Abstract
The retina of fish and amphibian contains genuine neural stem cells located at the most peripheral edge of the ciliary marginal zone (CMZ). However, their cell-of-origin as well as the mechanisms that sustain their maintenance during development are presently unknown. We identified Hes4 (previously named XHairy2), a gene encoding a bHLH-O transcriptional repressor, as a stem cell-specific marker of the Xenopus CMZ that is positively regulated by the canonical Wnt pathway and negatively by Hedgehog signaling. We found that during retinogenesis, Hes4 labels a small territory, located first at the pigmented epithelium (RPE)/neural retina (NR) border and later in the retinal margin, that likely gives rise to adult retinal stem cells. We next addressed whether Hes4 might impart this cell subpopulation with retinal stem cell features: inhibited RPE or NR differentiation programs, continuous proliferation, and slow cell cycle speed. We could indeed show that Hes4 overexpression cell autonomously prevents retinal precursor cells from commitment toward retinal fates and maintains them in a proliferative state. Besides, our data highlight for the first time that Hes4 may also constitute a crucial regulator of cell cycle kinetics. Hes4 gain of function indeed significantly slows down cell division, mainly through the lengthening of G1 phase. As a whole, we propose that Hes4 maintains particular stemness features in a cellular cohort dedicated to constitute the adult retinal stem cell pool, by keeping it in an undifferentiated and slowly proliferative state along embryonic retinogenesis. Stem Cells 2012;30:2784–2795
Keywords: Retina, Neural stem cells, Hes4/XHairy2, Wnt and Hedgehog signaling, Cell cycle kinetics
INTRODUCTION
Contrasting with the mammalian situation, the retina of adult fish and amphibians contains a population of neural stem cells, which allow continuous tissue growth throughout the animal life as well as regeneration following retinal damage [1]. These retinal stem cells (RSCs) reside in a permanently proliferating region located at the margin of the retina, known as the ciliary marginal zone (CMZ; [2]). The spatial organization of the CMZ mirrors the temporal sequence of retinal development with stem cells being found in its most peripheral part, successively followed more centrally by actively dividing progenitors and then by their postmitotic progeny [3, 4].
Much progress has been made these past few years in the characterization of CMZ neural stem cells. This includes the formal demonstration of their multipotency and self renewal ability [5, 6], advance in the description of their niche [7, 8] and the identification of candidate genes and signaling pathways to regulate their postembryonic activity [9–17]. However, several questions remain unresolved and in particular, that of their embryonic origin. Whether they arise from a discrete population of cells and how they escape from cell cycle exit and differentiation signals during retinal development is hitherto unknown. Comprehensive analysis of RSC ontogeny and properties necessitates reliable markers to formally identify these immature cells within proliferating heterogeneous cell populations in vivo. To gain insight into the molecular signature of RSCs, we recently performed a large-scale expression screen in the Xenopus CMZ [14]. Among identified RSC markers, we retrieved the Hes4 gene (previously known as XHairy2 in Xenopus and ortholog of zebrafish Her9 and chick cHairy1) that encodes a transcriptional repressor of the bHLH-O family.
Hes family genes are well known as Notch transcriptional targets that can regulate cellular differentiation, cell fate decisions, and embryonic patterning in various developmental systems [18, 19]. Notably, several members of this family, including Hes1, which is closely related to Hes4 in terms of sequence similarity [18], have been intensively studied in the developing vertebrate brain. In this context, several lines of evidence converge toward a role in boundary formation and maintenance of neural stem/progenitor cells, mainly through prevention of neuronal differentiation [20–27]. Hes1 is expressed as well in the embryonic retina, where it regulates distinct aspects of eye morphogenesis and is required for proper timing of neurogenesis [28–30]. Hes1 also emerged as a safeguard of cellular quiescence, through protection against terminal differentiation and permanent cell cycle arrest [31, 32]. In contrast to Hes1, the Hes4 gene was largely ignored in mammalian studies, presumably due to the absence of an ortholog in mouse. It is, however, expressed in humans and has been shown to be involved in several aspects of other vertebrate species development. In particular, Hes4 proved to play a significant role in maintaining the undifferentiated state of Xenopus neural crest cells [33–35] and zebrafish inner ear progenitors [36]. In addition, a recent publication by Kubo and Nakagawa identified the chick Hes4 ortholog, cHairy1 [37], as highly expressed in the CMZ and required for the maintenance of this structure downstream Wnt signaling [38]. We thus decided to gain further insights into Hes4 expression and function in the developing Xenopus retina.
We found that in contrast to mouse Hes1 [28, 30] and chick cHairy1 [39], Hes4 is not expressed in the neural retina (NR) at any stage examined but labels the presumptive retinal pigmented epithelium (RPE) and forming CMZ before being restricted to stem cells of the mature retina. Wnt and Hedgehog signaling pathways contribute to this dynamic expression pattern through positive and negative regulation, respectively. Finally, functional analysis revealed that Hes4 imparts retinal cells with stem-like properties: inhibited commitment toward RPE and neuronal fate, prolonged proliferative capacities, and slow cell cycle kinetics.
MATERIALS AND METHODS
Embryo Collection, Transgenic Line
Xenopus laevis embryos were obtained by conventional methods of in vitro fertilization. Xenopus tropicalis transgenic animals carrying the Wnt reporter pbin8LefdGFP construct have previously been described [40] and were obtained by natural fertilization between a wild-type female and a transgenic male carrying a single insertion of the transgene [41]. All experiments were approved by the Direction Départementale des Services Vétérinaires de l'Essonne, Evry, France.
Pharmacological Treatments
Cyclopamine (20 μM; LC Laboratories, Woburn, MA, http://lclabs.com), purmorphamine (100 μM; Calbiochem, San Diego, CA, http://www.emdbiosciences.com), DAPT (N– [N–(3,5–Difluorophenacetyl)-L-alanyl]-S-phenylglycine t-butyl ester; 100 μM; Sigma, St Louis, MO, http://www.sigma aldrich.com), and IWR1 (Inhibitor of Wnt Response 1; 50 μM; Sigma) were applied to the embryo culture medium from stage 25 to stage 35. BIO (6-bromoindirubin-3′-oxime; 20 μM; Sigma) was applied for 1 hour on stage 25 embryos, which were then rinsed and kept in drug-free medium for 15 or 24 hours [41]. Effectiveness of drug treatment was systematically assessed through whole mount in situ hybridization by checking the expression of known target genes of the considered pathway: Gli1 or Patched1 (Ptc1) for Hedgehog [12], CyclinD1 for Wnt [10], and HRT1 for Notch [42].
