Abstract
Precision measurement is a hallmark of physics but the small length scale (~ nanometer) of elementary biological components and thermal fluctuations surrounding them challenge our ability to visualize their action. Here, we highlight the recent developments in single molecule nanometry where the position of a single fluorescent molecule can be determined with nanometer precision, reaching the limit imposed by the shot noise, and the relative motion between two molecules can be determined with ~ 0.3 nm precision at ~ 1 millisecond time resolution, and how these new tools are providing fundamental insights on how motor proteins move on cellular highways. We will also discuss how interactions between three and four fluorescent molecules can be used to measure three and six coordinates, respectively, allowing us to correlate movements of multiple components. Finally, we will discuss recent progress in combining angstrom precision optical tweezers with single molecule fluorescent detection, opening new windows for multi-dimensional single molecule nanometry for biological physics.
1. Introduction
There are two types of motion in biology: diffusion and directed motion. Diffusion is thermally driven and is best characterized by the Brownian motion of molecules. Directed motion is typically driven by motor proteins that consume chemical energy. While being still intrinsically diffusive, the consumption of chemical energy aids to temporarily modulate the energy landscape, thus rectifying the motion. It carries out many cellular functions, including the transfer and manipulation of genetic information, transport of intracellular cargos, muscle contraction, and cell division. Growing measurement capabilities to manipulate and measure biological motions now make it possible to detect single photons, pico-Newton (pN) forces, and sub-nanometer distances and have extended our observation down to the smallest functional units in biology, a single biological molecule.
The functions and interactions of biomolecules are often too complex to be described by generalized rules. Their functions are usually not the mere sum of those of the building blocks but are often an emergent property of the integrated entity. Amino acid sequence of a protein and its atomic structure by themselves do not explicitly tell how it will function. Only when the structural information is combined with experimental analysis of the functions can we begin to comprehend the marvelous interworking of these molecules. In this review, we discuss several single molecule techniques for understanding their functions, with an emphasis on applications to the motor proteins.
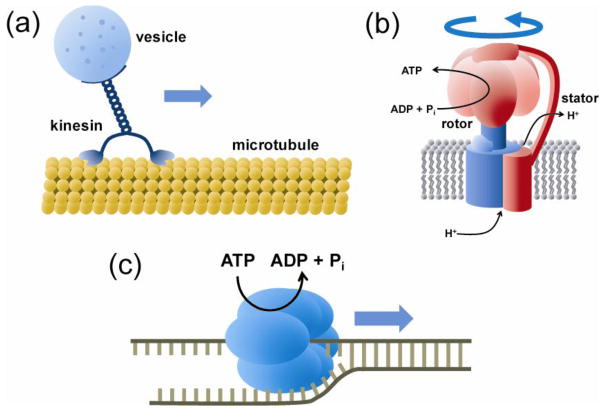
Motor proteins convert chemical energy, in many cases derived from the ATP molecule, the major source of cellular energy, into mechanical work. There are several classes of motor proteins (Figure 1). Cytoskeletal motors move along the cytoskeleton of the cell to perform muscle contraction, cell division, and shuttling cellular components (Vale and Milligan, 2000; Vale, 2003; Gennerich and Vale, 2009). They include myosins moving along the actin filaments and kinesins and dyneins moving along the microtubules. Rotary motors make rotating movement around certain axis. Bacterial flagellum (Kearns, 2010; Brown et al., 2011; Erhardt et al., 2010) and mitochondrial F0F1-ATP synthase (Jonckheere et al., 2012) are the best known examples in this category. Nucleic acid motors move along DNA or RNA. Included in this category are the helicases which are responsible for separating double stranded nucleic acids (Yodh et al., 2010), DNA polymerases that make double stranded DNA using a single stranded DNA template (Johnson and O’Donnell, 2005), RNA polymerases that synthesizes RNA by transcribing the genetic information of DNA (Selth et al., 2010), ribosomes that translate the information of messenger RNAs into proteins (Frank and Gonzalez, 2010; Schmeing and Ramakrishnan, 2009), topoisomerases that control the overwinding or underwinding of the DNA (Vos et al., 2011), and viral packaging motors that inject the genome into viral capsids (Rao and Feiss, 2008; Smith, 2011).
Figure 1.
Biological motors. (a) Cytoskeleton motors move along a one-dimensional filament of actins or tubulins. Shown is a kinesin carrying a vesicle cargo along a microtubule track. (b) Rotary motors couple biochemical reactions to rotary motion. For example, F0F1-ATP synthase converts a proton gradient across a membrane into the chemical energy by synthesizing ATP or vice versa. (c) Various motors, including DNA/RNA polymerase, helicase, and ribosome, move along nucleic acids to polymerize biopolymers or to change their geometric conformations. A helicase which unwinds double stranded DNA or RNA into single strands is shown.
Single molecule measurements avoid ensemble averaging and reveal full time trajectories of biological reactions. Therefore, they are ideally suited for studying the motor proteins which can move on inhomogeneous cellular tracks and exhibit non-equilibrium dynamics. In addition, motor proteins usually make conformational changes with multiple degrees of freedom and multiple proteins often function in concert. Therefore, it is necessary to observe multiple variables at once, in real time and at the single molecule resolution. Yet, their movements and the forces involved are very small. Motor proteins move in discrete steps that are only a few Å to nanometers in size and their movements are sensitive to pico-Newton forces. Owing to the advances in fluorescence microscopy and mechanical manipulation techniques during the last two decades, we can now observe and manipulate individual molecules for an extended period of time.
In order to detect single molecules through fluorescence, two requirements must be met. First, there has to be at most one molecule per detection volume. This can be achieved by making the sample dilute. Second, fluorescence signal from a single molecule has to be significantly larger than the background. Development of photochemically stable and bright fluorescent tags and biochemical methods to label the target molecules realized sensitive detection of single molecules. Fluorescent proteins, organic dyes, and inorganic quantum dots have been developed and successfully used for this purpose. Background can be reduced by minimizing the excitation or detection volume using various methods, including confocal detection, total internal reflection (TIR), physical confinement of the sample, and manipulation of the effective excitation volume.
In order to apply and measure forces on single molecules, various tools have been developed. Optical trap holds a dielectric bead in a local potential minimum and the applied force can be determined from the displacement of the bead. Magnetic tweezers trap paramagnetic beads and manipulate them but with the ability to control the orientation of the beads. Atomic force microscope tethers the apex of the probe to the target molecule in order to control or measure the force through the deformation of the cantilever. In this review, we will discuss the application of the above techniques and their combinations, in an increasing order of complexity, to various biological systems, mainly focusing on the motor proteins.
2. One color: localizing and tracking single molecules
Optical microscopy has a fundamental resolution limit, so called diffraction limit. It imposes the minimum distance between two point sources of light for them to be optically distinguished as d = λ/(2nsinθ), where λ is the wavelength of the light, n is the refractive index of the medium, and θ is the angular aperture of the microscope objective. However, when it comes to imaging a single fluorophore that can be approximated as a point light source, we can localize its position with arbitrarily high accuracy provided that we know the point spread function (PSF) of the optics accurately (Yildiz and Selvin, 2005). The following expression gives the localization accuracy as the standard error of the position (Thompson et al., 2002):
where s is the standard deviation of the PSF, a is the pixel size of the camera, b is the background photon count and N is the number of collected photons. With typical values of s = 200 nm, a = 130 nm, and b = 0.7 photons, the theoretical localization accuracy becomes 23 nm with only 100 photons and it goes down to 2 nm with 10,000 photons. Experimental localization uncertainty was shown to be only 30% greater than the theoretical value (Thompson et al., 2002). However, Thomson et al.’s work used an intensely fluorescent bead containing many dyes.
This principle was extended to the level of single fluorescent molecules to observe motor proteins’ movement with nanometer precision. Selvin and colleagues (Yildiz et al., 2003) observed the stepwise motion of the cytoskeleton motor, myosin V, which moves along actin filaments by localizing the position of its fluorescent label as a function of time (Figure 2a). Myosin V is a dimeric motor with two heads and its center moves 37 nm per ATP consumed. By analyzing the alternating step sizes, they were able to demonstrate that the motor moves in hand-over-hand manner, much like a bipedal walker (Figure 2b). The same method was used to show that several other cytoskeleton motors also move via the hand-over-hand mechanism where the two heads strictly coordinate to move forward (Yildiz et al., 2004b; Yildiz et al., 2004a; Okten et al., 2004). but that such strict coordination is absent for another cytoskeleton motor protein, dynein (DeWitt et al., 2012; Qiu et al., 2012).
Figure 2.
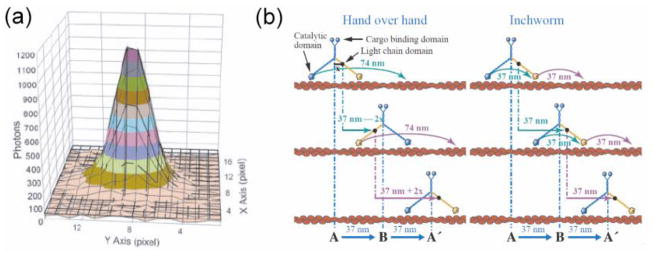
Localization of a single fluorophore and suggested mechanism for the stepping motion of myosin (Yildiz et al., 2003). (a) Gaussian fitting (solid lines) of the image of a single fluorophore immobilized on surface. While the width of the distribution is 287 nm, the center can be determined to 1.3 nm accuracy. (b) Two possible stepping mechanisms of myosin V. Both mechanisms give the same step size of the center of mass but individual arm will give different step sizes.
If we want to move beyond test tube experiments and observe single motor proteins in living cells, we need brighter and more photostable probes because of high cellular scattering and background fluorescence and because removal of oxygen molecules, a common trick used to extend the photobleaching lifetime, can be detrimental to the cell. Semiconductor quantum dots are excellent probes for this purpose because they are far more photostable than organic fluorophores or fluorescent proteins, and broadly tunable in color by engineering the synthesis (Michalet et al., 2005; Courty et al., 2006a; Bannai et al., 2007). Researchers developed methods to track single molecules tagged with quantum dots (Dubertret et al., 2002; Larson et al., 2003; Dahan et al., 2003). For example, Courty et al. tracked kinesin both in vitro and in vivo (Courty et al., 2006b) and measured kinesin’s velocity and processivity, which quantifies how far a motor protein moves along the track before it falls off. The unique ability of single molecule tracking enabled them to reveal multiple stages of directional motion intervened by diffusive phases (Figure 3).
Figure 3.
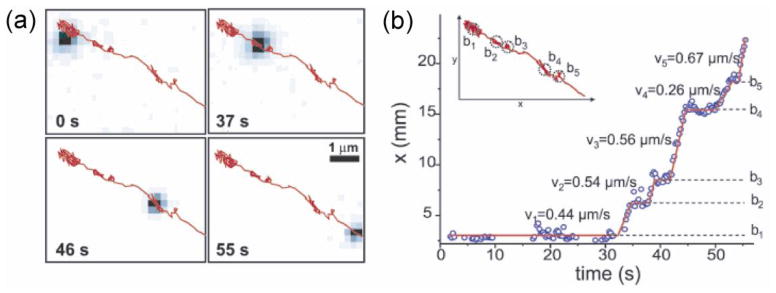
Tracking of kinesin by quantum-dot labels (Courty et al., 2006b). (a) Tracking of a quantum-dot-labeled kinesin inside a living cell. Images at different time points are shown with the overall trajectory overlaid in red. (b) Inset: trajectory of the kinesin shown in (a). The position along x axis vs time demonstrates successive directed movements (with corresponding velocities vi) and diffusive motions (marked by bi).
