productivity.
The deletion-mutant, 403G100, was specifically engineered and adapted for secondary heterotrophy because acetate is often a major byproduct during E. coli growth. Improvements in maximum specific growth rate after adaptation were consistent with previously reported adaptive evolution studies (Conrad et al., 2009; Fong and Palsson, 2004; Ibarra et al., 2002). While acetate served as the representative byproduct for metabolite exchange, other byproduct exchanges are possible. For instance, trace levels of lactate, succinate, fumarate and formate were observed and their exchange would further enhance the system as would the incorporation of additional byproduct metabolizing specialist strains. The engineered ecological role of the current study’s glucose negative strain is quite different than other recent uses of glucose negative strains in artificial consortia (Eiteman and Altman, 2006; Eiteman et al., 2008). In these studies, the glucose negative strain was utilized to co-metabolize a glucose and xlyose sugar mixture, the strain was not designed to metabolize inhibitory byproducts. The presented system is also distinct from a range of synthetic consortia that are built on auxotrophic complementation (Wintermute and Silver, 2010b). These systems are comprised of mutants typically unable to grow in minimal medium without a complementing strain and are arguably not designed for direct incorporation into bioprocess applications. The presented primary producer strains are not dependent on the consumer to grow.
The engineered consortium’s partitioning of metabolic functionality is analogous to cascading trophic levels observed in naturally occurring microbial communities as well as in highly adapted, long term E. coli chemostat cultures (Egland et al., 2004; Kinnersley et al., 2009; Rosenzweig et al., 1994). For instance, the primary producer/consumer relationship spontaneously establishes itself from monocultures during long term cultivation. The presented study demonstrates that it is possible to engineer these naturally occurring ecological patterns into E. coli cultures further establishing the possibility of metabolically and genetically defined model systems for testing microbial ecology phenomena. The consortia are also designed to remove complications associated with multisubstrate growth. The scavenging strain in Rosenzweig et al. utilizes a combination of both glucose and acetate while the current specialist strain cannot catabolize glucose.
While the engineered binary consortia members are designed to not compete for the same reduced carbon source, they may compete for the same electron acceptor (e.g. oxygen). The acetate specialist requires an external terminal electron acceptor to oxidize organic acids while the primary producer strains are facultative. Competition for terminal electron acceptor is a potential limitation of current binary systems. However, building a consortium with community members engineered for partitioned electron donor and electron acceptor (e.g. oxygen and nitrate) functionality would free the strains from resource competition. Again, this is a common theme in naturally occurring systems (Taffs et al., 2009).
The largest emergent increase in culture productivity was observed in biofilms. The culturing environment of the colony biofilm, with the microbes’ close physical proximity, differs considerably from the planktonic batch and chemostat cultures. Biofilms contain inherent chemical gradients including oxygen gradients which produce localized byproduct secretion and accumulation (Stewart and Franklin, 2008; Xu et al., 1998). Colony biofilms serve as an excellent test system for the current engineered binary consortia due to the immobile nature and proximity of cells. The presence of a dedicated acetate-consuming population increases overall population productivity likely by moderating position specific organic acid concentrations and pH while concurrently capturing byproduct carbon. The close physical localization and specialization of metabolism can be compared to interactions between subcellular compartments in eukaryotes. Eukaryotic cellular organization is analogous to microbial communities and the increased productivity observed in the current study could support competitive advantages associated with the eukaryote cellular organization and metabolic regulation.
This study establishes the design, construction and characterization of synthetic E. coli consortia based on productivity enhancing syntrophic metabolic exchange. The metabolic engineering blueprint was demonstrated using three distinct culturing systems including batch, chemostat and biofilms. The study presents proof of concept for a novel bioprocess strategy which mimics an ecological phenomenon found in most natural microbial communities; primary productivity enhancement through interactions with specialized, secondary heterotrophs. Integrating this global ecological theme with synthetic biology represents a powerful tool for optimizing bioprocesses and can be incorporated easily with most existing microbial bioprocess systems.
Highlights.
Ecological strategy, syntrophy, was built into artificial microbial consortia.
Consortia comprised of engineered, byproduct-crossfeeding E. coli strains.
