Abstract
The CH2Cl2-MeOH extract of a South African tunicate described as the new Synoicum globosum Parker-Nance sp. nov. (Ascidiacea, Aplousobranchia) was subjected to 1H NMR-guided fractionation. This resulted in the identification of new 3″-bromorubrolide F (1), 3′-bromorubrolide E (2), 3′-bromorubrolide F (3) and 3′, 3″-dibromorubrolide E (4), and reisolation of known rubrolides E (5) and F (6), based on NMR spectroscopic and mass spectrometric data. Biological testing of both new and known members of this reported antimicrobial family of halogenated, aryl-substituted furanones indicated moderate antibacterial properties for 3′-bromorubrolide E (2), 3′, 3″-dibromorubrolide E (4), and rubrolide F (6) against methicillin-resistant Staphylococcus aureus (MRSA) and S. epidermidis.
Marine organisms have proven to be a rich source of new chemical entities, and the first marine drugs have been approved, with others continuing in clinical trials.1,2 In particular, ascidians (tunicates) have provided a variety of clinical candidates and new carbon skeletons including non-nitrogenated, halogenated rubrolides, the first of which were isolated from British Columbian specimens of the ascidian Ritterella rubra.3 Rubrolides comprise a central furanone ring substituted at C-4 and C-5 with phenolic moieties. The phenolic moieties are differentially halogenated in the known rubrolides A-O. However, among them only rubrolide F is selectively methylated at C-4″.3-5 Attempts to synthesize new, more potent antimicrobials resulted in a suite of analogues having one or two phenolic moieties with varying halogenation and hydroxy group methylation.6,7 The biological activities of the known rubrolide natural products and their synthetic analogues include antibacterial activity against Staph. aureus and Bacillus subtilis (rubrolides A, B, C),3 selective inhibition of protein phosphatases 1 and 2A,3 weak to moderate activity against various cancer cell lines (rubrolides I, K, L, M),4 inhibition of human aldose reductase (rubrolide L),8,9 and moderate anti-inflammatory properties (rubrolide O).10 Interestingly, a synthetic series of analogues in which the furanone is substituted with an aryl group comprises potent inhibitors of bacterial cell wall biosynthesis, which have been patented for their potential antimicrobial applications.11
As part of our ongoing investigation of a 2004 collection of organic extracts from South African ascidians,12 1H NMR and MS screening identified a Synoicum CH2Cl2-MeOH extract that exhibited a complex aromatic 1H signature as well as isotopic patterns for halogenated mass ions. Successive fractionation of this extract (2.4 g) by RP SPE followed by RP HPLC yielded four new rubrolide analogues (1-4) together with the known rubrolides E (5) and F (6).
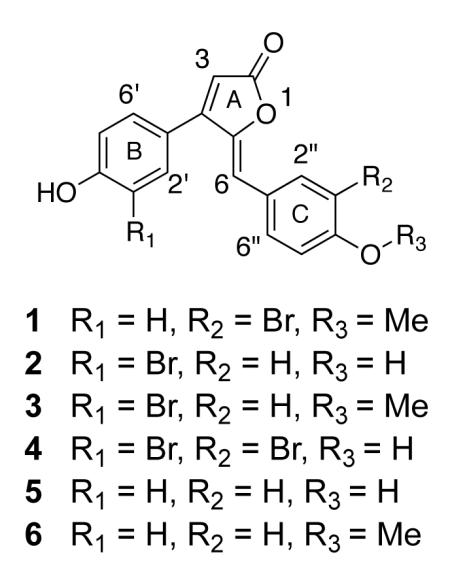