The cell is a three-dimensional (3D) object and we need to look into the third dimension to visualize the full trajectory of molecular motors. Several optical techniques have been developed to gain resolution on the z axis. Kao and Verkman used a cylindrical lens in the detection path to create asymmetry of the image deformation depending on the height of the fluorophore (Kao and Verkman, 1994). The astigmatism causes an ellipsoidal shape of a point light source and the direction of its major axis was used to retrieve information on the z position. Simple defocusing methods have also been used for 3D tracking. Speidel et al. acquired z resolution without special optical trick by analyzing the ring pattern and radii from defocused images (Speidel et al., 2003). Toprak et al. introduced a bifocal imaging method, simultaneously imaging the focused and defocused planes to track the 3D motion of the target (Toprak et al., 2007). Pavani et al. implemented a double-helix point spread function which rotates with the z position of the target and improved the localization of z position (Pavani et al., 2009).
One color × many
Another way of using one fluorophore to study cellular components is the super-resolution imaging, implemented by several different approaches. Despite the fundamental diffraction limit, there are ways to circumvent the limit utilizing the quantum nature of single fluorophores. If we can excite only a small population of fluorophores at a given moment so that they are more separated than the diffraction limit, we can determine the position of each fluorophore with high precision, as discussed above for a single fluorophore. Fluorophores under excitation exhibit diverse photophysical behaviors, including blinking, bleaching, and intensity fluctuation (Ha and Tinnefeld, 2012). This needs to be overcome when we want to use them as reliable reporters of the presence, position, or number of the tagged molecules. But, as a change of viewpoint, we can take advantage of such photophysics to switch them on and off randomly or under control by illumination (Bates et al., 2005). Combining the localization method and serial, stochastic excitation and imaging of single fluorophores, researchers have developed several super-resolution imaging techniques to overcome the diffraction limit (Rust et al., 2006; Hess et al., 2006; Betzig et al., 2006). For extensive review of the techniques, we redirect the readers to the review by Huang et al. (Huang et al., 2009). Even though the achievement of fast frame rate approaching the video rate currently remains challenging and the techniques have not been widely applied to study dynamic motor proteins so far, super-resolution imaging techniques provide promising ways to image and track large number of molecular motors in cellular environment.
3. Two colors: measuring sub-nanometer dynamics
When we have two fluorophores with different colors, we do not merely double the number of observables but also add a new kind of dimension. One remarkable phenomenon between two fluorophores is that they transfer energy by dipole-dipole interaction when there is a spectral overlap between the emission of one fluorophore and the absorption of the other. The efficiency of this process, known as fluorescence resonance energy transfer (FRET), is sharply dependent on the distance between the fluorophores and thus can be used as a measure of the distance (Figure 4a). FRET was implemented in the single molecule regime (Ha et al., 1996) and various studies showed that FRET can report the time dependent distance between two molecules or two positions within one molecule.
Figure 4.
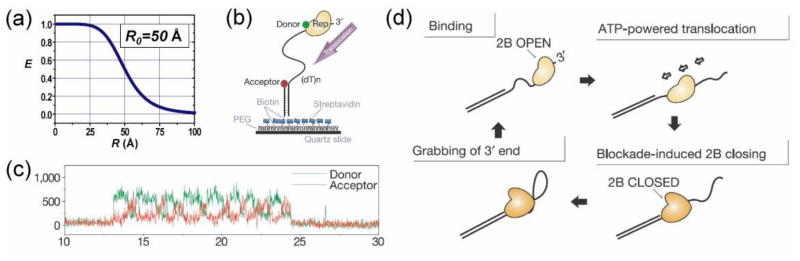
smFRET as a technique to study the dynamics of helicases (Myong et al., 2005). (a) Theoretical distance dependence of FRET efficiency. (b) Schematic of a donor-labeled helicase bound to a single stranded DNA. FRET between the donor and the acceptor on the end of the single stranded region can be used to study helicase translocation. (c) Fluorescence signal abruptly increases when the donor-labeled protein binds to the DNA (~13 s). This is followed by repetitive cycles of gradual FRET increase followed by an abrupt FRET decrease as indicated by anti-correlated changes of donor and acceptor intensities until the signal disappears due to donor photobleaching or protein dissociation. (d) Schematic model on the repetitive shuttling of the helicase on the same segment of single stranded DNA.
In contrast to the techniques based on the localization of a single fluorophore, single molecule FRET (smFRET) measures the relative distance between two positions and therefore is uncoupled from the Brownian motion of the whole complex. Through appropriate choice of dye pairs, we can probe distances ranging from 2 nm to 10 nm with a temporal resolution typically down to several milliseconds or sub-milliseconds if pushed harder (Chung et al., 2012). As FRET is based on the dipole-dipole interaction between the fluorophores, its efficiency depends on their orientations. The interpretation of smFRET data usually assumes random and fast reorientation of the fluorophores tethered to the molecules of interest and thus assumes the dipole orientation factor to be 2/3 from ideal isotropic distribution. When the fluorophore’s orientation is fixed relative to the biomolecule and is known, one can actually calculate the orientation factor for a more precise determination of distance (Iqbal et al., 2008). The fundamental limit of the temporal resolution of smFRET is estimated to be 1~10 μs due to the Brownian motion of the fluorophores and their tethering linkers (Badali and Gradinaru, 2011). Therefore, there is ample potential for the use of smFRET for molecular motors which typically undergo reactions in millisecond time scales. Indeed, smFRET has been applied to study many kinds of molecular motors: mostly nucleic acid motors including helicases (Myong et al., 2005; Joo et al., 2006; Myong et al., 2007; Park et al., 2010), RNA polymerases (Treutlein et al., 2012; Kim et al., 2011; Kapanidis et al., 2006; Andrecka et al., 2008; Tang et al., 2009), and ribosomes (Blanchard et al., 2004; Lee et al., 2007c; Fei et al., 2008; Cornish et al., 2008), and one important exception of kinesin which is a cytoskeletal motor (Rice et al., 1999).
Helicases are nucleic acid motors that catalytically unwind double stranded nucleic acids into single strands by consuming ATPs. Various single molecule techniques have been employed to study their mechanism including smFRET (Myong et al., 2005; Joo et al., 2006; Myong et al., 2007; Park et al., 2010), optical tweezer (Perkins et al., 2004), magnetic tweezer (Lionnet et al., 2007; Ramanathan et al., 2009), atomic force microscopy (Henn et al., 2001; Zhang et al., 2007), and flow-induced DNA stretching (Bianco et al., 2001; Lee et al., 2006) and these studies have provided unique insights from various perspectives. For a comprehensive review of helicases studied with various single molecule methods, we redirect the readers to (Yodh et al., 2010). smFRET method particularly proved to be successful in measuring their dynamics with high spatial precision down to single basepair (bp). In a smFRET study, Myong et al. proposed that NS3 helicase of Hepatitis C virus unwinds duplex DNA by actively unwinding the duplex rather than passively waiting for thermal fraying of the bps (Myong et al., 2007). They suggested that the accumulation of elastic strain in the complex from stepwise translocation triggers the active destabilization of the duplex. Fundamental step size of translocation is another key question for helicases. Myong et al. showed that NS3 helicase unwinds the DNA in distinct steps of 3 bps (Myong et al., 2007). Analysis of the duration at each step revealed the existence of 3 elemental steps, suggesting a mechanistic model that the motor translocates in increments of 1 nucleotide (nt) while breaking 3 bps at once using the accumulated tension.
This concept that a protein can store elastic energy through multiple cycles of chemical energy consumption and that such stored energy can be released in a single burst was further supported by Lee et al.’s work on yeast exoribonuclease, Rrp44 (Lee et al., 2012). In this work, the Rrp44 enzyme that can digest RNA strand one base at a time even in the presence of basepaired structures was studied by attaching the dyes to the RNA so that FRET would increase while the enzyme unwinds the RNA. The surprise was that the enzyme unwound RNA in large steps about 4 basepairs even though the motion is powered by RNA digestion in single base steps, strongly suggesting that the enzyme-RNA complex can be viewed as a spring that converts chemical energy into storable mechanical energy.
Compared to other single molecule methods, smFRET has the unique ability of probing the internal dynamics of molecular complexes, which is often obscured by the overall Brownian motion of the complex in other methods. This ability made it possible to reveal the “repetitive shuttling” behavior of helicases on partial DNA duplex (Myong et al., 2005) (Figure 4b–d). Such behavior was proposed to play a role in removing proteins, such as RecA filaments, from single stranded DNAs thus preventing unwanted recombination.
smFRET has been successful in another class of nucleic acid motor, RNA polymerase (RNAP). In cooperation with accessory proteins in most cases, RNA polymerase detects a promoter sequence on the duplex DNA and initiates, elongates, and terminates RNA synthesis. RNAPs switch from the initiation phase to the elongation phase by undergoing specific structural rearrangement and/or the dissociation of accessory proteins. smFRET measurements visualized this transition in the single-subunit T7 RNAP by controlling the length of the synthesized RNA strand and showing how it relates to the structural rearrangement (Tang et al., 2009). In ensemble experiments in which synchrony between different reaction steps is difficult to maintain, it is not easy to ascertain whether there exist multiple steps or it is only the relative populations in two states that changes. By examining multiple labeling sites, Tang et al. showed that the transcription initiation occurs through the scrunching of the DNA and the rotational motion of one domain of the polymerase (Tang et al., 2008). One step further in complexity, we studied mitochondrial RNAP from yeast, which comprises two subunits: the polymerase, Rpo41, and the specificity factor, Mtf1 (Kim et al., 2011). We revealed the repetitive opening–closing transitions of the transcription initiation complex that were shown to be modulated by Mtf1 and ATP. We also demonstrated multiple opening trials upon initial binding of the proteins to the promoter. With the unique ability of smFRET to measure fluctuations in real time, we suggested the roles of the opening-closing transitions in sensing how much ATP is present in the cell and in selecting the correct sequence from which RNA synthesis begins.
