Abstract
Prethrombin-2 is the immediate zymogen precursor of the clotting enzyme thrombin, which is generated upon cleavage at R15 and separation of the A chain and catalytic B chain. The X-ray structure of prethrombin-2 solved in the free form at 1.9 Å resolution shows the 215–217 segment collapsed into the active site. Remarkably, some of the crystals harvested from the same crystallization well diffract to 2.2 Å resolution in the same space group but produce a structure where the 215–217 segment does not hinder access to the active site. The two alternative conformations of prethrombin-2, open and collapsed, echo the active E and inactive E* forms of the mature enzyme. These findings validate the emerging paradigm that the allosteric E*-E equilibrium is a key property of the trypsin fold and demonstrate that the E and E* forms coexist under the same solution conditions and can be trapped by harvesting different crystals in the same crystallization well. Another unanticipated feature of prethrombin-2 is that R15 is buried within the protein in ionic interactions with E14e, D14l and E18 and its exposure is necessary for proteolytic attack and conversion to thrombin. Based on this structural observation, we constructed the E14eA/D14lA/E18A triple mutant to reduce electrostatic coupling with R15 and promote zymogen activation. The mutation causes prethrombin-2 to spontaneously convert to thrombin, without the need for the snake venom ecarin or the physiological prothrombinase complex.
Four protease families account for over 40% of all proteolytic enzymes in humans. These are the ubiquitin-specific proteases (1), the adamalysins (2), prolyl oligopeptidases (3) and the trypsin-like proteases responsible for digestion, blood coagulation, fibrinolysis, development, fertilization, apoptosis and immunity (4). Trypsin-like proteases utilize a canonical catalytic triad for activity, composed of the highly conserved residues H57, D102 and S195 (chymotrypsinogen numbering). Catalysis is assisted by the oxyanion hole, defined by the backbone N atoms of G193 and S195, the 215–217 segment shaping the western wall of the primary specificity pocket and residue D189 at the bottom of this pocket that engages the Arg residue at the P1 position of substrate (4–6). Nearly all members of the family are expressed as inactive zymogens that are irreversibly converted to the mature protease by proteolytic cleavage at R15, leading to generation of a new N-terminus that ion-pairs with the highly conserved D194 next to the catalytic S195 and organizes both the oxyanion hole and primary specificity pocket for substrate binding and catalysis. Few exceptions to this rule exist and involve mechanisms that mimic the important ionic interaction with D194 either via alternative residues within the protein, as seen in tissue-type plasminogen activator (7), or with the assistance of external activators, as seen in the interaction of streptokinase with plasminogen (8) or staphylocoagulase with prothrombin (9).
The paradigm based on the zymogen→protease conversion has been very useful to rationalize the onset of catalytic activity and its regulation, particularly for enzyme cascades (10–12). However, recent structural findings suggest that the paradigm is an oversimplification that overlooks the conformational plasticity of the trypsin fold (13). One of the most compelling cases supporting the emerging view of trypsin-like proteases as allosteric enzymes is provided by the clotting protease thrombin (14). In the absence of ligands, thrombin undergoes an equilibrium between two conformations: the active E form with the active site open and the inactive E* form where the active site is blocked by the side chain of W215 and collapse of the entire 215–217 segment (15). Both forms have recently been trapped crystallographically for the same thrombin construct (16). Whether the allosteric E*-E equilibrium is a prerogative of the mature enzyme, or is already present in the inactive zymogen has not been established conclusively and represents an unsolved issue of general relevance to the entire family of trypsin-like proteases.
Identification of alternative conformations for a trypsin-like protease or its zymogen requires crystallization of the free form. In the case of thrombin precursors, such crystals have so far been obtained only for prethrombin-1, that assumes a collapsed conformation similar to the E* form of the mature enzyme (17). There are currently no crystals of prothrombin and the other inactive precursor, prethrombin-2, has been crystallized only in the bound form (9, 18). Prethrombin-2 differs from thrombin only in the intact R15-I16 peptide bond (19). Recent findings indicate that prethrombin-2 may be the dominant intermediate along the thrombin generation pathway when prothrombin is proteolytically processed on the activated platelet surface (20). This provides additional motivation to solve the structure of prethrombin-2 in the physiologically relevant free form. Here we report X-ray crystal structures of prethrombin-2 in the free form that document two conformations of the 215–217 segment consistent with the E*-E equilibrium observed in the mature enzyme. Remarkably, the two conformations are trapped from crystals harvested from the same crystallization well under identical solution conditions. Furthermore, the side chain of R15 at the site of activation is not accessible to solvent and is buried into an anionic pocket defined by E14e, D14l and E18. When these three negatively charged residues are replaced by Ala, the resulting triple mutant E14eA/D14lA/E18A auto-activates to thrombin, which is a feature not present in the wild-type or any thrombin mutant reported to-date.
Materials and Methods
Prethrombin-2 wild-type and mutants S195A, E14eA/D14lA/E18A, E14eA/D14lA/E18A/S195A, W215A/E217A and E14eA/D14lA/E18A/W215A/E217A were expressed in E. coli, refolded and purified to homogeneity as previously described (21) with minor modifications. Inclusion bodies form 1 L of cells were solubilized by adding 7 M Gnd-HCl and 30 mM L-cysteine to a final concentration of 30–40 mg/ml. After 2–3 hr at room temperature, the unfolded protein was first diluted into 6 M Gnd-HCl, 0.6 M L-arginine HCl, 50 mM Tris, pH 8.3, 0.5 M NaCl, 1 mM EDTA, 10% glycerol, 0.2% Brij 58, 1 mM L-cysteine and then refolded by reverse dilution to a final concentration of 0.15–0.2 mg/ml. The refolded protein in 0.6 M L-arginine HCl, 50 mM Tris, pH 8.3, 0.5 M NaCl, 1 mM EDTA, 10% glycerol, 0.2% Brij 58, 1 mM L-cysteine was extensively dialyzed against 10 mM Tris, pH 7.4, 0.2 M NaCl, 2 mM EDTA, 0.1% PEG6000 for 24–30 h at room temperature and then, after centrifugation and filtration, loaded overnight onto a 5 ml Heparin-sepharose column. Finally, the correctly folded prethrombin-2 was eluted with a linear gradient of 0.2–0.95 M NaCl. When required, benzamidine was added to the refolding, dialysis and purification buffers at a final concentration of 10 mM. For crystallization purposes, the N-terminal T7-tag belonging to the pET21a vector (Novagen) was removed by incubating prethrombin-2 S195A with thrombin using a ratio 1:100 (w/w) for 24–30 h at room temperature. Proteolysis was monitored either by Coomassie-stained SDS-PAGE and by western blot analysis using anti-T7-tag antibodies. Prethrombin-2 S195A without T7-tag was further purified by heparin-affinity chromatography, dialyzed overnight against 0.05 M Na2HPO4, 0.35 M NaCl, pH 7.3, and concentrated up to 10 mg/ml. The homogeneity and chemical identity of the final preparations were verified by SDS-PAGE and by RP-HPLC-MS analysis, giving a purity grade > 98%. Activity of the auto-activated constructs was tested against chromogenic and physiological substrates as detailed elsewhere (21, 22).