A molecular formula of C18H13O4Br for rubrolide 1 was deduced from the HRESIMS ion cluster for [M+H]+ at m/z 373.0081/375.0070 (1:1). Inspection of the 1H NMR spectrum revealed nine olefinic or aromatic signals (δH 6.23 – 8.10, Table 1) and a 3H midfield singlet consistent with an aromatic O-methyl (δH 3.93) group. Multiplicity-edited HSQC and HMBC data indicated the presence of eight quaternary carbons (δC 120.5, 126.5, 133.5, 146.7, 156.9, 159.4, 159.9, 169.5), nine methine carbons (δC 111.1, 111.6, 111.7, 115.6, 115.6, 129.9, 129.9, 131.2, 134.6) and one O-methyl carbon (δC 55.3). HMBC correlations from a 1H doublet at δ 6.23 (H-3) to 13C resonances at δ 169.5 (C-2), 159.4 (C-4) and 146.7 (C-5) defined the furanone (ring A) of the rubrolide skeleton. A para-substituted phenolic moiety was indicated by two mutually coupled 2H doublets (δH 7.47 and 6.95), and confirmed by apparent HMBC correlations from these doublets to their same HSQC-correlated 13C resonances. This B-ring could be located at C-4 of ring A based on an HMBC correlation from the H-2′/6′ doublet (δH 7.47) to the C-4 resonance. The presence of COSY couplings between H-5″ and H-6″ 1H signals, in parallel with a long range coupling (2.1 Hz) between H-2″ and H-6″, led to assignment of ring C as a 1, 3, 4-trisubstitued aromatic moiety. Inspection of the HMBC data for 1 to establish the connectivity between rings A and C identified the olefinic linkage H-6 (δH 6.28, s). HMBC correlations were observed from the H-6 singlet to 13C resonances for C-4 and C-5 of ring A, and to C-2″ and C-6″ of ring C. Deshielded methoxy H3-7″ showed an HMBC correlation to C-4″, thus locating this substituent on ring C. A relatively shielded quaternary carbon resonance at δC 126.5, assigned as C-3″ (from HMBC correlations) in the 1, 3, 4-trisubstituted ring C, remained as the site of bromine substitution. Therefore, rubrolide 1 was assigned as 5-(3-bromo-4-methoxybenzylidene)-4-(4-hydroxyphenyl)furan-2(5H)-one, and given the trivial name 3″-bromorubrolide F.
Table 1.
1H (700 MHz) and 13C (175 MHz) NMR Spectroscopic Data (methanol-d4) for Rubrolides 1-4.
| position | 3″-Bromorubrolide F (1) | 3′-Bromorubrolide E (2) | 3′-Bromorubrolide F (3) | 3′, 3″-Dibromorubrolide E (4) | ||||
|---|---|---|---|---|---|---|---|---|
| δC, mult. | δH (mult., J in Hz) | δC, mult. | δH (mult., J in Hz) | δC, mult. | δH (mult., J in Hz) | δC, mult. | δH (mult., J in Hz) | |
| 2 | 169.5 (C) | 170.4 (C) | 171.5 (C) | 171.5 (C) | ||||
| 3 | 111.1 (CH) | 6.23 (d, 0.6) | 111.9 (CH) | 6.20 (s) | 111.4 (CH) | 6.16 (s) | 111.7 (CH) | 6.16 (s) |
| 4 | 159.4 (C) | 158.3 (C) | 159.5 (C) | 159.5 (C) | ||||
| 5 | 146.7 (C) | 146.3 (C) | 147.8 (C) | 147.5 (C) | ||||
| 6 | 111.6 (CH) | 6.28 (s) | 114.5 (CH) | 6.24 (s) | 115.0 (CH) | 6.31 (s) | 114.7 (CH) | 6.22 (s) |
| 1′ | 120.5 (C) | 123.2 (C) | 121.3 (C) | 122.3 (C) | ||||
| 2′ | 129.9 (CH) | 7.47 (d, 8.6) | 133.6 (CH) | 7.72 (d, 2.0) | 134.1 (CH) | 7.70 (d, 2.2) | 134.4 (CH) | 7.70 (d, 2.2) |
| 3′ | 115.6 (CH) | 6.95 (d, 8.7) | 110.9 (C) | 113.3 (C) | 113.1 (C) | |||
| 4′ | 159.9 (C) | 157.4 (C) | 161.6 (C) | 161.2 (C) | ||||
| 5′ | 115.6 (CH) | 6.95 (d, 8.7) | 116.8 (CH) | 7.03 (d, 8.3) | 118.9 (CH) | 6.94 (d, 8.3) | 118.8 (C) | 6.97 (d, 8.4) |
