Summary
New materials that can bind and deliver oligonucleotides such as short interfering RNA (siRNA) without toxicity are greatly needed to fulfill the promise of therapeutic gene silencing. Amphiphilic macromolecules (AMs) were functionalized with linear ethyleneimines to create cationic AMs capable of complexing with siRNA. Structurally, the parent AM is formed from a mucic acid backbone whose tetra-hydroxy groups are alkylated with 12-carbon aliphatic chains to form the hydrophobic component of the macromolecule. This alkylated mucic acid is then mono-functionalized with poly(ethylene glycol) (PEG) as a hydrophilic component. The resulting AM contains a free carboxylic acid within the hydrophobic domain. In this work, linear ethyleneimines were conjugated to the free carboxylic acid to produce an AM with one primary amine (1N) or one primary amine and four secondary amines (5N). Further, an AM with amine substitution both to the free carboxylic acid in the hydrophobic domain and also to the adjacent PEG was synthesized to produce a polymer with one primary amine and eight secondary amines (9N), four located on each side of the AM hydrophobic domain. All amine-functionalized AMs formed nanoscale micelles but only the 5N and 9N AMs had cationic zeta potentials, which increased with increasing number of amines. All AMs exhibited less inherent cytotoxicity than linear polyethyleneimine (L-PEI) at concentrations of 10 µM and above. By increasing the length of the cationic ethyleneimine chain and the total number of amines, successful siRNA complexation and cellular siRNA delivery was achieved in a malignant glioma cell line. In addition, siRNA-induced silencing of firefly luciferase was observed using complexes of siRNA with the 9N AM and comparable to L-PEI, yet showed better cell viability at higher concentrations (above 10 µM). This work highlights the promise of cationic AMs as safe and efficient synthetic vectors for siRNA delivery. Specifically, a novel polymer (9N) was identified for efficient siRNA delivery to cancer cells and will be further evaluated.
Keywords: amphiphiles, gene delivery, nanotechnology, siRNA
Introduction
The use of short interfering RNA (siRNA) molecules for gene silencing has enormous clinical potential for treating human disease, particularly for anticancer applications.[1, 2] Recent advances in siRNA delivery technology have led to the initiation of several human clinical trials using therapeutic siRNA.[3, 4] However, further development of safe, efficient siRNA delivery systems is required to advance siRNA therapeutics for routine clinical use and address diverse disease states. The delivery of siRNA and other nucleic acid molecules to malignant cells has been attempted, for example, with varying degrees of success with numerous non-viral molecules including proteins, peptides, and synthetic polymers.[5]
Self-assembled polymeric micelles have shown particular promise as drug and gene delivery vehicles due to their unique properties including steric stability, size suitable for passive tumor targeting, low cytotoxicity, high water solubility, and high drug encapsulation efficiency.[6–8] Polymeric micelle systems are currently being investigated as drug delivery vehicles in several Phase I and II clinical trials in the United States,[9–11] and are being evaluated in Phase III and IV studies internationally.[12, 13] More recently, several polymeric micelle systems have been evaluated for siRNA delivery,[9, 10, 14, 15] or for the co-delivery of siRNA and hydrophobic anticancer drugs.[16, 17] However, further development is needed for a delivery system that possesses increased complex stability, lowered toxicity, biodegradability, as well as the versatility for treating multiple disease states and ease of modification (e.g., targeting moieties) to increase specificity. Specifically, for polymeric micelles, improvements on existing systems are necessary to improve their drug loading capacity, stability in the blood stream, and ability to penetrate the cell membrane to make these systems viable for widespread clinical use.[18] [16]
Nanoscale amphiphilic macromolecules (AMs) are novel, polymeric micelles developed by Uhrich and colleagues for treatment of cardiovascular disease and drug delivery.[19–28] The unimers are composed of a branched hydrophobic component formed by the tetra-alkylation of mucic acid, a biocompatible sugar, which is further derivatized with linear, hydrophilic poly(ethylene glycol) (PEG) – all of which are linked via biodegradable bonds. In aqueous media, the unimers self-assemble to form nano-sized micelles at concentrations as low at 100 nM,[27] making them at least as stable as other polymeric micellar systems with CMC values on the order of 10−6 M.[29] Further, the polymers are biocompatible and capable of effectively delivering hydrophobic drugs intracellularly.[22–24, 26, 30]
AMs are attractive for multiple applications due to their facile tunability, with multiple means of synthetic modification on both the hydrophilic and hydrophobic portions of the unimer. In this work, the hydrophobic functionality was exploited to create non-viral vectors for siRNA delivery. Specifically, linear, cationic ethyleneimine groups were conjugated to the unimer’s hydrophobic backbone to facilitate electrostatic encapsulation and subsequent delivery of siRNA to malignant glioma cells. Ethyleneimines were chosen due to their similarity to the highly efficient non-viral vector, polyethyleneimine (PEI). However, PEI suffers from high cytotoxicity limiting its use for systemic in vivo applications where high polymer concentrations are required.[18, 31] The minimum number of amine groups necessary to efficiently deliver siRNA and elicit a gene-silencing response in malignant glioma cells, while maintaining the favorable structural properties and low cytotoxicity of the AM materials, was identified in this work. This proof-of-concept study outlines the rational design approach to siRNA delivery systems and identifies a promising new siRNA delivery system.
Experimental Part
Synthetic Materials
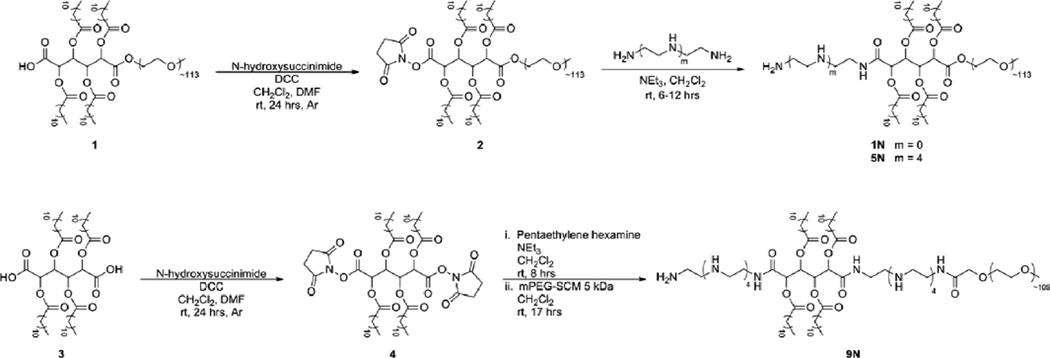
Unless otherwise stated, solvents and reagents were purchased from Fisher Scientific (Pittsburgh, PA) and Sigma-Aldrich (St. Louis, MO) and used as received. Poly(ethylene glycol) 5 kDa was purchased from Polysciences, Inc. (Warrington, PA) and dried by azeotropic distillation from toluene before use. N-hydroxysuccinimide(NHS)-functionalized PEG, Methoxy-PEG-succinimidyl carboxymethyl (MW 5 kDa) (mPEG-SCM). was purchased from Laysan Bio, Inc (Arab, AL) and used as received. 1,[26] 2,[22] and 3 [26] (Scheme 1) ere prepared as previously described.