Crystallization of prethrombin-2 S195A was achieved at 22 °C by the vapor diffusion technique using an Art Robbins Instruments Phoenix™ liquid handling robot and mixing equal volumes (0.3 µL) of protein and reservoir solution. Optimization of crystal growth was achieved by hanging drop vapor diffusion method mixing 3 µL of protein (11 mg/ml) with equal volumes of reservoir solution (Table 1). Crystals were grown in 0.1 M Tris, pH 8.5 and 11% PEG8000 after optimization. A total of eight diffraction quality crystals were harvested from the same crystallization well and were cryoprotected in the solution similar to mother liquid but containing 25% glycerol prior to flash freezing. X-ray diffraction data were collected with a home source (Rigaku 1.2 kW MMX007 generator with VHF optics) Rigaku Raxis IV++ detector and were indexed, integrated and scaled with the HKL2000 software package (23). Structures for the open and collapsed crystal forms were solved by molecular replacement using MOLREP from the CCP4 suite (24) and Protein Data Bank accession code 1HAG for wild type prethrombin-2 (18) as a search model. Refinement and electron density generation were performed with REFMAC5 from the CCP4 suite and 5% of the reflections were randomly selected as a test set for cross-validation. Model building and analysis of the structures were carried out using COOT (25). In the final stages of refinement for both structures, TLS tensors modeling rigid-body anisotropic temperature factors were calculated and applied to the model. Ramachandran plots were calculated using PROCHECK (26). Statistics for data collection and refinement are summarized in Table 1. Atomic coordinates and structure factors have been deposited in the Protein Data Bank (accession codes 3SQH for the open form and 3SQE for the collapsed form).
Table 1.
Crystallographic data for prethrombin-2
| Open form | Collapsed form | |
|---|---|---|
| Buffer/salt | 100 mM Tris, pH 8.5 | 100 mM Tris, pH 8.5 |
| PEG | 8000 (11%) | 8000 (11%) |
| PDB ID | 3SQH | 3SQE |
| Data collection: | ||
| Wavelength (Å) | 1.54 | 1.54 |
| Space group | P21 | P21 |
| Unit cell dimensions (Å) | a=44.4, b=60.0, c=49.7 | a=44.4, b=58.0, c=52.9 |
| β=96.5° | β=98.2° | |
| Molecules/asymmetric unit | 1 | 1 |
| Resolution range (Å) | 40–2.2 | 40–1.9 |
| Observations | 60302 | 132860 |
| Unique observations | 13204 | 20289 |
| Completeness (%) | 98.8 (97.9) | 95.7 (93.5) |
| Rsym (%) | 8.0 (32.0) | 7.9 (21.5) |
| I/σ(I) | 15.7 (3.5) | 21.3 (8.5) |
| Refinement: | ||
| Resolution (Å) | 40–2.2 | 40–1.9 |
| Rcryst, Rfree | 0.189, 0.244 | 0.171, 0.206 |
| Reflections (working/test) | 11782/661 | 18147/1046 |
| Protein atoms | 2304 | 2361 |
| Solvent molecules | 82 | 210 |
| Rmsd bond lengthsa (Å) | 0.012 | 0.011 |
| Rmsd anglesa (°) | 1.4 | 1.3 |
| Rmsd ΔB (Å2) (mm/ms/ss)b | 3.32/1.38/2.43 | 1.32/1.01/2.38 |
| <B> protein (Å2) | 43.8 | 31.0 |
| <B> solvent (Å2) | 47.8 | 42.8 |
| Ramachandran plot: | ||
| Most favored (%) | 99.6 | 99.6 |
| Generously allowed (%) | 0.4 | 0.4 |
| Disallowed (%) | 0.0 | 0.0 |
Root-mean-squared deviation (Rmsd) from ideal bond lengths and angles and Rmsd in B-factors of bonded atoms.
mm, main chain-main chain; ms, main chain-side chain; ss, side chain-side chain.
Results and Discussion
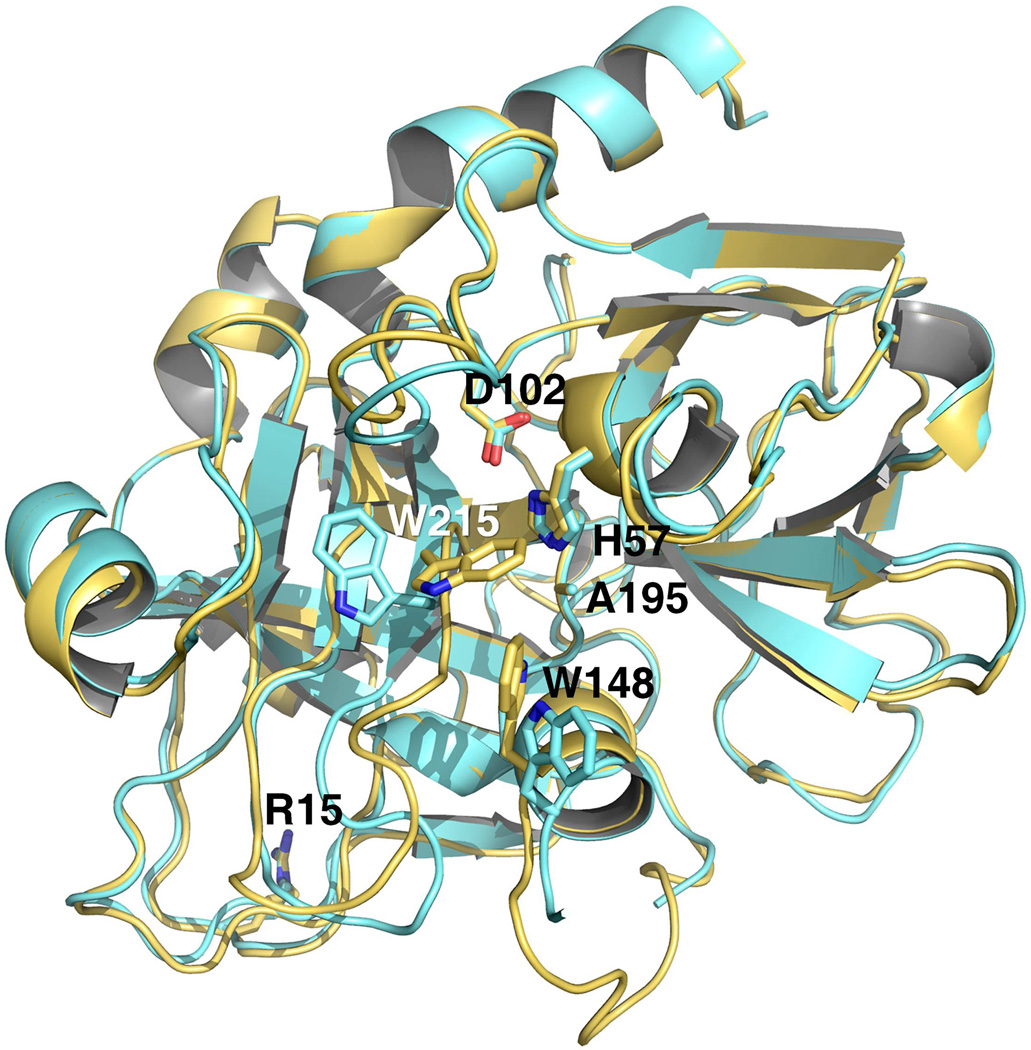
Prethrombin-2 was crystallized previously bound to hirugen and the active site inhibitor PPACK (18) or staphylocoagulase (9). The hirugen-bound structure is particularly relevant because the inhibitor engages exosite I and leaves the active site free. In this structure, the active site assumes an open conformation and R15 in the activation domain is fully exposed to solvent for proteolytic attack. Because exosite I and the active site are energetically linked in both the enzyme (27, 28) and zymogen (29, 30), the conformation of the active site in the prethrombin-2 structure bound to hirugen may be influenced by the liganded state of exosite I, as recently observed for the mature enzyme (31). Hence, the architecture of the active site of a zymogen such as prethrombin-2 must be established from a structure obtained in the absence of any ligands. The structure of the prethrombin-2 mutant S195A was solved in the free form at 1.9 Å resolution with a final Rfree=0.206 (Table 1). Overall, the structure differs (rmsd=0.362 Å) from that of prethrombin-2 bound to hirugen at exosite I (18) in