| 6′ | 129.9 (CH) | 7.47 (d, 8.6) | 129.5 (CH) | 7.42 (dd, 8.3, 2.0) | 130.2 (CH) | 7.38 (dd, 8.3, 2.2) | 130.5 (CH) | 7.39 (d, 8.4, 2.2) |
| 1″ | 133.5 (C) | 125.2 (C) | 127.4 (C) | 126.3 (C) | ||||
| 2″ | 134.6 (CH) | 8.10 (d, 2.1) | 133.2 (CH) | 7.70 (d, 8.7) | 133.5 (CH) | 7.79 (d, 8.6) | 136.7 (CH) | 8.03 (d, 2.1) |
| 3″ | 126.5 (C) | 116.0 (CH) | 6.83 (d, 8.7) | 115.2 (CH) | 6.98 (d, 8.9) | 112.7 (C) | ||
| 4″ | 156.9 (C) | 159.6 (C) | 162.1 (C) | 159.6 (C) | ||||
| 5″ | 111.7 (CH) | 7.10 (d, 8.6) | 116.0 (CH) | 6.83 (d, 8.7) | 115.2 (CH) | 6.98 (d, 8.9) | 118.3 (CH) | 6.87 (d, 8.6) |
| 6″ | 131.2 (CH) | 7.78 (dd, 8.6, 2.1) | 133.2 (CH) | 7.70 (d, 8.7) | 133.5 (CH) | 7.79 (d, 8.6) | 132.9 (CH) | 7.59 (dd, 8.6, 2.1) |
| 7″ | 55.3 (CH3) | 3.93 (s) | 55.8 (CH3) | 3.84 (s) | ||||
HRESIMS of rubrolide 2 gave an [M+H]+ ion cluster at m/z 358.9933/360.9916 (1:1) for a molecular formula of C17H11O4Br. Comparison of the 1H NMR spectra for 1 and 2 indicated significant changes in the shifts for all downfield 1H signals and the absence of the midfield 3H singlet (δ 3.93, Table 1). Assignment of COSY and HMBC correlations showed that in the case of 2, ring B is the 1, 3, 4-trisubstitued aromatic moiety, and ring C is the 1,4-disubstitued aromatic unit. Therefore, it could be concluded that in 2, the bromine is located at C-3′ of ring B. The lack of an O-methyl singlet indicated a free 4″-hydroxy group in 2 compared to 1. Thus, the structure of 2 was defined as 4-(3-bromo-4-hydroxyphenyl)-5-(4-hydroxybenzylidene)furan-2(5H)-one, and given the trivial name 3′-bromorubrolide E.
The HRESIMS data for rubrolide 3 gave an [M+H]+ ion cluster at m/z 373.0060/375.0048 (1:1), for the same molecular formula of C18H13O4Br as that for 1. Comparison of the 1H chemical shifts for 2 and 3 showed a slight upfield shift for B-ring 1H resonances (H-2′, H-5′ and H-6′) and a large downfield shift for C-ring 1H resonances (H-2″, H-3″, H-5″ and H-6″, Table 1). The larger variations in δH values for ring C could be explained by methylation of OH-4″ in 3, as confirmed by an HMBC correlation from a 3H singlet at δ 3.84 (H3-7″) to the C-4″ resonance (δC 162.1). Therefore, the structure of rubrolide 3 was assigned as 4-(3-bromo-4-hydroxyphenyl)-5-(4-methoxybenzylidene)furan-2(5H)-one, and given the trivial name 3′-bromorubrolide F.
Rubrolide 4 has a molecular formula of C17H10O4Br2 as assigned from the [M−H]− ion cluster at m/z 434.8873/436.8929/438.8940 (1:2:1). Examination of the 1H and COSY NMR data indicated that the 1H shifts for ring B protons (H-2′, H-5′ and H-6′) were similar to those for ring B of 3′-bromorubrolide E (2, Table 1). Additionally, COSY-coupled H-2″/H-6″ and H-5″/H-6″ established ring C as a 1, 3, 4-trisubstitued benzene. The assignment of quaternary C-3′ (δC 113.1) and C-3″ (δC 112.7), as deduced from HMBC, localized bromine substituents at these positions. Thus, rubrolide 4 was assigned as 5-(3-bromo-4-hydroxybenzylidene)-4-(3-bromo-4-hydroxyphenyl)furan-2(5H)-one, and given the trivial name 3′,3″-dibromorubrolide E.