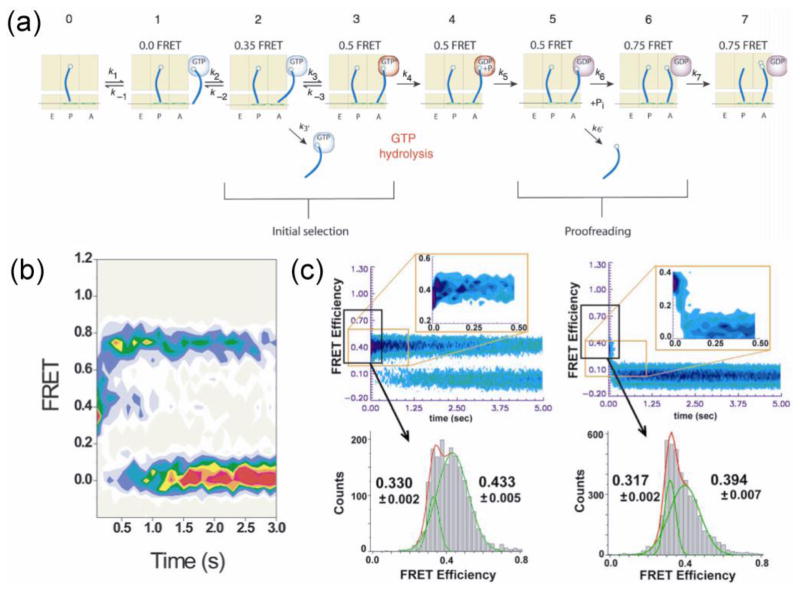
Another example of nucleic acid motor extensively studied by single molecule methods is the ribosome. The ribosome is the protein factory that translates genetic information on the messenger RNA (mRNA) into a chain of amino acids to synthesize proteins. It recruits the transfer RNA (tRNA) which brings an amino acid and a 3 nt sequence, called the anti-codon, in the tRNA that basepairs with a corresponding 3 nt codon sequence on the mRNA, thereby ensuring an accurate ‘translation’ of the genetic information. Because of ~ 7 nm distance between the decoding site and the amino-acid binding site of the tRNA, it is remarkable that the ribosome is able to signal a match to a codon across its body to allow bond formation between amino acids. In some of the earliest single molecule studies of the ribosome, smFRET was used to detect the movements between two tRNA molecules within the ribosome (Blanchard et al., 2004; Lee et al., 2007c; Kim et al., 2007), providing new insights into how correct tRNA molecules are accommodated and incorrect ones are rejected. Figure 5a–b shows the stepwise accommodation of a tRNA that has entered the ribosome revealed by smFRET. Given the small difference of basepairing energy between the matching and nearly matching codons, it remained elusive how the ribosome gains such low error rate of 10−4 in decoding the mRNA. By smFRET, Lee et al. detected a minute difference in the distance between two adjacent tRNAs caused by a single mismatch in the codon and proposed that the ribosome utilizes it as a discriminating tool for tRNA selection (Lee et al., 2007c) (Figure 5c). Another interesting question is how the ribosome ‘translocates’ on the mRNA in 3 nt steps without falling off prematurely. smFRET analyses showed highly coordinated global and local motions of the ribosome itself, painting a picture of the ribosome as a Brownian ratchet (Frank and Gonzalez, 2010).
Figure 5.
Model and experimental observation of tRNA accommodation in the ribosome. (a) A revised model of two-staged tRNA selection (initial selection and proofreading step that follows GTP hydrolysis) based on smFRET experiments (Blanchard et al., 2004). (b) Time evolution of FRET after the initial binding of cognate correct tRNA when GTP hydrolysis is allowed so that both stages of tRNA selection can occur (Blanchard et al., 2004). (c) Time evolution of FRET comparing the delivery of correct (left) and nearly correct (right) tRNAs stalled by non-hydrolizable GTP analog, GDPNP, so that only the initial selection can occur. Both the mid-FRET and low-FRET efficiencies are higher for the correct tRNA by significant amount, suggesting that the tRNA selection has high fidelity even before GTP hydrolysis (Lee et al., 2007c).
Single molecule methods based on detecting one fluorophore (single particle tracking, noted as “ONE” below) and those based on two fluorophores (single molecule FRET, noted as “TWO” below) are complementary tools with contrasting pros and cons (Table 1). While ONE can determine the position down to ~ 2 nm accuracy with 10,000 photons with practically unlimited range of displacement, TWO can easily detect distance changes as small as ~ 0.5 nm with only 100 photons but with a narrow detection range of 2 ~ 10 nm. For ONE, we measure the absolute position of each fluorophore in the laboratory frame and thus the suppression of drift and vibration is critical. In contrast, for TWO, we only measure the relative distance between two fluorophores and, accordingly, the method is robust against drift or vibration. Considering the above differences, ONE is better suited for observing the behavior of a motor protein over a long distance, whereas TWO is well suited for a detailed view of the local dynamics of a single molecule or a complex of molecules. TWO has also been used to detect the one-dimensional diffusion of proteins on nucleic acids (Roy et al., 2009; Liu et al., 2008).
Table 1.
ONE vs TWO. Single molecule fluorescence methods based on one dye and two dyes are compared. They are complementary to each other regarding the precision or range of distance required for the measurement.
| ONE | TWO | |
|---|---|---|
| Distance precision | > 1.5 nm | < 0.5 nm |
| Photon budget | 10,000 photons | 100 photons |
| Range of distance | unlimited | 2 ~ 8 nm |
| Frame of reference | absolute position (two or three dimensional) | relative distance or orientation (one dimensional) |
| Requirement for low drift and low vibration equipment | very high | low |
One color for one distance
Despite the popularity and successes of smFRET method, the requirement of two labels per measurable can limit its application because labeling a protein at a specific location with a high yield can be challenging and may disrupt the protein’s function. In addition, FRET is insensitive to changes that involve distances below 2 ~ 3 nm.
An alternative approach is to probe a distance change with only one dye, taking advantage of the fact that the fluorescence properties of a dye can be strongly influenced by its local environment. For example, Yang et al. deduced the hidden internal conformational dynamics of an enzyme, flavin reductase, by measuring the fluorescence quenching of its substrate, flavin, by photoinduced electron transfer to a nearby aromatic amino acid tyrosine (Yang et al., 2003). Yet another example, suitable for studying DNA-protein and RNA-protein interactions, is an effect called protein-induced fluorescence enhancement (PIFE), first utilized in ensemble experiments (Fischer et al., 2004) and more recently in single molecule experiments (Luo et al., 2007; Myong et al., 2009). Cy3 dye attached to DNA or RNA becomes brighter when it is proximal to a protein, likely due to steric hindrance that inhibits a non-radiative decay pathway involving the twisting motion of the dye around its own axis. Hwang et al. calibrated the distance dependence of the fluorescence enhancement using a variety of DNA/RNA-protein systems to demonstrate its wide applicability as well as its unique ability to detect short-range movements that are not readily accessible by FRET (Hwang et al., 2011) (Figure 6). Fluorescence decay lifetime measured by time-correlated single photon counting technique can improve the reliability of the data and also reveal additional details if there exist more than one decay lifetime (Sorokina et al., 2009). Finally, self-quenching between two identical dyes that form a dimer could be used to follow the conformational changes of a single protein between 1.5 nm and 1 nm distances, again a range inaccessible to FRET (Zhou et al., 2011b), and to monitor the distance changes between two kinesin monomers (Toprak et al., 2009).
Figure 6.
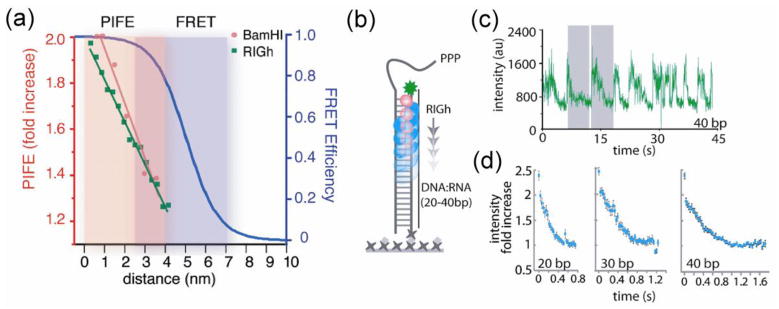
Measuring distance changes with one dye (Hwang et al., 2011). (a) Calibration of PIFE (protein induced fluorescence enhancement) effect as a function of distance obtained from exemplary proteins: restriction enzyme, BamHI, and a motor protein RIG-I, and comparison to FRET vs distance. (b) As RIG-I translocates on RNA-DNA duplex, fluorescence signal of a fluorophore attached to one end of the duplex changes, decreasing as the protein moves away from the fluorophore. (c) A single molecule fluorescence intensity time trace showing repetitive translocation of RIG-I observed via PIFE. (d) Time traces averaged over about 50 translocation cycles over duplexes of 20, 30, and 40 base pairs in length.
4. Three and four colors: exploring complex correlations
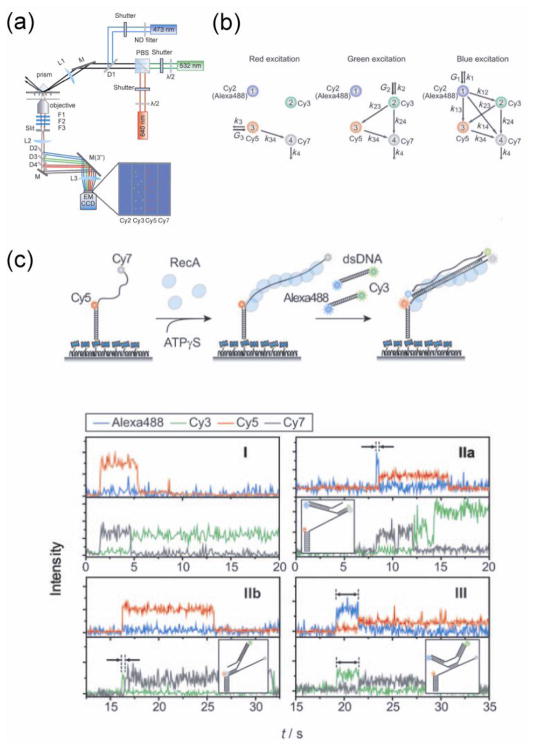
The structural complexity of molecular motors and the existence of multiple correlated components within the motor complexes demand that we probe multiple observables at the same time in order to fully understand their working mechanisms. It is now possible to detect three or four different colors at the single molecule level (Hohng et al., 2004; Lee et al., 2010a; Clamme and Deniz, 2005; Lee et al., 2007a; Eid et al., 2009; DeRocco et al., 2010) (Figure 7a). Three color FRET can measure up to three different pairwise distances simultaneously and four color FRET can measure six different pairwise distances. By alternating different lasers to excite different subsets of the dyes, it became possible to deconvolve all of the distances (Kapanidis et al., 2004; Lee et al., 2010a) (Figure 7b).
Figure 7.
Three and four color FRET (Lee et al., 2010a). (a) Schematic of 4-color FRET setup with shutters for alternating excitation. (b) Diagram of energy transfer between 4 dyes. With alternating excitation, it is possible to determine all six pairwise distances between the dyes. (c) Strand exchange by RecA filament demonstrated in real time by 4-color FRET. Simultaneously observing two pairs of dyes revealed various types of exchanging mechanism, depending on which part of the DNA strand initializes the exchange.
The capability of multi-color fluorescence technique to probe complex dynamics of proteins and nucleic acids has been demonstrated in several examples (Stein et al., 2011; Lee et al., 2007a; Lee et al., 2007b; Lee et al., 2010c; DeRocco et al., 2010; Roy et al., 2009; Lee et al., 2010a; Munro et al., 2010a; Munro et al., 2010b; Clamme and Deniz, 2005). DeRocco et al. measured the conformation of a mismatched DNA by smFRET while monitoring the colocalization of the mismatch repair protein, MutSα, with additional fluorescence color (DeRocco et al., 2010). Simultaneous observation of three dyes enabled them to correlate the MutSα binding with the DNA bending. By three-color FRET, Roy et al. showed the spontaneous diffusional migration of single-stranded DNA binding protein (SSB) by observing its motion between two ends of the ssDNA (Roy et al., 2009). Such diffusional motion can facilitate the melting of short DNA hairpins and stimulate RecA filament elongation during DNA recombination and repair. While two-color FRET between two ends of the ssDNA was enough to prove the existence of such dynamics, three-color FRET with additional label on SSB made it clear if the dynamics represents the partial unwrapping of the ssDNA or diffusional motion of the protein as it could reveal the correlated change of FRET between SSB and each end of the ssDNA.