the region defining access to the active site and the activation domain. Because of the intact R15-I16 bond, no new N-terminus is present in the B chain ready to engage the carboxylate of D194. Yet residues of the catalytic triad assume a correct orientation (Figure 1) with the Cβ atom of the mutated A195 in the same position as the Cβ atom of S195 in the mature enzyme and within 3.7 Å of the Nε2 atom of H57, and the Oδ2 atom of D102 within 2.7 Å of the Nδ1 atom of H57, as seen in the structures of other zymogens in the free form (13, 32). The side chain of D194 repositions and finds new H-bonding partners in the backbone N atoms of W141, G142 and N143. The entire 141–144 segment shifts upward toward the adjacent segment carrying E192 and G193, the highly conserved H-bonding interaction between the backbone N atom of N143 and the backbone O atom of E192 is disrupted and a new H-bond forms between the backbone O atom of G193 and the backbone N atom of L144. As a result, the 191–193 segment changes direction relative to the mature enzyme, the oxyanion hole defined by the backbone N atoms of residues is disrupted and the backbone N atom of G193 engages the Oδ atom of N143 in a H-bonding interaction. These are features recently documented in prethrombin-1 (17).
Figure 1. Crystal structures of prethrombin-2 in the open (yellow) and collapsed (orange) conformations.
The two structures have a similar overall fold (rmsd=0.307 Å) and arrangement of the catalytic triad, but differ in the position of the 215–217 segment. In the open form, this segment is positioned away from the active site with W215 in contact with F227. In the collapsed form, the 215–217 collapses and occludes the active site with W215 clustering with the catalytic H57, W60d from the 60-loop and W148 from the autolysis loop, as observed in the recent structure of prethrombin-1 (17). The two conformations of prethrombin-2, open and collapsed, resemble the E and E* forms of the mature enzyme (16). Note the position of R15 in both structures pointing away from the solvent and into a cavity lined up by negatively charged residues (see also Figure 3).
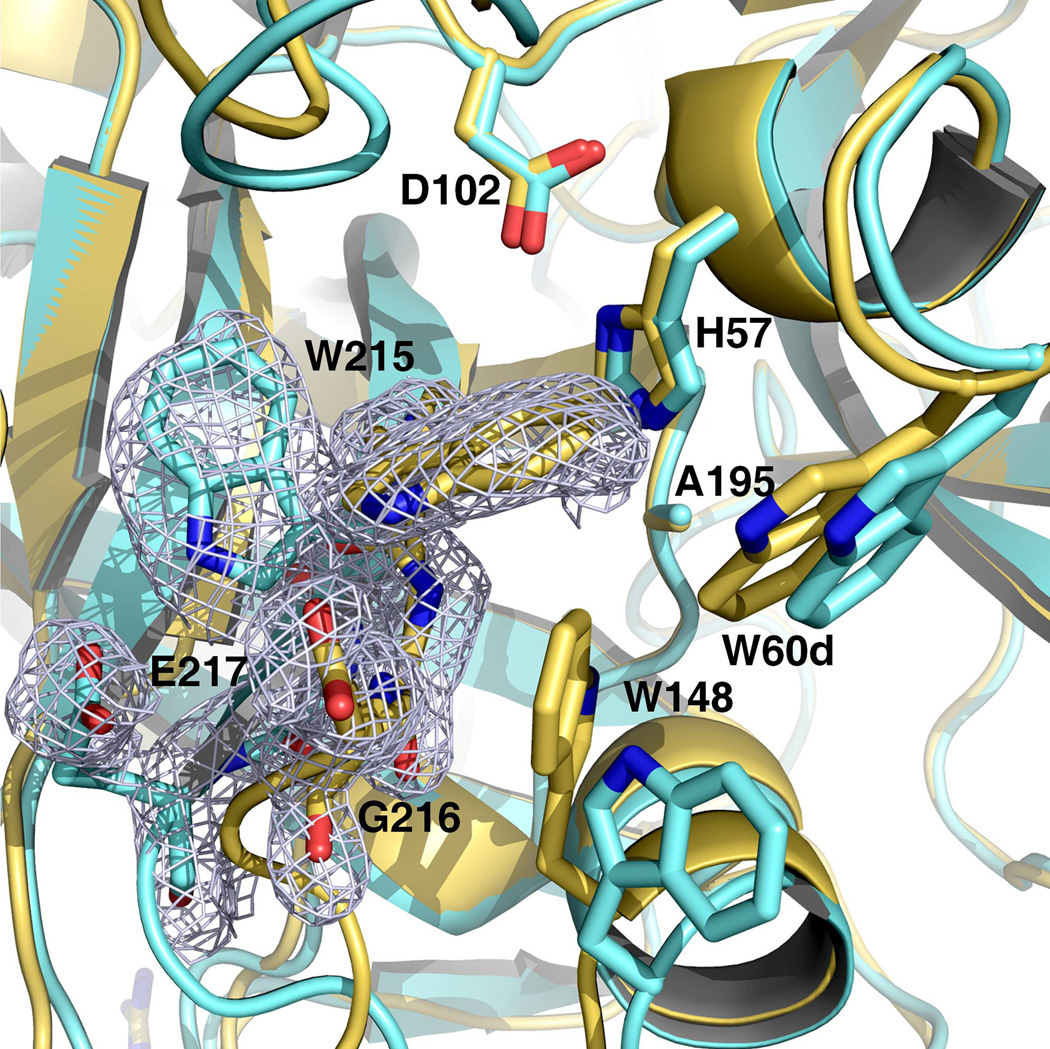
The 215–217 segment of prethrombin-2 in the free form is collapsed into the active site (Figure 1) and W215 forms a cluster of hydrophobic residues together with W60d in the adjacent 60-loop and the indole of W148 from the autolysis loop that assumes a helical conformation (Figure 2), as observed in the structure of prethrombin-1 (17). Remarkably, some of the crystals harvested from the same crystallization well diffracted to 2.2 Å resolution in the same space group P21 but with slightly different cell parameters and produced a structure with a conformation of the 215–217 segment similar to that observed in the hirugen-bound form (18). In this open conformation, the 215–217 segment moves 6.1 Å away from the active site and W215 comes into van der Waals interaction with F227 after relinquishing the hydrophobic interaction with W60d and W148 (Figure 1). The autolysis loop, however, retains its unusual helical conformation and enables W148 to partially screen the active site. The two conformations of prethrombin-2 differing in the collapsed and open arrangement of the 215–217 segment have an overall rmsd=0.307 Å. They differ in accessibility of the active site and are significantly populated under the same solution conditions: both forms can be trapped crystallographically in a 3:5 ratio (open:collapsed) by harvesting crystals grown in the same crystallization well. The conformational plasticity of prethrombin-2 is consistent with the results from a recent analysis of the entire Protein Data Bank database (13). Trypsin-like enzymes exist in equilibrium between two forms, E* and E, that differ in the position of the 215–217 segment. In the E form, the segment assumes an open conformation and poses no steric hindrance to substrate binding. In the E* form, the segment is collapsed into the active site and precludes substrate binding. The same equilibrium is present in the zymogen form. The collapsed conformation of prethrombin-1 (17) has already been mentioned. Chymotrypsinogen (33, 34) adopts a collapsed form for the 215–217 segment, but in a high resolution structure an alternative open conformation of the 215–217 segment was documented in the second molecule in the asymmetric unit (32). However, the