Synthetic consortia demonstrated emergent property of enhanced biomass productivity.
Consortia interactions produced predictable, self-assembling microlaminated biofilms.
New synthetic biology strategy highlights potential biocatalyst platform technology.
Acknowledgments
The authors would like to acknowledge the helpful discussions and assistance of Betsey Pitts and Trevor Zuroff. The authors would also like to thank an anonymous reviewer for insightful comments and suggestions. This work was funded by the National Institutes of Health (NIH) grants (EB006532 and P20 RR16455) and by the National Science Foundation IGERT program in Geobiological Systems (DGE 0654336).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alexeeva S, et al. Quantitative assessment of oxygen availability: perceived aerobiosis and its effect on flux distribution in the respiratory chain of Escherichia coli. J Bacteriol. 2002;184:1402–6. doi: 10.1128/JB.184.5.1402-1406.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderl JN, et al. Role of antibiotic penetration limitation in Klebsiella pneumoniae biofilm resistance to ampicillin and ciprofloxacin. Antimicrob Agents Chemother. 2000;44:1818–24. doi: 10.1128/aac.44.7.1818-1824.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderl JN, et al. Role of nutrient limitation and stationary-phase existence in Klebsiella pneumoniae biofilm resistance to ampicillin and ciprofloxacin. Antimicrob Agents Chemother. 2003;47:1251–6. doi: 10.1128/AAC.47.4.1251-1256.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aristidou AA, et al. Modification of central metabolic pathway in escherichia coli to reduce acetate accumulation by heterologous expression of the bacillus subtilis acetolactate synthase gene. Biotechnol Bioeng. 1994;44:944–51. doi: 10.1002/bit.260440810. [DOI] [PubMed] [Google Scholar]
- Baba T, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2:2006 0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateson MM, Ward DM. Photoexcretion and Fate of Glycolate in a Hot-Spring Cyanobacterial Mat. Applied and Environmental Microbiology. 1988;54:1738–1743. doi: 10.1128/aem.54.7.1738-1743.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner K, Arnold FH. Self-organization, layered structure, and aggregation enhance persistence of a synthetic biofilm consortium. PLoS One. 2011;6:e16791. doi: 10.1371/journal.pone.0016791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner K, et al. Engineering microbial consortia: a new frontier in synthetic biology. Trends Biotechnol. 2008;26:483–9. doi: 10.1016/j.tibtech.2008.05.004. [DOI] [PubMed] [Google Scholar]
- Calhoun MW, et al. Energetic efficiency of Escherichia coli: effects of mutations in components of the aerobic respiratory chain. J Bacteriol. 1993;175:3020–5. doi: 10.1128/jb.175.10.3020-3025.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson RP, Taffs RL. Molecular-level tradeoffs and metabolic adaptation to simultaneous stressors. Curr Opin Biotechnol. 2010;21:670–6. doi: 10.1016/j.copbio.2010.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad TM, et al. Whole-genome resequencing of Escherichia coli K-12 MG1655 undergoing short-term laboratory evolution in lactate minimal media reveals flexible selection of adaptive mutations. Genome Biol. 2009;10:R118. doi: 10.1186/gb-2009-10-10-r118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–5. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Mey M, et al. Minimizing acetate formation in E-coli fermentations. Journal of Industrial Microbiology & Biotechnology. 2007;34:689–700. doi: 10.1007/s10295-007-0244-2. [DOI] [PubMed] [Google Scholar]
- Dittrich CR, et al. Redistribution of metabolic fluxes in the central aerobic metabolic pathway of E. coli mutant strains with deletion of the ackA-pta and poxB pathways for the synthesis of isoamyl acetate. Biotechnol Prog. 2005;21:627–31. doi: 10.1021/bp049730r. [DOI] [PubMed] [Google Scholar]