With four color FRET, Lee et al. analyzed the DNA strand exchange reaction mediated by RecA filament (Lee et al., 2010a) (Figure 7c). They grouped the dyes into two pairs and traced the distances at both ends of the DNA strand being exchanged. By probing two distances simultaneously, they identified subsets of molecules starting the strand exchange process from different positions, i.e. either from the ends or the middle of the strand. Another demonstration of multi-color FRET was on the ribosome. During translation elongation, tRNAs have to pass through a small gap between two subunits of the ribosome. This ‘translocation’ process involves multiple conformational changes, including the formation of tRNA hybrid states, closure of the ribosomal L1 stalk domain, and ratcheting between the subunits of the ribosome. By simultaneously probing dynamics of the tRNA and the L1 stalk, Munro et al. observed that two dynamics are not tightly coupled and they can be regulated by the elongation factor EF-G (Munro et al., 2010a; Munro et al., 2010b).
5. Two colors + one force: force-fluorescence nanometry
Motor proteins convert biochemical energy to mechanical work and as such the process can be modulated by force. By applying forces of precisely determined values, we can reveal the hidden reaction parameters by either slowing down the process to the detection time scale or probing the force-dependence of the dynamics. Several techniques have been developed to mechanically probe single molecules, including optical tweezers, magnetic tweezers, and atomic force microscopy. For a general overview and comparison of these techniques, refer to (Neuman and Nagy, 2008). In particular, optical tweezers have proven to be a powerful tool with high spatial and force resolutions. They can measure the mechanical properties over a force range of 0.1 to 100 pN (Moffitt et al., 2008).
Optical tweezers can achieve sub-nanometer precision down to a single angstrom but only at relatively high forces. At low forces, the tether connecting the trapped bead to the biomolecule, often a DNA molecule, is not fully stretched and cannot transmit small movements of the biomolecule to the bead. Additionally, in a system of bound molecules, a protein and a DNA for example, a considerably large conformational change of one molecule (protein) does not necessarily induce a detectable structural change of another molecule (DNA) coupled to the tweezers, which is the only readout in the tweezers setup. Combining the optical tweezers with single molecule fluorescence provides a solution to this problem (Figure 8).
Figure 8.
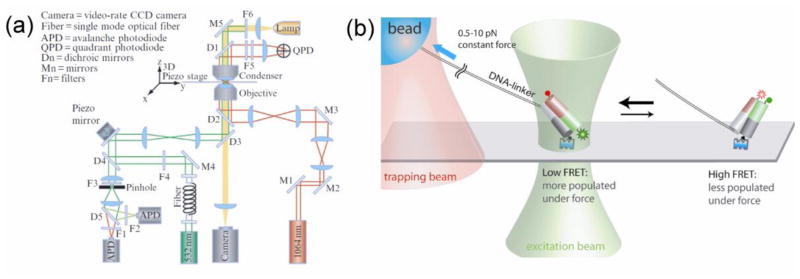
Combination of fluorescence and force measurements. (a) Schematic of the instrument design combining optical tweezers and confocal fluorescence measurement of two colors (Zhou et al., 2010). (b) Experimental scheme for measuring force-dependence of conformational dynamics of a DNA structure via FRET (Joo et al., 2008).
Combined single molecule force and fluorescence spectroscopy was first demonstrated to study myosins. Ishijima et al. simultaneously measured the ATPase activity and the mechanical action of the motor protein (Ishijima et al., 1998). They showed that the force generation does not always coincide with the release of the hydrolyzed ADP. There was often a delay until the force generation, suggesting that myosin has a hysteresis or memory state storing chemical energy from ATP hydrolysis. Simultaneous measurement of the ADP release (by fluorescence) and the mechanical event (by optical tweezers) enabled them to reveal the temporal correlation between the events. Later, the optical tweezers instrument was combined with fluorescence quenching between two fluorophores (Lang et al., 2004) and smFRET (Hohng et al., 2007; Tarsa et al., 2007) and revealed the detailed energy landscape of Holliday junctions. This hybrid approach was then applied to study the SSB protein (Zhou et al., 2011a). The combined force-fluorescence measurement demonstrated how mechanical force modulates SSB diffusion along the DNA and the unraveling of the DNA, thus providing evidence of reptation mechanism for SSB diffusion. They suggested that SSB can act as a sliding platform to recruit its interacting proteins for use in DNA replication, recombination, and repair.
In order to achieve the highest possible resolution in optical tweezers, researchers have developed dual trap optical tweezers that can decouple the trapped objects from the surface, which is the major source of drift and noise (Shaevitz et al., 2003; Moffitt et al., 2006). Combing such ultrahigh resolution optical tweezers with single molecule fluorescence (Comstock et al., 2011) raised a number of challenges: for example, faster photobleaching induced by the intense trapping laser. Long tethers can be used to spatially separate the beams (Ishijima et al., 1998; Hohng et al., 2007) but this inevitably sacrifices the spatial or force resolution. As an alternative approach, interlacing the trapping and excitation beams prevents simultaneous exposure to both beams (Brau et al., 2006) and solves the problem without sacrificing the resolution (Comstock et al., 2011).
Magnetic tweezers have also been combined with fluorescence detection (De Vlaminck and Dekker, 2012; van Mameren et al., 2008). Magnetic tweezers have several advantages over optical tweezers or atomic force microscope. First of all, they realize the least interfering force manipulation as they do not interact at all with non-magnetic sample. They do not suffer from sample heating or photo-destruction like normal optical tweezers. Due to the small gradient of magnetic field, they do not require complex feedback system for performing measurements at a constant force. Neither do they require sophisticated design to deal with the sources of noise found in optical tweezers. They provide precise control of the torque applied on the tether, which is possible but not very straightforward with optical tweezers. Another advantage is that they enable parallel measurements of many molecules, although the force experience by individual beads are not precisely the same. Several groups of researchers have presented the combination of magnetic tweezers with single molecule fluorescence techniques to study the intrinsic properties of DNAs (Lee et al., 2010b; Shroff et al., 2005) and the DNA packaging motor of bacteriophage ϕ29 (Hugel et al., 2007). This type of approach will become powerful especially when we need to assess the torsional strain generated by molecular motors or manipulate a large number of them simultaneously.
6. Theoretical and computational approaches to molecular motors
Due to the recent advances in the theory for understanding the dynamics of individual molecules, we can relate the observed single molecule dynamics to the underlying statistical mechanics and thermodynamics of the molecular motors. Be it fluctuations over poorly resolved group of states or well-defined transition kinetics between distinct states, we can statistically analyze the dynamics to describe the free energy landscape of the system along the observed reaction coordinate. Such approach is largely based on the energy landscape theory developed in the protein folding community (Bryngelson and Wolynes, 1989; Thirumalai et al., 2010). The framework of the energy landscape theory has been extended to nucleic acids and also to the functions of molecular motors which are often accompanied by global motions of the proteins (Schug and Onuchic, 2010).
Researchers have developed rigorous formulations to extract the underlying conformational dynamics from the distribution of observed smFRET efficiency or fluorescence lifetime of diffusing or immobilized molecules (Gopich and Szabo, 2007, 2009, 2012; Yang and Xie, 2002). It was shown that, if we measure the color and lifetime of each photon simultaneously, we can maximally extract the information on the connectivity between the underlying conformational states (Gopich and Szabo, 2012). On the other hand, models to describe the biological molecules under mechanical tension also have been developed and applied to various systems (Dudko et al., 2006; Dudko et al., 2007; Suzuki and Dudko, 2011). Using these models, we can extract intrinsic parameters of the molecular complex from single molecule pulling experiments, including the intrinsic rate of rupturing, the distance to the transition state, and the intrinsic activation free energy (Dudko et al., 2006).
Computational approaches have extensively been applied in describing and understanding the mechanisms of molecular motors. It used to be limited due to their large size and long time scale associated with their functions. However, due to the hierarchical structure of dynamics in various levels from atoms to large domains, we can coarse-grain the atomistic details and focus on the global motions which are directly related to their functions. Either all-atom or coarse-grained simulations have been performed to many of the molecular motors discussed in this review, including kinesin (Hyeon and Onuchic, 2007a, b; Hyeon and Onuchic, 2011), myosin (Tehver and Thirumalai, 2010; Takano et al., 2010), F1-ATPase (Koga and Takada, 2006), RNAP polymerase (Chen et al., 2010; Zamft et al., 2012), helicases (Yu et al., 2006), and ribosomes (Whitford et al., 2011), and extended our understanding of their functioning mechanisms. Hyeon and Onuchic developed a novel computational strategy to build the structural model of kinesin dimers attached to microtubules, for which the crystal structure does not exist, and explained the processivity and directionality of the kinesin motor by resolving the mechanism of nucleotide binding regulation between the two motor domains, for which the internal strain plays a key role (Hyeon and Onuchic, 2007a). In a following study, they focused on the stepping dynamics of kinesins and resolved the experimental debates on the existence of substeps (Hyeon and Onuchic, 2007b). They described the stepping of kinesin as rectified diffusion by viewing the power stroke as modulating the boundary condition for its diffusion. Integrating their results from coarse-grained modeling, they constructed the full thermodynamic cycle of the kinesin as a heat engine and demonstrated the relation between structural changes and work production in this molecular motor (Hyeon and Onuchic, 2011). Similar approach has been employed for another class of stepping motor, myosins. Tehver and Thirumalai applied their structural perturbation method to the actomyosin system and revealed the communication pathway in myosin from the ATP-binding site to the actin-binding region (Tehver and Thirumalai, 2010).
Nucleic acid motors make another example where computational approaches have proven to be successful. Chen et al. constructed coarse-grained self-organized polymer models of RNAP on promoter DNA and performed Brownian dynamics simulation of the promoter opening (Chen et al., 2010). They were able to structurally identify multiple stages in the process: DNA melting, scrunching, and bending. With the advancement of computational technology as well as its integration with crystallographic, cryo-EM, and single molecule methods, it became feasible to model and simulate the intricate machinery of the ribosome, which comprises multi-million atoms (Whitford et al., 2011). Sanbonmatsu et al. performed all-atom molecular dynamics simulation of tRNA accommodation in the ribosome (Sanbonmatsu et al., 2005; Whitford et al., 2010). They identified the bases of the ribosomal RNA that interact with the tRNA during its accommodation, some of which working as a gate to impede its translocation, and demonstrated that the flexibility of tRNA is essential for the selection of cognate tRNA (Sanbonmatsu et al., 2005). In more recent work, through an integrated approach of simulation and smFRET, they revealed the presence of reversible structural fluctuations and parallel pathways during tRNA accommodation (Whitford et al., 2010). Researchers have also simulated the interaction of the nascent polypeptide chain with the ribosome and shown how the nascent chain regulates its own translation at the atomic level (Trabuco et al., 2010a; Ishida and Hayward, 2008). Gumbart et al., through a method named molecular dynamics flexible fitting (Trabuco et al., 2008), constructed the ribosome model in complex with the membrane channel protein SecY, thus complementing the low resolution cryo-EM data (Gumbart et al., 2009). Remarkably, the same method was applied to reveal the interaction between tRNA and L1 stalk, that had been studied by smFRET (Cornish et al., 2009; Fei et al., 2008; Fei et al., 2009; Munro et al., 2010a), and the simulation provided a structural basis for interpreting and explaining the smFRET observations (Trabuco et al., 2010b).