alternative conformations are not associated with the drastic relocation of the side chain of W215 within the active site observed in the structure of prethrombin-2. Plasminogen features W215 in a “foot-in-mouth” conformation that occludes access to the primary specificity pocket (35, 36). The steric hindrance is partially relieved upon binding of streptokinase (8), as observed for prethrombin-2 when hirugen binds to exosite I (18). In complement profactor D the active site is screened by a collapse of the 215–217 segment (37) and a similar collapse is observed in prokallikrein 6 (38) and complement profactor C1r (39). Collapse of residue 215 against the catalytic H57 is observed in progranzyme K (40). The inactive α-subunit of the 7S nerve growth factor is a zymogen with a collapsed W215 (41). Several zymogens assume an open conformation of the 215–217 segment. They are trypsinogen (42, 43), complement profactors B (44) and MASP-2 (45), and coagulation factor XI (46). The E* and E forms have been trapped crystallographically in the same protein construct only recently and for a mature enzyme (16). The structures of prethrombin-2 reported here now document the two alternative conformations for the 215–217 segment in the same zymogen and under identical solution conditions, thereby validating the recent proposal that the E*-E equilibrium is a basic property of the trypsin fold for both the enzyme and zymogen forms (13). This emphasizes the need and importance to crystallize other members of the trypsin family in the free form to capture directly their allosteric properties. Crystallographers should be aware that the E and E* forms coexist under the same solution conditions and can be trapped by harvesting different crystals in the same crystallization well.
Figure 2. Active site accessibility in prethrombin-2.
The collapsed form of prethrombin-2 (orange) features a cluster of hydrophobic/aromatic residues that completely occludes access to the active site, as observed in the recent structure of prethrombin-1 (17). The cluster is formed by the collapse of W215 and W148 into the active site against W60d, with the indole ring of W215 moving >10 Å relative to its position in the open form (yellow). The electron density 2F0-Fc map (green mesh) outlines the alternative positions of W215 and is contoured at 1 σ.
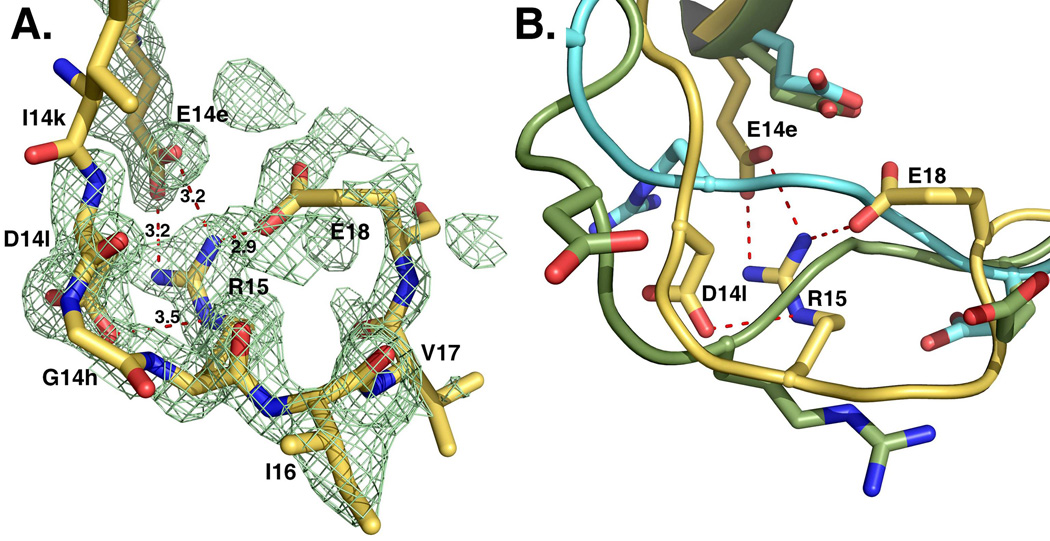
An unanticipated feature of the structure of prethrombin-2 is revealed by inspection of the activation domain around the site of cleavage at R15 that is engaged in specific interactions with the side chains of E14e, D14l and E18 (Figure 3). This region of the protein is either highly disordered or fully exposed to solvent in all existing structures of free zymogens deposited in the Protein Data Bank. Disorder of R15 is not linked to the conformation of the 215–217 segment in the active site and is observed in the structures of chymotrypsinogen (33), complement profactor D (37), complement profactor C1r (39) and the inactive α-subunit of the 7S nerve growth factor (41) where the 215–217 segment is collapsed, but also in trypsinogen (42, 43) and complement profactors B (44) where the 215–217 segment is in the open form. Likewise, exposure of R15 to solvent does not correlate with the conformation of the 215–217 segment and is documented in chymotrypsinogen (32, 34), plasminogen (35, 36), prokallikrein 6 (38), progranzyme K (40) and prethrombin-1 (17) where the 215–217 segment is collapsed, but also in the zymogen of MASP-2 (45) and chymotrypsinogen (32) where the 215–217 segment is in the open form. Coagulation factor XI deserves attention because it shows the guanidinium group of R369, equivalent to R15, partially exposed to solvent and also in polar interactions with the backbone O atoms of R378 and K367 (46). The buried position of R15 in prethrombin-2 in electrostatic interaction with three negatively charged residues is unprecedented for the zymogen of a trypsin-like protease and implies that activation of prethrombin-2 to thrombin would require a conformational change exposing R15 to solvent. A number of steps in the coagulation and complement cascades involve cofactor-assisted proteolytic activation of a zymogen (10–12) and the cofactor is assumed to cause conformational changes in the enzyme that facilitate substrate cleavage. Complement factors B and C2 are mostly inactive until binding of complement