Expression Constructs
pCS2-Flag-Hes4 (previously named XHairy2), pCS2-Hes4-myc-GR (inducible construct fused to the dexamethasone-responsive hormone-binding domain of the human glucocorticoid receptor (GR); referred as Hes4-GR in the text [34]), pCS2-Hes2 [43], pCS2-Xgadd-45γ [44], pCS2-GFP (a gift from David Turner), and pCS2-LacZ (a gift from Nancy Papalopulu) were previously described. pCS2-Hes4-myc-VP16-GR encodes a Hes4 glucocorticoid-inducible antimorphic variant where the VP16 transactivation domain is fused to the carboxylterminus of Hes4. It was generated by subcloning the Hes4 coding sequence into a pCS2-myc-VP16-GR vector after polymerase chain reaction (PCR) amplification. Details on the cloning procedure are available upon request.
Microinjections and In Vivo DNA Lipofection
Xenopus laevis Hes4 gene exists as two alloalleles, Hes4a and Hes4b, and the later was used in all overexpression experiments. Capped sense RNAs were transcribed using the mMessage mMachine SP6 kit (Ambion, Austin, TX, http://www.ambion.com). 500 pg of mRNA was injected animally into one or two blastomeres at the two-cell stage, together with GFP or β-galactosidase mRNA (250 pg). Protein activity of GR chimeric constructs was induced by incubating the embryos in 4 μg/ml dexamethasone (Sigma) from stage 12. Treating embryos before this stage lead to severe developmental defects and frequently to developmental arrest. Morpholino oligonucleotides against both alloaleles (“Hes4 Mo”; [34]) were injected into one blastomere at the four- or eight-cell stage (8–20 ng). Lipofection experiments were performed at stage 17/18 as previously described [45].
In Situ Hybridization
Digoxigenin-labeled antisense RNA probe synthesis and whole-mount in situ hybridization were performed as previously described [14]. Embryos were then vibratome sectioned (50 μM). Following image capture, labeling area was manually delineated and quantified (in Pixel2) in both the dorsal and ventral CMZ using Adobe Photoshop CS4 software. Shown in graphs are percentage of staining area increase/decrease compared to the control.
BrdU/EdU Incorporation and Immunohistochemistry
Embryos were injected intra-abdominally with BrdU (5-bromo-2′-deoxyuridine; 10 mM; Sigma) or EdU (5-ethynyl-2′-deoxyuridine; 1 mM; Invitrogen, Carlsbad, CA, http://www. invitrogen.com) for various durations, depending on the experiment. For birthdating analyses, embryos were injected every 10–12 hours from stage 34 to stage 41. Immunohistochemistry was performed on 12 μm-cryostat or -paraffin sections as previously described [12, 41]. Antibodies used are listed in Supporting Information Table 1. Cell nuclei were counterstained with Hoechst (Sigma). Detection of EdU-labeled cells was carried out with the Click-iT EdU Imaging Kit (Molecular Probes, Eugene, OR, http://probes.invitrogen.com). Fluorescent staining was visualized with a M2 Zeiss microscope. Images were captured using a digital Axiocam MRc camera and processed with AxioVision REL 7.8 and Adobe Photoshop CS4 softwares. Retinal area was manually delineated on transverse sections and quantified (in Pixel2) using Adobe Photoshop CS4 software.
Analysis of Cell-Cycle Parameters
Growth fraction (GF; proportion of proliferative cells), total cell cycle length (TC), and S-phase length (TS) were determined following EdU cumulative labeling using the Excel sheet provided by Dr R. Nowakowski [46]. Mitotic index and percentages of EdU-labeled mitosis were measured as previously described [12]. The time required for half-maximal appearance of EdU labeling in the mitotic population was taken as an estimation of the average G2 length (TG2; [47]). M-phase duration (TM) was calculated after determining the proportion of cells that were in mitosis (% M), taking into account TC and GF, as determined by cumulative EdU labeling: TM = % M × GF × TC. Finally, G1 duration (TG1) was deduced from the above values as TG1 = TC − (TS + TG2 + TM).
Reverse Transcription and Quantitative Real Time PCR
Reverse transcription of retinal mRNA and quantitative real time PCR (qPCR) were performed as described previously [41]. PCR primer sequences are listed in Supporting Information Table 2.
Quantification and Statistical Analysis
All experiments were performed at least in duplicate. Shown in figures are results from one representative experiment. In each histogram, values are given as mean ± SEM. In experiments involving two conditions, statistical analysis was performed by Student's t test, while in those involving the comparison of more than two treatments one-way ANOVA followed by Tukey's post hoc tests were performed (NS: not significant; *, p < .05; **, p < .01; ***, p < .001).
RESULTS
Hes4 Expression Likely Reveals the Embryonic Cell-of-Origin of Adult RSCs
The embryonic lineage that gives rise to adult RSCs in Xenopus is currently unknown. Gene expression profile within the CMZ has proved to mirror the temporal genetic sequence of retinal development [3, 4, 48]. As Hes4 labels the most peripheral stem cell-containing part of the CMZ ([14] and Figure 1Q, 1R), we hypothesized that establishing its expression profile during retinogenesis might retrospectively give clues about RSC ontogenesis. We thus analyzed the distribution of its transcripts at key stages of eye development and compared it with that of optic field (Pax6), NR (Rx), optic stalk (OS; Pax2), and RPE (Mitf1A) markers (Fig. 1; Supporting Information Fig. S1). At late neurula stage, Hes4 expression was observed in a restricted proximo-dorsal region of the optic field, clearly contrasting with that of Pax6 (Fig. 1A, 1B). In line with this, analysis on retinal sections at the optic vesicle stage revealed that Hes4 stained the Mitf1A-expressing presumptive RPE and was not detected in the Rx-positive retinal neuroepithelium, except in its most dorsal part at the border with the RPE (Fig. 1K, 1L). Of note, from stage 24–25, Hes4 transcripts were additionally observed in the OS (coinciding with Pax2 expression; Fig. 1L). During optic cup formation, Hes4 expression progressively declined in the differentiating central RPE while remaining high in its peripheral part and in the NR margins (Fig. 1M–1O). Staining in the margin progressively superimposed with the forming CMZ to finally get restricted to its most peripheral region (Fig. 1P–1R). As a whole, Hes4 expression delineates a territory clearly distinct from both the differentiating NR and RPE, that is first located in the dorsal part of the optic vesicle, then at the peripheral margin of the optic cup, and that finally coincides with RSC location following completion of embryonic retinogenesis. Importantly, such a dynamic expression pattern strongly resembles that of Gli3 and Smo, two previously described stem-cell specific markers [49]. In addition, we found that the same held true for two other genes that we recently identified as being expressed at the extreme tip of the CMZ, namely Id2 and Wnt8b (Supporting Information Fig. S2 and [41]). Altogether, these data thus suggest that adult RSCs originate from a discrete population of cells located at the RPE/NR border of the optic vesicle.
