Abstract
Over the past decade immuno-spin trapping (IST) has been used to detect and identify protein radical sites in numerous heme and metalloproteins. To date, however, the technique has had little application toward non-metalloproteins. In this study, we demonstrate the successful application of IST in a system free of transition metals and present the first conclusive evidence of ·NO-mediated protein radical formation in the HRas GTPase. HRas is a non-metalloprotein that plays a critical role in regulating cell growth control. Protein radical formation in Ras GTPases has long been suspected of initiating premature release of bound guanine nucleotide. This action results in altered Ras activity both in vitro and in vivo. As described herein, successful application of IST may provide a means for detecting and identifying radical-mediated Ras activation in many different cancers and disease states where Ras GTPases play an important role.
Keywords: Ras GTPase, radical-mediated activation, protein radical, immuno-spin trapping
Introduction
Immuno-spin trapping (IST), a technique pioneered by the Mason lab at the National Institute of Environmental Health Sciences (NIEHS), allows for detection of protein radicals via immunological-based techniques [1]. To date, IST has been limited in its applications and has been used nearly exclusively to detect protein radicals in metalloproteins [1-5]. Recent efforts demonstrate IST can be used to detect protein radicals in various non-metalloproteins [6, 7]. These studies, however, still required the active site of a separate metalloprotein to generate the free radical oxidizing species used to produce the protein radical on the target protein [6, 7]. Herein, we demonstrate the first successful application of IST on a system free of transition metals and present evidence supporting transient protein radical formation in a model Ras GTPase under conditions of nitrosative stress. These findings may alter how we test for protein radicals in non-metalloproteins and also promotes the practical application of IST in proteins where radical-mediated processes are suspected.
IST is a simple, yet novel, technique that consists of using an oxidizing species, such as ·NO2, to generate a protein radical. The 5,5′-dimethyl-1-pyrroline N-oxide (DMPO) spin trap is subsequently used to trap the protein radical intermediate. As DMPO traps the protein radical, a covalently-bound DMPO-nitroxide radical adduct is formed. This paramagnetic species can undergo one-electron reduction to the corresponding hydroxylamine or one-electron oxidation to the diamagnetic DMPO-nitrone. Of these redox states, only the nitrone species is thermodynamically stable [1]. The stability of this species permits the use of recently developed anti-DMPO antibodies to detect bound DMPO-nitrone protein adducts using highly sensitive enzyme-linked immunosorbent assays (ELISAs) or simple Western blotting [1]. One major advantage IST offers is the ability to detect protein radicals under various experimental conditions on the bench top without the need for an electron spin resonance (ESR) spectrometer. IST can also be conducted at much lower protein concentrations than required for typical spin trapping ESR experiments (μg vs. mg quantities). Moreover, as DMPO adduction adds ~111 Da to the molecular weight of the protein, tandem mass spectrometry (MS/MS) approaches can be utilized to unambiguously identify protein radical sites [3-5, 8, 9]. Perhaps the greatest advantage of immuno-spin trapping is its suitability for detecting protein radical events in living cells (i.e., cell cultures and/or in vivo) [10].
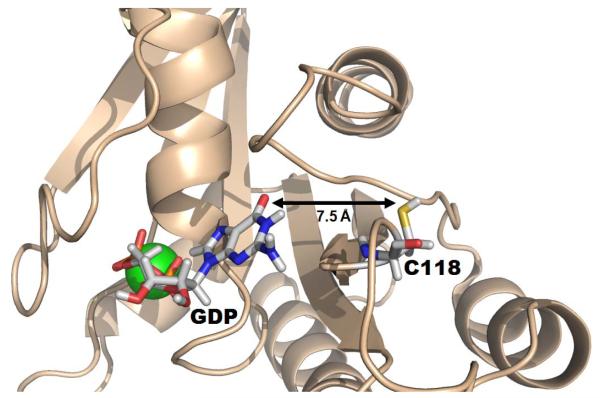
The HRas GTPase was chosen for the current study for two main reasons. First, numerous studies demonstrate small free radical oxidants can alter Ras activity both in vitro and in vivo [11-15]. Small free radical oxidants, such as nitrogen dioxide (·NO2), are hypothesized to promote guanine nucleotide exchange through generation of a transient Ras thiyl protein radical centered at Cys118 [12-15]. We have suggested elsewhere that electron transfer between the thiyl protein radical and bound guanine nucleotide initiates premature release of the nucleotide. This process can result in exchange of GTP for GDP and activation of the Ras protein in vivo [12-15]. As seen in Figure 1, the nearest distance between the Cys118 sulfhydryl and bound GDP is ~ 7.5 Å, according to the 1CRR NMR structure [16]. Electron transfer over such a distance is common given a suitable pathway for the transfer exists. Currently, only indirect evidence supports thiyl radical formation of Ras Cys118 in the presence of a free radical oxidant.
Figure 1. NMR solution structure (pdb 1CRR) of GDP-bound HRas.
Bound GDP and the Cys118 side chain are highlighted in sticks (Mg2+ is shown in green). Approximately 7.5 Å separates bound GDP from the sulfhydryl on Cys118.
Second, Ras GTPases are considered one of the most prevalent oncoproteins in human cancer. Mutations in Ras proteins are present at high levels in pancreatic (~90 %), colorectal (35-45 %), and lung (~30 %) cancers [17]. Recent studies have also linked endogenous nitric oxide (·NO), released from active endothelial nitric oxide synthase (eNOS), to enhanced tumor initiation and maintenance in oncogenic Ras-driven pancreatic cancer [18]. Previous in vitro studies from our lab demonstrated that S-nitrosation of Ras at Cys118 does not affect Ras activity [19]. These observations, suggest that thiyl radical production at Cys118, rather than Cys118 S-nitrosation, may be a key factor for ·NO-mediated regulation of Ras activity. We hypothesize the autoxidation product of ·NO, ·NO2, may contribute to Ras activation during eNOS-enhanced pancreatic tumorigenesis through production of a transient Ras protein radical. Successful detection of the Ras protein radical using IST-based approaches in vitro may lay the groundwork for future tests in cancer cell lysates and/or animal models.