7. Challenges ahead
Despite many accomplishments of single molecule approaches to the biological motors, we are facing technical challenges in exploring complex interactions and dynamics of biomolecules in close to physiological or in vivo environments. In general, fluorescence-based single molecule methods require sparsely distributed molecules and low background fluorescence. The cell is a crowded medium; (1) many biological processes require concentrations of participating molecules that are several orders of magnitude higher than what conventional single molecule measurements can deal with and (2) a diverse set of molecules are involved in any given process, some of which are often unknown or only transiently interacting, making it very difficult to recapitulate their action in vitro. Therefore, we need (1) a method to detect single molecules in a high background of diffusing fluorescent molecules and (2) an experimental scheme to observe single molecules under more physiologically relevant conditions even when we do not exactly know what kinds of interactions are present in vivo. (3) Further ahead, we want to measure single molecule dynamics within living cells, not to be compromised by the experimental scheme of in vitro measurements.
For the purpose of suppressing high background of diffusing molecules, there are three established techniques: stimulated emission depletion (STED) microscopy, vesicle encapsulation, and zero-mode waveguide. STED microscopy modulates the PSF by using a depleting beam with a singularity at the center that is combined with the excitation beam, leaving only the molecules within the small center volume available for fluorescence detection (Hell and Wichmann, 1994). This technique confines the excitation volume ~ 100 fold smaller compared to conventional confocal microscopy (Harke et al., 2008) and enables fluorescence detection close to the single molecule level at up to ~ 100 nM concentration. Vesicle encapsulation (Boukobza et al., 2001; Cisse et al., 2007) captures one set of molecules within lipid vesicles such that their effective concentrations reach up to ~ 100 μM due to the confinement. But the technique is especially useful for studying weak and transient interaction between pairs of molecules (Cisse et al., 2007; Benítez et al., 2008). Cisse et al. demonstrated fast annealing and melting of short DNA oligos which can only be measured at such high concentration (Cisse et al., 2012). Another approach is to use nano-fabricated wells in a metallic thin film as an optical blockade to the background, named as zero-mode waveguide (Levene et al., 2003). The smallest zero-mode waveguide enables us to detect immobilized single fluorophore in the presence of diffusing fluorophores at ~ 200 μM. Low concentration does not only slow down biological processes but often kinetically prevents certain reactions especially when the reaction requires the formation of multimeric protein complexes or recruiting multiple substrates in succession. Single molecule measurements at high concentration can uncover previously unaddressed details of such biochemical processes (Uemura et al., 2010) and offer a new way to sequence single DNA molecules in real time (Eid et al., 2009).
Single molecule measurements in vitro provide detailed mechanistic views of individual molecular motors but they are often different from the actual pictures in vivo, due to missing cofactors, unknown chemical modifications, or different solution environment. Super-resolution cellular imaging reveals more realistic pictures but its spatial or temporal resolutions in addressing individual molecules are more or less limited compared to well-controlled in vitro measurements. In order to bridge the gap, a group of researchers have presented a new approach by performing single molecule measurements with the cell lysate directly without further purification procedures (Jain et al., 2011; Yeom et al., 2011). They combined the conventional immunoprecipitation assay with single molecule fluorescence microscopy. In contrast to measurements with purified components, this method preserves the constitution of a complex in the cell and still addresses individual complexes so that we can reveal the in vivo stoichiometry of the complexes. Jain et al. specifically captured fresh protein complexes through their antibodies immobilized on surface and detected their binding partners through genetically encoded fluorescent tags, thus directly measuring their functionality and stoichiometry (Jain et al., 2011). Yeom et al. took a slightly different approach by enriching the complexes of interest using antibody beads before immobilizing them on the surface (Yeom et al., 2011). These approaches will provide an alternative platform to study single molecule dynamics in more in vivo-like context. For example, first applications to reveal new stoichiometric information were published shortly after (Means et al., 2011; Shen et al., 2012). Combined with cell sorting, the technique can be pushed toward single-cell-based systems biology.
Observing molecules within the cellular environment will show the most accurate and undisturbed picture of their interactions and dynamics. Single molecule imaging in live cell has been highly challenging but the recent advances in imaging technology are making breakthroughs in several directions. Super-resolution imaging techniques, which used to provide snapshots of fixed cells, have advanced toward higher spatio-temporal resolution in live cells. Jones et al. demonstrated 2D or 3D super-resolution imaging of cellular organelles with 25–50 nm spatial resolution and sub-second temporal resolution by labeling target proteins with fast switching and photostable dyes (Jones et al.). STED microscopy achieved video rate imaging of micron-sized field of view with ~ 60 nm lateral resolution (Westphal et al., 2008). Due to the limited number of fluorescence emission cycles that organic dyes or fluorescent proteins can undergo, there always is a trade-off among the temporal resolution, spatial resolution, and the maximum observation time. As the excitation beam excites any fluorophore on its path, we are further wasting the limited photon budget during 3D imaging. Planchon et al. developed a plane-illumination microscopy to minimize the unwanted excitation and also the out-of-focus background (Planchon et al., 2011). They combined scanned Bessel beam illumination with structured illumination or two-photon excitation to create thin light sheets and demonstrated rapid 3D imaging of living cells for an extended period.
In addition to the technical challenges discussed above, we are facing fundamental problems in interpreting single molecule data. Molecular motors combine large-scale conformational dynamics with biochemical processes happening in atomic scale in order to perform their functions. We can measure the former, at least for a few degrees of freedom with limited temporal resolution, but we often lack understanding of its causal relation to the biochemistry involved. This relation can be revealed in detail by computational approach. As discussed in this review, the computational capability has been rapidly growing, enabling us to simulate macromolecules up to millisecond time scale, in case of coarse-grained modeling, which is within the experimentally accessible regime. As the gap of the time scale between experimental and computational approaches is rapidly collapsing from both ends, we will eventually be able to explore the whole range of molecular motor dynamics, from the atoms to the cell.
Acknowledgments
We thank Jejoong Yoo for helpful discussions. This work was supported by U.S. National Science Foundation grants 0646550 and 0822613 and U.S. National Institutes of Health grant GM065367 to T.H. T.H. is an employee of the Howard Hughes Medical Institute.
References
- Andrecka J, Lewis R, Brückner F, Lehmann E, Cramer P, Michaelis J. Single-molecule tracking of mRNA exiting from RNA polymerase II. Proceedings of the National Academy of Sciences. 2008;105:135–40. doi: 10.1073/pnas.0703815105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badali D, Gradinaru CC. The effect of Brownian motion of fluorescent probes on measuring nanoscale distances by F[o-umlaut]rster resonance energy transfer. The Journal of Chemical Physics. 2011;134:225102–11. doi: 10.1063/1.3598109. [DOI] [PubMed] [Google Scholar]
- Bannai H, Levi S, Schweizer C, Dahan M, Triller A. Imaging the lateral diffusion of membrane molecules with quantum dots. Nat Protocols. 2007;1:2628–34. doi: 10.1038/nprot.2006.429. [DOI] [PubMed] [Google Scholar]
- Bates M, Blosser TR, Zhuang X. Short-Range Spectroscopic Ruler Based on a Single-Molecule Optical Switch. Physical Review Letters. 2005;94:108101. doi: 10.1103/PhysRevLett.94.108101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benítez JJ, Keller AM, Ochieng P, Yatsunyk LA, Huffman DL, Rosenzweig AC, Chen P. Probing Transient Copper Chaperone – Wilson Disease Protein Interactions at the Single-Molecule Level with Nanovesicle Trapping. Journal of the American Chemical Society. 2008;130:2446–7. doi: 10.1021/ja7107867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF. Imaging Intracellular Fluorescent Proteins at Nanometer Resolution. Science. 2006;313:1642–5. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- Bianco PR, Brewer LR, Corzett M, Balhorn R, Yeh Y, Kowalczykowski SC, Baskin RJ. Processive translocation and DNA unwinding by individual RecBCD enzyme molecules. Nature. 2001;409:374–8. doi: 10.1038/35053131. [DOI] [PubMed] [Google Scholar]
- Blanchard SC, Gonzalez RL, Kim HD, Chu S, Puglisi JD. tRNA selection and kinetic proofreading in translation. Nat Struct Mol Biol. 2004;11:1008–14. doi: 10.1038/nsmb831. [DOI] [PubMed] [Google Scholar]
- Boukobza E, Sonnenfeld A, Haran G. Immobilization in Surface-Tethered Lipid Vesicles as a New Tool for Single Biomolecule Spectroscopy. The Journal of Physical Chemistry B. 2001;105:12165–70. [Google Scholar]
- Brau RR, Tarsa PB, Ferrer JM, Lee P, Lang MJ. Interlaced Optical Force-Fluorescence Measurements for Single Molecule Biophysics. Biophysical journal. 2006;91:1069–77. doi: 10.1529/biophysj.106.082602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MT, Delalez NJ, Armitage JP. Protein dynamics and mechanisms controlling the rotational behaviour of the bacterial flagellar motor. Current Opinion in Microbiology. 2011;14:734–40. doi: 10.1016/j.mib.2011.09.009. [DOI] [PubMed] [Google Scholar]
- Bryngelson JD, Wolynes PG. Intermediates and barrier crossing in a random energy model (with applications to protein folding) The Journal of Physical Chemistry. 1989;93:6902–15. [Google Scholar]
