Abstract
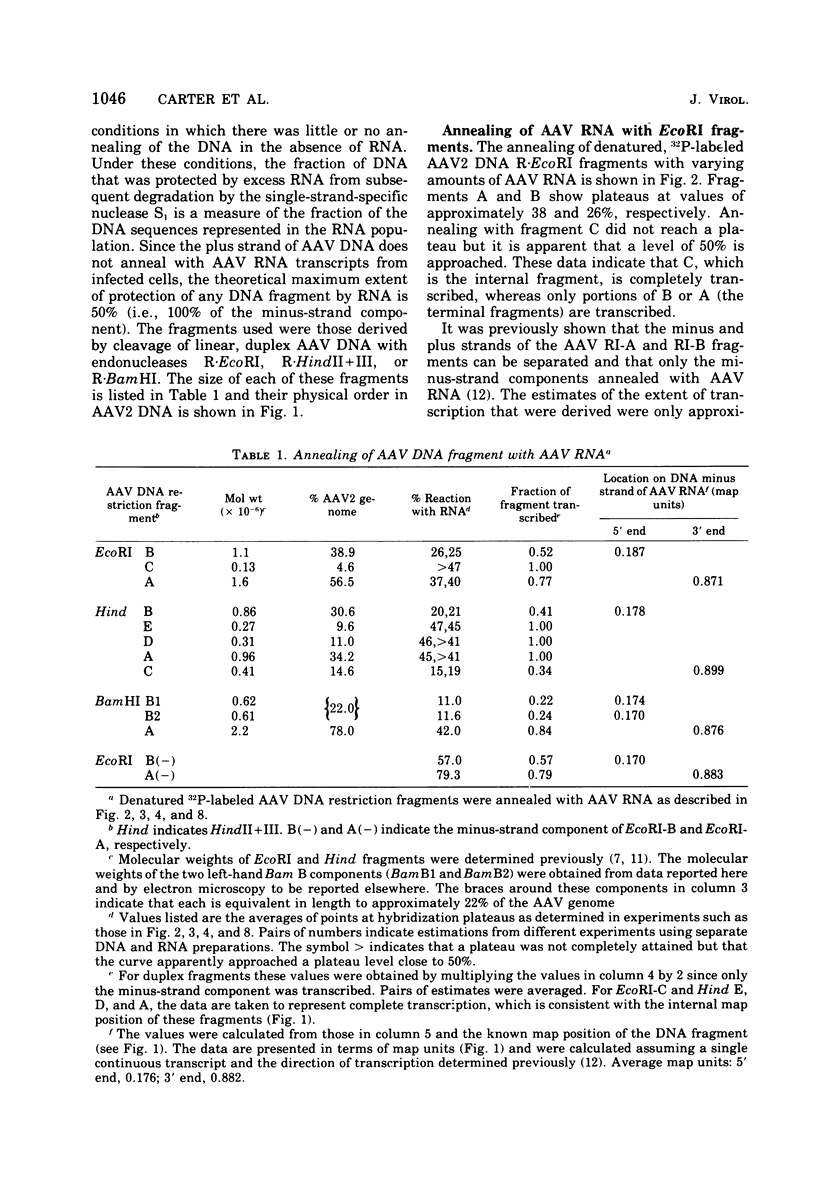
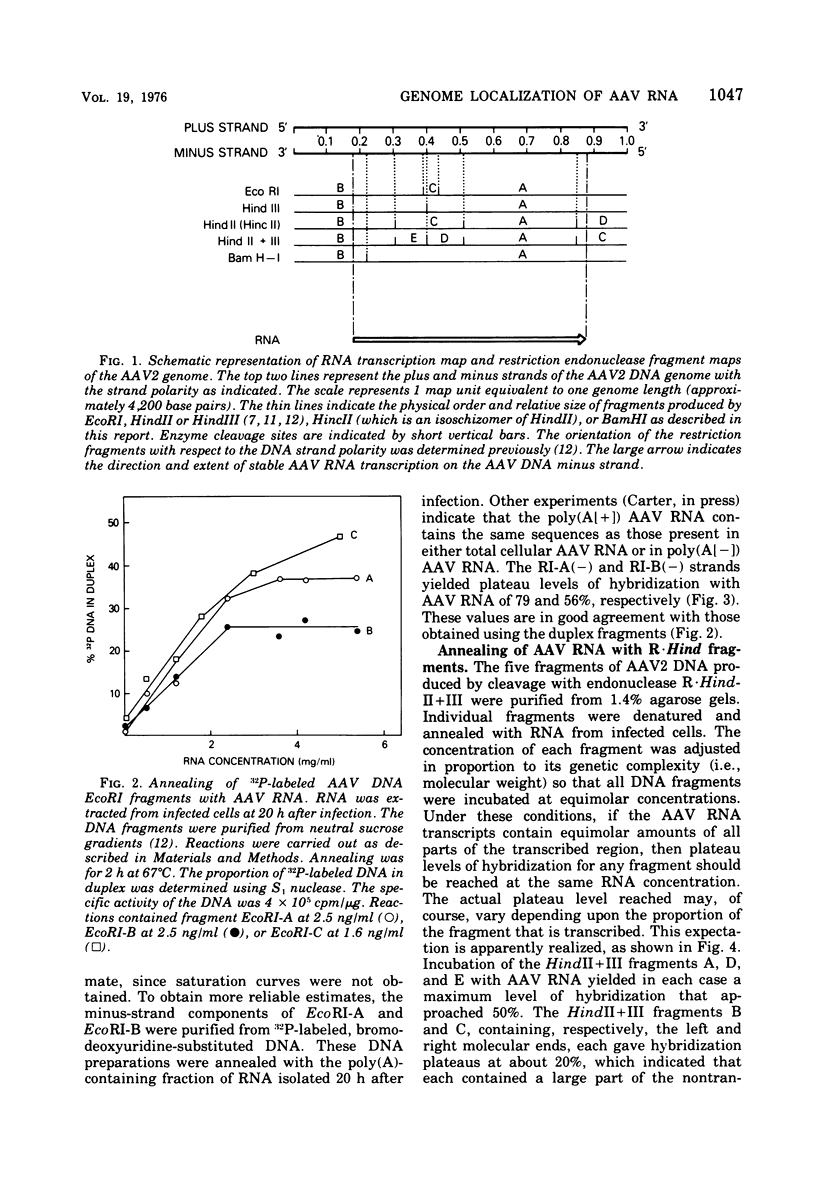
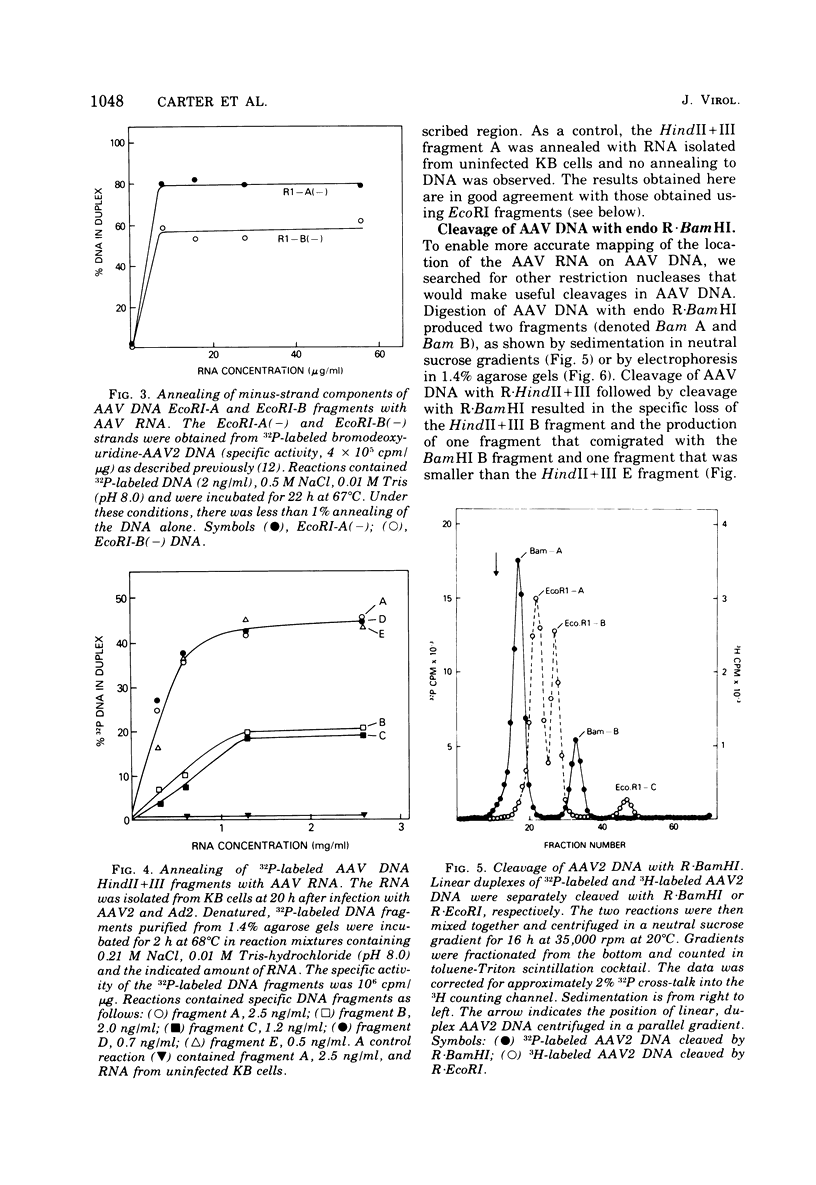
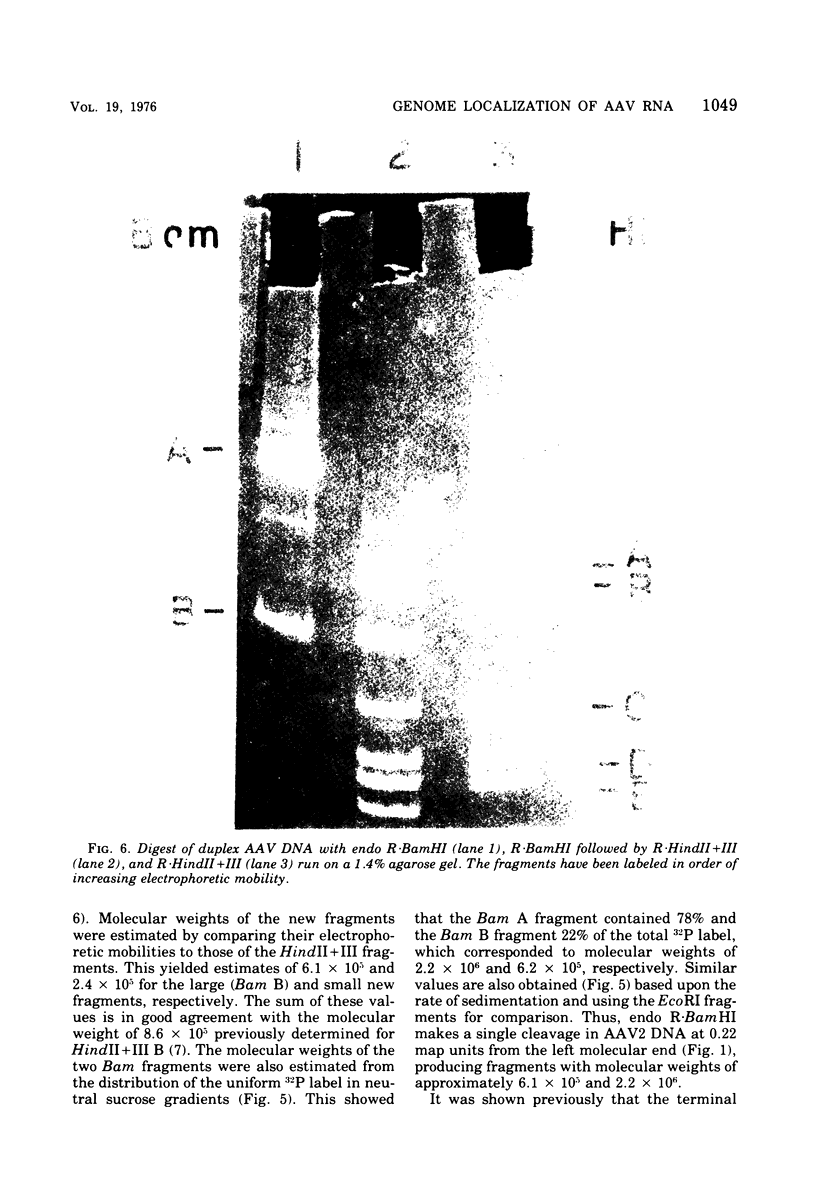
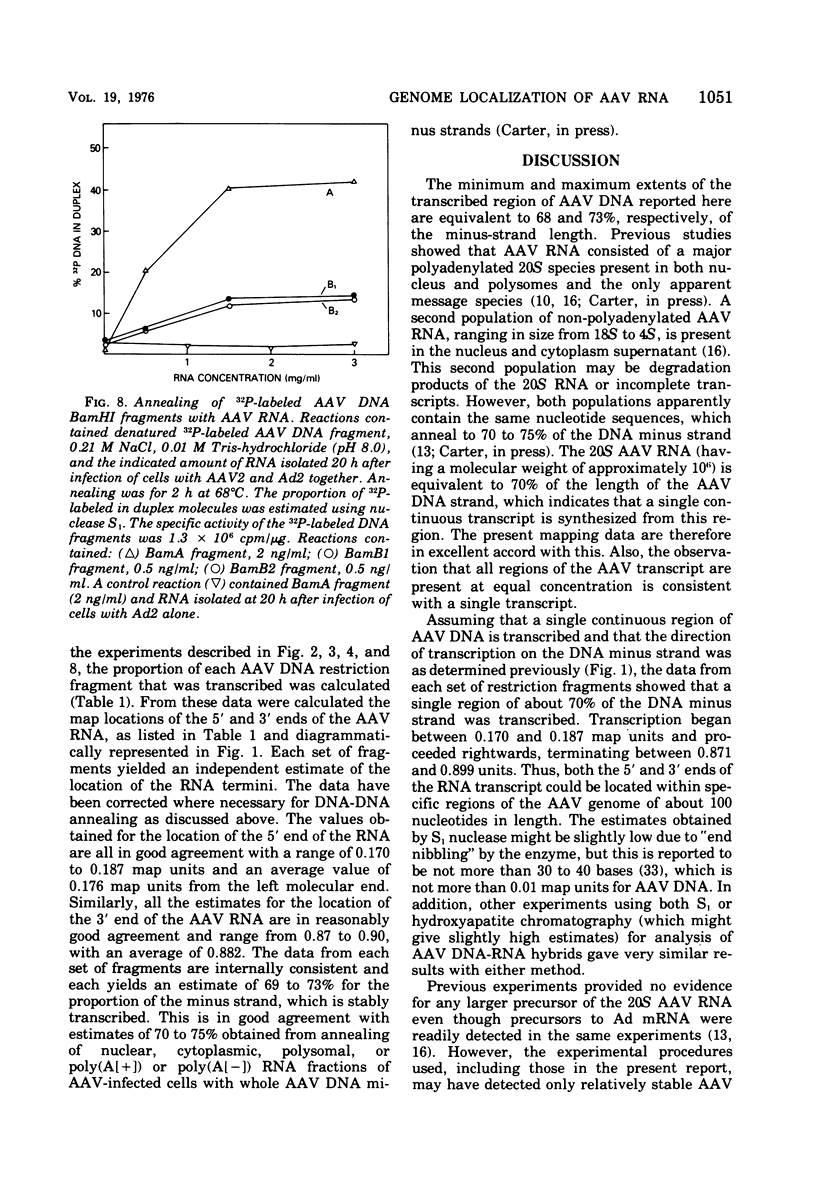
In previous work, linear duplex molecules of adeno-associated virus, type 2 (AAV2), DNA were cleaved with the restriction endonucleases R-EcoRI, R-HindII, and R-HindIII. The physical order of the specific fragments obtained was deduced and oriented with respect to the DNA strand polarity and the direction of transcription. Stable AAV RNA is transcribed only from 70% of the minus DNA strand. We report here RNA-DNA hybridization experiments using these restriction fragments to obtain a more accurate map of the portion of the AAV genome represented in stable RNA. The data obtained with several sets of restriction fragments annealed to either whole-cell RNA or poly(A)-containing RNA were internally consistent. The AAV RNA annealed with a continuous region of the AAV DNA, beginning at 0.18 map units (18%) from the left end of the molecule and ending at 0.88 map units. In addition, the restriction endonuclease BamHI was found to make one specific cleavage in AAV2 DNA at 0.22 map units, which is 0.04 map units (i.e., 160 nucleotides) to the right (""down stream'') of the point corresponding to the 5' end of the viral mRNA.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ando T. A nuclease specific for heat-denatured DNA in isolated from a product of Aspergillus oryzae. Biochim Biophys Acta. 1966 Jan 18;114(1):158–168. doi: 10.1016/0005-2787(66)90263-2. [DOI] [PubMed] [Google Scholar]