Scheme 1.
Synthesis of cationic-AMs; (top) synthesis of 1N and 5N from NHS-activation of 1 to yield 2 [21], (bottom) synthesis of 9N via di-activation of 3 with NHS to yield 4.
Polymer Characterization Methods
Proton nuclear magnetic resonance (1H-NMR) spectra of the products were obtained using a Varian 400 MHz or 500 MHz spectrophotometer. Samples were dissolved in chloroform-d, with a few drops of dimethyl sulfoxide-d6 if necessary, with tetramethylsilane as an internal reference. Molecular weights (Mw) and polydispersity indices (PDI) were determined using gel permeation chromatography (GPC) with respect to PEG (Sigma-Aldrich) on a Waters Stryagel® HR 3 THF column (7.8 × 300 mm). The Waters LC system (Milford, MA) was equipped with a 2414 refractive index detector, a 1515 isocratic HPLC pump, and 717plus autosampler. An IBM ThinkCentre computer with Waters Breeze Version 3.30 software installed was used for collection and processing of data. Samples were prepared at a concentration of 10 mg/mL in tetrahydrofuran, filtered using 0.45 µm pore size nylon or polytetrafluoroethylene syringe filters (Fisher Scientific) and placed in sample vials to be injected into the system. Melting points were determined by differential scanning calorimetry (DSC) on a TA DSC Q200. TA Universal Analysis 2000 software was used for data collection on a Dell Dimension 3000 computer. Samples (3–5 mg) were heated under dry nitrogen gas. Data were collected at heating and cooling rates of 10 °C /min with a two-cycle minimum.
Polymer Synthesis
1N: Ethylenediamine (50 µL, 0.75 mmol) was dissolved in HPLC-grade CH2Cl2 (3 mL) and triethylamine (0.15 mL, 1.1 mmol). In a separate vessel, 2 (0.51 g, 0.085 mmol) was dissolved in HPLC-grade CH2Cl2 (9 mL) and subsequently added to the solution of ethylenediamine dropwise via syringe pump at a rate of 1.0 mL/hr. The reaction was stirred overnight (~ 18 hrs). The reaction solution was then diluted with CH2Cl2 and subsequently washed with 0.1 N HCl/brine (1×) and brine (2×). The combined aqueous portions were extracted with CH2Cl2 and the combined organics dried over MgSO4, and concentrated to a yellow oil. The desired product was precipitated from the oil dissolved in CH2Cl2 (5 mL) by addition of 10-fold diethyl ether and cooling over dry ice for 1 hr. The solid was then collected by centrifugation at 3000 rpm for 5 min and the supernatant removed by decanting. The resulting white solid was dried under ambient atmosphere (12 hrs) and under high vacuum (12 hrs). Yield: 0.41 g, 80 %. 1H-NMR (CDCl3): δ 5.67 (m, 2H, CH), 5.14 (m, 2H, CH), 4.24 (m, 3H, CH2), 3.60 (m, ~0.45 kH, CH2O), 3.37 (s, 3H, OCH3), 2.37 (m, 8H, CH2), 2.29 (m, 4H, CH2), 1.81 (b, 4H, CH2), 1.60 (m, 8H, CH2), 1.26 (m, 64H, CH2), 0.87 (t, 12H, CH3). Tm = 58 °C GPC: Mw: 6.3 kDa; PDI: 1.1.
5N: Pentaethylenehexamine (0.15 mL, 0.64 mmol) was dissolved in HPLC-grade CH2Cl2 (10 mL) and triethylamine (0.33 mL, 2.4 mmol). In a separate vessel, 2 (0.48 g, 0.079 mmol) was dissolved in HPLC-grade CH2Cl2 (10 mL) and subsequently added to the solution of ethylenediamine dropwise via syringe pump at a rate of 1.0 mL/hr. The reaction was stirred overnight (~ 17 hrs). The bright yellow reaction solution was diluted with CH2Cl2 and subsequently washed with 0.1 N HCl/brine (1×) and brine (2×). The combined aqueous portions were extracted with CH2Cl2 and the combined organics dried over MgSO4, and concentrated to a cloudy yellow oil. The desired product was precipitated from the oil dissolved in CH2Cl2 (5 mL) by addition of 10-fold diethyl ether and cooling over dry ice for 1 hr. The solid was then collected by centrifugation at 3000 rpm for 5 min and the supernatant removed by decanting. The resulting white solid was dried under ambient atmosphere (12 hrs) and under high vacuum (12 hrs). Yield: 0.42 g, 86 %. 1H-NMR (DMSO): δ 5.50 (m, 2H, CH), 5.11 (m, 2H, CH), 3.41 (m, ~0.45 kH, CH2O), 3.24 (s, 3H, OCH3), 2.89 (m, 13 H, CH2), 2.80 (bs, 2H, CH2), 2.76 (bs, 7H, CH2), 2.64 (bs, 6H, CH2), 1.49 (m, 8H, CH2), 1.24 (m, 64H, CH2), 0.84 (t, 12H, CH3). Tm = 59 °C. GPC: Mw: 6.4 kDa; PDI: 1.1.