factors C3b and C4b (11, 47, 48). Complement factor D assumes an inactive conformation (37, 49) until binding to C3b and factor B promote substrate binding and catalysis (50, 51). Coagulation factor VIIa acquires full catalytic upon binding to tissue factor (52, 53). However, in the case of prethrombin-2, it is difficult to envision how factor Va within the prothrombinase complex may promote conversion to thrombin by acting exclusively on the enzyme factor Xa (19) and without causing exposure of R15 in the substrate to enable proteolytic attack. An alternative scenario should be considered where the cofactor Va acts directly on the substrate prethrombin-2 rather than, or in addition to the enzyme factor Xa. The enzyme itself can partake in this mechanism by promoting a conformational transition of the substrate mediated by binding to specific exosites (30). Whether R15 is also buried in prothrombin, for which no crystal structure is available, remains to be demonstrated. Prethrombin-1 differs from prothrombin for the lack of the Gla domain and kringle 1 and from prethrombin-2 for the presence of kringle 2. The structure of prethrombin-1 has been solved recently (17) and shows R15 fully exposed to solvent, suggesting that the presence of kringles may influence the conformation of the activation domain long-range.
Figure 3. Activation domain of prethrombin-2.
The segment around the cleavage site at R15 defines the activation domain and shows an intact R15-I16 peptide bond. R15 is buried inside the protein in electrostatic interaction with the side chains of E14e, D14l and E18. These interactions are shown for the open conformation, but are equivalent to those observed in the collapsed form. The orientation of R15 in the free form of prethrombin-2 is unprecedented among existing crystal structures of zymogens of trypsin-like proteases, including structures of prethrombin-2 bound to hirugen, PPACK or staphylocoagulase (9, 18). The electron density 2F0-Fc map (green mesh) is contoured at 1 σ.
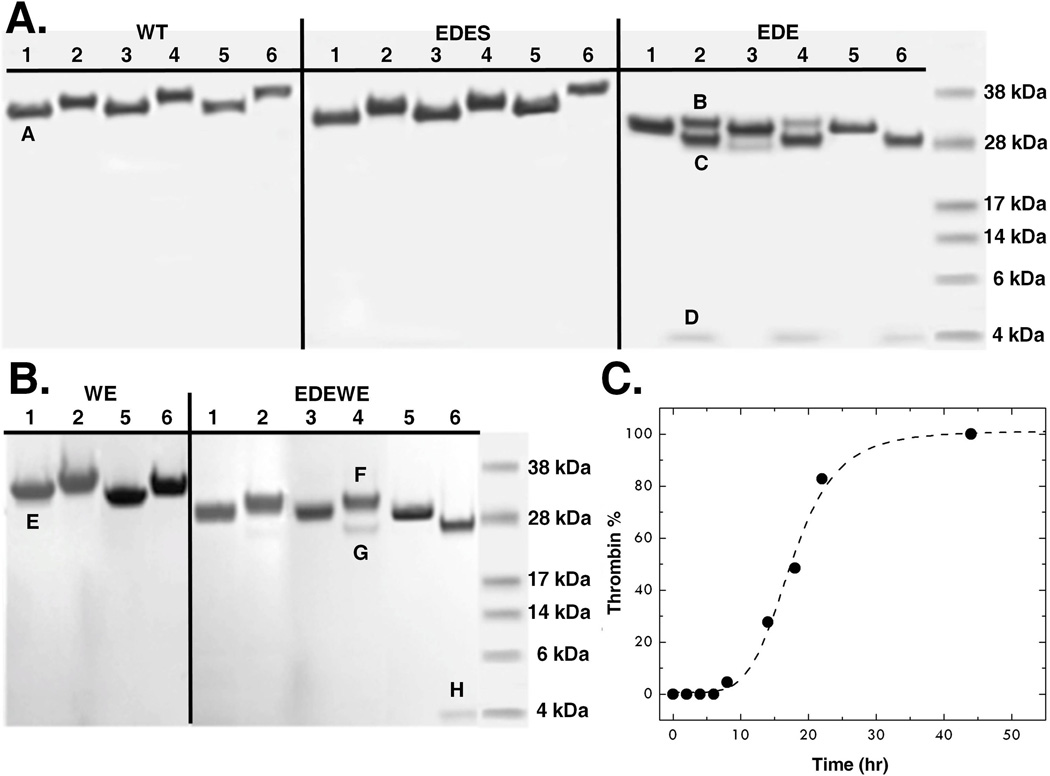
The unusual conformation of R15 and its specific interactions with the side chains of E14e, D14l and E18 raise important questions on the role of these residues in prethrombin-2 activation. Guided by the crystal structure, we expressed the prethrombin-2 mutant E14eA/D14lA/E18A with the expectation that it would be activated more efficiently than wild-type and would crystallize with R15 exposed to solvent. Surprisingly, the prethrombin-2 mutant E14eA/D14lA/E18A was found to convert spontaneously to thrombin (Figure 4), without the need of specific activators such as the snake venom ecarin or the physiological prothrombinase complex. This property is not present in the wild-type or any prethrombin-2 mutant of thrombin reported to-date. Auto-activation is complete in 90 hr and is specifically abrogated by the additional mutation S195A (Figure 4). Introduction of the triple mutation E14eA/D14lA/E18A in the prethrombin-2 mutant W215A/E217A also causes spontaneous conversion to the mature enzyme, but requires significantly higher concentrations and a longer time scale (Figure 4), consistent with the impaired catalytic activity of the thrombin mutant W215A/E217A (54). These observations support the conclusion that the E14eA/D14lA/E18A substitution promotes activity toward R15 to generate a mature enzyme. The auto-activation is likely initiated by prethrombin-2 itself, by analogy with the well known minuscule activity detected in other zymogens such as chymotrypsinogen (32), and then propagated by the mature enzyme. N-terminal sequencing confirms the presence of a new N-terminus at I16 for the catalytic B chain, along with the N-terminus of the A chain at T1h, for both constructs E14eA/D14lA/E18A and E14eA/D14lA/E18A/W215A/E217A (Figure 4). The products of auto-activation, E14eA/D14lA/E18A and E14eA/D14lA/E18A/W215A/E217A, cleave synthetic and physiological substrates with values of kcat/Km comparable to those of wild-type and the anticoagulant thrombin mutant W215A/E217A, respectively (Table 2). This is consistent with the modest perturbation of catalytic activity reported for single Ala mutants of the A chain (56). The remarkable properties introduced by the E14eA/D14lA/E18A mutation have substantial practical implications and could simplify production of recombinant thrombin wild-type and W215A/E215A for clinical use (55).