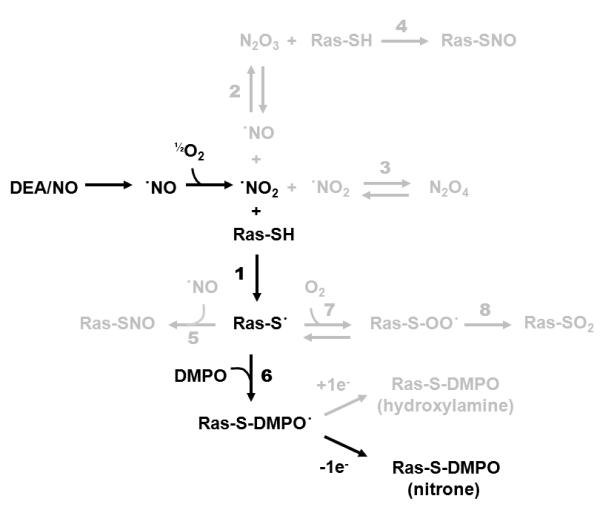
For the current study, ·NO2 oxidant was generated by autoxidation of ·NO liberated from the compound 2-(N,N-diethylamino)-diazenolate-2-oxide diethylammonium salt (DEA/NO). As opposed to bolus addition, the slow release of ·NO from DEA/NO is expected to be more representative of cellular ·NO production by active eNOS. As shown in Figure 2 (black pathway) and Table 1, detection of DMPO-nitrone adducts by IST involves a multitude of kinetic steps beginning with the autoxidation of liberated ·NO to produce ·NO2 and other higher NO-oxides [15, 20-29]. Slow release of ·NO not only simulates active eNOS, but also helps limit formation of the non-radical oxidant dinitrogen trioxide (N2O3) [30]. Competing reactions (grey pathways in Figure 2), unfavorable reaction rates, and low-yields of DMPO adduction highlight the challenge of applying IST in non-metalloproteins. The reactions and associated kinetic parameters for all pathways are listed in Table 1.
Figure 2. Ras immuno-spin trapping reaction diagram.
The black pathway shows the primary reaction steps involved in ·NO-mediated Ras immuno-spin trapping experiments. The grey pathways highlight competing reactions associated with the experiment. Reactions and kinetic parameters associated with all reaction steps are shown in Table 1.
Table 1.
Reaction and kinetic parameters associated with the Ras immuno-spin trapping pathways illustrated in Figure 2
| Reaction Number |
Reaction | kfor | krev | K | Ref. |
|---|---|---|---|---|---|
| 1 | ·NO2 + Ras-SH → Ras-S· | ≥ 2 × 107 M−1·s−1a | n/a | n/a | [15,20] |
| 2 | ·NO2 + ·NO ↔ N2O3 | 1.1 × 109 M−1 ·s−1 | 4.3 × 106 s−1 | 2.6 × 102 M−1 | [20,21] |
| 3 | ·NO2 + ·NO2 ↔ N2O4 | 4.5 × 108 M−1·s−1 | -- | 7 × 104 M−1 | [21-23] |
| 4 | N2O3 + Ras-SH → Ras-SNO | 1.6 × 103 M−1·s−1a | n/a | n/a | [15,24] |
| 5 | Ras-S· + ·NO → Ras-SNO | 3 × 109 M−1·s−1a | n/a | n/a | [25,26] |
| 6 | Ras-S· + DMPO → Ras-S-DMPO· | 2.6 × 108 M−1·s−1a | n/a | n/a | [15,27] |
| 7 | Ras-S· + O2 ↔ Ras-SOO· | 2.2 × 109 M−1·s−1a | 6.2 × 105 s−1a | 3.3 × 103 M−1a | [28,29] |
| 8 | Ras-SOO· → Ras-SO2 | 2 × 103 s−1a | n/a | n/a | [28] |
Kinetic rates reported are based on reactions using reduced glutathione (GSH).
As previously stated, IST has traditionally been used to detect protein radicals in heme proteins. The high oxidation potential of the compound I π-cation radical, formed post addition of H2O2, drives protein radical formation and DMPO-nitroxide adduction in heme proteins, such as myoglobin (Mb) [1]. The high-valence oxoferryl (FeIV=O) compound II intermediate then serves as a second oxidizing equivalent to further oxidize the DMPO-nitroxide to the DMPO-nitrone. This zwitterionic DMPO-nitrone species acts as the specific epitope recognized by the anti-DMPO antibodies [1-5, 8, 9]. Needless to say, it becomes difficult to match the efficiency of DMPO adduction and nitrone conversion observed in heme proteins to that of a non-metalloprotein, such as HRas.
Material and Methods
Protein Expression and Purification
All studies were conducted using a truncated form of human HRas (1-166), which lacks the C-terminal hyper-variable region. Previous studies have shown that the C-terminus of HRas is unstructured and its deletion does not affect nucleotide binding, release, or hydrolysis [31]. While removal of the hyper-variable region does not affect activity in vitro, it is required for posttranslational lipid modification and membrane localization in vivo [32]. As lipid modification does not occur in E. coli, inclusion of this region could lead to the exposure and potential oxidation of cysteines near the C-terminus that does not normally occur. HRas (1-166) was inserted into the pQLinkH vector containing a cleavable 6X N-terminal His-tag [33] and transformed into BL21 (DE3)-RIPL E. coli cells (Stratagene). The RIPL cells were used to supplement tRNAs for poorly expressed E. coli codons. The cells were plated onto LB agar plates containing 100 μg/mL ampicillin (Amp) and allowed to grow overnight at 37 °C. Colonies were isolated and a 250 mL LB broth (100 μg/mL Amp) starter growth was allowed to grow overnight at 37 °C with shaking. Twenty mL of the starter growth were then added to 1 L of LB broth (100 μg/mL Amp) and grown at 37 °C with shaking until an OD600 of 0.5 was reached. Isopropyl β-D-1-thiogalactopyranoside (IPTG) was added to a final concentration of 1 mM and the cells were grown for an additional 18 hrs at 18 °C before collection via centrifugation. Resuspension of the cells in lysis buffer (50 mM Hepes, 150 mM NaCl, 10 mM imidazole, 5 mM MgCl2, 100 μM tris(2-carboxyethyl)phosphine (TCEP), 50 μM GDP, pH 7.75) was performed prior to sonication using a Fisher Scientific 550 Sonic Dismembrator. Centrifugation was used to separate soluble HRas from cell debris. Soluble HRas was loaded onto a Ni-NTA agarose column (Qiagen), treated with wash buffer (50 mM Hepes, 1 M NaCl, 20 mM imidazole, 5 mM MgCl2, 100 μM TCEP, 50 μM GDP, pH 7.75), and eluted using elution buffer (50 mM Hepes, 150 mM NaCl, 300 mM imidazole, 5 mM MgCl2, 100 μM TCEP, 50 μM GDP, pH 7.75). The 6X His-tag was cleaved from purified HRas by overnight incubation with 6X His-tagged tobacco etch virus (TEV) protease (~2-3 mg/mL) at 4 °C. His-tagged TEV was removed by a second pass through a Ni-NTA agarose column. The concentration of GDP-bound Ras was determined using Abs280 with a molar absorptivity of 13530 M−1 cm−1. The final yield of purified HRas was found to be ~18 mg/L broth with purity greater than 95 % as determined by SDS-PAGE.