For new rubrolides 1-4, determination of the C-5/C-6 double bond geometry relied on NOE data. NOEs between H-6 and both H-2′/H-6′ and H-2″/H-6″ (where one of the pair was always distinguishable) were observed, placing H-6 in close proximity to both phenyl rings and concluding a Z geometry of the exocyclic double bond.10
The HRESIMS data for compounds 5 and 6 suggested their identities as rubrolides E and F, respectively. 1D and 2D NMR data were acquired in methanol-d4 for compound 5, and readily confirmed its identity as rubrolide E, although previously reported data were acquired in CDCl3 and DMSO-d6.3 The initially acquired NMR data in methanol-d4 for compound 6 were consistent with previously published 1H NMR data for rubrolide F.3 However, after additional purification of compound 6, significant changes in 1H shifts for the A and B rings were observed, while 2D NMR data provided the same rubrolide F structure as initially assigned. One explanation for this observation was the interconversion of 6 between phenoxy and phenol forms. The addition of NaOH to the NMR tube containing 6 caused a shift in ring A and B 1H resonances, which was reversed after the addition of formic acid (Figures S25-27). Although the addition of NaOH did not reproduce the exact chemical shifts previously reported,3 or observed for the initially isolated natural product here, it could be concluded that rubrolide F was first isolated as a salt (other than Na+) of the phenoxy (B-ring) anion in both cases. Our subsequent re-purification performed under neutral conditions resulted in protonation to yield the phenol form of 6 (Table S6).
Finally, we tested in vitro activities of rubrolides 1-4 as well as 5 and 6 against a panel of pathogenic bacteria including MRSA, S. epidermidis, gentamycin and vancomycin resistant Enterococcus faecalis and Escherichia coli, (Table 2). Overall, 3′-bromo-rubrolide F (3) and rubrolide F (6) showed significantly higher IC50 values and a lower percentage of growth inhibition than their non-methylated analogues. This trend was previously observed for synthetic rubrolide analogues tested against NCI-H460, MCF-7 and SF-268 cell lines,7 and may indicate a differential cell permeability.
Table 2.
Antibacterial Properties of Rubrolides 1-6.
| Compound | MRSA | S. epidermidis | E. faecalis | E. coli |
|---|---|---|---|---|
|
|
||||
| aIC50 μM (SEM) | aIC50 μM (SEM) | Percent growth inhibition at 100 μg/mL (SEM) | ||
|
|
||||
| 3″-Bromorubrolide F (1) | 256 (1.3) | 98 (1.2) | 43 (8.3) | 22 (1.9) |
| 3′-Bromorubrolide E (2) | 82 (1.4) | 38 (1.7) | 16 (14.1) | 0 (26.2) |
| 3′-Bromorubrolide F (3) | 360 (1.3) | 42 (1.2) | 2 (3.1) | 14 (14.4) |
| 3′, 3″-Dibromorubrolide E (4) | 89 (1.3) | 28 (1.4) | 2 (12.9) | 25 (4.6) |
| Rubrolide E (5) | 105 (1.5) | 21 (1.5) | 89 (3.6) | 16 (2.8) |
| Rubrolide F (6) | 1006 (1.3) | 79 (1.1) | 47 (10.4) | 15 (5.5) |
IC50 values represent the mean of 3 independent experiments, derived using nonlinear regression analysis; SEM = standard error of the mean
EXPERIMENTAL SECTION
General Experimental Procedures
UV spectra were acquired on a JASCO J-815 CD spectrometer. NMR data were acquired in methanol-d4 referenced to residual CH3OH chemical shifts (δC 49.2, δH 3.31) on a Bruker Avance III 700 MHz spectrometer equipped with a 5mm 13C cryogenic probe. HRESIMS data were acquired in positive mode on a Waters Micromass LCT Premier and AB SCIEX Triple TOF 5600. HPLC purifications were performed using a Shimadzu dual LC-20AD solvent delivery system with a Shimadzu SPD-M20A UV/VIS photodiode array detector.