- Chen J, Darst SA, Thirumalai D. Promoter melting triggered by bacterial RNA polymerase occurs in three steps. Proceedings of the National Academy of Sciences. 2010;107:12523–8. doi: 10.1073/pnas.1003533107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung HS, McHale K, Louis JM, Eaton WA. Single-Molecule Fluorescence Experiments Determine Protein Folding Transition Path Times. Science. 2012;335:981–4. doi: 10.1126/science.1215768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cisse I, Okumus B, Joo C, Ha T. Fueling protein – DNA interactions inside porous nanocontainers. Proceedings of the National Academy of Sciences. 2007;104:12646–50. doi: 10.1073/pnas.0610673104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cisse II, Kim H, Ha T. A rule of seven in Watson-Crick base-pairing of mismatched sequences. Nat Struct Mol Biol. 2012;19:623–7. doi: 10.1038/nsmb.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clamme J-P, Deniz AA. Three-Color Single-Molecule Fluorescence Resonance Energy Transfer. ChemPhysChem. 2005;6:74–7. doi: 10.1002/cphc.200400261. [DOI] [PubMed] [Google Scholar]
- Comstock MJ, Ha T, Chemla YR. Ultrahigh-resolution optical trap with single-fluorophore sensitivity. Nat Meth. 2011;8:335–40. doi: 10.1038/nmeth.1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornish PV, Ermolenko DN, Noller HF, Ha T. Spontaneous Intersubunit Rotation in Single Ribosomes. Molecular Cell. 2008;30:578–88. doi: 10.1016/j.molcel.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornish PV, Ermolenko DN, Staple DW, Hoang L, Hickerson RP, Noller HF, Ha T. Following movement of the L1 stalk between three functional states in single ribosomes. Proceedings of the National Academy of Sciences. 2009;106:2571–6. doi: 10.1073/pnas.0813180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courty S, Bouzigues C, Luccardini C, Ehrensperger MV, Bonneau S, Dahan M, James I. Methods in Enzymology. Academic Press; 2006a. pp. 211–28. [DOI] [PubMed] [Google Scholar]
- Courty S, Luccardini C, Bellaiche Y, Cappello G, Dahan M. Tracking Individual Kinesin Motors in Living Cells Using Single Quantum-Dot Imaging. Nano Letters. 2006b;6:1491–5. doi: 10.1021/nl060921t. [DOI] [PubMed] [Google Scholar]
- Dahan M, Levi S, Luccardini C, Rostaing P, Riveau B, Triller A. Diffusion Dynamics of Glycine Receptors Revealed by Single-Quantum Dot Tracking. Science. 2003;302:442–5. doi: 10.1126/science.1088525. [DOI] [PubMed] [Google Scholar]
- De Vlaminck I, Dekker C. Recent Advances in Magnetic Tweezers. Annual Review of Biophysics. 2012;41:453–72. doi: 10.1146/annurev-biophys-122311-100544. [DOI] [PubMed] [Google Scholar]
- DeRocco V, Anderson T, Piehler J, Erie DA, Weninger K. Four-color single-molecule fluorescence with noncovalent dye labeling to monitor dynamic multimolecular complexes. Biotechniques. 2010;49:807–16. doi: 10.2144/000113551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeWitt MA, Chang AY, Combs PA, Yildiz A. Cytoplasmic Dynein Moves Through Uncoordinated Stepping of the AAA+ Ring Domains. Science. 2012;335:221–5. doi: 10.1126/science.1215804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubertret B, Skourides P, Norris DJ, Noireaux V, Brivanlou AH, Libchaber A. In Vivo Imaging of Quantum Dots Encapsulated in Phospholipid Micelles. Science. 2002;298:1759–62. doi: 10.1126/science.1077194. [DOI] [PubMed] [Google Scholar]
- Dudko OK, Hummer G, Szabo A. Intrinsic Rates and Activation Free Energies from Single-Molecule Pulling Experiments. Physical Review Letters. 2006;96:108101. doi: 10.1103/PhysRevLett.96.108101. [DOI] [PubMed] [Google Scholar]
- Dudko OK, Mathé J, Szabo A, Meller A, Hummer G. Extracting Kinetics from Single-Molecule Force Spectroscopy: Nanopore Unzipping of DNA Hairpins. Biophysical journal. 2007;92:4188–95. doi: 10.1529/biophysj.106.102855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eid J, Fehr A, Gray J, Luong K, Lyle J, Otto G, Peluso P, Rank D, Baybayan P, Bettman B, Bibillo A, Bjornson K, Chaudhuri B, Christians F, Cicero R, Clark S, Dalal R, deWinter A, Dixon J, Foquet M, Gaertner A, Hardenbol P, Heiner C, Hester K, Holden D, Kearns G, Kong X, Kuse R, Lacroix Y, Lin S, Lundquist P, Ma C, Marks P, Maxham M, Murphy D, Park I, Pham T, Phillips M, Roy J, Sebra R, Shen G, Sorenson J, Tomaney A, Travers K, Trulson M, Vieceli J, Wegener J, Wu D, Yang A, Zaccarin D, Zhao P, Zhong F, Korlach J, Turner S. Real-Time DNA Sequencing from Single Polymerase Molecules. Science. 2009;323:133–8. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- Erhardt M, Namba K, Hughes KT. Bacterial Nanomachines: The Flagellum and Type III Injectisome. Cold Spring Harbor Perspectives in Biology. 2010:2. doi: 10.1101/cshperspect.a000299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei J, Bronson JE, Hofman JM, Srinivas RL, Wiggins CH, Gonzalez RL. Allosteric collaboration between elongation factor G and the ribosomal L1 stalk directs tRNA movements during translation. Proceedings of the National Academy of Sciences. 2009;106:15702–7. doi: 10.1073/pnas.0908077106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei J, Kosuri P, MacDougall DD, Gonzalez RL., Jr Coupling of Ribosomal L1 Stalk and tRNADynamics during Translation Elongation. Molecular Cell. 2008;30:348–59. doi: 10.1016/j.molcel.2008.03.012. [DOI] [PubMed] [Google Scholar]
- Fischer CJ, Maluf NK, Lohman TM. Mechanism of ATP-dependent Translocation of E. coli UvrD Monomers Along Single-stranded DNA. Journal of Molecular Biology. 2004;344:1287–309. doi: 10.1016/j.jmb.2004.10.005. [DOI] [PubMed] [Google Scholar]
- Frank J, Gonzalez RL. Structure and Dynamics of a Processive Brownian Motor: The Translating Ribosome. Annual Review of Biochemistry. 2010;79:381–412. doi: 10.1146/annurev-biochem-060408-173330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gennerich A, Vale RD. Walking the walk: how kinesin and dynein coordinate their steps. Current Opinion in Cell Biology. 2009;21:59–67. doi: 10.1016/j.ceb.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopich IV, Szabo A. Single-Molecule FRET with Diffusion and Conformational Dynamics. The Journal of Physical Chemistry B. 2007;111:12925–32. doi: 10.1021/jp075255e. [DOI] [PubMed] [Google Scholar]
- Gopich IV, Szabo A. Decoding the Pattern of Photon Colors in Single-Molecule FRET. The Journal of Physical Chemistry B. 2009;113:10965–73. doi: 10.1021/jp903671p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopich IV, Szabo A. Theory of the energy transfer efficiency and fluorescence lifetime distribution in single-molecule FRET. Proceedings of the National Academy of Sciences. 2012;109:7747–52. doi: 10.1073/pnas.1205120109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumbart J, Trabuco LG, Schreiner E, Villa E, Schulten K. Regulation of the Protein-Conducting Channel by a Bound Ribosome. Structure. 2009;17:1453–64. doi: 10.1016/j.str.2009.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha T, Enderle T, Ogletree DF, Chemla DS, Selvin PR, Weiss S. Probing the interaction between two single molecules: fluorescence resonance energy transfer between a single donor and a single acceptor. Proceedings of the National Academy of Sciences. 1996;93:6264–8. doi: 10.1073/pnas.93.13.6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha T, Tinnefeld P. Photophysics of fluorescent probes for single-molecule biophysics and super-resolution imaging. Annual Review of Physical Chemistry. 2012;63:595–617. doi: 10.1146/annurev-physchem-032210-103340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harke B, Ullal CK, Keller J, Hell SW. Three-Dimensional Nanoscopy of Colloidal Crystals. Nano Letters. 2008;8:1309–13. doi: 10.1021/nl073164n. [DOI] [PubMed] [Google Scholar]
- Hell SW, Wichmann J. Breaking the diffraction resolution limit by stimulated emission: stimulated-emission-depletion fluorescence microscopy. Opt Lett. 1994;19:780–2. doi: 10.1364/ol.19.000780. [DOI] [PubMed] [Google Scholar]
- Henn A, Medalia O, Shi S-P, Steinberg M, Franceschi F, Sagi I. Visualization of unwinding activity of duplex RNA by DbpA, a DEAD box helicase, at single-molecule resolution by atomic force microscopy. Proceedings of the National Academy of Sciences. 2001;98:5007–12. doi: 10.1073/pnas.071372498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess ST, Girirajan TPK, Mason MD. Ultra-High Resolution Imaging by Fluorescence Photoactivation Localization Microscopy. Biophysical journal. 2006;91:4258–72. doi: 10.1529/biophysj.106.091116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohng S, Joo C, Ha T. Single-Molecule Three-Color FRET. Biophysical journal. 2004;87:1328–37. doi: 10.1529/biophysj.104.043935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohng S, Zhou R, Nahas MK, Yu J, Schulten K, Lilley DMJ, Ha T. Fluorescence-Force Spectroscopy Maps Two-Dimensional Reaction Landscape of the Holliday Junction. Science. 2007;318:279–83. doi: 10.1126/science.1146113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Bates M, Zhuang X. Super-Resolution Fluorescence Microscopy. Annual Review of Biochemistry. 2009;78:993–1016. doi: 10.1146/annurev.biochem.77.061906.092014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugel T, Michaelis J, Hetherington CL, Jardine PJ, Grimes S, Walter JM, Falk W, Anderson DL, Bustamante C. Experimental Test of Connector Rotation during DNA Packaging into Bacteriophage ?29 Capsids. PLoS Biol. 2007;5:e59. doi: 10.1371/journal.pbio.0050059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang H, Kim H, Myong S. Protein induced fluorescence enhancement as a single molecule assay with short distance sensitivity. Proceedings of the National Academy of Sciences. 2011;108:7414–8. doi: 10.1073/pnas.1017672108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyeon C, Onuchic J. A Structural Perspective on the Dynamics of Kinesin Motors. Biophysical journal. 2011;101:2749–59. doi: 10.1016/j.bpj.2011.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyeon C, Onuchic JN. Internal strain regulates the nucleotide binding site of the kinesin leading head. Proceedings of the National Academy of Sciences. 2007a;104:2175–80. doi: 10.1073/pnas.0610939104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyeon C, Onuchic JN. Mechanical control of the directional stepping dynamics of the kinesin motor. Proceedings of the National Academy of Sciences. 2007b;104:17382–7. doi: 10.1073/pnas.0708828104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal A, Arslan S, Okumus B, Wilson TJ, Giraud G, Norman DG, Ha T, Lilley DMJ. Orientation dependence in fluorescent energy transfer between Cy3 and Cy5 terminally attached to double-stranded nucleic acids. Proceedings of the National Academy of Sciences. 2008;105:11176–81. doi: 10.1073/pnas.0801707105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida H, Hayward S. Path of Nascent Polypeptide in Exit Tunnel Revealed by Molecular Dynamics Simulation of Ribosome. Biophysical journal. 2008;95:5962–73. doi: 10.1529/biophysj.108.134890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishijima A, Kojima H, Funatsu T, Tokunaga M, Higuchi H, Tanaka H, Yanagida T. Simultaneous Observation of Individual ATPase and Mechanical Events by a Single Myosin Molecule during Interaction with Actin. Cell. 1998;92:161–71. doi: 10.1016/s0092-8674(00)80911-3. [DOI] [PubMed] [Google Scholar]
- Jain A, Liu R, Ramani B, Arauz E, Ishitsuka Y, Ragunathan K, Park J, Chen J, Xiang YK, Ha T. Probing cellular protein complexes using single-molecule pull-down. Nature. 2011;473:484–8. doi: 10.1038/nature10016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A, O’Donnell M. Cellular DNA replicases: components and dynamics at the replication fork. Annual Review of Biochemistry. 2005;74:283–315. doi: 10.1146/annurev.biochem.73.011303.073859. [DOI] [PubMed] [Google Scholar]