Figure 4. Auto-activation of prethrombin-2.
(A) The prethrombin-2 mutant E14eA/D14lA/E18A (EDE) shows evidence of auto-activation, which is not seen in the wild-type (WT) and is selectively abrogated by the additional mutation S195A (EDES). After heparin-sepharose purification, the concentration of each protein was adjusted to 0.27 mg/ml and auto-activation was followed at room temperature for 0 (lanes 1, 2), 4 (lanes 3, 4) and 90 (lanes 5, 6) hr. (B) Auto-activation is also observed when the E14eA/D14lA/E18A mutation is introduced in the prethrombin-2 mutant W215A/E217A (WE) to yield the construct E14eA/D14lA/E18A/W215A/E217A (EDEWE). In this case, the concentration was adjusted to 3 mg/ml and the reaction was followed at room temperature for 0 (lanes 1, 2), 3 (lanes 3, 4) and 7 (lanes 5, 6) days. No evidence of auto-activation is detected for WE over the same time scale. Samples were analyzed under non-reducing (lanes 1, 3, 5) and reducing (lanes 2, 4, 6) conditions. In the case of EDE and EDEWE, the two bands pertaining to the A and B chains of the mature enzyme are easily detected under reducing conditions and conversion to thrombin is complete after 90 h or 7 days, respectively. The chemical identity of the A and B chains was confirmed by N-terminal sequencing. Bands in the gel are labeled as follows: A and E mapped to N-terminal sequence GRGSE and refer to prethrombin-2 constructs with the T7tag from the expression vector partially cleaved and then processed during E. coli expression as reported (57, 58); B and F mapped to N-terminal sequence TFGSG and refer to prethrombin-2 with a single N-terminus starting at T1h; C and G mapped to N-terminal sequence IVAGS and refer to the B chain of thrombin with the N-terminus I16 and the mutation E18A introduced in the EDE and EDEWE constructs; D and H mapped to N-terminal sequence TFGSG and refer to the A chain of thrombin with the N-terminus T1h.
Table 2.
Values of kcat/Km (µM−1s−1) for thrombin wild-type and mutants toward synthetic and physiological substrates
| Enzyme | FPR | FpA | PAR1 | Protein Ca |
|---|---|---|---|---|
| Wt | 37±1 | 17±1 | 27±1 | 0.22±0.01 |
| EDE | 19±1 | 7.2±0.5 | 9.1±0.6 | 0.11±0.01 |
| WE | 0.0014±0.0001 | 0.00027±0.00001 | 0.0099±0.0003 | 0.027±0.001 |
| EDEWE | 0.0015±0.0001 | 0.00026±0.00001 | 0.016±0.001 | 0.031±0.001 |
EDE: E14eA/D14lA/E18A; EDEWE: E14eA/D14lA/E18A/W215A/E217A; FpA: fibrinopeptide A; FPR: H-D-Phe-Pro-Arg-p-nitroanilide; WE: W215A/E217A; Experimental conditions are: 5 mM Tris, 0.1% PEG 8K, pH 7.4, 37 °C, 145 mM NaCl.
In the presence of 100 nM thrombomodulin and 5 mM CaCl2.
Acknowledgments
We are grateful to Ms. Tracey Baird for her help with illustrations.
Abbreviations used
- PDB
Protein Data Bank
- PEG
polyethyleneglycol
Footnotes
This work was supported in part by the National Institutes of Health Research Grants HL49413, HL58141, HL73813 and HL95315 (to E.D.C.).
References
- 1.Ciechanover A, Iwai K. The ubiquitin system: from basic mechanisms to the patient bed. IUBMB Life. 2004;56:193–201. doi: 10.1080/1521654042000223616. [DOI] [PubMed] [Google Scholar]
- 2.Gomis-Ruth FX. Structural aspects of the metzincin clan of metalloendopeptidases. Mol Biotechnol. 2003;24:157–202. doi: 10.1385/MB:24:2:157. [DOI] [PubMed] [Google Scholar]
- 3.Rea D, Fulop V. Structure-function properties of prolyl oligopeptidase family enzymes. Cell Biochem Biophys. 2006;44:349–365. doi: 10.1385/CBB:44:3:349. [DOI] [PubMed] [Google Scholar]
- 4.Page MJ, Di Cera E. Serine peptidases: classification, structure and function. Cell Mol Life Sci. 2008;65:1220–1236. doi: 10.1007/s00018-008-7565-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hedstrom L. Serine protease mechanism and specificity. Chem Rev. 2002;102:4501–4524. doi: 10.1021/cr000033x. [DOI] [PubMed] [Google Scholar]
- 6.Perona JJ, Craik CS. Structural basis of substrate specificity in the serine proteases. Protein Sci. 1995;4:337–360. doi: 10.1002/pro.5560040301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ranby M, Bergsdorf N, Nilsson T. Enzymatic properties of the one- and two-chain form of tissue plasminogen activator. Thromb Res. 1982;27:175–183. doi: 10.1016/0049-3848(82)90197-9. [DOI] [PubMed] [Google Scholar]
- 8.Wakeham N, Terzyan S, Zhai P, Loy JA, Tang J, Zhang XC. Effects of deletion of streptokinase residues 48–59 on plasminogen activation. Protein Eng. 2002;15:753–761. doi: 10.1093/protein/15.9.753. [DOI] [PubMed] [Google Scholar]
- 9.Friedrich R, Panizzi P, Fuentes-Prior P, Richter K, Verhamme I, Anderson PJ, Kawabata S, Huber R, Bode W, Bock PE. Staphylocoagulase is a prototype for the mechanism of cofactor-induced zymogen activation. Nature. 2003;425:535–539. doi: 10.1038/nature01962. [DOI] [PubMed] [Google Scholar]
- 10.Krem MM, Di Cera E. Evolution of enzyme cascades from embryonic development to blood coagulation. Trends Biochem Sci. 2002;27:67–74. doi: 10.1016/s0968-0004(01)02007-2. [DOI] [PubMed] [Google Scholar]
- 11.Gros P, Milder FJ, Janssen BJ. Complement driven by conformational changes. Nat Rev Immunol. 2008;8:48–58. doi: 10.1038/nri2231. [DOI] [PubMed] [Google Scholar]
- 12.Davie EW, Fujikawa K, Kisiel W. The coagulation cascade: initiation, maintenance, and regulation. Biochemistry. 1991;30:10363–10370. doi: 10.1021/bi00107a001. [DOI] [PubMed] [Google Scholar]