4: Product 3 (5.10 g, 5.43 mmol) and NHS (5.38 g, 46.8 mmol) were dissolved in anhydrous CH2Cl2 (100 mL) and anhydrous DMF (18 mL) under argon. Once a clear solution was obtained, N,N’-dicyclohexylcarbodiimide (17 mL, 17 mmol) was added and the reaction stirred at room temp under argon for 24 hours. The resulting solution with white suspension was stored at −4 °C overnight. The dicyclohexyl urea (DCU) byproduct was then removed by vacuum filtration and the filtrate washed with 0.1 N HCl and 50:50 brine/H2O, dried over MgSO4, and concentrated. The resulting white solid was then dissolved in a small amount of CH2Cl2 (5–10 mL) and stored at −4 °C for 2–3 hours. The resulting white suspension was filtered to remove residual DCU. The filtrate was then concentrated to dryness and the white solid dried under high vacuum overnight. Yield = 4.5 g, 73 %. 1H-NMR (CDCl3): δ = 5.96 (s, 2H, CH), 5.57 (s, 1H, CH), 2.81 (s, 8H, CH2), 2.49 (m, 6H, CH2), 2.37 (m, 2H, CH2), 1.64 (m, 8H, CH2), 1.27 (m, 64H, CH2), 0.89 (t, 12H, CH3).
9N: Pentaethylenehexamine (0.05 mL, 0.2 mmol) was dissolved in CH2Cl2 (3 mL) and triethylamine (0.15 mL, 1.1 mmol). In a separate vessel, 4 (0.10 g, 0.090 mmol) was dissolved in HPLC-grade CH2Cl2 (3 mL) and subsequently added to the solution of ethylenediamine dropwise via syringe pump at a rate of 1.0 mL/hr. The reaction was stirred at room temperature a total of 8 hrs. mPEG-SCM (0.45 g, 0.090 mmol) dissolved in CH2Cl2 (7 mL) was then added to the yellow reaction solution dropwise via syringe pump at a rate of 1.0 mL/hr. The reaction was stirred at room temperature overnight (~ 17 hrs). The solvent was then removed from the reaction solution by rotary evaporation. The oil/solid was then redispersed in CH2Cl2 and filtered to remove the solid NHS-byproduct. The filtrate was concentrated to an oil and product precipitated from the oil dissolved in CH2Cl2 (5 mL) by addition of 10-fold diethyl ether. The solid was then collected by centrifugation at 3000 rpm for 5 min and the supernatant removed by decanting. The resulting white solid was washed with diethyl ether (1×) and dried under ambient atmosphere (12 hrs) and under high vacuum (12 hrs). Yield: 0.45 g, 87 %. 1H-NMR (CDCl3): δ 7.26 (s, 4H, CH), 3.69 (m, ~0.44 kH, CH2O), 3.38 (s, 3H, OCH3), 3.05 (bm, 15H, CH2), 2.55 (bm, 16H, CH2), 2.07 (bm, 40H, CH2), 1.65 (bs, 7H, CH2), 1.48 (t, 5H, CH2), 1.26 (m, 37H, CH2), 0.88 (t, 12H, CH3). Tm = 59 °C. GPC: Mw: 5.5 kDa; PDI = 1.1.
Size and Zeta Potential of AMs and AM/siRNA complexes
Dynamic light scattering (DLS) and zeta potential analyses were performed using a Malvern Instruments Zetasizer Nano ZS-90 instrument (Southboro, MA). DLS measurements were performed at a 90° scattering angle at 25°C. Size distributions by volume of measurements were collected in triplicate, averaged and reported. Zeta potential measurements were collected in triplicate, averaged and the Z-average charges reported. For all measurements, error bars represent peak widths of the average value.
Sample Preparation
AMs alone: Polymer solutions at a concentration of 1.0 mg/mL were prepared using picopure water and filtered with a 0.45 µM Nylon syringe filter (Fischer Scientific, Pittsburgh, PA).
AM/siRNA complexes: Complexes were prepared in picopure water at various nitrogen/phosphate (N/P) ratios. For size and zeta potential measurements, 2mL of solutions containing AM/siRNA complexes were prepared at polymer concentrations sufficient for detection by the zetasizer instrument (1 mg/mL for 1 and 1N, and 2 mg/mL for 5N and 9N). Solutions were briefly vortexed and incubated for at least 60 min at room temperature to allow for complex formation prior to size and zeta potential analysis.
Gel Electrophoresis
Polymer/siRNA (Dharmacon, Lafayette, CO) complexes were first prepared at the desired nitrogen to phosphorous (N/P) ratios by mixing solutions of polymers (stocks maintained in DI water) and siRNA in PBS (final siRNA concentration of 12.5 µg/mL). Since polymer 1 does not contain primary amines, the mass of polymer 1 added for the gel electrophoresis experiments was equivalent to the mass of polymer 1N added at the N/P ratios indicated in Figure 2. Solutions were briefly vortexed, and incubated for 60 min at room temperature to allow for complex formation. Polymer/siRNA complexes were loaded into 1% agarose gels run in an electrophoresis chamber at 70 V for 40 minutes. Following electrophoresis, gels were stained with SYBR Green II RNA gel stain (Invitrogen, Carlsbad, CA) for 30 minutes prior to imaging on a Bio-Rad Molecular Imager FX (Bio-Rad Laboratories, Hercules, CA) to visualize unbound siRNA. The fluorescence intensities of bands were quantified using Quantity One Quantitation software (Bio-Rad Laboratories, Hercules, CA).
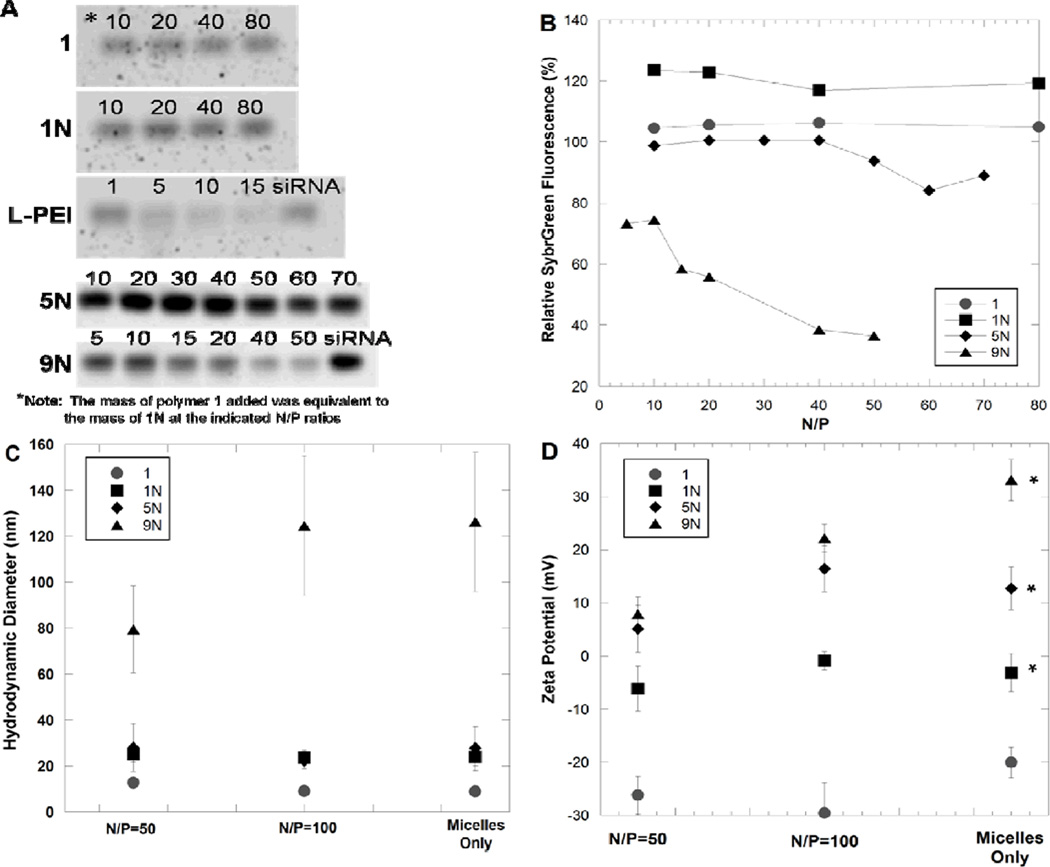
Figure 2.