- Jonckheere A, Smeitink J, Rodenburg R. Mitochondrial ATP synthase: architecture, function and pathology. Journal of Inherited Metabolic Disease. 2012;35:211–25. doi: 10.1007/s10545-011-9382-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SA, Shim S-H, He J, Zhuang X. Fast, three-dimensional super-resolution imaging of live cells. Nat Meth. 8:499–505. doi: 10.1038/nmeth.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo C, Balci H, Ishitsuka Y, Buranachai C, Ha T. Annual Review of Biochemistry. Palo Alto: Annual Reviews; 2008. pp. 51–76. [DOI] [PubMed] [Google Scholar]
- Joo C, McKinney SA, Nakamura M, Rasnik I, Myong S, Ha T. Real-Time Observation of RecA Filament Dynamics with Single Monomer Resolution. Cell. 2006;126:515–27. doi: 10.1016/j.cell.2006.06.042. [DOI] [PubMed] [Google Scholar]
- Kao HP, Verkman AS. Tracking of single fluorescent particles in three dimensions: use of cylindrical optics to encode particle position. Biophysical journal. 1994;67:1291–300. doi: 10.1016/S0006-3495(94)80601-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapanidis AN, Lee NK, Laurence TA, Doose S, Margeat E, Weiss S. Fluorescence-aided molecule sorting: Analysis of structure and interactions by alternating-laser excitation of single molecules. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:8936–41. doi: 10.1073/pnas.0401690101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapanidis AN, Margeat E, Ho SO, Kortkhonjia E, Weiss S, Ebright RH. Initial Transcription by RNA Polymerase Proceeds Through a DNA-Scrunching Mechanism. Science. 2006;314:1144–7. doi: 10.1126/science.1131399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearns DB. A field guide to bacterial swarming motility. Nat Rev Micro. 2010;8:634–44. doi: 10.1038/nrmicro2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Tang G-Q, Patel SS, Ha T. Opening – closing dynamics of the mitochondrial transcription pre-initiation complex. Nucleic Acids Research. 2011;40:371–80. doi: 10.1093/nar/gkr736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HD, Puglisi JD, Chu S. Fluctuations of Transfer RNAs between Classical and Hybrid States. Biophysical journal. 2007;93:3575–82. doi: 10.1529/biophysj.107.109884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga N, Takada S. Folding-based molecular simulations reveal mechanisms of the rotary motor F1–ATPase. Proceedings of the National Academy of Sciences. 2006;103:5367–72. doi: 10.1073/pnas.0509642103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang MJ, Fordyce PM, Engh AM, Neuman KC, Block SM. Simultaneous, coincident optical trapping and single-molecule fluorescence. Nat Meth. 2004;1:133–9. doi: 10.1038/nmeth714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson DR, Zipfel WR, Williams RM, Clark SW, Bruchez MP, Wise FW, Webb WW. Water-Soluble Quantum Dots for Multiphoton Fluorescence Imaging in Vivo. Science. 2003;300:1434–6. doi: 10.1126/science.1083780. [DOI] [PubMed] [Google Scholar]
- Lee G, Bratkowski MA, Ding F, Ke A, Ha T. Elastic Coupling Between RNA Degradation and Unwinding by an Exoribonuclease. Science. 2012;336:1726–9. doi: 10.1126/science.1216848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J-B, Hite RK, Hamdan SM, Sunney Xie X, Richardson CC, van Oijen AM. DNA primase acts as a molecular brake in DNA replication. Nature. 2006;439:621–4. doi: 10.1038/nature04317. [DOI] [PubMed] [Google Scholar]
- Lee J, Lee S, Ragunathan K, Joo C, Ha T, Hohng S. Single-Molecule Four-Color FRET. Angewandte Chemie. 2010a;122:10118–21. doi: 10.1002/anie.201005402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M, Kim SH, Hong S-C. Minute negative superhelicity is sufficient to induce the B-Z transition in the presence of low tension. Proceedings of the National Academy of Sciences. 2010b;107:4985–90. doi: 10.1073/pnas.0911528107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee NK, Kapanidis AN, Koh HR, Korlann Y, Ho SO, Kim Y, Gassman N, Kim SK, Weiss S. Three-Color Alternating-Laser Excitation of Single Molecules: Monitoring Multiple Interactions and Distances. Biophysical journal. 2007a;92:303–12. doi: 10.1529/biophysj.106.093211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee NK, Koh HR, Han KY, Kim SK. Folding of 8–17 Deoxyribozyme Studied by Three-Color Alternating-Laser Excitation of Single Molecules. Journal of the American Chemical Society. 2007b;129:15526–34. doi: 10.1021/ja0725145. [DOI] [PubMed] [Google Scholar]
- Lee S, Lee J, Hohng S. Single-Molecule Three-Color FRET with Both Negligible Spectral Overlap and Long Observation Time. PLoS ONE. 2010c;5:e12270. doi: 10.1371/journal.pone.0012270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T-H, Blanchard SC, Kim HD, Puglisi JD, Chu S. The role of fluctuations in tRNA selection by the ribosome. Proceedings of the National Academy of Sciences. 2007c;104:13661–5. doi: 10.1073/pnas.0705988104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levene MJ, Korlach J, Turner SW, Foquet M, Craighead HG, Webb WW. Zero-Mode Waveguides for Single-Molecule Analysis at High Concentrations. Science. 2003;299:682–6. doi: 10.1126/science.1079700. [DOI] [PubMed] [Google Scholar]
- Lionnet T, Spiering MM, Benkovic SJ, Bensimon D, Croquette V. Real-time observation of bacteriophage T4 gp41 helicase reveals an unwinding mechanism. Proceedings of the National Academy of Sciences. 2007;104:19790–5. doi: 10.1073/pnas.0709793104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Abbondanzieri EA, Rausch JW, Grice SFJL, Zhuang X. Slide into Action: Dynamic Shuttling of HIV Reverse Transcriptase on Nucleic Acid Substrates. Science. 2008;322:1092–7. doi: 10.1126/science.1163108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo G, Wang M, Konigsberg WH, Xie XS. Single-molecule and ensemble fluorescence assays for a functionally important conformational change in T7 DNA polymerase. Proceedings of the National Academy of Sciences. 2007;104:12610–5. doi: 10.1073/pnas.0700920104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Means CK, Lygren B, Langeberg LK, Jain A, Dixon RE, Vega AL, Gold MG, Petrosyan S, Taylor SS, Murphy AN, Ha T, Santana LF, Tasken K, Scott JD. An entirely specific type I A-kinase anchoring protein that can sequester two molecules of protein kinase A at mitochondria. Proceedings of the National Academy of Sciences. 2011;108:E1227–E35. doi: 10.1073/pnas.1107182108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalet X, Pinaud FF, Bentolila LA, Tsay JM, Doose S, Li JJ, Sundaresan G, Wu AM, Gambhir SS, Weiss S. Quantum Dots for Live Cells, in Vivo Imaging, and Diagnostics. Science. 2005;307:538–44. doi: 10.1126/science.1104274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffitt J, Chemla Y, Smith S, Bustamante C. Recent Advances in Optical Tweezers. Annual Review of Biochemistry. 2008;77:205–28. doi: 10.1146/annurev.biochem.77.043007.090225. [DOI] [PubMed] [Google Scholar]
- Moffitt JR, Chemla YR, Izhaky D, Bustamante C. Differential detection of dual traps improves the spatial resolution of optical tweezers. Proceedings of the National Academy of Sciences. 2006;103:9006–11. doi: 10.1073/pnas.0603342103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro JB, Altman RB, Tung C-S, Cate JHD, Sanbonmatsu KY, Blanchard SC. Spontaneous formation of the unlocked state of the ribosome is a multistep process. Proceedings of the National Academy of Sciences. 2010a;107:709–14. doi: 10.1073/pnas.0908597107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro JB, Altman RB, Tung C-S, Sanbonmatsu KY, Blanchard SC. A fast dynamic mode of the EF-G-bound ribosome. EMBO J. 2010b;29:770–81. doi: 10.1038/emboj.2009.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myong S, Bruno MM, Pyle AM, Ha T. Spring-Loaded Mechanism of DNA Unwinding by Hepatitis C Virus NS3 Helicase. Science. 2007;317:513–6. doi: 10.1126/science.1144130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myong S, Cui S, Cornish PV, Kirchhofer A, Gack MU, Jung JU, Hopfner K-P, Ha T. Cytosolic Viral Sensor RIG-I Is a 5′-Triphosphate–dependent Translocase on Double-Stranded RNA. Science. 2009;323:1070–4. doi: 10.1126/science.1168352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myong S, Rasnik I, Joo C, Lohman TM, Ha T. Repetitive shuttling of a motor protein on DNA. Nature. 2005;437:1321–5. doi: 10.1038/nature04049. [DOI] [PubMed] [Google Scholar]
- Neuman KC, Nagy A. Single-molecule force spectroscopy: optical tweezers, magnetic tweezers and atomic force microscopy. Nat Meth. 2008;5:491–505. doi: 10.1038/nmeth.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okten Z, Churchman LS, Rock RS, Spudich JA. Myosin VI walks hand-over-hand along actin. Nat Struct Mol Biol. 2004;11:884–7. doi: 10.1038/nsmb815. [DOI] [PubMed] [Google Scholar]
- Park J, Myong S, Niedziela-Majka A, Lee KS, Yu J, Lohman TM, Ha T. PcrA Helicase Dismantles RecA Filaments by Reeling in DNA in Uniform Steps. Cell. 2010;142:544–55. doi: 10.1016/j.cell.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavani SRP, Thompson MA, Biteen JS, Lord SJ, Liu N, Twieg RJ, Piestun R, Moerner WE. Three-dimensional, single-molecule fluorescence imaging beyond the diffraction limit by using a double-helix point spread function. Proceedings of the National Academy of Sciences. 2009;106:2995–9. doi: 10.1073/pnas.0900245106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins TT, Li H-W, Dalal RV, Gelles J, Block SM. Forward and Reverse Motion of Single RecBCD Molecules on DNA. Biophysical journal. 2004;86:1640–8. doi: 10.1016/S0006-3495(04)74232-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planchon TA, Gao L, Milkie DE, Davidson MW, Galbraith JA, Galbraith CG, Betzig E. Rapid three-dimensional isotropic imaging of living cells using Bessel beam plane illumination. Nat Meth. 2011;8:417–23. doi: 10.1038/nmeth.1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu W, Derr ND, Goodman BS, Villa E, Wu D, Shih W, Reck-Peterson SL. Dynein achieves processive motion using both stochastic and coordinated stepping. Nat Struct Mol Biol. 2012;19:193–200. doi: 10.1038/nsmb.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanathan SP, van Aelst K, Sears A, Peakman LJ, Diffin FM, Szczelkun MD, Seidel R. Type III restriction enzymes communicate in 1D without looping between their target sites. Proceedings of the National Academy of Sciences. 2009;106:1748–53. doi: 10.1073/pnas.0807193106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao VB, Feiss M. The bacteriophage DNA packaging motor. Annual Review of Genetics. 2008:647–81. doi: 10.1146/annurev.genet.42.110807.091545. [DOI] [PubMed] [Google Scholar]
- Rice S, Lin AW, Safer D, Hart CL, Naber N, Carragher BO, Cain SM, Pechatnikova E, Wilson-Kubalek EM, Whittaker M, Pate E, Cooke R, Taylor EW, Milligan RA, Vale RD. A structural change in the kinesin motor protein that drives motility. Nature. 1999;402:778–84. doi: 10.1038/45483. [DOI] [PubMed] [Google Scholar]