- 13.Gohara DW, Di Cera E. Allostery in trypsin-like proteases suggests new therapeutic strategies. Trends Biotechnol. 2011 doi: 10.1016/j.tibtech.2011.06.001. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Di Cera E. Thrombin. Mol Aspects Med. 2008;29:203–254. doi: 10.1016/j.mam.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bah A, Garvey LC, Ge J, Di Cera E. Rapid kinetics of Na+ binding to thrombin. J Biol Chem. 2006;281:40049–40056. doi: 10.1074/jbc.M608600200. [DOI] [PubMed] [Google Scholar]
- 16.Niu W, Chen Z, Gandhi PS, Vogt AD, Pozzi N, Pelc LA, Zapata FJ, Di Cera E. Crystallographic and kinetic evidence of allostery in a trypsin-like protease. Biochemistry. 2011;50:6301–6307. doi: 10.1021/bi200878c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Z, Pelc LA, Di Cera E. Crystal structure of prethrombin-1. Proc Natl Acad Sci U S A. 2010;107:19278–19283. doi: 10.1073/pnas.1010262107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vijayalakshmi J, Padmanabhan KP, Mann KG, Tulinsky A. The isomorphous structures of prethrombin2, hirugen-, and PPACK-thrombin: changes accompanying activation and exosite binding to thrombin. Protein Sci. 1994;3:2254–2271. doi: 10.1002/pro.5560031211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mann KG, Butenas S, Brummel K. The dynamics of thrombin formation. Arterioscler Thromb Vasc Biol. 2003;23:17–25. doi: 10.1161/01.atv.0000046238.23903.fc. [DOI] [PubMed] [Google Scholar]
- 20.Wood JP, Silveira JR, Maille NM, Haynes LM, Tracy PB. Prothrombin activation on the activated platelet surface optimizes expression of procoagulant activity. Blood. 2010;117:1710–1718. doi: 10.1182/blood-2010-09-311035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marino F, Pelc LA, Vogt A, Gandhi PS, Di Cera E. Engineering thrombin for selective specificity toward protein C and PAR1. J. Biol. Chem. 2010;285:19145–19152. doi: 10.1074/jbc.M110.119875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pozzi N, Chen R, Chen Z, Bah A, Di Cera E. Rigidification of the autolysis loop enhances Na+ binding to thrombin. Biophys Chem. 2011;159:6–13. doi: 10.1016/j.bpc.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Otwinowski Z, Minor W. Processing of x-ray diffraction data collected by oscillation methods. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 24.Bailey S. The CCP4 suite. Programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 25.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 26.Morris AL, MacArthur MW, Hutchinson EG, Thornton JM. Stereochemical quality of protein structure coordinates. Proteins. 1992;12:345–364. doi: 10.1002/prot.340120407. [DOI] [PubMed] [Google Scholar]
- 27.Ayala Y, Di Cera E. Molecular recognition by thrombin. Role of the slow-->fast transition, site-specific ion binding energetics and thermodynamic mapping of structural components. J Mol Biol. 1994;235:733–746. doi: 10.1006/jmbi.1994.1024. [DOI] [PubMed] [Google Scholar]
- 28.Liu LW, Vu TK, Esmon CT, Coughlin SR. The region of the thrombin receptor resembling hirudin binds to thrombin and alters enzyme specificity. J Biol Chem. 1991;266:16977–16980. [PubMed] [Google Scholar]
- 29.Kroh HK, Tans G, Nicolaes GAF, Rosing J, Bock PE. Expression of allosteric linkage between the sodium ion binding site and exosite I of thrombin during prothrombin activation. J Biol Chem. 2007;282:16095–16104. doi: 10.1074/jbc.M610577200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bock PE, Panizzi P, Verhamme IM. Exosites in the substrate specificity of blood coagulation reactions. J Thromb Haemost. 2007;5(Suppl 1):81–94. doi: 10.1111/j.1538-7836.2007.02496.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gandhi PS, Chen Z, Mathews FS, Di Cera E. Structural identification of the pathway of long-range communication in an allosteric enzyme. Proc Natl Acad Sci USA. 2008;105:1832–1837. doi: 10.1073/pnas.0710894105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang D, Bode W, Huber R. Bovine chymotrypsinogen A X-ray crystal structure analysis and refinement of a new crystal form at 1.8 A resolution. J Mol Biol. 1985;185:595–624. doi: 10.1016/0022-2836(85)90074-9. [DOI] [PubMed] [Google Scholar]
- 33.Freer ST, Kraut J, Robertus JD, Wright HT, Xuong NH. Chymotrypsinogen: 2.5-angstrom crystal structure, comparison with alpha-chymotrypsin, and implications for zymogen activation. Biochemistry. 1970;9:1997–2009. doi: 10.1021/bi00811a022. [DOI] [PubMed] [Google Scholar]
- 34.Pjura PE, Lenhoff AM, Leonard SA, Gittis AG. Protein crystallization by design: chymotrypsinogen without precipitants. J Mol Biol. 2000;300:235–239. doi: 10.1006/jmbi.2000.3851. [DOI] [PubMed] [Google Scholar]
- 35.Peisach E, Wang J, de los Santos T, Reich E, Ringe D. Crystal structure of the proenzyme domain of plasminogen. Biochemistry. 1999;38:11180–11188. doi: 10.1021/bi991130r. [DOI] [PubMed] [Google Scholar]
- 36.Wang X, Terzyan S, Tang J, Loy JA, Lin X, Zhang XC. Human plasminogen catalytic domain undergoes an unusual conformational change upon activation. J Mol Biol. 2000;295:903–914. doi: 10.1006/jmbi.1999.3397. [DOI] [PubMed] [Google Scholar]
- 37.Jing H, Macon KJ, Moore D, DeLucas LJ, Volanakis JE, Narayana SV. Structural basis of profactor D activation: from a highly flexible zymogen to a novel self-inhibited serine protease, complement factor D. Embo J. 1999;18:804–814. doi: 10.1093/emboj/18.4.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gomis-Rüth FX, Bayés A, Sotiropoulou G, Pampalakis G, Tsetsenis T, Villegas V, Avilés FX, Coll M. The structure of human prokallikrein 6 reveals a novel activation mechanism for the kallikrein family. J Biol Chem. 2002;277:27273–27281. doi: 10.1074/jbc.M201534200. [DOI] [PubMed] [Google Scholar]