Characterization of AM/siRNA complexes by gel electrophoresis (A and B), dynamic light scattering (C), and zeta potential (D). Relative SybrGreen fluorescence corresponds to unbound siRNA detected in an electrophoresis band normalized by a band of free siRNA (B). Images of gel electrophoresis bands are shown with the N/P ratio for each band denoted above the band, where the mass of polymer 1 added was equivalent to 1N at the indicated N/P ratios (A). Gel electrophoresis siRNA complexation studies were performed at least three times for each polymer, and one representative gel image and band quantification is shown here. The zeta potential measurements of each polymer were compared to each other at varying N/P ratios, and asterisks in D indicate polymers whose zeta potentials differed as a function of N/P ratio (p<0.05).
Cell Culture
All cell culture products were obtained from Invitrogen (Carlsbad, CA). U87 MG cells (ATCC HTB-14) were maintained in DMEM medium supplemented with 10% fetal bovine serum (FBS), L-glutamine, and penicillin-streptomycin. A U87 cell line containing a stably integrated destabilized EGFP (d1EGFP) transgene (U87-GFP) was generated as described previously,[33] and was maintained under constant selective pressure by G418 (500 µg/mL), and the growth medium was supplemented with sodium pyruvate and nonessential amino acids. U87-Luc, a human glioblastoma cell line with constitutive expression of firefly luciferase, was generously provided by Dr. Xu-Li Wang (Department of Pharmaceutics and Pharmaceutical Chemistry, University of Utah). U87-Luc cells were maintained in minimal essential medium supplemented with 10% FBS, penicillin-streptomycin, and maintained under selective pressure by G418.
Cytotoxicity Assay
U87 glioma cells were seeded into 96 well plates (Corning, Corning, NY) at 10,000 cells per well in DMEM supplemented with 10% FBS and 1 % penicillin-streptomycin and incubated overnight at 37 °C, with 5 % CO2. The media was removed by aspiration and replaced with 200 µL aminated-AM or PEI dissolved in media at desired concentrations (n=4 per condition). Untreated control wells received media only. After 72 hours, cells were harvested by trypsinization (75 µL trypsin-EDTA followed with 75 µL complete media to neutralize trypsin) and 50 µL of staining solution (48:1:1 media:DMSO:Guava ViaCount Flex reagent (Guava Technologies, Hayward, CA) was added to each well. Cells were counted using a Guava EasyCyte Plus (Guava Technologies, Haywood CA) instrument with an original volume of 0.2 mL and a dilution factor of one.
siRNA Delivery Assay
U87-Luc cells were plated at a density of 5000 cells/well in 96-well plates approximately 20 hours prior to transfection. Immediately prior to transfection, polymer/siRNA complexes were prepared in 20 µL of PBS (N/P=50 for the AMs, and N/P=15 for linear PEI). Linear polyethyleneimine (Polysciences, Inc., Warrington, PA) commonly used polymeric transfection reagent, was used as a positive control. An irrelevant siRNA sequence not targeted against firefly luciferase was delivered as a negative control. The polyplexes were brought to a total volume of 100 µL in OptiMEM medium to obtain a final siRNA concentration of 100 nM. The serum-containing culture medium was aspirated from the cells, and each well treated with 100 µL of the polyplexes in OptiMEM medium. Each treatment was performed in triplicate. After a 4 hr incubation period, the transfection mixture was replaced with serum-containing growth medium and maintained under normal growth conditions until the cells were assayed for firefly luciferase expression 24 hours after the initial treatment.
For fluorescence imaging, a similar transfection protocol was performed on U87-GFP cells seeded onto an 8-well LabTek coverglass chamber (Nalge Nunc, Naperville, IL) at a density of 5000 cells/well. U87-GFP cells were treated with a Cy5-labeled siRNA to facilitate imaging of cellular localization of siRNA.
Fluorescence Microscopy
Uptake of a fluorescently labeled siRNA (Dharmacon, Lafayette, CO) sequence into U87-GFP cells was evaluated using fluorescence microscopy. Imaging was performed 24 hours after siRNA transfection using an Olympus IX81 model fluorescent microscope (Olympus, Center Valley, PA). Imaging was performed at 20X magnification. The following excitation and emission wavelengths were used: GFP (excitation=482 nm, emission=536 nm) and Cy5 siRNA (excitation=628 nm, emission= 692 nm).
Luciferase Detection Assay
Cells were prepared for firefly luciferase detection using the Luciferase Assay System (Promega, Madison, WI) according to the manufacturer’s protocol. Firefly luciferase was quantified using The Reporter microplate luminometer (Turner Biosystems, Sunnyvale, CA). Following luciferase quantification, cell lysates were assayed for total protein content using the BCA Protein Assay kit (Pierce, Rockford, IL) according to the manufacturer’s protocol.
Statistics
Statistical comparisons for zeta potential measurements, luciferase silencing and polymer cytotoxicity were performed using a one-way ANOVA test with a Fisher’s all-pairs post hoc comparison test.