Ascidian Collection
Synoicum globosum Parker-Nance sp. nov. was collected on July 20, 2004 by hand using SCUBA at a depth of 18 m in the White Sands Reef in Algoa Bay, Eastern Cape Province, South Africa (33:59.916S, 25:42.573W). The type specimen (SAF2004-54) for this new ascidian species is housed at the South African Institute for Aquatic Biodiversity (SAIAB), Grahamstown, South Africa.
Ascidian Taxonomy
A descriptive synopsis of the new species Synoicum globosum Parker-Nance sp. nov. (Ascidiacea, Aplousobranchia) relative to known congeners and a detailed morphological description with figures are provided in the Supporting Information. In summary, colonies are gelatinous but firm and consist of egg-shaped heads on a short cone-shaped stalk with irregular basal mass. Externally, the surface of the head is free of sand, although a dense accumulation of fine sand particles is visible in the surface layer of the stalk and the wider irregular basal attachment mass is heavily encrusted with larger sand particles and epifauna. Zooids are red and arranged in circular systems clearly distinguishable against the mustard yellow test material of freshly collected material. The internal test is transparent with fine sand, shell pieces, as well as other foreign material that decreases in density towards the central part of the colony. The slender zooids (8-20 mm) are closely packed together in a distinct layer, and comprise a thorax (1.0-2.7 mm), abdomen (0.9-2.0 mm) and posterior abdomen (6.0-14.8 mm). The branchial opening has six small lobes and the atrial opening is small with a single ribbonlike tongue with small distal teeth. The branchial sac has 11-14 rows of stigmata with 10-11 stigmata in each of the rows. One to two larvae are present in developmental sequence in the atrial cavity. Mature larvae (trunk 0.56-0.66 mm in length and 0.14-0.40 mm in width) have 12 or more lateral ampullae arranged in 2-3 rows with epidermal vesicles present dorsally and ventrally and tail wound about halfway around the trunk.
Extraction and Isolation
The organism was extracted with CH2Cl2-MeOH 2:1 to yield 1.62 g of organic extract that was subjected to reversed phased solid phase extraction (RP18-SPE) using a stepped gradient of 50-100% MeOH in H2O. The 75% MeOH-H2O and 100% MeOH fractions were subsequently separated by RP18-HPLC (Synergi Fusion-RP, Phenomenex, 250 mm × 10 mm, 65 % MeOH-H2O) to yield 3″-bromorubrolide F (1, 3.0 mg), 3′-bromorubrolide E (2, 7.4 mg), 3′-bromo-rubrolide F (3, 2.8 mg) and 3′, 3″-dibromorubrolide E (4, 1.4 mg), rubrolide E (5, 13.5 mg) and rubrolide F (6, 4.3 mg).
Antibacterial Assay
Antibacterial activity was determined using the microtiter dilution method in a 96 well plate format.13 Crude extracts were tested at 125, 12.5, and 1.25 μg/mL against a panel of clinically relevant human pathogens including methicillin-resistant Staphylococcus aureus (ATCC® BAA-1720TM), S. epidermidis (ATCC® 35984™), gentamycin and vancomycin resistant Enterococcus faecalis (ATCC® 700802™), and Escherichia coli (O157:H7)14 to determine the appropriate concentration range for testing of pure rubrolides. Bacterial strains were cultured as follows: MRSA and S. epidermidis in tryptic soy broth (Becton, Dickinson and Co.), E. faecalis in brain heart infusion medium (Becton, Dickinson and Co.), and all Gramnegative bacteria in Luria-Bertani liquid medium (Becton, Dickinson and Co). For all experiments, the cultures were inoculated with 5 × 105 cfu/mL in a final volume of 200 μL in 96-well tissue culture treated microtiter plates (Greiner Bio-One) and incubated without shaking at 37 °C for 16 h, at which point bacterial growth was determined turbidimetrically (OD600) using a BioTek Synergy HT multimode plate reader. Each microtiter plate contained a positive control (chloramphenicol at a final concentration of 30 μg/mL), a negative control (bacteria grown in the presence of DMSO), and a contamination control (medium only). IC50 values for rubrolides 1-6 were determined against MRSA and S. epidermidis as described above, using a concentration range of 130 - 0.3 μg/mL. Additionally, the susceptibility of E. faecalis and E. coli to pure compounds was tested at 100 μg/mL. Test samples were inoculated in technical duplicate on each plate and assays were performed in biological triplicate. Percentage growth inhibition was determined using the plate-based microtiter dilution assay, and IC50 values were derived using nonlinear regression analysis.