- Aviv H., Leder P. Purification of biologically active globin messenger RNA by chromatography on oligothymidylic acid-cellulose. Proc Natl Acad Sci U S A. 1972 Jun;69(6):1408–1412. doi: 10.1073/pnas.69.6.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachenheimer S., Darnell J. E. Adenovirus-2 mRNA is transcribed as part of a high-molecular-weight precursor RNA. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4445–4449. doi: 10.1073/pnas.72.11.4445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berns K. I., Adler S. Separation of two types of adeno-associated virus particles containing complementary polynucleotide chains. J Virol. 1972 Feb;9(2):394–396. doi: 10.1128/jvi.9.2.394-396.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berns K. I., Kelly T. J., Jr Letter: Visualization of the inverted terminal repetition in adeno-associated virus DNA. J Mol Biol. 1974 Jan 15;82(2):267–271. doi: 10.1016/0022-2836(74)90344-1. [DOI] [PubMed] [Google Scholar]
- Berns K. I., Kort J., Fife K. H., Grogan E. W., Spear I. Study of the fine structure of adeno-associated virus DNA with bacterial restriction endonucleases. J Virol. 1975 Sep;16(3):712–719. doi: 10.1128/jvi.16.3.712-719.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berns K. I. Molecular biology of the adeno-associated viruses. Curr Top Microbiol Immunol. 1974;65:1–20. doi: 10.1007/978-3-642-65875-4_1. [DOI] [PubMed] [Google Scholar]