Results and Discussion
The goal of this study was to create novel synthetic vectors that exploit the structural properties of PEI beneficial for siRNA delivery while reducing the inherent cytotoxicity associated with PEI. AMs were modified with two different lengths of ethyleneimine chains to yield three novel polymer systems: ethylenediamine to yield 1N, or pentaethylenehexamine to yield 5N and 9N polymers. The polymers were synthesized as shown in Scheme 1 from the amine-specific N-hydroxysuccinimide (NHS)-activated polymer 1, which has been the focus of a previous publication.[23][21]
The parent compound, 3, served as the basic building block for the polymer modifications. Specifically, the carboxylic acid on the mucic acid backbone was activated with N-hydroxysuccinimide to functionalize the polymers with linear ethyleneimines, systematically increasing the total number of amines in the final polymers from one, 1N, up to nine, 9N. For 1N and 5N, an excess of the diamines coupled with their slow addition to the polymer solution via syringe pump were utilized to control for the disubstitution of polymer to both primary amines. For 9N, a 2:1 molar ratio of compound 4 to pentaethylene hexamine coupled with the slow addition of 4 to the diamine via syringe pump were used to limit the formation of undesired oligomers. Subsequently, a 1:1 molar ratio of the NHS-PEG with respect to 4 coupled with its slow addition to the diaminated 4 via syringe pump were utilized to limit the coupling of PEG to both sides of the 4. For all aminated polymers, Isolation of cationic AMs with amines conjugated to, rather than associated with, the polymer was insured by precipitation from diethyl ether; this process precipitates the AM products but not the ethyleneimine starting materials. Amine conjugation was further verified by 1H NMR spectroscopy. In addition to monitoring 1H NMR spectra for the disappearance of protons associated with the NHS activating group (~ 2.8 ppm), new peaks assigned to the ethyleneamine protons were observed resonating at 1.3 and 1.8 ppm for 1N and from 2.5–3.0 ppm for 5N and 9N. For all cationic AMs, the integrations of the 1H NMR are consistent with mono PEG substitution to produce the desired, cationic AMs. The molecular weights of the cationic AMs were determined by GPC relative to PEG standards. As shown in Table 1, 1N and 5N have similar molecular weights while the molecular weight of 9N is approximately 1 kDa less. This difference can be attributed to the use of different PEG starting materials from different vendors with varying peak molecular weights. In addition, due to the incorporation of amines between the hydrophobic and hydrophilic component in the structure of 9N, the polymer may associate more with the column than the other cationic polymers, thereby making the molecular weight appear lower than it actually is. For all cationic AMs, the absence of a high molecular weight peak in the GPC corresponding to ~ 10 kDa suggests there was little-to-no PEG di-substitution in any of the resulting polymers.
Table 1.
Molecular weights, poydispersity indices (PDI), and melting temperature (Tm) ethyleneimine-modified AMs.
| Cationic AM | MW (kDa) | PDI | Tm (° C) |
|---|---|---|---|
| 1N | 6.3 | 1.1 | 58 |
| 5N | 6.4 | 1.1 | 59 |
| 9N | 5.5 | 1.1 | 59 |
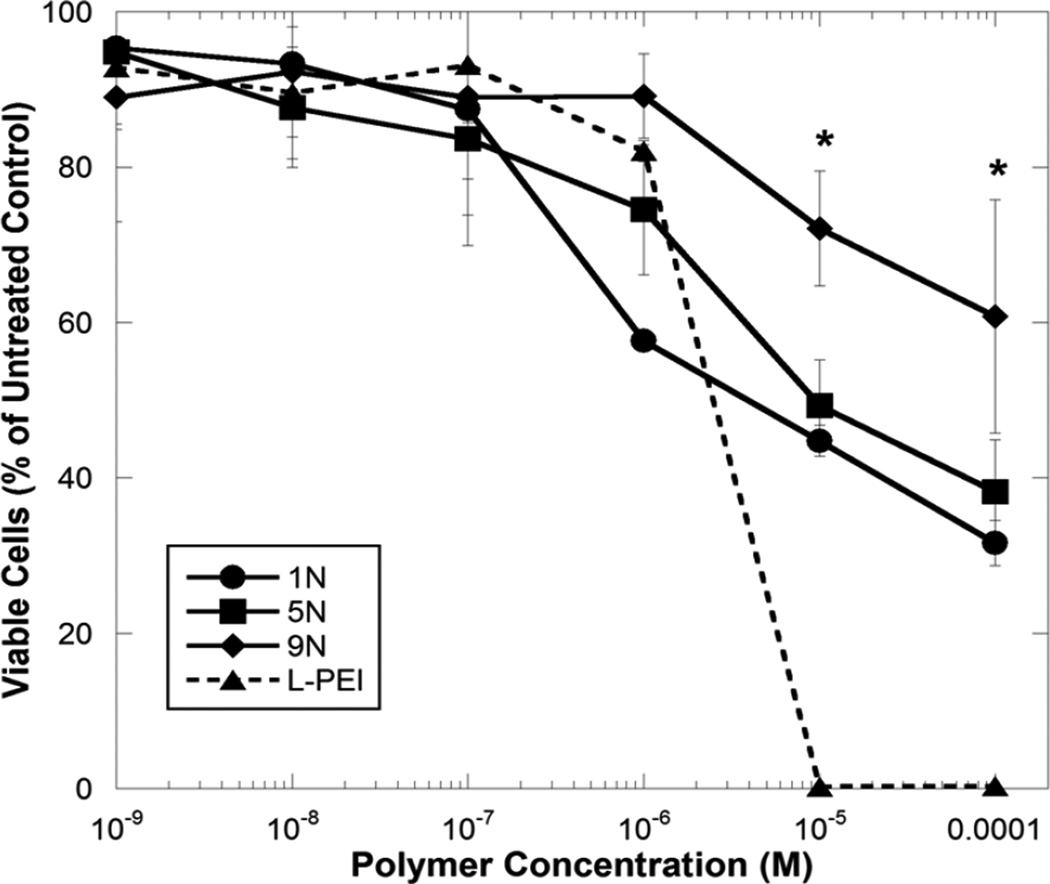
The toxicity of the aminated-AMs compared to linear PEI 25 kDa (L-PEI) was assessed in U87 glioma cells. A dose response curve was generated for all samples by counting viable cells remaining after a 72-hour exposure to the polymers (Figure 1). A significant decrease in cytotoxicity (p<0.05) was observed for all AMs compared to L-PEI at the highest concentrations tested (10−5 and 10−4 M). Interestingly, when comparing the AMs, the 9N material exhibited the lowest cytotoxicity compared to the AMs containing fewer amine groups. One explanation for this observation is that the surface charge density of the 9N micelles is lower as their hydrodynamic diameter (shown in Figure 2c) has increased by a factor of approximately five and, therefore, their area is increased by a factor of about 25 compared to the 1N and 5N micelles.