Figure 1.
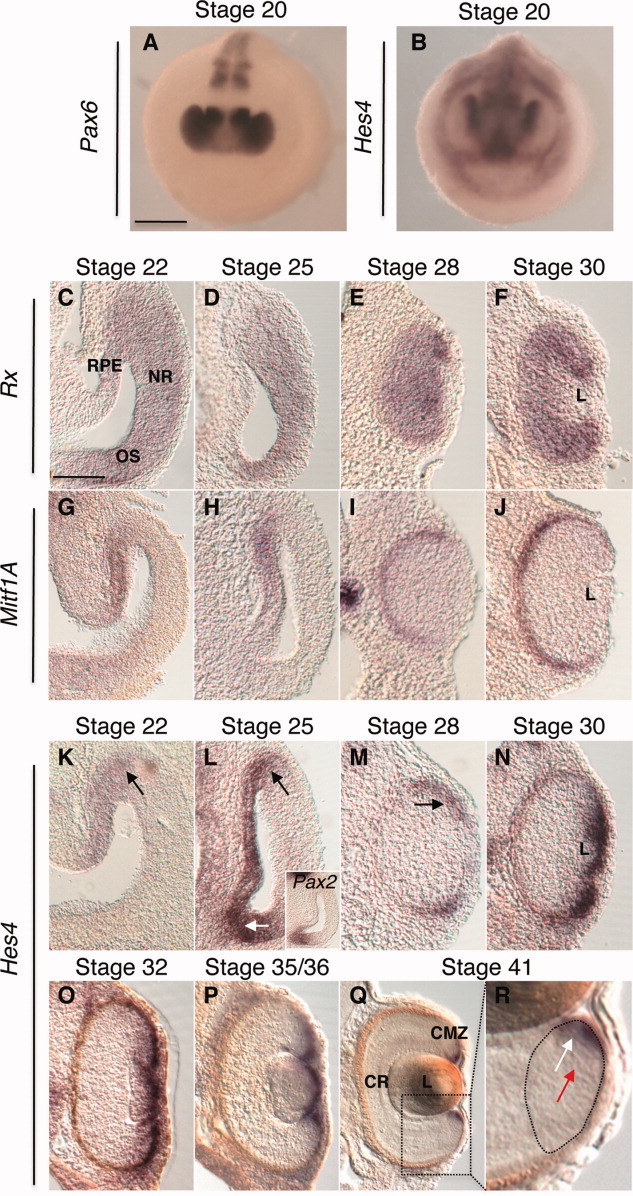
Hes4 expression during retinogenesis. Comparative in situ hybridization analysis of Hes4 expression profile (B, K–R) with that of Pax6(A), Rx(C–F)Mitf1A(G–J), and Pax2 (inset in L). White arrow in (L) point to the OS. Black arrows in (K–M) indicate the RPE/NR border (K, L) and the retinal margin (M). (Q) corresponds to a magnification of the CMZ delineated in (R). White and red arrows, respectively, indicate stem cell and progenitor zones of the CMZ. Scale bars = 300 μm (A, B) or 50 μm (C–R). Abbreviations: CMZ, ciliary marginal zone; CR, central retina; L: lens; NR, neural retina; OS, optic stalk; RPE, retinal pigmented epithelium.
Hes4 Expression Is Positively Regulated by Wnt Signaling and Negatively by the Hedgehog Pathway
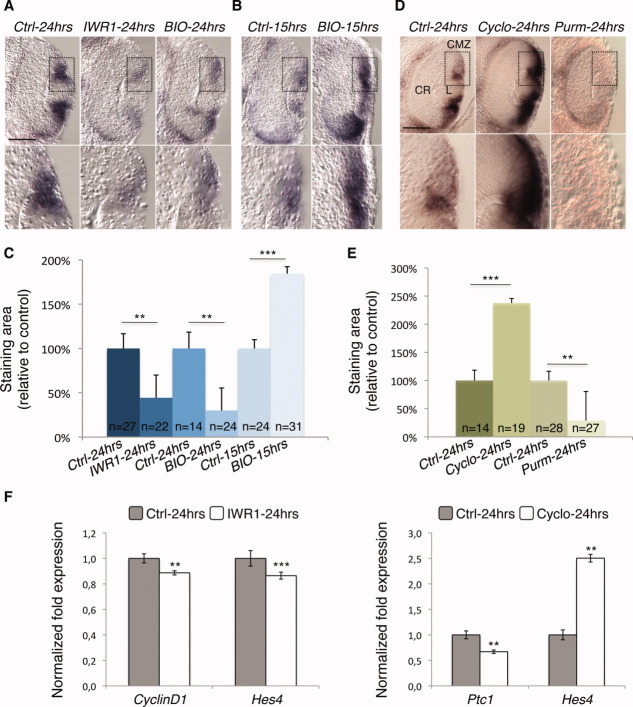
We then aimed at identifying the signaling pathways regulating Hes4 embryonic expression. A potential candidate is the canonical Wnt pathway since it proved to be active in the peripheral retina of various species [10, 50, 51] and was shown to regulate CMZ expression of cHairy1, the Hes4 ortholog in chick [38]. However, the status of Wnt activity during embryonic retinogenesis of Xenopus has never been investigated in detail. Taking advantage of a Xenopus tropicalis reporter line carrying a destabilized eGFP downstream a synthetic Wnt-responsive promoter [40], we thus analyzed the profile of Wnt activity in the developing retina. Except in the presumptive RPE and OS of the optic vesicle where it was not detected, eGFP staining strikingly superimposed with that of Hes4 (Supporting Information Fig. S3). Consistent with a role in promoting Hes4 expression, we found that pharmacological blockade of Wnt signaling from stage 25 to stage 35 (24-hour IWR1 treatment) significantly lowered Hes4 levels in the CMZ, as inferred from both in situ hybridization and qPCR analyses (Fig. 2A, 2C, 2F). Unexpectedly, similar inhibition was observed upon a 24-hour treatment with BIO, an activator of the Wnt pathway (Fig. 2A, 2C). Shortening the treatment to 15 hours, however, led to a consistent increase of Hes4 labeling (Fig. 2B, 2C). As Hes1 is known to repress its own expression [52], these results led us to investigate whether this might also be the case for Hes4. In line with this, we found that misexpression of the Hes4 antimorphic variant Hes4VP16-GR (which transforms the Hes4 repressor into a transcriptional activator) led to ectopic Hes4 staining in the epidermis of injected embryos (Supporting Information Fig. S4A). Additionally, morpholino-mediated Hes4 knockdown significantly enhanced Hes4 expression in its endogenous domain at the neurula stage (Supporting Information Fig. S4B). As a whole, these results suggest that Hes4 levels in the forming CMZ are positively regulated by the Wnt pathway and modulated by a negative feedback loop.