- Berns K. I., Rose J. A. Evidence for a single-stranded adenovirus-associated virus genome: isolation and separation of complementary single strands. J Virol. 1970 Jun;5(6):693–699. doi: 10.1128/jvi.5.6.693-699.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blin N., von Gabain A., Bujard H. Isolation of large molecular weight DNA from agarose gels for further digestion by restriction enzymes. FEBS Lett. 1975 Apr 15;53(1):84–86. doi: 10.1016/0014-5793(75)80688-0. [DOI] [PubMed] [Google Scholar]
- Carter B. J. Analysis of parvovirus mRNA by sedimentation and electrophoresis in aqueous and nonaqueous solution. J Virol. 1974 Oct;14(4):834–839. doi: 10.1128/jvi.14.4.834-839.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter B. J., Khoury G., Denhardt D. T. Physical map and strand polarity of specific fragments of adenovirus-associated virus DNA produced by endonuclease R-EcoRI. J Virol. 1975 Sep;16(3):559–568. doi: 10.1128/jvi.16.3.559-568.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter B. J., Khoury G., Rose J. A. Adenovirus-associated virus multiplication. IX. Extent of transcription of the viral genome in vivo. J Virol. 1972 Dec;10(6):1118–1125. doi: 10.1128/jvi.10.6.1118-1125.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter B. J., Khoury G. Specific cleavage of adenovirus-associated virus DNA by restriction endonuclease R-EcoRI--characterization of cleavage products. Virology. 1975 Feb;63(2):523–538. doi: 10.1016/0042-6822(75)90325-6. [DOI] [PubMed] [Google Scholar]