Figure 1.
Cytotoxicity of cationic-AMs and L-PEI to U87 glioma cells after 72 hours of exposure. Data represent mean ± standard deviation (n=4). Astericks represent concentrations at which cationic-AMs elicited a significantly lower cytotoxicity than L-PEI (p < 0.05).
The ability of cationic AMs to complex anionic siRNA was evaluated using gel electrophoresis. Complexes were formed at a range of N/P ratios and run on an electrophoresis gel to separate un-complexed siRNA from the AM/siRNA complexes. The decrease in fluorescence intensity of the band corresponding to un-complexed siRNA verifies the siRNA complexation efficiency of the cationic AMs. Polymers containing zero or one cationic amine group (i.e., 1 and 1N) displayed no complexation of siRNA at charge ratios tested up to nitrogen/phosphorus (N/P) 80 (Figure 2b). By increasing the number of amine groups to five (5N), a modest extent of siRNA complexation (approximately 20%) was observed at N/P ratios of 60 and higher. Significantly improved siRNA complexation efficiency was observed by using the AM containing nine amine groups, 9N, where most of the siRNA was encapsulated by N/P=50. The ability of AMs to complex siRNA was compared to L-PEI, where nearly complete siRNA complexation was observed by gel electrophoresis at N/P≥15. Hence, for subsequent physical and biological characterization studies, AM/siRNA complexes were formed at N/P≥50.
All cationic AMs formed micelles in the nanoscale size range as determined by dynamic light scattering (Figure 2c). 1N and 5N formed micelles of approximately the same size as polymer, 1, while micelles formed from 9N were much larger (~125 nm), presumably due to charge repulsion of the highly cationic ethyleneimine units. Once complexed with siRNA (Figure 2c, N/P=50 and 100), all cationic AMs maintained the nanoscale size of the AMs alone. Self-assembled polymeric micelles are known to have stable sizes that are dictated primarily by the architecture of the amphiphilic polymer segments[6]. Especially at high N/P ratios, the size of polymer micelles often remains unchanged in the presence of nucleic acids as the presence of relatively small amounts of siRNA does not change the properties of the stable polymer micelles[6, 14]. Maintaining sizes of less than ~100 nm is desirable for improved circulation time, passive tumor targeting by the enhanced permeation and retention (EPR) effect, and optimal cellular uptake.[34, 35]
Successful conjugation of the amines was shown by the disappearance of N-hydroxysuccinimide in the 1H NMR as well as the increase in the zeta potential from negative (for polymer 1), to less negative (polymer 1N), and positive (polymers 5N and 9N), as shown in Figure 2d (micelles only). The zeta potential increased with increasing ethyleneimine length, further indicating the successful incorporation of amine groups. When siRNA was complexed with the cationic AMs at N/P ratios of 50 and 100, the zeta potentials for the aminated polymers 5N and 9N significantly changed compared to the native polymer in the absence of siRNA (p<0.05). Specifically, at N/P 50, the zeta potentials for complexes of siRNA and 5N decreased from 12.7 mV of the 5N alone to 5.3 mV when complexed with siRNA. Likewise, the zeta potential of 9N decreased from 33.1 mV alone to 7.89 mV when complexed with siRNA. The zeta potentials for both AMs increased at N/P 100 – back to that for the vehicle alone for 5N but only to 22.2 mV for 9N. This data suggests that 9N complexed most efficiently with siRNA at the N/P ratios evaluated in this study, as the decrease in zeta potential is a result of charge neutralization when the negatively charged siRNA complexes with the cationic AMs. These results are in agreement with the gel electrophoresis data. Based on the physical characterization of AM/siRNA complexes by gel electrophoresis, dynamic light scattering, and zeta potential, 9N was expected to be the most effective siRNA delivery vehicle.
The ability of AMs to facilitate cellular delivery of siRNA and elicit silencing of the reporter gene, firefly luciferase, in U87 cells was evaluated. Polyplexes of AMs and anti-luciferase siRNA were formed (siRNA concentration: 100 nM, N/P=50, AM concentration: ~10−5 M) and delivered to U87-Luc cells which were subsequently assayed for luciferase expression. To visually evaluate the cellular uptake of siRNA, a fluorescently labeled siRNA sequence was delivered separately to U87-GFP cells which were then imaged using fluorescent microscopy.
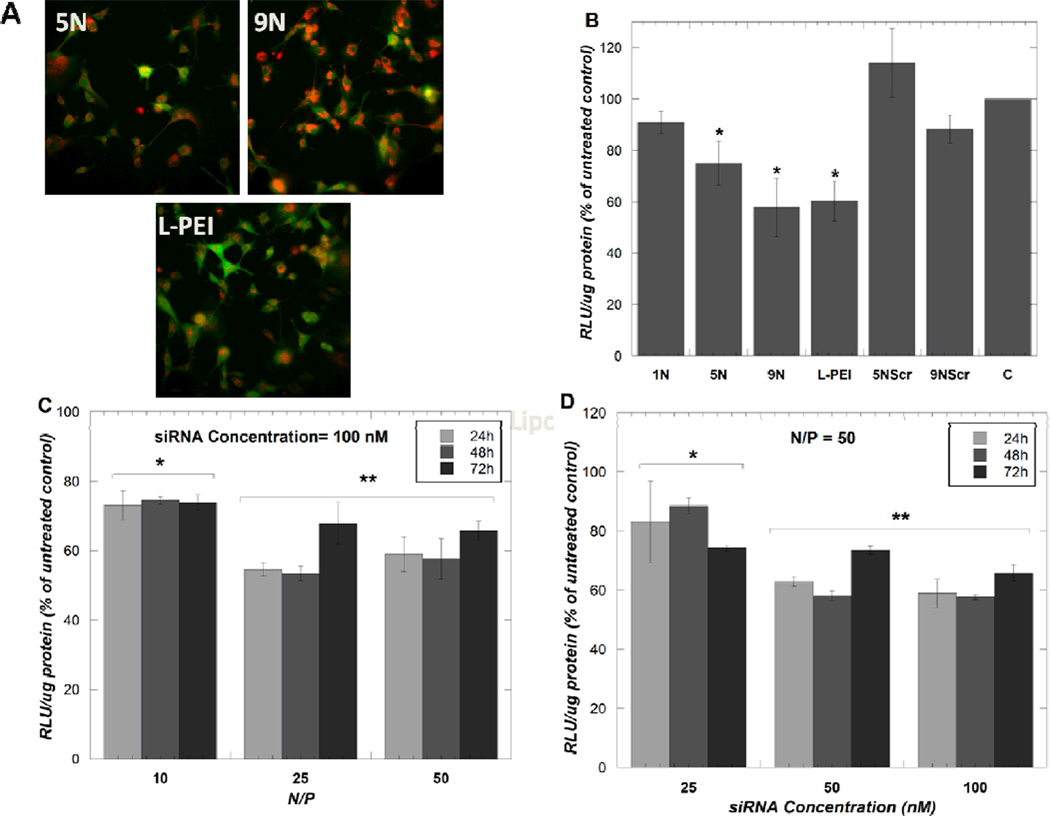
Successful cellular association of siRNA delivered by 5N and 9N was observed using fluorescent microscopy and was qualitatively comparable to siRNA delivered by L-PEI (Figure 3A). Significant luciferase silencing (p <0.05) was observed using the AMs containing five or nine amines (5N and 9N), but not observed using the AMs containing just one amine group (1N) (Figure 3B). A similar luciferase silencing response was observed between the 9N and L-PEI, a widely-studied polymeric system for nucleic acid delivery (Figure 3B). Delivering a scrambled siRNA sequence did not elicit luciferase silencing, demonstrating that the AMs alone do not induce off-target silencing effects.