Figure 2.
Wnt and Hedgehog signalings affect Hes4 expression in an opposite manner. (A–E):In situ hybridization analysis of Hes4 expression on retinal sections, following a 24-hour (from stage 25 to stage 35; A, C–E) or a 15-hour (from stage 25 to stage 32; B, C) treatment with the indicated drug. Shown beneath each retinal section is a higher magnification of the dorsal CMZ delineated with dotted lines. (C, E): Quantification of Hes4 staining area in the dorsal CMZ in each condition. (F): Quantitative polymerase chain reaction analysis of retinal Hes4 expression at stage 35 following 24 hours of Hedgehog or Wnt signaling inhibition. Ptc1 and CyclinD1 serve as controls of drug efficiency. Scale bar = 50 μm. Abbreviations: BIO, 6-bromoindirubin-3′-oxime; CMZ, ciliary marginal zone; CR, central retina; Cyclo, cyclopamine; IWR1, inhibitor of wnt response 1; L, lens; N, number of analyzed sections; Purm, purmorphamine.
We next investigated the impact of Hedgehog signaling based on previous data in mouse retinal explants showing that Hes1 expression is stabilized by the Shh effector Gli2 [53]. As a first attempt to address this issue, we compared Hes4 expression pattern during retinogenesis with that of Gli1 as a readout of Hedgehog signaling activity (Supporting Information Fig. S3). As previously described [12, 49], Gli1 transcripts were detected at high levels in the presumptive RPE and OS of stage 25 embryos and at low levels in the NR and NR/RPE border. Strikingly, upon optic cup formation, Gli1 and Hes4 clearly exhibited complementary expression patterns, with Gli1 being mainly detectable in the differentiating RPE and Hes4 restricted to the peripheral margin of the retina. Finally, in the mature retina, Gli1 labeling was detected in the central CMZ and was faint in its most peripheral Hes4-expressing part. These results suggest that in contrast to mouse Hes1, Hes4 might be downregulated by Hedgehog signaling. In line with this, we found that Hedgehog pathway inhibition with cyclopamine significantly enhanced Hes4 expression in the CMZ (Fig. 2D–2F). Conversely, Hedgehog activation with purmorphamine strongly reduced Hes4 staining (Fig. 2D, 2E). Altogether, it is likely that Hedgehog-mediated inhibition of Hes4 contributes to restrain its expression domain to the peripheral margin of the optic cup and to the stem cell compartment of the CMZ.
Although Hes genes are recognized as canonical targets of Notch signaling, a growing wealth of data indicate that they may function independently in several contexts, including the retina [38, 53]. Considering the above data, we investigated whether this might be the case for Hes4 as well. We thus blocked the Notch pathway with DAPT, a chemical compound that inhibits the γ-secretase-dependent release of the Notch intracellular domain and was shown to repress Hes1/5 expression in the chick retina [54]. Contrasting with the effects of Wnt and Hedgehog signaling perturbations in the same time window, Hes4 levels were not significantly lowered by a 24-hour DAPT exposure (Supporting Information Fig. S5B, S5E, S5F) and were only affected when treatment duration was extended to 48 hours (Supporting Information Fig. S5D, S5E). Such a delay suggests that the Notch pathway does not directly contribute to Hes4 regulation in the forming CMZ.
Hes4 Loss of Function Severely Impairs Eye Formation
We next aimed at addressing Hes4 function during retinal development through a knockdown experiment. Morphological analysis of stage 37 tadpoles injected with Hes4 Morpholinos (Hes4 Mo) revealed severe dose-dependent eye defects ranging from a strong reduction of eye size to a complete absence of eye (Fig. 3A). No discernable eye malformations were observed upon injection of a standard Morpholino (Ctrl Mo; data not shown). In order to exclude a potential toxicity of the Hes4 Mo, we tested whether the eye-absent morphant phenotype could be rescued upon coinjection of Hes4 Mo with an inducible Hes4 construct (Hes4-GR) devoid of Hes4 Mo target sequence. The percentage of eye-absent embryos was scored in the different conditions, and we found that it dropped from 78% in Hes4 Mo-injected embryos to 12.5% in Hes4 Mo plus Hes4-GR-injected ones (Fig. 3B). Such a rescue clearly suggests that the loss of eye is a specific phenotype of Hes4 knockdown. Consistent with this, severe eye defects were observed as well following overexpression in the presumptive neurectoderm of the antimorphic Hes4 variant Hes4VP16-GR (data not shown).
Figure 3.