Immuno-spin Trapping Experiments
For the immuno-spin trapping experiments, F28L (1-166), or F28L/C118S HRas (1-166), were exchanged and concentrated using chelexed 100 mM potassium phosphate (pH 7.4) buffer containing 5 mM MgCl2, 50 μM GDP, and 25 μM diethylene triamine pentaacetic acid (DTPA). In addition, 100 mM DMPO and 0.25 - 1 mM DEA/NO were added to ~350 μM protein (500 μL total volume) and reactions were allowed to proceed for 2-10 mins at 37 °C in 1.5 mL closed Eppendorf tubes. After the brief incubation at 37 °C, the Eppendorf tubes were un-capped and the samples were exposed to open air at 25 °C for 1 hr. The samples were diluted to ~20 μM in H2O, and SDS-PAGE was performed under non-reducing conditions (i.e., no β-mercaptoethanol present in the loading buffer). Electrophoretic transfer to a nitrocellulose membrane (12 V, 1 hr) was performed using a Genie blotter (Idea Scientific) prior to overnight blocking using 4% fish gelatin in phosphate buffered saline (PBS; pH 7.4). The blot was washed for 2 min in wash buffer (0.2 % fish gelatin, 0.05 % Tween in PBS, pH 7.4) before being subjected to 1 hr treatment with 1 : 5000 rabbit polyclonal anti-DMPO antibodies in wash buffer [2]. The membrane was subsequently washed for 5 min 4X in wash buffer and treated with alkaline phosphatase conjugated anti-rabbit IgG (1 : 5000) for 1 hr. After a second 4X wash, the membrane was treated with 5 mL of pH 9.6 TRIS buffered saline (TBS) containing CDP-Star (50 μL, Roche) and Tropix Nitro-Block II™ (250 μL, Applied Biosystems). The resulting chemiluminescent product was captured using radiographic film. Band intensities of SDS-PAGE and Western blots were quantified using ImageJ software.
MS Data Collections
Acquisition of MS data were performed using a hybrid Qe-Fourier Transform Ion Cyclotron Resonance Mass Spectrometer equipped with a 12.0 Tesla actively shielded magnet (12.0 T AS, Apex Qe-FTICR-MS, Bruker Daltonics, Billerica, MA, USA) and an Apollo II microelectrospray (μESI) source. The voltages on the μESI spray capillary, spray shield, capillary exit, deflector, ion funnel and skimmer were set at +4.2 kV, +3.6 kV, +340 V, +310 V, +185 V, and +25 V, respectively. Temperature of the μESI source was maintained at 180 °C. Desolvation was carried out using a nebulization gas flow (2.0 bar) and a countercurrent drying gas flow (4.5 L/s). HRas samples were prepared by re-suspending lyophilized protein in a mixture of acetonitrile/water/acetic acid (49.0 : 49.0 : 2.0 v/v/v) at a concentration of ~0.2 μg/μL. The samples were directly infused using a syringe pump (Harvard Apparatus, Holliston, MA, USA) and a 100 μL syringe (Hamilton, Reno, NV, USA), and electrosprayed at an infusion flow rate of 120 μL/hr. Before transfer, ion packets were accumulated inside the collision cell for 1.0 second, and 100 scans per spectrum were acquired in the ICR cell with a resolution of 580,000 at m/z 400 Da.
The primary method used to characterize the site of Ras DMPO adduction was Fourier transform ion cyclotron resonance mass spectrometry (FTICR-MS) coupled with electron-capture dissociation capability (ECD-MS/MS). Precursor ions of HRas were isolated in a quadrupole (Q1) and subjected to ECD-MS/MS in the ICR cell directly. A precursor ion isolation window width was fixed at 2.0 Da on the ion packets that were accumulated inside the collision cell for 10 seconds. Low energy electrons were generated by the heated hollow dispenser cathode with a bias voltage of -2.5 V, and the ECD lens voltage was set at +15.0 V. The electrons, produced by the hollow dispenser cathode (operated at 1.7 A), were pulsed into the ICR cell for a period of 3.0 ms, which led to dissociation of the precursor protein ions that were trapped in the ICR cell. To maximize the ion population before irradiation, the ICR cell was filled with 5 iterations of ion accumulation from the external collision cell, and 100 MS/MS scans per spectrum were acquired with a resolution of 580,000 at m/z 400 Da.
Results
Immuno-spin trapping allows for detection of protein radicals via immunological-based techniques. However, the technique has been used almost exclusively for detection of protein radicals in metalloproteins. Herein, we employed IST to detect thiyl radical formation in the non-metalloprotein HRas. In an effort to overcome competing reactions (Figure 2, grey pathways) and enhance HRas protein radical detection, two adjustments were made in our experimental design. First, a phenylalanine-to-leucine point mutation was introduced at site 28 in our HRas (1-166) construct. The F28L mutation destabilizes nucleotide binding and results in faster rates of intrinsic guanine nucleotide dissociation in both HRas and Cdc42 (a Ras-related GTPase) [13, 34-36]. Structural studies indicate this mutation creates small perturbations that are localized to the nucleotide binding site and does not alter the secondary structure of the protein [34-36]. Given the critical role of Cys118 in ·NO-mediated Ras guanine nucleotide exchange [11, 12], we anticipate that treatment of wt Ras with ·NO2 will initially generate a thiyl radical. Electron transfer between the thiyl radical and guanine nucleotide base will be extremely fast and ultimately limits our ability to detect Ras thiyl radical formation. The faster rate of intrinsic nucleotide exchange implies a decrease in the amount of bound nucleotide available for electron transfer events. We, therefore, anticipate this mutant will enhance the lifetime of the Ras thiyl protein radical and increase our ability to trap the species. Second, to facilitate the reaction between HRas and ·NO2, relatively high concentrations of protein were used (~350 μM). The reaction kinetics associated with the HRas IST experiments are listed in Table 1.