Figure 3.
Fluorescent microscope images of siRNA distribution (red) in U87-d1EGFP cells (green) when delivered by the indicated polymers (A) and luciferase silencing in U87-Luc cells 24 hours post-transfection (B). L-PEI was used at N/P=15. The samples 5NScr and 9NScr indicate treatments with a scrambled siRNA sequence. Time-course and dose titrations were performed of 9N /siRNA complexes to U87-Luc cells (C and D). Data represent mean ± standard deviation (n=3). Asterisks indicate treatments that elicited statistically significant luciferase silencing compared to the untreated control, C (B) or treatments that elicited statistically different luciferase silencing compared to the other treatment groups in the experiment (C and D) (p<0.05).
To study the dynamics and dose-dependence of luciferase silencing by 9N, the most promising aminated-AM, siRNA transfection experiments were performed at various time points, polymer concentrations (Figure 3C) and siRNA concentrations (Figure 3D). The minimum N/P ratio required for a maximum luciferase silencing response was N/P=25 (siRNA concentration: 100 nM, AM concentration: 1.2×10−5 M), (Figure 3C), and the minimum siRNA concentration required for optimal silencing response was 50 nM (Figure 3D).
Studying the dose response and dynamics of siRNA delivery by AMs provides insights into the mechanisms governing siRNA delivery by these novel molecules. Our results suggest that using polymer 9N at N/P>25 is not biologically beneficial as similar extents of gene silencing were observed using 9N at N/P=25 and N/P=50. The goal in polymeric delivery systems is to identify the lowest possible polymer concentration that can achieve optimal siRNA delivery, as having excessively high polymer concentrations can elicit undesirable cytotoxicity and may result in insolubility of polymers in aqueous media. Further, we observed that using siRNA concentrations of 50 nM was sufficient to achieve maximal luciferase gene silencing with 9N. Presumably, having siRNA in excess of the minimum effective concentrations is unnecessary as the number of target mRNAs present in the cell is limited; the cells are sufficiently targeted by siRNA at 50 nM.
We also observed trends in siRNA silencing as a function of time, where maximum siRNA silencing was observed at 48 hours, and decreased after 72 hours. This trend in gene silencing dynamics is consistent with previous work evaluating the gene silencing dynamics of antisense oligonucleotides delivered by branched PEIs where the silencing of green fluorescent protein (GFP) became less pronounced after 24 hours[36]. This decrease in gene silencing activity after 48 hours can likely be attributed to intracellular degradation of siRNA molecules by nucleases over time.
Interestingly, the trends observed in the quantitative luciferase silencing assay differed somewhat from the qualitative observations of cellular association of a fluorescently labeled siRNA sequence into U87GFP cells. It appeared that 9N delivered more siRNA to the cells than L-PEI in the fluorescent images, however, this trend was not observed in the luciferase silencing assay where both 9N and L-PEI elicited similar extents of luciferase silencing. This observation suggests that while 9N may be capable of delivering siRNA to cells, other intracellular barriers such as siRNA unpackaging or endosomal escape may be affecting gene silencing activity by 9N. This phenomenon will be investigated further in subsequent work.
Conclusions
Amphiphilic macromolecules that self-assemble to form nanoscale micellar assemblies were functionalized with linear ethyleneimines to render them positively charged for improved siRNA complexation. By increasing the number of secondary amines from one up to nine (i.e., from 1N to 9N, respectively), increased zeta potential and stable complexation with siRNA was achieved. All cationic AMs were less cytotoxic to U87 cells than L-PEI at polymer concentrations of 10 µM or greater. The cationic AM with nine total amines, 9N, successfully delivered siRNA molecules to U87 cells and elicited silencing of the reporter gene, firefly luciferase. This work highlights the promise of AMs for siRNA delivery and specifically identified a novel AM molecule, 9N, that displays low cytotoxicity compared to L-PEI, stable complexes with siRNA while maintaining a nanoscale size, and efficiently delivers siRNA delivery to malignant glioma cells.
Acknowledgements
We acknowledge the gift of U87-Luc cells from Xu-Li Wang at the University of Utah and financial support from Rutgers University, the NIH (2R01EB008278-07, P.I. CR), a Schering-Plough fellowship (SMS), and an NSF IGERT (DGE-0504497) fellowship (CLW).