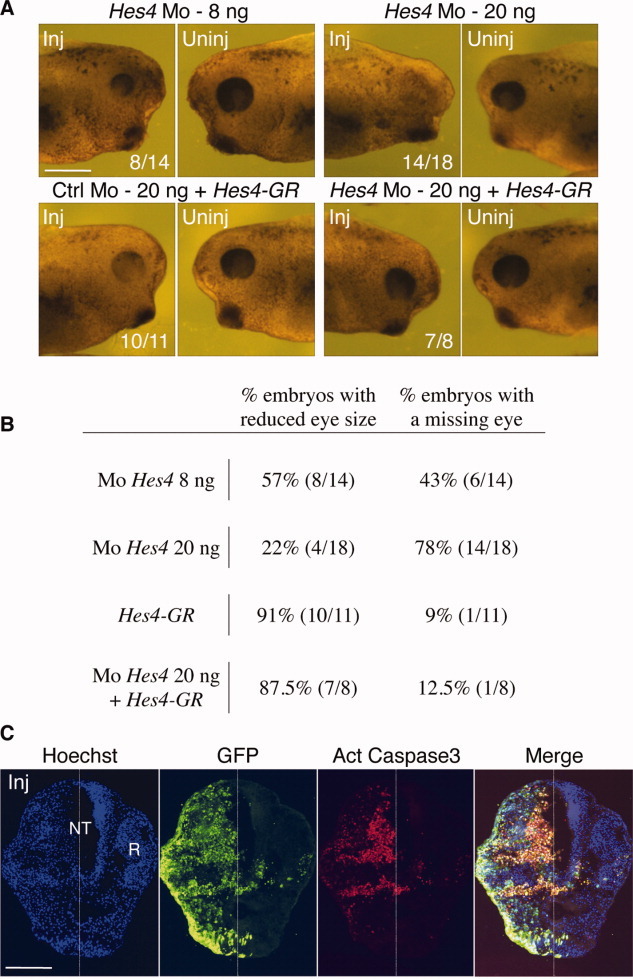
Hes4 is required for proper eye formation. (A, B): Analysis of Hes4 loss of function. Hes4 or Ctrl Mo were injected at the indicated dose with or without Hes4-GR mRNA. (A): Typical pictures of stage 37 injected embryos (lateral view of the head). Note that embryos injected with Hes4-GR mRNA alone exhibit a defective RPE pigmentation and a slightly smaller eye. (B): Quantification of morphant and rescued embryos exhibiting the indicated phenotypes. (C): Transverse sections of a stage 25 morphant embryo (injected with 8 ng Hes4 Mo) immunostained with anti-active caspase3 and anti-GFP (to visualize the injected side). Scale bars = 300 μm (A) or 100 μm (C). Abbreviations: GR, glucocorticoid receptor; GFP, green fluorescent protein; Inj, injected side; NT, neural tube; R, retina; Uninj, uninjected side.
Since two previous studies in Xenopus reported increased apoptosis at neurula stages following Hes4 loss of function [33, 35], we wondered whether cell death might account as well for the observed eye malformations. We thus performed anti-active caspase3 immunostaining on stage 25 retinal sections from Hes4 Mo injected embryos. Massive apoptosis was evident on the injected side (Fig. 3C). Thus, as previously proposed in neural crest stem cells [33, 35], Hes4 might be required for cell survival in the retina. Of note, cell death was not restricted to Hes4-expressing tissues and spread in particular to the NR, which might be a secondary consequence of the loss of surrounding tissues.
Hes4 Misexpression Prevents RPE Differentiation and Neuronal Commitment of Retinal Precursor Cells
Since the dramatic phenotype of Hes4 knockdown precludes any further functional investigation, we turned to a gain of function strategy using the Hes4-GR inducible construct. Morphological analysis at stage 37 revealed a defective RPE pigmentation (Fig. 3A) and a slight reduction of retinal size in Hes4-GR-injected embryos (23% ± 2.5% decrease of retinal surface compared to controls, as measured on 19 and 37 sections, respectively; p < .001). Effects on retinal cell type differentiation were then monitored using specific markers (Fig. 4A). In line with the aforementioned defects in retinal pigmentation, Hes4 overexpressing retinas displayed a virtual absence of XAR-1 expression, a marker of the differentiated RPE. In addition, substantial perturbations in laminar organization were observed in the central retina (CR). In line with this, both rhodopsin (data not shown) and calbindin stainings were severely reduced suggesting that rods and cones were profoundly missing. Syntaxin expression was strongly decreased in the inner and outer plexiform layers as well as in the optic nerve, consistent with defective interneuron and ganglion cell production. Thus, Hes4 misexpression drastically impairs both RPE and neuronal differentiation.
Figure 4.
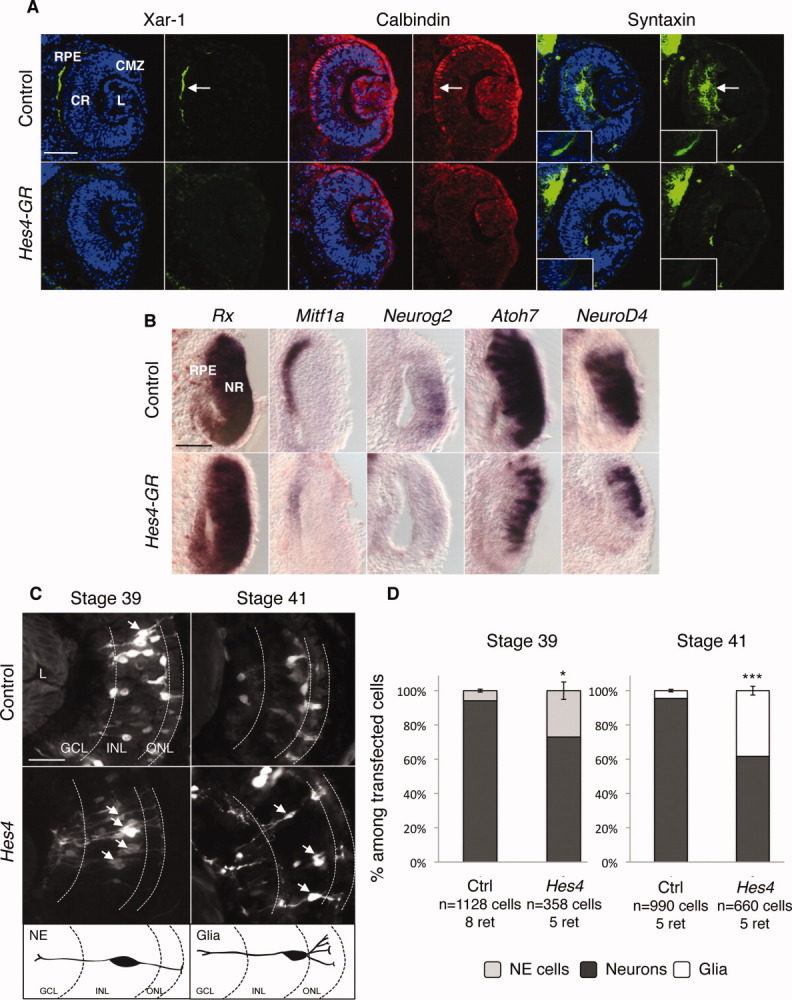
Hes4 misexpression inhibits neuronal and RPE differentiation. (A): Immunofluorescence analysis of cell-type-specific marker expression in stage 37 retinas, following Hes4-GR mRNA injection. Arrows point to the staining in the RPE (Xar-1), photoreceptors (calbindin) or interneuron and ganglion cell fibers (syntaxin). The optic nerve is shown in insets. (B):In situ hybridization analysis with the indicated probe in stage 25 retinas, following Hes4-GR mRNA injection. (C, D): Analysis of cell type distribution in stage 39 or 41 retinas, following Hes4 lipofection. (C) Typical sections of retinas transfected with GFP alone (control) or GFP plus Hes4. Arrows indicate NE at stage 39 or Müller glial cells at stage 41. Respective morphologies of these cells are illustrated on the schematics below. (D) Quantification of NE, neurons, and glia among transfected cells at stage 39 or 41. Scale bars = 50 μm (A, B) or 25 μm (C). Abbreviations: CMZ, ciliary marginal zone; CR, central retina; GR, glucocorticoid receptor; GCL, ganglion cell layer; INL/ONL, inner/outer nuclear layer; L, lens; NE, neuroepithelial cell; NR, neural retina; RPE, retinal pigmented epithelium.