- Egland PG, et al. Interspecies communication in Streptococcus gordonii-Veillonella atypica biofilms: signaling in flow conditions requires juxtaposition. Proc Natl Acad Sci U S A. 2004;101:16917–22. doi: 10.1073/pnas.0407457101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiteman MA, Altman E. Overcoming acetate in Escherichia coli recombinant protein fermentations. Trends Biotechnol. 2006;24:530–6. doi: 10.1016/j.tibtech.2006.09.001. [DOI] [PubMed] [Google Scholar]
- Eiteman MA, et al. A co-fermentation strategy to consume sugar mixtures effectively. J Biol Eng. 2008;2:3. doi: 10.1186/1754-1611-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiteman MA, et al. A substrate-selective co-fermentation strategy with Escherichia coli produces lactate by simultaneously consuming xylose and glucose. Biotechnol Bioeng. 2009;102:822–7. doi: 10.1002/bit.22103. [DOI] [PubMed] [Google Scholar]
- Fong SS, Palsson BO. Metabolic gene-deletion strains of Escherichia coli evolve to computationally predicted growth phenotypes. Nat Genet. 2004;36:1056–8. doi: 10.1038/ng1432. [DOI] [PubMed] [Google Scholar]
- Girvan MS, et al. Bacterial diversity promotes community stability and functional resilience after perturbation. Environ Microbiol. 2005;7:301–13. doi: 10.1111/j.1462-2920.2005.00695.x. [DOI] [PubMed] [Google Scholar]
- Gross R, et al. Microbial biofilms: new catalysts for maximizing productivity of long-term biotransformations. Biotechnol Bioeng. 2007;98:1123–34. doi: 10.1002/bit.21547. [DOI] [PubMed] [Google Scholar]
- Hamilton M. The Biofilm Laboratory Step-By-Step Protocols for Experimental Design, Analysis, and Data Interpretation. Cytergy Publishing; Bozeman MT: 2003. [Google Scholar]
- Herigstad B, et al. How to optimize the drop plate method for enumerating bacteria. J Microbiol Methods. 2001;44:121–9. doi: 10.1016/s0167-7012(00)00241-4. [DOI] [PubMed] [Google Scholar]
- Hua Q, et al. Responses of the central metabolism in Escherichia coli to phosphoglucose isomerase and glucose-6-phosphate dehydrogenase knockouts. J Bacteriol. 2003;185:7053–67. doi: 10.1128/JB.185.24.7053-7067.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarra RU, et al. Escherichia coli K-12 undergoes adaptive evolution to achieve in silico predicted optimal growth. Nature. 2002;420:186–9. doi: 10.1038/nature01149. [DOI] [PubMed] [Google Scholar]
- Kato S, et al. Network relationships of bacteria in a stable mixed culture. Microb Ecol. 2008;56:403–11. doi: 10.1007/s00248-007-9357-4. [DOI] [PubMed] [Google Scholar]
- Kinnersley MA, et al. E Unibus Plurum: genomic analysis of an experimentally evolved polymorphism in Escherichia coli. PLoS Genet. 2009;5:e1000713. doi: 10.1371/journal.pgen.1000713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein AH, et al. The intracellular concentration of acetyl phosphate in Escherichia coli is sufficient for direct phosphorylation of two-component response regulators. Journal of Bacteriology. 2007;189:5574–5581. doi: 10.1128/JB.00564-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kneitel JM, Chase JM. Trade-offs in community ecology: linking spatial scales and species coexistence. Ecology Letters. 2004;7:69–80. [Google Scholar]
- Kondo S, Miura T. Reaction-Diffusion Model as a Framework for Understanding Biological Pattern Formation. Science. 2010;329:1616–1620. doi: 10.1126/science.1179047. [DOI] [PubMed] [Google Scholar]
- Lasko DR, et al. Bacterial response to acetate challenge: a comparison of tolerance among species. Applied Microbiology and Biotechnology. 2000;54:243–247. doi: 10.1007/s002530000339. [DOI] [PubMed] [Google Scholar]
- Majewski RA, Domach MM. Simple Constrained-Optimization View of Acetate Overflow in Escherichia-Coli. Biotechnology and Bioengineering. 1990;35:732–738. doi: 10.1002/bit.260350711. [DOI] [PubMed] [Google Scholar]