3″-Bromorubrolide F (1)
Yellow amorphous solid; UV (MeOH) λmax (log ε) 248 (3.8), 356 (4.0) nm; HRESIMS [M+H]+ (50%) m/z 373.0081, (calcd for C18H14O479Br, 373.0075), [M+H]+ (50%) m/z 375.0070, (calcd for C18H14O481Br, 375.0055); 1H and 13C NMR data (Table 1); COSY, HMBC, NOESY data (Table S1).
3′-Bromorubrolide E (2)
Yellow amorphous solid; UV (MeOH) λmax (log ε) 250 (4.1), 371 (4.3) nm; HRESIMS [M+H]+ (50%) m/z 358.9933, (calcd for C17H12O479Br, 358.9919), [M+H]+ (50%) m/z 360.9916, (calcd for C17H12O481Br, 360.9899); 1H and 13C NMR data (Table 1); COSY, HMBC, NOESY data (Table S2).
3′-Bromorubrolide F (3)
Yellow amorphous solid; UV (MeOH) λmax (log ε) 248 (3.8), 365 (3.9) nm; HRESIMS [M+H]+ (50%) m/z 373.0060, (calcd for C18H14O479Br, 373.0051), [M+H]+ (50%) m/z 375.0048, (calcd for C18H14O481Br, 375.0055); 1H and 13C NMR data (Table 1); COSY, HMBC, NOESY data (Table S3).
3′, 3”-Dibromorubrolide E (4)
Yellow amorphous solid; UV (MeOH) λmax (log ε) 254 (4.0), 366 (4.1), 444 (3.6) nm; HRESIMS [M−H]− (25%) m/z 434.8872, (calcd for C17H9O479Br81Br, 434.8868), [M−H]− (50%) m/z 436.8853, (calcd for C17H9O479Br81Br, 436.8847), [M−H]− (25%) m/z 438.8835, (calcd for C17H9O481Br2, 438.8827); 1H and 13C NMR data (Table 1); COSY, HMBC, NOESY data (Table S4).
Rubrolide E (5)
Yellow amorphous solid; UV (MeOH) λmax (log ε) 226 (3.9), 247 (3.9), 368 (4.0) nm; HRESIMS [M+H]+ m/z 281.0821, (calcd for C17H13O4, 281.0814); 1H NMR (400 MHz, CD3OD) 7.69 (2H, d, J = 8.8 Hz, H-2″, H-6″), 7.44 (2H, d, J = 8.6 Hz, H-2′, H-6′), 6.91 (2H, d, J = 8.5 Hz, H-3′, H-5′) 6.81 (2H, d, J = 8.8 Hz, H-2″, H-6″), 6.30 (1H, s, H-5), 6.11 (1H, s, H-2); 13C NMR (100 MHz, methanol-d4) 171.5 (C, C-1), 161.0 (C, C-3), 159.7 (C, C-4′, C-4″), 145.0 (C, C-4), 133.7 (CH, C-2″, C-6″), 131.2 (CH, C-2′, C-6′), 126.5 (CH, C-3″, C-5″), 125.0 (C, C-1″), 120.6 (C, C-1′), 117.2 (CH, C-3′, C-5′), 115.9 (CH, C-5), 110.7 (CH, C-2), see Table S5 for 2D data and literature for CDCl315 and DMSO-d36 NMR data.