To get further insights into this phenotype, we checked the status of several markers at earlier stages by in situ hybridization (Fig. 4B). We observed that Rx expression was not affected suggesting that Hes4-overexpressing cells retain a retinal identity. However, Mif1A staining was strongly reduced compared to controls, consistent with an impairment of RPE determination. Besides, expression of the proneural genes Neurog2, Atoh7, and NeuroD4 (previously called XNgnr-1, Xath5, and Xath3, respectively) was also decreased, suggesting that Hes4 inhibits neurogenesis by preventing commitment of precursor cells toward a neuronal fate. Consistent with this, we found that the antimorphic Hes4 variant Hes4VP16-GR was able to induce ectopic neurogenesis, as inferred by N-tubulin staining following mRNA injection in the presumptive epidermis (Supporting Information Fig. S4A).
We next followed the fate of Hes4-overexpressing cells in a clonal analysis using in vivo lipofection. At stage 39, most cells in control clones were already differentiated into neurons as judged by their position and morphology. In contrast, an important proportion of Hes4-overexpressing cells still exhibited a neuroepithelial morphology characteristic of undifferentiated neural precursors (Fig. 4C, 4D). Analysis at stage 41 revealed that this delayed differentiation eventually ended up with an excessive production of Müller glial cells at the expense of neurons (Fig. 4C, 4D). In addition to their typical morphology, Müller cell identity was further confirmed by immunostaining using an anti-CRALBP antibody (data not shown). These data indicate that Hes4 overexpression cell autonomously delays differentiation and reduces neurogenesis. As a whole, these results suggest that Hes4 maintains cells of the retinal margin in an undifferentiated state during embryonic retinogenesis.
Hes4 Misexpression Maintains Retinal Precursors in a Proliferative State
We next examined whether Hes4-dependent blockade of differentiation was accompanied by prolonged proliferative capacities. We first analyzed BrdU incorporation at stage 37 following Hes4-GR mRNA injection. At this point of retinal histogenesis, the majority of cells in the wild-type retina are postmitotic and proliferation is restricted to the CMZ. In contrast, a dramatic increase in BrdU-positive cell number was observed in the CR of Hes4-overexpressing embryos (Fig. 5A, 5B).
Figure 5.
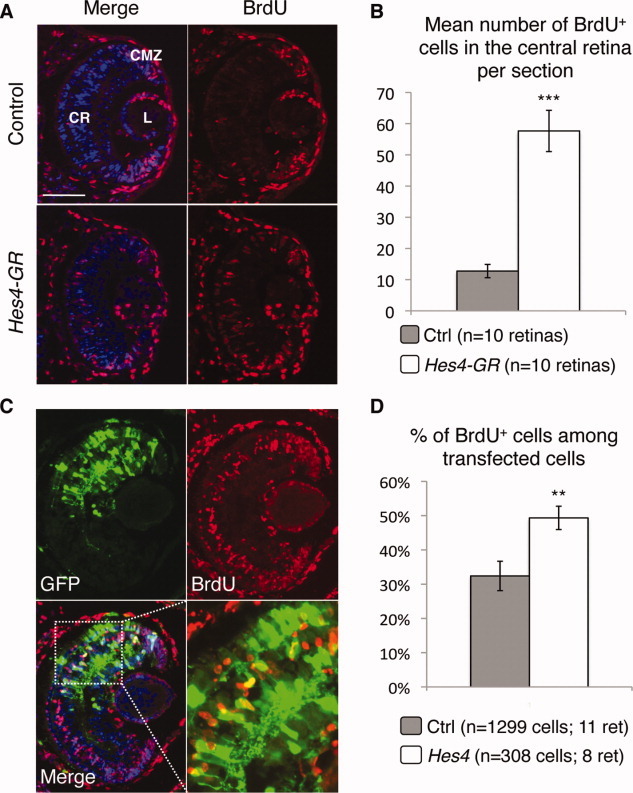
Hes4 overexpression delays cell cycle exit. (A, B): BrdU incorporation assay (3-hour pulse) at stage 37, following Hes4-GR mRNA injection. (A): Typical retinal sections immunostained with anti-BrdU. (B): Corresponding quantification. (C, D): Birthdating experiments in Hes4 lipofected retinas. (C): Typical retinal section immunostained for both GFP and BrdU, following continuous BrdU exposure from stage 34 to 41. (D): Percentage of BrdU+ cells among transfected cells. Scale bar = 50 μm. Abbreviations: BrdU, 5-bromo-2′-deoxyuridine; CMZ, ciliary marginal zone; CR, central retina; GR, glucocorticoid receptor; GFP, green fluorescent protein; L, lens.