- McCleary WR, et al. Is acetyl phosphate a global signal in Escherichia coli? J Bacteriol. 1993;175:2793–8. doi: 10.1128/jb.175.10.2793-2798.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInerney MJ, et al. Syntrophy in anaerobic global carbon cycles. Current opinion in biotechnology. 2009;20:623–32. doi: 10.1016/j.copbio.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon KD, et al. Integrating ecology into biotechnology. Curr Opin Biotechnol. 2007;18:287–92. doi: 10.1016/j.copbio.2007.04.007. [DOI] [PubMed] [Google Scholar]
- Miller JH. A Short Course in Bacterial Genetics: A Laboratory Manual and Handbook for Escherichia coli and Related Bactera. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1992. [Google Scholar]
- Minty JJ, Lin X. Engineering A Synthetic Microbial Consortium for Efficient Production of Biofuels. AICHE Annual meeting; AICHE, Salt Lake City. 2010. [Google Scholar]
- Nicolaou SA, et al. A comparative view of metabolite and substrate stress and tolerance in microbial bioprocessing: From biofuels and chemicals, to biocatalysis and bioremediation. Metab Eng. 2010;12:307–31. doi: 10.1016/j.ymben.2010.03.004. [DOI] [PubMed] [Google Scholar]
- Rosche B, et al. Microbial biofilms: a concept for industrial catalysis? Trends in Biotechnology. 2009;27:636–643. doi: 10.1016/j.tibtech.2009.08.001. [DOI] [PubMed] [Google Scholar]
- Rosenzweig RF, et al. Microbial Evolution in a Simple Unstructured Environment - Genetic Differentiation in Escherichia-Coli. Genetics. 1994;137:903–917. doi: 10.1093/genetics/137.4.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart PS, Franklin MJ. Physiological heterogeneity in biofilms. Nat Rev Microbiol. 2008;6:199–210. doi: 10.1038/nrmicro1838. [DOI] [PubMed] [Google Scholar]
- Taffs R, et al. In silico approaches to study mass and energy flows in microbial consortia: a syntrophic case study. BMC Syst Biol. 2009;3:114. doi: 10.1186/1752-0509-3-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinh CT, et al. Design, construction and performance of the most efficient biomass producing E-coli bacterium. Metabolic Engineering. 2006;8:628–638. doi: 10.1016/j.ymben.2006.07.006. [DOI] [PubMed] [Google Scholar]
- Trinh CT, et al. Minimal Escherichia coli cell for the most efficient production of ethanol from hexoses and pentoses. Applied and Environmental Microbiology. 2008;74:3634–3643. doi: 10.1128/AEM.02708-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters MC, et al. Contributions of antibiotic penetration, oxygen limitation, and low metabolic activity to tolerance of Pseudomonas aeruginosa biofilms to ciprofloxacin and tobramycin. Antimicrobial Agents and Chemotherapy. 2003;47:317–323. doi: 10.1128/AAC.47.1.317-323.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wintermute EH, Silver PA. Dynamics in the mixed microbial concourse. Genes Dev. 2010a;24:2603–14. doi: 10.1101/gad.1985210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wintermute EH, Silver PA. Emergent cooperation in microbial metabolism. Mol Syst Biol. 2010b;6:407. doi: 10.1038/msb.2010.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe AJ. The acetate switch. Microbiology and Molecular Biology Reviews. 2005;69:12. doi: 10.1128/MMBR.69.1.12-50.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu KD, et al. Spatial physiological heterogeneity in Pseudomonas aeruginosa biofilm is determined by oxygen availability. Appl Environ Microbiol. 1998;64:4035–9. doi: 10.1128/aem.64.10.4035-4039.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YT, et al. Metabolic flux analysis of Escherichia coli deficient in the acetate production pathway and expressing the Bacillus subtilis acetolactate synthase. Metab Eng. 1999;1:26–34. doi: 10.1006/mben.1998.0103. [DOI] [PubMed] [Google Scholar]
- Zuroff TR, et al. Robustness analysis of culturing perturbations on Escherichia coli colony biofilm beta-lactam and aminoglycoside antibiotic tolerance. BMC Microbiol. 2010;10:185. doi: 10.1186/1471-2180-10-185. [DOI] [PMC free article] [PubMed] [Google Scholar]