References
- 1.Aagaard L, Rossi J. Advanced Drug Delivery Review. 2007;59:75. doi: 10.1016/j.addr.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gartel A, Kandel E. Biomolecular Engineering. 2006;23:17. doi: 10.1016/j.bioeng.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 3.Edelstein ML, Abedi MR, Wixon J. The Journal of Gene Medicine. 2007;9:833. doi: 10.1002/jgm.1100. [DOI] [PubMed] [Google Scholar]
- 4.Edelstein ML, Abedi MR, Wixon J, Edelstein RM. The Journal of Gene Medicine. 2004;6:597. doi: 10.1002/jgm.619. [DOI] [PubMed] [Google Scholar]
- 5.Tokatlian T, Segura T. Nanomedicine and Nanobiotechnology. 2010;2:305. doi: 10.1002/wnan.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kataoka K, Harada A, Nagasaki Y. Advanced Drug Delivery Review. 2001;47:113. doi: 10.1016/s0169-409x(00)00124-1. [DOI] [PubMed] [Google Scholar]
- 7.Nishiyama N, Kataoka K. Pharmacology & Therapeutics. 2006;112:630. doi: 10.1016/j.pharmthera.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 8.Osada K, Kataoka K. Advances in Polymer Science. 2006;202:113. [Google Scholar]
- 9.Itaka K, Kanayama N, Nishiyama N, Jang W, Yamasaki Y, Nakamura K, Kawaguchi H, Kataoka K. Journal of the American Chemical Society. 2005;126:13612. doi: 10.1021/ja047174r. [DOI] [PubMed] [Google Scholar]
- 10.Xiong X, Uludag H, Lavasanifar A. Biomaterials. 2009;30:242. doi: 10.1016/j.biomaterials.2008.09.025. [DOI] [PubMed] [Google Scholar]
- 11.Yokoyama M. Expert Opinion on Drug Delivery. 2010;7:145. doi: 10.1517/17425240903436479. [DOI] [PubMed] [Google Scholar]
- 12.Samyang. ClinicalTrials.gov [Internet] National Library of Medicine (US); 2009. A Trial to Evaluate the Efficacy and Safety of Genexol®-PM Compared to Genexol® in Subjects With Recurrent or Metastatic Breast Cancer. p. NCT00876486/. [Google Scholar]
- 13.Korean Breast Cancer Study Group. ClinicalTrials.gov [Internet] National Library of Medicine (US); 2009. A Clinical Trial of Paclitaxel Loaded Polymeric Micelle in Patients With Taxane-Pretreated Recurrent Breast Cancer. p. NCT00912639/. [Google Scholar]
- 14.Matsumoto S, Christie R, Nishiyama N, Miyata K, Ishii A, Oba M, Koyama H, Yamasaki Y, Kataoka K. Biomacromolecules. 2009;10:119. doi: 10.1021/bm800985e. [DOI] [PubMed] [Google Scholar]
- 15.Sun T, Du J, Yan L, Mao H, Wang J. Biomaterials. 2008;29:4348. doi: 10.1016/j.biomaterials.2008.07.036. [DOI] [PubMed] [Google Scholar]
- 16.Beh C, Seow W, Wang Y, Zhang Y, Ong Z, Ee P, Yang Y. Biomacromolecules. 2009;10:41. doi: 10.1021/bm801109g. [DOI] [PubMed] [Google Scholar]
- 17.Zhu C, Jung S, Luo S, Meng F, Zhu X, Park T, Zhong Z. Biomaterials. 2010;31:2408. doi: 10.1016/j.biomaterials.2009.11.077. [DOI] [PubMed] [Google Scholar]
- 18.Kim W, Kim S. Pharmaceutical Research. 2009;26:657. doi: 10.1007/s11095-008-9774-1. [DOI] [PubMed] [Google Scholar]
- 19.Chnari E, Lari H, Tian L, Uhrich K, Moghe P. Biomaterials. 2005;26:3749. doi: 10.1016/j.biomaterials.2004.09.038. [DOI] [PubMed] [Google Scholar]
- 20.Chnari E, Nikitczuk J, Uhrich K, Moghe P. Biomacromolecules. 2006;7:597. doi: 10.1021/bm0506905. [DOI] [PubMed] [Google Scholar]
- 21.Chnari E, Nikitczuk J, Wang J, Uhrich K, Moghe P. Biomacromolecules. 2006;7:1796. doi: 10.1021/bm0600872. [DOI] [PubMed] [Google Scholar]
- 22.Djordjevic J, Barch M, Uhrich K. Pharmaceutical Research. 2005;22:24. doi: 10.1007/s11095-004-9005-3. [DOI] [PubMed] [Google Scholar]
- 23.Djordjevic J, Del Rosario L, Wang J, Uhrich K. Journal of Bioactive and Compatible Polymers. 2008;23:532. [Google Scholar]
- 24.Iverson N, Plourde N, Sparks S, Wang J, Patel E, Nackman G, Uhrich K, Moghe P. under review. 2010 [Google Scholar]
- 25.Iverson NM, Sparks SM, Demirdirek B, Uhrich KE, Moghe PV. Acta Biomaterialia. 2010;6:3081. doi: 10.1016/j.actbio.2010.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tao L, Uhrich K. Journal of Colloid and Interface Science. 2006;298:102. doi: 10.1016/j.jcis.2005.12.018. [DOI] [PubMed] [Google Scholar]
- 27.Tian L, Yam L, Zhou N, Tat H, Uhrich K. Macromolecules. 2004;37:538. [Google Scholar]
- 28.Wang J, Plourde N, Iverson N, Moghe P, Uhrich K. International Journal of Nanomedicine. 2007;2:697. [PMC free article] [PubMed] [Google Scholar]
- 29.Torchilin V. Pharmaceutical Research. 2007;24:1. doi: 10.1007/s11095-006-9132-0. [DOI] [PubMed] [Google Scholar]
- 30.Harmon A, Uhrich K. Journal of Bioactive and Compatible Polymers. 2009;24:185. [Google Scholar]
- 31.Zintchenko A, Philipp A, Dehshahri A, Wagner E. Bioconjugate Chemistry. 2008;19:1448. doi: 10.1021/bc800065f. [DOI] [PubMed] [Google Scholar]
- 32.Djordjevic J, Del Rosario L, Wang J, Uhrich KE. Journal of Bioactive and Compatible Polymers. 2008;23:532. [Google Scholar]
- 33.Waite C, Sparks S, Uhrich K, Roth C. BMC Biotechnology. 2009;9 doi: 10.1186/1472-6750-9-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brannon-Peppas L, Blanchette JO. Advance Drug Delivery Reviews. 2004;56:1649. doi: 10.1016/j.addr.2004.02.014. [DOI] [PubMed] [Google Scholar]
- 35.Davis ME, Chen Z, Shin DM. Nature Reviews Druge Discovery. 2008;7:771. doi: 10.1038/nrd2614. [DOI] [PubMed] [Google Scholar]
- 36.Sundaram S, Lee LK, Roth CM. Nucleic Acids Res. 2007;35:4396. doi: 10.1093/nar/gkm450. [DOI] [PMC free article] [PubMed] [Google Scholar]