We next assayed in lipofection experiments whether these effects on proliferation were cell autonomous (Supporting Information Fig. S6). To challenge the specificity of the Hes4 phenotype, transfection of another Hes family member, Hes2, was performed in a parallel batch of embryos. In contrast to Hes4, Hes2 is not expressed in stem cells but restricted to retinal progenitors where it acts as a gliogenic factor. We previously showed that it does so at least in part by delaying cell cycle exit of late precursor cells, while having no effect on early progenitor cell proliferation [43]. In line with this, we found that Hes2 overexpression increased the percentage of BrdU-positive cells among transfected cells at stage 35 but not at stage 32. In contrast, proliferation rates of Hes4-misexpressing cells were elevated at both stages with respect to the control situation (Supporting Information Fig. S6). This suggests that both early and late retinal precursors are maintained longer in the cell cycle upon Hes4 overexpression.
To examine the timing of cell cycle exit more directly, we performed birthdating experiments (Fig. 5C). BrdU was injected at regular intervals so that it would be constantly available from stage 34 to stage 41, and therefore mark all cells born in that period. We found that the proportion of BrdU-labeled cells among transfected cells was significantly increased following Hes4 transfection, compared to the control situation (Fig. 5D). This clearly indicates that Hes4 delays cell cycle withdrawal of precursor cells.
Finally, we wondered whether such prolonged proliferation might account for the aforementioned imbalance between neuronal and glial cell production (Fig. 4D). We addressed this issue by counteracting Hes4-dependent maintenance in the cell cycle through colipofection with Xgadd-45γ. This cell cycle inhibitor was previously shown to accelerate retinal cell birthdate in similar assays [44]. We found that Xgadd-45γ indeed concomitantly rescued the Hes4-induced delayed cell cycle exit and the deficit in neuronal cells (Supporting Information Fig. S7), strongly suggesting a tight coupling between differentiation and proliferation defects.
Hes4 Misexpression Slows Down Cell Cycle Kinetics of Retinal Precursor Cells
As stated above, Hes4 misexpression leads to a reduction of retinal size, an apparently contradictory phenotype in view of the delayed cell cycle withdrawal of retinal precursor cells. As no increase in apoptosis could be detected following Hes4-GR mRNA injection (data not shown), we thus wondered whether this paradox might be explained by slower cell cycle kinetics of Hes4-overexpressing progenitors compared to controls.
To address this possibility, we first evaluated the mitotic index in the retinal neuroepithelium of stage 25 embryos, using the mitotic marker phospho-Histone H3 (P-H3; [12]). P-H3 positive cell proportion was calculated among all nuclei and further corrected by the GF (percentage of proliferative cells) as determined by EdU cumulative labeling experiments (see below; Fig. 6B; Supporting Information Table 3). We found a significantly lower mitotic index in Hes4-GR-injected optic vesicles compared to the control (Fig. 6A), suggesting that Hes4 overexpression slows down cell cycle speed of retinal precursors.
Figure 6.
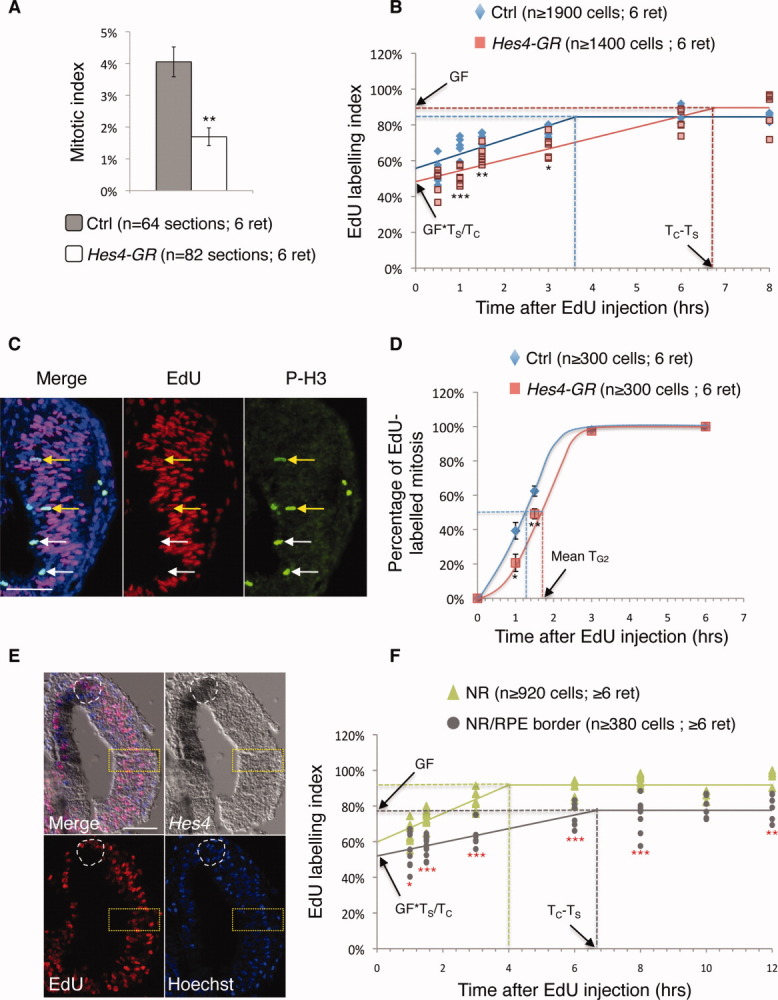
Hes4 overexpression slows down cell cycle speed of retinal precursors. (A): Quantification of the mitotic index in the retinal neuroepithelium at stage 25, following Hes4-GR mRNA injection. (B): Evaluation of the EdU labeling index in the NR of control and Hes4-GR-injected embryos, after cumulative EdU labeling starting from stage 25. (C, D): G2 length evaluation in vivo, following Hes4-GR mRNA injection. (C) Typical retinal section (stage 25) stained for both phospho-Histone H3 (P-H3) and EdU, following a 1.5-hour EdU pulse. White and yellow arrows point to P-H3+/EdU− and to P-H3+/EdU+ cells, respectively. (D) Proportion of EdU-labeled mitosis along with increasing EdU exposure times. (E, F): Proportion of EdU-labeled nuclei after cumulative EdU labeling, starting from stage 22. EdU detection was performed following in situ hybridization with a Hes4 probe. Shown in (E) are typical retinal sections stained for both Hes4 and EdU, following a 1-hour EdU pulse. Labeling index was measured in the Hes4-expressing NR/RPE border (delineated by white dotted lines in E) or in the Hes4-negative NR (yellow dotted lines) of control embryos. Scale bar = 50 μm. Abbreviations: EdU, 5-ethynyl-2′-deoxyuridine; GR, glucocorticoid receptor; NR, neural retina; RPE, retinal pigmented epithelium.