- Carter B. J., Koczot F. J., Garrison J., Rose J. A., Dolin R. Separate helper functions provided by adenovirus for adenovirus-associated virus multiplication. Nat New Biol. 1973 Jul 18;244(133):71–73. doi: 10.1038/newbio244071a0. [DOI] [PubMed] [Google Scholar]
- Carter B. J., Rose J. A. Adenovirus-associated virus multiplication. 8. Analysis of in vivo transcription induced by complete or partial helper viruses. J Virol. 1972 Jul;10(1):9–16. doi: 10.1128/jvi.10.1.9-16.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter B. J., Rose J. A. Transcription in vivo of a defective parvovirus: sedimentation and electrophoretic analysis of RNA synthesized by adenovirus-associated virus and its helper adenovirus. Virology. 1974 Sep;61(1):182–199. doi: 10.1016/0042-6822(74)90253-0. [DOI] [PubMed] [Google Scholar]
- Crawford L. V., Follett E. A., Burdon M. G., McGeoch D. J. The DNA of a minute virus of mice. J Gen Virol. 1969 Jan;4(1):37–46. doi: 10.1099/0022-1317-4-1-37. [DOI] [PubMed] [Google Scholar]
- Danna K. J., Sack G. H., Jr, Nathans D. Studies of simian virus 40 DNA. VII. A cleavage map of the SV40 genome. J Mol Biol. 1973 Aug 5;78(2):363–376. doi: 10.1016/0022-2836(73)90122-8. [DOI] [PubMed] [Google Scholar]