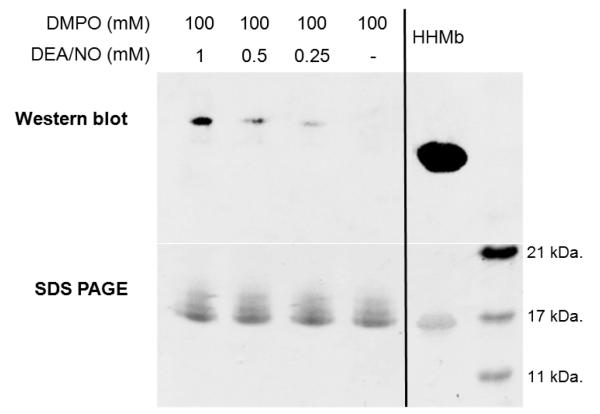
HRas, containing a cysteine-to-serine point (C118S) mutation, was used as a negative control for the IST experiments. This mutation does not alter the structure or biochemical properties of Ras and has used been extensively as a negative control for numerous studies of Ras involving reactive nitrogen species [15]. For the IST reactions, buffered F28L and F28L/C118S HRas (1-166) were allowed to react with varying concentrations of DEA/NO and DMPO in an open reaction vial for 1 hr. Best results were obtained when reacting ~350 μM protein with 1 mM DEA/NO and 100 mM DMPO in chelexed phosphate buffer (pH 7.4). The F28L HRas immunoblot obtained post-reaction using an anti-DMPO polyclonal antibody, and its corresponding SDS-PAGE gel, is shown in Figure 3. Treatment of F28L HRas with sub-stoichiometric to ~3-fold excess DEA/NO resulted in increasing amounts of Ras-DMPO nitrone adducts. In the absence of DEA/NO, Ras-DMPO adducts were not observed. These analyses were performed in triplicate with comparable results in each run.
Figure 3. Immunoblot and SDS-PAGE gel of the Ras IST experiment.
Immunoblot (top) and SDS-PAGE (bottom) gels are shown for DEA/NO treated F28L HRas in the presence of DMPO. For the IST treatment, ~350 μM of Ras was exposed to varying concentrations of DEA/NO (0, 0.25, 0.5, and 1.0 mM) in the presence of 100 mM DMPO. The Ras protein was allowed to react with DEA/NO and DMPO for 2-10 min at 37 °C in a closed Eppendorf tube. After 10 min, the Eppendorf tube containing the reaction mixture was opened and left at 25 °C for 1 hr. As the concentration of DEA/NO is increased, a concomitant increase in the Ras-DMPO nitrone adduct is observed. For the positive control, 10 μM HHMb was treated with 100 μM H2O2 in the presence of 100 mM DMPO in a closed Eppendorf tube at 25 °C for 1 hr. Here, 3.2 μg of Ras were loaded into each lane, and 1.0 μg of the positive HHMb control was loaded into the last lane (far right).
Quantification of the Western band intensities in Figure 3 show that addition of 0.25 mM and 0.5 mM DEA/NO yields bands that are 13.8 and 36.4 % the respective intensity of the 1 mM DEA/NO band. While DEA/NO concentration greatly affects the Western band intensity, only a 6.5 % relative standard deviation (% RSD) is observed in the SDS-PAGE band intensities for the 1, 0.5, 0.25, and 0 mM DEA/NO samples. As seen in Figure 3, the disparity between Ras-DMPO adduction and the positive control horse heart myoglobin (HHMb) is significant although the concentration of Ras in the blot is over 3-fold higher than that of HHMb (3.2 μg Ras/lane; 1 μg HHMb/lane). Densitrometry quantification corroborates these observations as the SDS-PAGE band intensity of the HHMb control was found to be 71 % less intense than the average of the Ras SDS-PAGE bands. As discussed in the introduction, IST is much more efficient for heme proteins, like HHMb, due to the high oxidation potential of the reactive compound I intermediate formed after addition of H2O2. Therefore, a much higher concentration of the positive control HHMb-DMPO adduct is to be expected.
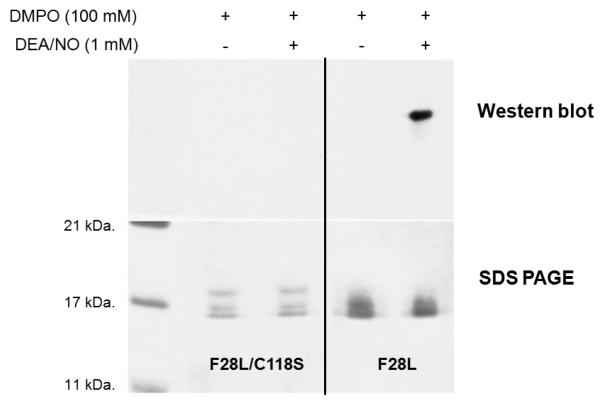
To establish ·NO2 as a one-electron oxidation source, we performed the reaction under identical conditions (~350 μM protein, 100 mM DMPO, pH 7.4) in a sealed, deoxygenated reaction vial containing ·NO2 (g) at ~30:1 excess over Ras. Samples exposed to ·NO2 (g) gave rise to observable Ras-DMPO adducts in the subsequent immunoblot (Supporting Information). Ras-DMPO adducts were not detected in the absence of ·NO2 (g) under identical conditions. To determine if Ras protein radical formation is centered at redox active Cys118, IST experiments were also conducted on a F28L/C118S HRas double mutant. In a side-by-side comparison, F28L/C118S HRas and F28L HRas were treated with identical concentrations of DEA/NO (1 mM) and DMPO (100 mM). DMPO adducts were not observed in the F28L/C118S HRas samples, while positive adducts were observed in the F28L HRas samples (Figure 4). These data were collected in duplicate with comparable results. The % RSD for the F28L/C118S HRas bands were found to 7.4 %, while an 11.4 % RSD was found for the F28L HRas bands in the SDS-PAGE gel.
Figure 4. Immunoblot and SDS-PAGE gel of an IST experiment utilizing a C118S redox-inactive Ras mutant.
Immunoblot (top) and SDS-PAGE gels (bottom) are shown comparing identical reactions of F28L/C118S and F28L HRas using 1 mM DEA/NO and 100 mM DMPO. For both reactions, ~350 μM of F28L/C118S and F28L HRas were exposed to 1.0 mM DEA/NO in the presence of 100 mM DMPO. Both reactions were allowed to proceed for 2-10 min at 37 °C in a closed Eppendorf tube and for an additional 1 hr in an open Eppendorf tube at 25 °C. For the gel and immunoblot, 3.2 μg of F28L HRas and 3.0 μg of F28L/C118S HRas were used for loading. The F28L/C118S HRas mutant shows no evidence of DMPO adduction in the Western blot, while treatment of F28L HRas under the same conditions results in observable DMPO adducts (top panel).