- Roy R, Kozlov AG, Lohman TM, Ha T. SSB protein diffusion on single-stranded DNA stimulates RecA filament formation. Nature. 2009;461:1092–7. doi: 10.1038/nature08442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rust MJ, Bates M, Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat Meth. 2006;3:793–6. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanbonmatsu KY, Joseph S, Tung C-S. Simulating movement of tRNA into the ribosome during decoding. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15854–9. doi: 10.1073/pnas.0503456102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeing TM, Ramakrishnan V. What recent ribosome structures have revealed about the mechanism of translation. Nature. 2009;461:1234–42. doi: 10.1038/nature08403. [DOI] [PubMed] [Google Scholar]
- Schug A, Onuchic JN. From protein folding to protein function and biomolecular binding by energy landscape theory. Current Opinion in Pharmacology. 2010;10:709–14. doi: 10.1016/j.coph.2010.09.012. [DOI] [PubMed] [Google Scholar]
- Selth LA, Sigurdsson S, Svejstrup JQ. Transcript Elongation by RNA Polymerase II. Annu Rev Biochem. 2010;79:271–93. doi: 10.1146/annurev.biochem.78.062807.091425. [DOI] [PubMed] [Google Scholar]
- Shaevitz JW, Abbondanzieri EA, Landick R, Block SM. Backtracking by single RNA polymerase molecules observed at near-base-pair resolution. Nature. 2003;426:684–7. doi: 10.1038/nature02191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Z, Chakraborty A, Jain A, Giri S, Ha T, Prasanth KV, Prasanth SG. Dynamic association of ORCA with pre-RC components regulates DNA replication initiation. Molecular and Cellular Biology. 2012 doi: 10.1128/MCB.00362-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shroff H, Reinhard BM, Siu M, Agarwal H, Spakowitz A, Liphardt J. Biocompatible force sensor with optical readout and dimensions of 6 nm3. Nano Letters. 2005;5:1509–14. doi: 10.1021/nl050875h. [DOI] [PubMed] [Google Scholar]
- Smith DE. Single-molecule studies of viral DNA packaging. Current Opinion in Virology. 2011;1:134–41. doi: 10.1016/j.coviro.2011.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorokina M, Koh H-R, Patel SS, Ha T. Fluorescent Lifetime Trajectories of a Single Fluorophore Reveal Reaction Intermediates During Transcription Initiation. Journal of the American Chemical Society. 2009;131:9630–1. doi: 10.1021/ja902861f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speidel M, Jon A, Florin E-L. Three-dimensional tracking of fluorescent nanoparticles with subnanometer precision by use of off-focus imaging. Opt Lett. 2003;28:69–71. doi: 10.1364/ol.28.000069. [DOI] [PubMed] [Google Scholar]
- Stein IH, Steinhauer C, Tinnefeld P. Single-Molecule Four-Color FRET Visualizes Energy-Transfer Paths on DNA Origami. Journal of the American Chemical Society. 2011;133:4193–5. doi: 10.1021/ja1105464. [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Dudko OK. Biomolecules under mechanical stress: A simple mechanism of complex behavior. The Journal of Chemical Physics. 2011;134:065102. doi: 10.1063/1.3533366. [DOI] [PubMed] [Google Scholar]
- Takano M, Terada TP, Sasai M. Unidirectional Brownian motion observed in an in silico single molecule experiment of an actomyosin motor. Proceedings of the National Academy of Sciences. 2010;107:7769–74. doi: 10.1073/pnas.0911830107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G-Q, Roy R, Bandwar RP, Ha T, Patel SS. Real-time observation of the transition from transcription initiation to elongation of the RNA polymerase. Proceedings of the National Academy of Sciences. 2009;106:22175–80. doi: 10.1073/pnas.0906979106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G-Q, Roy R, Ha T, Patel SS. Transcription Initiation in a Single-Subunit RNA Polymerase Proceeds through DNA Scrunching and Rotation of the N-Terminal Subdomains. Molecular Cell. 2008;30:567–77. doi: 10.1016/j.molcel.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarsa PB, Brau RR, Barch M, Ferrer JM, Freyzon Y, Matsudaira P, Lang MJ. Detecting Force-Induced Molecular Transitions with Fluorescence Resonant Energy Transfer. Angewandte Chemie International Edition. 2007;46:1999–2001. doi: 10.1002/anie.200604546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tehver R, Thirumalai D. Rigor to Post-Rigor Transition in Myosin V: Link between the Dynamics and the Supporting Architecture. Structure. 2010;18:471–81. doi: 10.1016/j.str.2010.01.019. [DOI] [PubMed] [Google Scholar]
- Thirumalai D, O’Brien EP, Morrison G, Hyeon C. In: Annual Review of Biophysics. Rees DC, et al., editors. Vol. 39. Palo Alto: Annual Reviews; 2010. pp. 159–83. [DOI] [PubMed] [Google Scholar]
- Thompson RE, Larson DR, Webb WW. Precise Nanometer Localization Analysis for Individual Fluorescent Probes. Biophysical journal. 2002;82:2775–83. doi: 10.1016/S0006-3495(02)75618-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toprak E, Balci H, Blehm BH, Selvin PR. Three-Dimensional Particle Tracking via Bifocal Imaging. Nano Letters. 2007;7:2043–5. doi: 10.1021/nl0709120. [DOI] [PubMed] [Google Scholar]
- Toprak E, Yildiz A, Hoffman MT, Rosenfeld SS, Selvin PR. Why kinesin is so processive. Proceedings of the National Academy of Sciences. 2009;106:12717–22. doi: 10.1073/pnas.0808396106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabuco LG, Harrison CB, Schreiner E, Schulten K. Recognition of the Regulatory Nascent Chain TnaC by the Ribosome. Structure. 2010a;18:627–37. doi: 10.1016/j.str.2010.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabuco LG, Schreiner E, Eargle J, Cornish P, Ha T, Luthey-Schulten Z, Schulten K. The Role of L1 Stalk-tRNA Interaction in the Ribosome Elongation Cycle. Journal of Molecular Biology. 2010b;402:741–60. doi: 10.1016/j.jmb.2010.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabuco LG, Villa E, Mitra K, Frank J, Schulten K. Flexible Fitting of Atomic Structures into Electron Microscopy Maps Using Molecular Dynamics. Structure. 2008;16:673–83. doi: 10.1016/j.str.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treutlein B, Muschielok A, Andrecka J, Jawhari A, Buchen C, Kostrewa D, Hög F, Cramer P, Michaelis J. Dynamic Architecture of a Minimal RNA Polymerase II Open Promoter Complex. Molecular Cell. 2012;46:136–46. doi: 10.1016/j.molcel.2012.02.008. [DOI] [PubMed] [Google Scholar]
- Uemura S, Aitken CE, Korlach J, Flusberg BA, Turner SW, Puglisi JD. Real-time tRNA transit on single translating ribosomes at codon resolution. Nature. 2010;464:1012–7. doi: 10.1038/nature08925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale RD. The Molecular Motor Toolbox for Intracellular Transport. Cell. 2003;112:467–80. doi: 10.1016/s0092-8674(03)00111-9. [DOI] [PubMed] [Google Scholar]
- Vale RD, Milligan RA. The Way Things Move: Looking Under the Hood of Molecular Motor Proteins. Science. 2000;288:88–95. doi: 10.1126/science.288.5463.88. [DOI] [PubMed] [Google Scholar]
- van Mameren J, Peterman EJG, Wuite GJL. See me, feel me: methods to concurrently visualize and manipulate single DNA molecules and associated proteins. Nucleic Acids Research. 2008;36:4381–9. doi: 10.1093/nar/gkn412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos SM, Tretter EM, Schmidt BH, Berger JM. All tangled up: how cells direct, manage and exploit topoisomerase function. Nat Rev Mol Cell Biol. 2011;12:827–41. doi: 10.1038/nrm3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westphal V, Rizzoli SO, Lauterbach MA, Kamin D, Jahn R, Hell SW. Video-Rate Far-Field Optical Nanoscopy Dissects Synaptic Vesicle Movement. Science. 2008;320:246–9. doi: 10.1126/science.1154228. [DOI] [PubMed] [Google Scholar]
- Whitford PC, Altman RB, Geggier P, Terry DS, Munro JB, Onuchic JN, Spahn CMT, Sanbonmatsu KY, Blanchard SC, Rodnina MV, Wintermeyer W, Green R. Springer; Vienna: 2011. pp. 303–19. [Google Scholar]
- Whitford PC, Geggier P, Altman RB, Blanchard SC, Onuchic JN, Sanbonmatsu KY. Accommodation of aminoacyl-tRNA into the ribosome involves reversible excursions along multiple pathways. RNA. 2010;16:1196–204. doi: 10.1261/rna.2035410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Luo G, Karnchanaphanurach P, Louie T-M, Rech I, Cova S, Xun L, Xie XS. Protein Conformational Dynamics Probed by Single-Molecule Electron Transfer. Science. 2003;302:262–6. doi: 10.1126/science.1086911. [DOI] [PubMed] [Google Scholar]
- Yang H, Xie XS. Probing single-molecule dynamics photon by photon. The Journal of Chemical Physics. 2002;117:10965. [Google Scholar]
- Yeom K-H, Heo I, Lee J, Hohng S, Kim VN, Joo C. Single-molecule approach to immunoprecipitated protein complexes: insights into miRNA uridylation. EMBO Rep. 2011;12:690–6. doi: 10.1038/embor.2011.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz A, Forkey JN, McKinney SA, Ha T, Goldman YE, Selvin PR. Myosin V Walks Hand-OverHand: Single Fluorophore Imaging with 1.5-nm Localization. Science. 2003;300:2061–5. doi: 10.1126/science.1084398. [DOI] [PubMed] [Google Scholar]
- Yildiz A, Park H, Safer D, Yang Z, Chen L-Q, Selvin PR, Sweeney HL. Myosin VI Steps via a Hand-over-Hand Mechanism with Its Lever Arm Undergoing Fluctuations when Attached to Actin. Journal of Biological Chemistry. 2004a;279:37223–6. doi: 10.1074/jbc.C400252200. [DOI] [PubMed] [Google Scholar]
- Yildiz A, Selvin PR. Fluorescence Imaging with One Nanometer Accuracy: ??Application to Molecular Motors. Accounts of Chemical Research. 2005;38:574–82. doi: 10.1021/ar040136s. [DOI] [PubMed] [Google Scholar]
- Yildiz A, Tomishige M, Vale RD, Selvin PR. Kinesin Walks Hand-Over-Hand. Science. 2004b;303:676–8. doi: 10.1126/science.1093753. [DOI] [PubMed] [Google Scholar]
- Yodh JG, Schlierf M, Ha T. Insight into helicase mechanism and function revealed through single-molecule approaches. Quarterly Reviews of Biophysics. 2010;43:185–217. doi: 10.1017/S0033583510000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Ha T, Schulten K. Structure-Based Model of the Stepping Motor of PcrA Helicase. Biophysical journal. 2006;91:2097–114. doi: 10.1529/biophysj.106.088203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamft B, Bintu L, Ishibashi T, Bustamante C. Nascent RNA structure modulates the transcriptional dynamics of RNA polymerases. Proceedings of the National Academy of Sciences. 2012;109:8948–53. doi: 10.1073/pnas.1205063109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Dillingham MS, Thomas CD, Allen S, Roberts CJ, Soultanas P. Directional Loading and Stimulation of PcrA Helicase by the Replication Initiator Protein RepD. Journal of Molecular Biology. 2007;371:336–48. doi: 10.1016/j.jmb.2007.05.050. [DOI] [PubMed] [Google Scholar]
- Zhou R, Kozlov A, Roy R, Zhang J, Korolev S, Lohman T, Ha T. SSB Functions as a Sliding Platform that Migrates on DNA via Reptation. Cell. 2011a;146:222–32. doi: 10.1016/j.cell.2011.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Kunzelmann S, Webb MR, Ha T. Detecting Intramolecular Conformational Dynamics of Single Molecules in Short Distance Range with Subnanometer Sensitivity. Nano Letters. 2011b;11:5482–8. doi: 10.1021/nl2032876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Schlierf M, Ha T, Nils GW. Methods in Enzymology. Academic Press; 2010. pp. 405–26. [DOI] [PubMed] [Google Scholar]