- Denhardt D. T., Eisenberg S., Bartok K., Carter B. J. Multiple structures of adeno-associated virus DNA: analysis of terminally labeled molecules with endonuclease R-Hae III. J Virol. 1976 May;18(2):672–684. doi: 10.1128/jvi.18.2.672-684.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerry H. W., Kelly T. J., Jr, Berns K. I. Arrangement of nucleotide sequences in adeno-associated virus DNA. J Mol Biol. 1973 Sep 15;79(2):207–225. doi: 10.1016/0022-2836(73)90001-6. [DOI] [PubMed] [Google Scholar]
- Glynn I. M., Chappell J. B. A simple method for the preparation of 32-P-labelled adenosine triphosphate of high specific activity. Biochem J. 1964 Jan;90(1):147–149. doi: 10.1042/bj0900147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward G. S., Smith M. G. The chromosome of bacteriophage T5. I. Analysis of the single-stranded DNA fragments by agarose gel electrophoresis. J Mol Biol. 1972 Feb 14;63(3):383–395. doi: 10.1016/0022-2836(72)90435-4. [DOI] [PubMed] [Google Scholar]
- Koczot F. J., Carter B. J., Garon C. F., Rose J. A. Self-complementarity of terminal sequences within plus or minus strands of adenovirus-associated virus DNA. Proc Natl Acad Sci U S A. 1973 Jan;70(1):215–219. doi: 10.1073/pnas.70.1.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillehaug J. R., Kleppe K. Kinetics and specificity of T4 polynucleotide kinase. Biochemistry. 1975 Mar 25;14(6):1221–1225. doi: 10.1021/bi00677a020. [DOI] [PubMed] [Google Scholar]
- Mayor H. D., Torikai K., Melnick J. L., Mandel M. Plus and minus single-stranded DNA separately encapsidated in adeno-associated satellite virions. Science. 1969 Dec 5;166(3910):1280–1282. doi: 10.1126/science.166.3910.1280. [DOI] [PubMed] [Google Scholar]