As DMPO adduction results in a mass increase of ~111 Da, complementary MS experiments were employed for site-specific determination of DMPO modification sites. MS experiments on DEA/NO / DMPO treated F28L HRas (~350 μM protein, 1 mM DEA/NO, 100 mM DMPO) were carried out using μESI FTICR-MS ECD on a Bruker Daltonics spectrometer. FTICR-MS with ECD is a top-down mass spectrometry approach that provides sub-nanomolar detection limits on intact proteins without the need for sample proteolysis [37-39]. This approach was advantageous for our current effort given the expected low yield of DMPO adduction.
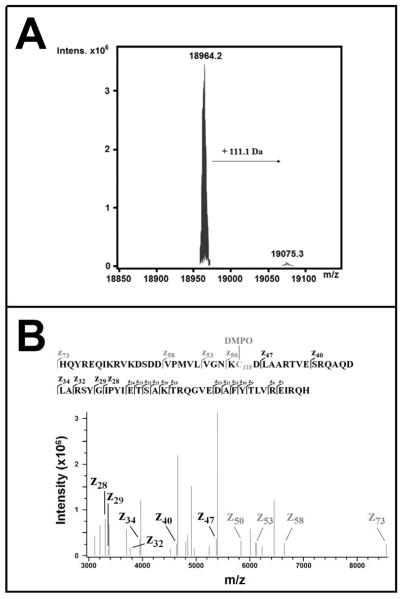
According to its amino acid sequence (Supporting Information), the theoretical average mass of F28L HRas (1-166) ([M+H]+) is 18964.2 Da. Using μESI-FTICR-MS, exact mass measurements of DEA/NO / DMPO treated F28L HRas identified a major peak at the theoretical average mass (18964.2 Da, with mass error < 1 ppm in the undeconvoluted spectrum). As seen in Figure 5A, a second peak was observed that exactly matched the theoretical average mass of F28L HRas with adduction of a single DMPO molecule (19075.3 Da, with mass error < 1 ppm in the undeconvoluted spectrum). This second peak displayed approximately 2 – 3 % the intensity of the non-modified F28L HRas peak and was consistent with estimated levels of DMPO adduction observed in the IST experiments (Figure 3).
Figure 5. Mass spectral analysis of HRas after IST treatment.
Panel A: Top-down μESI-FTICR-MS analysis (100 scans) on F28L HRas obtained post reaction with 1 mM DEA/NO and 100 mM DMPO. Exact mass measurements of F28L HRas identify non-modified protein (18964.2 kDa, mass error < 1 ppm in the undeconvoluted spectrum) and DMPO-adducted F28L HRas (19075.3 kDa, mass error < 1 ppm in the undeconvoluted spectrum). The + 111.1 mass shift is consistent with the mass addition of 1 DMPO molecule. The modified protein is observed at ~2 - 3 % the intensity of unmodified protein. Panel B: ECD-MS/MS spectrum of the resulting mass fragments of DMPO-modified F28L HRas. The sequence and fragmentation pattern for the z73 – z5 fragment ions is shown at the top. In the ECD-MS/MS spectrum (bottom, mass error < 1 ppm), +111.1 Da mass shifts are observed in the z50, z53, z58, and z73 fragment ions (grey) indicating DMPO adduction only in the fragments containing Lys117, Cys118, and Asp119.
To identify the specific DMPO adduction site(s), precursor ions (m/z 830.3 Da, 23+ charge state) corresponding to DMPO-modified protein were isolated for top-down mass spectrometry experiments using ECD-MS/MS (Figure 5B). Inspection of the c- and z-type fragment ions in the ECD-MS/MS spectrum revealed +111.1 Da mass shifts in the z50, z53, z58, and z73 ions (Figure 5B, grey). No other c- or z-type fragment ions (c3 – c35; z5 – z47) showed mass shifts consistent with DMPO adduction (see Supporting Information for the full ECD-MS/MS spectrum). These data indicate DMPO adduction is localized to one of the three amino acids differentiating the z47 and z50 fragment ions (K117, C118, and D119). As DMPO adducts were not observed in the C118S IST control experiments (Figure 4), and because the oxidizing potential of Asp and Lys residues are very high in comparison to Cys, we confidently assign Cys118 as the site of DMPO adduction under our experimental conditions (~350 μM protein, 1 mM DEA/NO, 100 mM DMPO, pH 7.4).
Discussion
This report highlights the first conclusive evidence of protein radical formation in the non-metalloprotein HRas. A combination of IST (Figure 4) and FTICR-ECD-MS/MS (Figure 5A and 5B) approaches confirm DMPO adduction occurs at redox active Cys118. While in vitro IST data provide qualitative evidence of Ras protein radical formation, particularly at Cys118, it does not eliminate the possibility of secondary protein radicals formed through intramolecular electron transfer of the initial Cys118 thiyl radical. This hypothesis is reasonable given the propensity of protein radicals to undergo electron transfer, sometimes even in the presence of DMPO [4, 40, 41]. Nonetheless, the combination of IST and MS data in this report demonstrate protein radical formation in HRas in the presence of nitric oxide in vitro. The question remains whether or not IST will be efficient enough to detect Ras protein radicals in cell cultures or animal models. For the current study, an F28L HRas mutant was chosen because of its fast intrinsic guanine nucleotide exchange rate. In vivo, Ras GTPases will interact with numerous guanine nucleotide exchange factors (GEFs) that enhance nucleotide exchange rates. These interactions should facilitate detection of Ras protein radical intermediates.
A caveat of using more traditional routes of protein radical detection, such as ESR, lies in the percent of Ras-DMPO adduction. Typically ESR requires much higher protein concentrations, particularly when spin trapping experiments are involved. This alone makes spectroscopic techniques, like ESR, less attractive when studying proteins that are susceptible to aggregation or precipitation at higher concentrations (> 1 mM). Moreover, a greater emphasis is being placed on techniques that allow detection of protein radical events in cell cultures or in vivo systems. Under typical physiological conditions, the low levels of molecular oxygen and high reducing environment of cells would appear to be an issue for translation of IST into a cellular context. Cancer cells, however, exhibit a well-established altered redox environment and often contain altered levels of antioxidant enzymes, small molecule oxidants, and small molecule antioxidants [42, 43]. Coupling the altered redox environment with the additional stimuli of ·NO released from active eNOS may create a scenario where trapping Ras protein radicals in cancer cells using IST becomes feasible. Recent studies indicate that oncogenic KRas activates the phosphatidylinositol-3-kinase (PI3K)/Akt pathway, which leads to phosphorylation of eNOS in oncogenic KRas-driven pancreatic cancer [18]. The ·NO released from active eNOS subsequently activates endogenous (non-mutated) H- and NRas by S-nitrosation at Cys118. Therefore, activated H- and NRas could stimulate PI3K and may contribute to positive feedback activation of the PI3K/Akt/eNOS pathway [44]. The importance of this cannot be understated, as the PI3K/Akt/eNOS pathway is a primary pathway contributing to tumor maintenance in oncogenic KRas-driven pancreatic cancer [44].