Rubrolide F (6)
Yellow amorphous solid; UV (MeOH) λmax (log ε) 248 (3.8), 356 (4.0) nm; HRESIMS [M+H]+ m/z 295.0985, (calcd for C18H15O4, 295.0970); 1H and 13C NMR data (Table S6).
Supplementary Material
ACKNOWLEDGMENTS
We thank Professor Bill Fenical of Scripps Institution of Oceanography, San Diego, USA for the generous donation of funding that enabled the first large-scale SCUBA collection of marine ascidians in Algoa Bay, South Africa. We are also grateful to Jeff Morré of the Environmental Health Sciences Center at OSU for MS data acquisition (NIEHS P30 ES00210). The National Science Foundation (CHE-0722319) and the Murdock Charitable Trust (2005265) are acknowledged for their support of the OSU Natural Products and Small Molecule Nuclear Magnetic Resonance Facility. Additional funding was provided by the OSU College of Pharmacy.
Footnotes
Supporting Information. Tabulated 1D and 2D NMR data and spectra for rubrolides 1-6 in methanol-d4. Synopsis of the genus Synoicum and detailed morphological description of the new species Synoicum globosum Parker-Nance sp. nov. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
REFERENCES
- (1).Kong D-X, Jiang Y-Y, Zhang H-Y. Drug Discov. Today. 2010;15:884–886. doi: 10.1016/j.drudis.2010.09.002. [DOI] [PubMed] [Google Scholar]
- (2).Montaser R, Luesch H. Future Med. Chem. 2011;3:1475–1489. doi: 10.4155/fmc.11.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Miao S, Andersen RJ. J. Org. Chem. 1991;56:6275–6280. [Google Scholar]
- (4).Ortega MJ, Zubía E, Ocaña JM, Naranjo S, Salvá J. Tetrahedron. 2000;56:3963–3967. [Google Scholar]
- (5).Pearce AN, Chia EW, Berridge MV, Maas EW, Page MJ, Webb VL, Harper JL, Copp BR. J. Nat. Prod. 2006;70:111–113. doi: 10.1021/np060188l. [DOI] [PubMed] [Google Scholar]
- (6).Bellina F, Anselmi C, Viel S, Mannina L, Rossi R. Tetrahedron. 2001;57:9997–10007. [Google Scholar]
- (7).Bellina F, Anselmi C, Martina F, Rossi R. Eur. J. Org. Chem. 2003;12:2290–2302. [Google Scholar]
- (8).Manzanaro S, Salvá J, de la Fuente JÁ. J. Nat. Prod. 2006;69:1485–1487. doi: 10.1021/np0503698. [DOI] [PubMed] [Google Scholar]
- (9).Boukouvalas J, McCann LC. Tetrahedron Lett. 2010;51:4636–4639. [Google Scholar]
- (10).Pearce AN, Chia EW, Berridge MV, Maas EW, Page MJ, Webb VL, Harper JL, Copp BR. J. Nat. Prod. 2006;70:111–113. doi: 10.1021/np060188l. [DOI] [PubMed] [Google Scholar]
- (11).Caufield CE, Antane SA, Morris KM, Naughton SM, Quagliato DA, Andrae PM, Enos A, Chiarello JF. In: A61K 31/341. Office USP, editor. Wyeth, Madison, NJ (US): USA: 2009. p. 35. [Google Scholar]
- (12).Sikorska J, Hau AM, Anklin C, Parker-Nance S, Davies-Coleman MT, Ishmael JE, McPhail KL. J. Org. Chem. 2012;77:6066–6075. doi: 10.1021/jo3008622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Smith SM, Beattie AJ, Gillings MR, Holley MP, Stow AJ, Turnbull CL, Wilson PD, Briscoe DA. J. Microbiol. Methods. 2008;72:103–106. doi: 10.1016/j.mimet.2007.10.003. [DOI] [PubMed] [Google Scholar]
- (14).Sikora AE, Beyhan S, Bagdasarian M, Yildiz FH, Sandkvist M. J. Bacteriol. 2009;191:5398–5408. doi: 10.1128/JB.00092-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Tale NP, Shelke AV, Tiwari GB, Thorat PB, Karade NN. Helv. Chim. Acta. 2012;95:852–857. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