- Middleton J. H., Edgell M. H., Hutchison C. A., 3rd Specific fragments of phi X174 deoxyribonucleic acid produced by a restriction enzyme from Haemophilus aegyptius, endonuclease Z. J Virol. 1972 Jul;10(1):42–50. doi: 10.1128/jvi.10.1.42-50.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson C. C. Phosphorylation of nucleic acid by an enzyme from T4 bacteriophage-infected Escherichia coli. Proc Natl Acad Sci U S A. 1965 Jul;54(1):158–165. doi: 10.1073/pnas.54.1.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts R. J., Breitmeyer J. B., Tabachnik N. F., Myers P. A. A second specific endonuclease from Haemophilus aegyptius. J Mol Biol. 1975 Jan 5;91(1):121–123. doi: 10.1016/0022-2836(75)90375-7. [DOI] [PubMed] [Google Scholar]
- Rose J. A., Berns K. I., Hoggan M. D., Koczot F. J. Evidence for a single-stranded adenovirus-associated virus genome: formation of a DNA density hybrid on release of viral DNA. Proc Natl Acad Sci U S A. 1969 Nov;64(3):863–869. doi: 10.1073/pnas.64.3.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose J. A., Koczot F. Adenovirus-associated virus multiplication. VI. Base compostion of the deoxyribonucleic acid strand species and strand-specific in vivo transcription. J Virol. 1971 Nov;8(5):771–777. doi: 10.1128/jvi.8.5.771-777.