While successful detection of ·NO-mediated protein radical formation in HRas has now been reported, the exact mechanism through which the Ras DMPO-nitroxide is oxidized to the corresponding DMPO-nitrone remains a question. Efforts to identify the exact oxidation mechanism will be the focus of future work. However, several critical insights are available from this report and other previous efforts. As Williams et al. have shown, stable S-nitrosation of HRas at Cys118 does not alter guanine nucleotide cycling and does not alter Ras activity [19]. Rather, the causative factor of Ras activation lies in the mechanism of S-nitrosation itself. As seen in Figure 2, S-nitrosation of Ras can occur through different pathways. One involves a one-electron oxidation of Cys118 (via reaction with ·NO2) followed by a radical recombination with ·NO (Figure 2, Reactions 1 and 5). Another involves a non-radical pathway involving attack of an electrophilic N2O3 (Figure 2, Reaction 4). This latter scenario is unlikely given the rate of N2O3 hydrolysis is nearly an order of magnitude greater than its reaction toward thiols [15, 24]. Therefore, a pathway which involves a Ras Cys118 radical must be considered, given data indicating high levels of endogenous H and NRas S-nitrosation upon activation of eNOS in KRas-driven pancreatic cancer [18]. Successful implementation of IST to trap intermediate Ras protein radicals during this process in vivo would greatly advance our knowledge of how ·NO mechanistically drives Ras activation during pancreatic tumor initiation and maintenance.
Conclusion
In conclusion, we have presented direct evidence of protein radical formation in a model Ras GTPase and confirmed DMPO adduction is centered at redox active Cys118 using immuno-spin trapping and FTICR-MS approaches. This study also demonstrates the successful application of IST on a non-metalloprotein where transition metals or separate metalloproteins were not used to generate an oxidizing species. The findings may have far reaching implications for detecting protein radical events in other non-metalloproteins, such as phosphatases [45], caspases [46, 47] and protein kinases [48], where ·NO-mediated processes are known to influence protein activity via redox active cysteines. The in vitro detection of Ras GTPase protein radicals by IST, even as a proof-of-principle, also sets the groundwork for integration of the technique in future cellular-based investigations. While Ras GTPases are known to be mutated in numerous different cancers, including cancer of the pancreas, colon, and lung [17], little emphasis has been placed on understanding the role of radical-mediated Ras activation in these disease states. This remains true today, even as evidence clearly implies a radical-mediated pathway contributes to tumor initiation and maintenance in Ras-driven pancreatic cancer [18]. We anticipate IST may become a useful method to test for radical-mediated Ras activation in many different cancers or disease states where Ras GTPases play a significant role. Successful detection of Ras protein radicals in cancer cell cultures or animal models may pave important directions toward the development of new anti-Ras therapeutic agents targeting radical-mediated activation pathways.
Supplementary Material
Highlights.
Evidence of ·NO-mediated protein radical formation in HRas is presented.
Immuno-spin trapping (IST) and tandem MS/MS indicate a Cys118 Ras protein radical.
IST may be used to detect thiyl radical formation in the Ras oncoprotein.
The IST approach described can be used for other non-metalloproteins.
Acknowledgement
We gratefully acknowledge research support by the National Institute of Health (Grant # RO1GM75431, RO1CA089614), and the Lineberger Comprehensive Cancer Center postdoctoral training grant at the University of North Carolina, Chapel Hill (Grant # 5T32CA009156-35).
Abbreviations
- DEA/NO
2-(N,N-diethylamino)-diazenolate-2-oxide diethylammonium salt
- DMPO
5,5-dimethyl-1-pyrroline N-oxide
- eNOS
endothelial nitric oxide synthase
- FT-ICR
Fourier transform ion cyclotron resonance
- GAPs
GTPase activating proteins
- GDP
guanosine-5′-diphosphate
- GEFs
guanine exchange factors
- GTP
guanosine-5′-triphosphate
- IST
immuno-spin trapping
- N2O3
dinitrogen trioxide
- ·NO
nitric oxide
- ·NO2
nitrogen dioxide
- PI3K
phosphatidylinositol-3-kinase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supporting Information Available Supporting material is available, including F28L and F28L/C118S immunoblots for IST experiments, full μESI-FTICR-MS and μESI-FTICR-ECD MS/MS spectra, amino acid sequence, and full fragmentation pattern of F28L HRas.