- 39.Budayova-Spano M, Lacroix M, Thielens NM, Arlaud GJ, Carlos Fontecilla-Camps J, Gaboriaud C. The crystal structure of the zymogen catalytic domain of complement protease C1r reveals that a disruptive mechanical stress is required to trigger activation of the C1 complex. EMBO J. 2002;21:231–239. doi: 10.1093/emboj/21.3.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hink-Schauer C, Estebanez-Perpina E, Wilharm E, Fuentes-Prior P, Klinkert W, Bode W, Jenne DE. The 2.2-A crystal structure of human pro-granzyme K reveals a rigid zymogen with unusual features. J Biol Chem. 2002;277:50923–50933. doi: 10.1074/jbc.M207962200. [DOI] [PubMed] [Google Scholar]
- 41.Bax B, Blundell TL, Murray-Rust J, McDonald NQ. Structure of mouse 7S NGF: a complex of nerve growth factor with four binding proteins. Structure. 1997;5:1275–1285. doi: 10.1016/s0969-2126(97)00280-3. [DOI] [PubMed] [Google Scholar]
- 42.Fehlhammer H, Bode W, Huber R. Crystal structure of bovine trypsinogen at 1–8 A resolution. II. Crystallographic refinement, refined crystal structure and comparison with bovine trypsin. J Mol Biol. 1977;111:415–438. doi: 10.1016/s0022-2836(77)80062-4. [DOI] [PubMed] [Google Scholar]
- 43.Kossiakoff AA, Chambers JL, Kay LM, Stroud RM. Structure of bovine trypsinogen at 1.9 A resolution. Biochemistry. 1977;16:654–664. doi: 10.1021/bi00623a016. [DOI] [PubMed] [Google Scholar]
- 44.Milder FJ, Gomes L, Schouten A, Janssen BJC, Huizinga EG, Romijn RA, Hemrika W, Roos A, Daha MR, Gros P. Factor B structure provides insights into activation of the central protease of the complement system. Nat Struct Mol Biol. 2007;14:224–228. doi: 10.1038/nsmb1210. [DOI] [PubMed] [Google Scholar]
- 45.Gál P, Harmat V, Kocsis A, Bián T, Barna L, Ambrus G, Végh B, Balczer J, Sim RB, Náray-Szabó G, Závodszky P. A true autoactivating enzyme. Structural insight into mannose-binding lectin-associated serine protease-2 activations. J Biol Chem. 2005;280:33435–33444. doi: 10.1074/jbc.M506051200. [DOI] [PubMed] [Google Scholar]
- 46.Papagrigoriou E, McEwan PA, Walsh PN, Emsley J. Crystal structure of the factor XI zymogen reveals a pathway for transactivation. Nat Struct Mol Biol. 2006;13:557–558. doi: 10.1038/nsmb1095. [DOI] [PubMed] [Google Scholar]
- 47.Arlaud GJ, Barlow PN, Gaboriaud C, Gros P, Narayana SV. Deciphering complement mechanisms: the contributions of structural biology. Mol Immunol. 2007;44:3809–3822. doi: 10.1016/j.molimm.2007.06.147. [DOI] [PubMed] [Google Scholar]
- 48.Ponnuraj K, Xu Y, Macon K, Moore D, Volanakis JE, Narayana SV. Structural analysis of engineered Bb fragment of complement factor B: insights into the activation mechanism of the alternative pathway C3-convertase. Mol Cell. 2004;14:17–28. doi: 10.1016/s1097-2765(04)00160-1. [DOI] [PubMed] [Google Scholar]
- 49.Narayana SV, Carson M, el-Kabbani O, Kilpatrick JM, Moore D, Chen X, Bugg CE, Volanakis JE, DeLucas LJ. Structure of human factor, DA complement system protein at 2.0 A resolution. J Mol Biol. 1994;235:695–708. doi: 10.1006/jmbi.1994.1021. [DOI] [PubMed] [Google Scholar]
- 50.Forneris F, Ricklin D, Wu J, Tzekou A, Wallace RS, Lambris JD, Gros P. Structures of C3b in complex with factors B and D give insight into complement convertase formation. Science. 2010;330:1816–1820. doi: 10.1126/science.1195821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jing H, Babu YS, Moore D, Kilpatrick JM, Liu XY, Volanakis JE, Narayana SV. Structures of native and complexed complement factor D: implications of the atypical His57 conformation and self-inhibitory loop in the regulation of specific serine protease activity. J Mol Biol. 1998;282:1061–1081. doi: 10.1006/jmbi.1998.2089. [DOI] [PubMed] [Google Scholar]
- 52.Banner DW, D'Arcy A, Chene C, Winkler FK, Guha A, Konigsberg WH, Nemerson Y, Kirchhofer D. The crystal structure of the complex of blood coagulation factor VIIa with soluble tissue factor. Nature. 1996;380:41–46. doi: 10.1038/380041a0. [DOI] [PubMed] [Google Scholar]
- 53.Eigenbrot C, Kirchhofer D, Dennis MS, Santell L, Lazarus RA, Stamos J, Ultsch MH. The factor VII zymogen structure reveals reregistration of beta strands during activation. Structure. 2001;9:627–636. doi: 10.1016/s0969-2126(01)00624-4. [DOI] [PubMed] [Google Scholar]
- 54.Cantwell AM, Di Cera E. Rational design of a potent anticoagulant thrombin. J Biol Chem. 2000;275:39827–39830. doi: 10.1074/jbc.C000751200. [DOI] [PubMed] [Google Scholar]
- 55.Erdmann J. Engineered thrombin aims to take on heparin. Chem Biol. 2010;17:1267–1268. doi: 10.1016/j.chembiol.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 56.Papaconstantinou ME, Bah A, Di Cera E. Role of the A chain in thrombin function. Cell Mol Life Sci. 2008;65:1943–1947. doi: 10.1007/s00018-008-8179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Frottin F, Martinez A, Peynot P, Mitra S, Holz RC, Giglione C, Meinnel T. The proteomics of N-terminal methionine cleavage. Mol Cell Proteomics. 2006;5:2336–2349. doi: 10.1074/mcp.M600225-MCP200. [DOI] [PubMed] [Google Scholar]
- 58.Xiao Q, Zhang F, Nacev BA, Liu JO, Pei D. Protein N-terminal processing: substrate specificity of Escherichia coli and human methionine aminopeptidases. Biochemistry. 2010;49:5588–5599. doi: 10.1021/bi1005464. [DOI] [PMC free article] [PubMed] [Google Scholar]