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. A., Sugden B., Sambrook J. Detection of two restriction endonuclease activities in Haemophilus parainfluenzae using analytical agarose--ethidium bromide electrophoresis. Biochemistry. 1973 Jul 31;12(16):3055–3063. doi: 10.1021/bi00740a018. [DOI] [PubMed] [Google Scholar]
- Shenk T. E., Rhodes C., Rigby P. W., Berg P. Biochemical method for mapping mutational alterations in DNA with S1 nuclease: the location of deletions and temperature-sensitive mutations in simian virus 40. Proc Natl Acad Sci U S A. 1975 Mar;72(3):989–993. doi: 10.1073/pnas.72.3.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H. O., Nathans D. Letter: A suggested nomenclature for bacterial host modification and restriction systems and their enzymes. J Mol Biol. 1973 Dec 15;81(3):419–423. doi: 10.1016/0022-2836(73)90152-6. [DOI] [PubMed] [Google Scholar]
- Sutton W. D. A crude nuclease preparation suitable for use in DNA reassociation experiments. Biochim Biophys Acta. 1971 Jul 29;240(4):522–531. doi: 10.1016/0005-2787(71)90709-x. [DOI] [PubMed] [Google Scholar]
- Weiss B., Live T. R., Richardson C. C. Enzymatic breakage and joining of deoxyribonucleic acid. V. End group labeling and analysis of deoxyribonucleic acid containing single straned breaks. J Biol Chem. 1968 Sep 10;243(17):4530–4542. [PubMed] [Google Scholar]
- Wilson G. A., Young F. E. Isolation of a sequence-specific endonuclease (BamI) from Bacillus amyloliquefaciens H. J Mol Biol. 1975 Sep 5;97(1):123–125. doi: 10.1016/s0022-2836(75)80028-3. [DOI] [PubMed] [Google Scholar]