This project was supported by NIH grant 5R01 GM075431-04 and the Lineberger Comprehensive Cancer Center postdoctoral training grant 5T32CA009156-35
References
- [1].Mason RP. Using anti-5,5-dimethyl-1-pyrroline N-oxide (anti-DMPO) to detect protein radicals in time and space with immuno-spin trapping. Free Radic Biol Med. 2004;36:1214–1223. doi: 10.1016/j.freeradbiomed.2004.02.077. [DOI] [PubMed] [Google Scholar]
- [2].Detweiler CD, Deterding LJ, Tomer KB, Chignell CF, Germolec D, Mason RP. Immunological identification of the heart myoglobin radical formed by hydrogen peroxide. Free Radic Bio Med. 2002;33:364–369. doi: 10.1016/s0891-5849(02)00895-x. [DOI] [PubMed] [Google Scholar]
- [3].Detweiler CD, Lardinois OM, Deterding LJ, de Montellano PR, Tomer KB, Mason RP. Identification of the myoglobin tyrosyl radical by immuno-spin trapping and its dimerization. Free Radic Bio Med. 2005;38:969–976. doi: 10.1016/j.freeradbiomed.2004.12.031. [DOI] [PubMed] [Google Scholar]
- [4].Bhattacharjee S, Deterding LJ, Jiang J, Bonini MG, Tomer KB, Ramirez DC, Mason RP. Electron transfer between a tyrosyl radical and a cysteine residue in hemoproteins: spin trapping analysis. J Am Chem Soc. 2007;129:13493–13501. doi: 10.1021/ja073349w. [DOI] [PubMed] [Google Scholar]
- [5].Lardinois OM, Detweiler CD, Tomer KB, Mason RP, Deterding LJ. Identifying the site of spin trapping in proteins by a combination of liquid chromatography, ELISA, and off-line tandem mass spectrometry. Free Radic Bio Med. 2008;44:893–906. doi: 10.1016/j.freeradbiomed.2007.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Ranguelova K, Bonini MG, Mason RP. (Bi)sulfite oxidation by copper, zinc-superoxide dismutase: sulfite-derived, radical-initiated protein radical formation. Environ Health Persp. 2010;118:970–975. doi: 10.1289/ehp.0901533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Ranguelova K, Chatterjee S, Ehrenshaft M, Ramirez DC, Summers FA, Kadiiska MB, Mason RP. Protein radical formation resulting from eosinophil peroxidase-catalyzed oxidation of sulfite. J Biol Chem. 2010;285:24195–24205. doi: 10.1074/jbc.M109.069054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Deterding LJ, Ramirez DC, Dubin JR, Mason RP, Tomer KB. Identification of free radicals on hemoglobin from its self-peroxidation using mass spectrometry and immuno-spin trapping: observation of a histidinyl radical. J Biol Chem. 2004;279:11600–11607. doi: 10.1074/jbc.M310704200. [DOI] [PubMed] [Google Scholar]
- [9].Lardinois OM, Tomer KB, Mason RP, Deterding LJ. Identification of protein radicals formed in the human neuroglobin-H2O2 reaction using immuno-spin trapping and mass spectrometry. Biochemistry. 2008;47:10440–10448. doi: 10.1021/bi800771k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Chatterjee S, Ehrenshaft M, Bhattacharjee S, Deterding LJ, Bonini MG, Corbett J, Kadiiska MB, Tomer KB, Mason RP. Immuno-spin trapping of a post-translational carboxypeptidase B1 radical formed by a dual role of xanthine oxidase and endothelial nitric oxide synthase in acute septic mice. Free Radic Biol Med. 2009;46:454–461. doi: 10.1016/j.freeradbiomed.2008.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Lander HM, Hajjar DP, Hempstead BL, Mirza UA, Chait BT, Campbell S, Quilliam LA. A molecular redox switch on p21(ras). Structural basis for the nitric oxide p21(ras) interaction. J Biol Chem. 1997;272:4323–4326. doi: 10.1074/jbc.272.7.4323. [DOI] [PubMed] [Google Scholar]
- [12].Heo J, Campbell SL. Mechanism of p21Ras S-nitrosylation and kinetics of nitric oxide-mediated guanine nucleotide exchange. Biochemistry. 2004;43:2314–2322. doi: 10.1021/bi035275g. [DOI] [PubMed] [Google Scholar]
- [13].Heo J, Prutzman KC, Mocanu V, Campbell SL. Mechanism of free radical nitric oxide-mediated Ras guanine nucleotide dissociation. J Mol Biol. 2005;346:1423–1440. doi: 10.1016/j.jmb.2004.12.050. [DOI] [PubMed] [Google Scholar]
- [14].Heo J, Campbell SL. Superoxide anion radical modulates the activity of Ras and Ras related GTPases by a radical-based mechanism similar to that of nitric oxide. J Biol Chem. 2005;280:12438–12445. doi: 10.1074/jbc.M414282200. [DOI] [PubMed] [Google Scholar]
- [15].Raines KW, Bonini MG, Campbell SL. Nitric oxide cell signaling: S-nitrosation of Ras superfamily GTPases. Cardiovasc Res. 2007;75:229–239. doi: 10.1016/j.cardiores.2007.04.013. [DOI] [PubMed] [Google Scholar]
- [16].Kraulis PJ, Domaille PJ, Campbell-Burk SL, Van Aken T, Laue ED. Solution structure and dynamics of ras p21.GDP determined by heteronuclear three- and four-dimensional NMR spectroscopy. Biochemistry. 1994;33:3515–3531. doi: 10.1021/bi00178a008. [DOI] [PubMed] [Google Scholar]
- [17].Bos JL. Ras oncogenes in human cancer: a review. Cancer Res. 1989;49:4682–4689. [PubMed] [Google Scholar]
- [18].Lim KH, Ancrile BB, Kashatus DF, Counter CM. Tumour maintenance is mediated by eNOS. Nature. 2008;452:646–649. doi: 10.1038/nature06778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Williams JG, Pappu K, Campbell SL. Structural and biochemical studies of p21Ras S-nitrosylation and nitric oxide-mediated guanine nucleotide exchange. Proc Natl Acad Sci USA. 2003;100:6376–6381. doi: 10.1073/pnas.1037299100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Ford E, Hughes MN, Wardman P. Kinetics of the reactions of nitrogen dioxide with glutathione, cysteine, and uric acid at physiological pH. Free Radic Biol Med. 2002;32:1314–1323. doi: 10.1016/s0891-5849(02)00850-x. [DOI] [PubMed] [Google Scholar]
- [21].Augusto O, Bonini MG, Amanso AM, Linares E, Santos CC, De Menezes SL. Nitrogen dioxide and carbonate radical anion: two emerging radicals in biology. Free Radic Biol Med. 2002;32:841–859. doi: 10.1016/s0891-5849(02)00786-4. [DOI] [PubMed] [Google Scholar]
- [22].Grätzel M, Henglein A, Lilie J, Beck G. Pulsradiolytische untersuchung einigerelementarprozesse der oxidation und reduction des nitritions. Ber Bunsenges Phys Chem. 1969;73:646–653. [Google Scholar]
- [23].Broszkiewicz RK. The pulse radiolysis study of NaNO2 and NaNO3 solutions. Bull Acad Pol Sci Sedr Sci Chim. 1976;24:221–229. [Google Scholar]
- [24].Herold S, Rock G. Mechanistic studies of S-nitrosothiol formation by NO/O2 and by NO/methemoglobin. Arch Biochem Biophys. 2005;436:386–96. doi: 10.1016/j.abb.2005.02.013. [DOI] [PubMed] [Google Scholar]
- [25].Keshive M, Singh S, Wishnok JS, Tannenbaum SR, Deen WM. Kinetics of S-nitrosation of thiols in nitric oxide solutions. Chem Res Toxicol. 1996;9:988–993. doi: 10.1021/tx960036y. [DOI] [PubMed] [Google Scholar]
- [26].Madej E, Folkes LK, Wardman P, Czapski G, Goldstein S. Thiyl radicals react with nitric oxide to form S-nitrosothiols with rate constants near the diffusion-controlled limit. Free Radic Biol Med. 2008;44:2013–2018. doi: 10.1016/j.freeradbiomed.2008.02.015. [DOI] [PubMed] [Google Scholar]
- [27].Davies MJ, Forni LG, Shuter SL. Electron spin resonance and pulse radiolysis studies on the spin trapping of sulphur-centered radicals. Chem Biol Interact. 1987;61:177–188. doi: 10.1016/0009-2797(87)90038-x. [DOI] [PubMed] [Google Scholar]
- [28].Zhang X, Zhang N, Schuchmann H-P, von Sonntag C. Pulse radiolysis of 2-mercaptoethanol in oxygenated aquesous solutions. Generation of the thiylperoxyl radical. J Phys Chem. 1994;98:6541–6547. [Google Scholar]
- [29].Nauser T, Pelling J, Schöneich C. Thiyl radical reaction with amino acid side chains: rate constants for hydrogen transfer and relevance for posttranslational protein modification. Chem Res Toxicol. 2004;17:1323–1328. doi: 10.1021/tx049856y. [DOI] [PubMed] [Google Scholar]
- [30].Jourd’heuil D, Jourd’heuil FL, Feelisch M. Oxidation and nitrosation of thiols at low micromolar exposure to nitric oxide. Evidence for a free radical mechanism. J Biol Chem. 2003;278:15720–15726. doi: 10.1074/jbc.M300203200. [DOI] [PubMed] [Google Scholar]
- [31].John J, Schlichting I, Schiltz E, Wittinghofer A. C-terminal truncation of p21H preserves crucial kinetic and structural properties. J Biol Chem. 1989;264:13086–13092. [PubMed] [Google Scholar]
- [32].Silvius J. Mechanisms of ras protein targeting in mammalian cells. J Membr Biol. 2002;190:83–92. doi: 10.1007/s00232-002-1026-4. [DOI] [PubMed] [Google Scholar]
- [33].Scheich C, Kümmel D, Soumailakakis D, Heinemann U, Büssow K. Vectors for co-expression of an unrestricted number of proteins. Nucleic Acids Res. 2007;35:1–7. doi: 10.1093/nar/gkm067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Schlichting I, John J, Frech M, Chardin P, Wittinghofer A, Zimmerman H, Rösch P. Proton NMR studies of transforming and nontransforming H-ras p21 mutants. Biochemistry. 1990;29:504–511. doi: 10.1021/bi00454a026. [DOI] [PubMed] [Google Scholar]
- [35].Adams PD, Loh AP, Oswald RE. Backbone dynamics of an oncogenic mutant of Cdc42Hs shows increased flexibility at the nucleotide-binding site. Biochemistry. 2004;43:9968–9977. doi: 10.1021/bi0490901. [DOI] [PubMed] [Google Scholar]
- [36].Adams PD, Oswald RE. Solution structure of an oncogenic mutant of Cdc42Hs. Biochemistry. 2006;45:2577–2583. doi: 10.1021/bi051686g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Kelleher NL, Lin HY, Valaskovic GA, Aaseruud DJ, Fridriksson EK, McLafferty FW. Top down versus bottom up protein characterization by tandem high resolution mass spectrometry. J Am Chem Soc. 1999;121:806–812. [Google Scholar]
- [38].Reid GE, McLuckey SA. ‘Top down’ protein characterization via tandem mass spectrometry. J Mass Spectrom. 2002;37:663–675. doi: 10.1002/jms.346. [DOI] [PubMed] [Google Scholar]
- [39].Siuti N, Kelleher NL. Decoding protein modifications using top-down mass spectrometry. Nat Methods. 2007;4:817–821. doi: 10.1038/nmeth1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Shang H, Xu Y, Kalyanaraman B. Intramolecular electron transfer between tyrosyl radical and cysteine residue inhibits tyrosine nitration and induces thiyl radical formation in model peptides treated with MPO, H2O2, and NO2−: EPR spin-trapping studies. J Biol Chem. 2005;280:40684–40698. doi: 10.1074/jbc.M504503200. [DOI] [PubMed] [Google Scholar]
- [41].Aubert C, Vos MH, Mathis P, Eker AP, Brettel K. Intraprotein radical transfer during photoactivation of DNA photolyase. Nature. 2000;405:586–590. doi: 10.1038/35014644. [DOI] [PubMed] [Google Scholar]
- [42].Valko M, Rhodes CJ, Moncol J, Izakovic M, Mazur M. Free radicals, metals and antioxidants in oxidative stress-induced cancer. Chem Biol Interact. 2006;160:1–40. doi: 10.1016/j.cbi.2005.12.009. [DOI] [PubMed] [Google Scholar]
- [43].McEligot AJ, Yang S, Meyskens FL., Jr. Redox regulation by intrinsic species and extrinsic nutrients in normal and cancer cells. Annu Rev Nutr. 2005;25:261–295. doi: 10.1146/annurev.nutr.25.050304.092633. [DOI] [PubMed] [Google Scholar]
- [44].Davis MF, Vigil D, Campbell SL. Regulation of ras proteins by reactive nitrogen species. Free Rad Biol Med. 2011;51:565–575. doi: 10.1016/j.freeradbiomed.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Tomko RJ, Jr., Lazo JS. Multimodal control of cdc25a by nitrosative stress. Cancer Res. 2008;68:7457–7465. doi: 10.1158/0008-5472.CAN-08-0625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Li J, Billiar TB, Talanian RV, Kim YM. Nitric oxide reversibly inhibits seven members of the caspase family via S-nitrosylation. Biochem Biophys Res Commun. 1997;240:419–424. doi: 10.1006/bbrc.1997.7672. [DOI] [PubMed] [Google Scholar]
- [47].Kim YM, Talanian RV, Billiar TR. Nitric oxide inhibits apoptosis by preventing increases in caspase-3-like activity via two distinct mechanisms. J Biol Chem. 1997;49:31138–31148. doi: 10.1074/jbc.272.49.31138. [DOI] [PubMed] [Google Scholar]
- [48].Carvalho-Filho MA, Ueno M, Hirabara SM, Seabra AB, Carvalheira JBC, de Oliveira MG, Velloso LA, Curi R, Saad MJA. S-Nitrosation of the insulin receptor, insulin receptor substrate 1, and protein kinase B/Akt: a novel mechanism of insulin resistance. Diabetes. 2005;54:959–967. doi: 10.2337/diabetes.54.4.959. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.