We next aimed at directly measuring total cell cycle (TC) and S-phase (TS) lengths in the NR through a cumulative EdU labeling assay. As shown in Figure 6B and Supporting Information Table 3, the duration of G2 + M + G1 (TC − TS), as inferred from the time point at which the EdU labeling index reached the plateau, was longer in Hes4-misexpressing cells compared to wild-type ones. Calculation of TS and TC using the Nowakowski excel sheet [46] confirmed the hypothesis of extended cell cycle duration following Hes4-GR overexpression (+39% compared to the control situation; 14.73 vs. 10.62 hours) and showed a slight increase of S-phase length (+13%; 7.94 vs. 7.00 hours; Supporting Information Table 3).
Finally, in order to determine whether a specific phase of the cell cycle was particularly affected, we individually estimated G2 (TG2), M (TM), and G1 (TG1) durations. G2 length was evaluated using the percentage of labeled mitoses (PLM) paradigm [12]. Noticeably, the PLM was consistently lower in Hes4-GR-overexpressing retinas compared to control ones, indicating a delayed S- to M-phase progression (Fig. 6C, 6D). Mean TG2 was estimated at 1.6 hours and 1.2 hours, respectively (+33%; Fig. 6D; Supporting Information Table 3). Taking into account data gathered from the measurement of the mitotic index shown previously (Fig. 6A), we next determined TM (see Materials and Methods section for calculation details) and finally deduced TG1 as being equal to TC − (TS + TG2 + TM). This revealed that increased time spent in G1 is the main parameter that accounts for the longer cell cycle observed upon Hes4 overexpression (+148%; 4.94 vs. 1.99 hours), S and G2 phases being lengthened as well although to a lesser extent (Supporting Information Table 3).
The above data suggest that Hes4 transcription factor acts as a regulator of cell cycle speed. In line with this, the stem cell compartment of the CMZ where Hes4 is expressed was recently shown to exhibit slow cell cycle kinetics [6]. As stated above, Hes4 expression at the NR/RPE border of the optic vesicle likely prefigures the subpopulation of cells dedicated to form the adult RSC cohort. We thus wondered whether cells in this territory might be endowed with slow cell cycle kinetics as well. To address this issue, we performed EdU cumulative labeling in stage 22–24 wild-type embryos and analyzed EdU incorporation in two distinct territories: the Hes4-negative NR and the Hes4-expressing NR/RPE border (Fig. 6E). Calculations of TS and TC at the NR/RPE border yielded values of 13.57 and 20.26 hours compared to 7.47 and 11.47 hours in the NR (Fig. 6F; Supporting Information Table 3). Altogether these results strongly suggest that Hes4 contributes to the maintenance of slow cell cycle kinetics at the NR/RPE border.
DISCUSSION
New Insights into the Embryonic Cell-of-Origin of Adult Xenopus RSCs
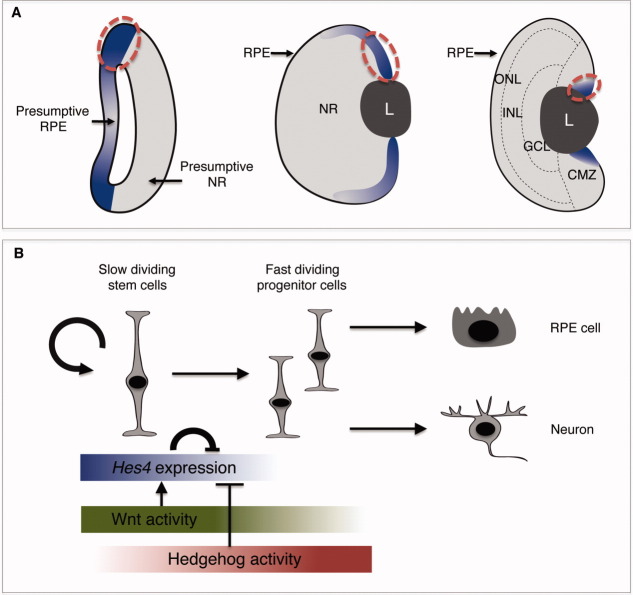
Elegant tracing experiments recently allowed to identify the offspring of a single CMZ stem cell during fish postnatal life [5]. However, the embryonic lineage of adult RSCs has never been investigated so far. Our retrospective analysis of Hes4 expression pattern suggests that these cells originate from the RPE/NR border of the optic vesicle. This hypothesis is substantiated by our finding that other stem cell-specific markers of the postembryonic CMZ display similar expression dynamic during retinogenesis: first labeling the presumptive RPE at optic vesicle stage, then the retinal margin of the optic cup, and finally the most peripheral stem-cell containing part of the CMZ in the mature retina. Importantly, it also fits with recent results from four dimensional manual cell tracking in zebrafish, suggesting that part of the optic vesicle medial layer (classically considered as prospective RPE) contributes to the CMZ [55]. Finally, such an early restricted expression of RSC markers challenges the classical schematic representation of retinal histogenesis, which implies that RSCs are initially found throughout the NR of the newly emerged optic vesicle and thereafter persist in its periphery as central cells differentiate. It rather suggests that a presumptive RSC cohort is already segregated from both the NR and RPE lineages as early as the optic vesicle stage (Fig. 7A).
Figure 7.
A proposed model for retinal stem cell (RSC) cell-off-origin and Hes4 function during embryonic retinogenesis. (A): Schema highlighting the dynamics of Hes4 expression during retinogenesis. Adult RSCs likely originate from the most dorsal Hes4-expressing part of the optic vesicle (red dotted lines). (B): Schematic illustration of Hes4 regulation and function in the presumptive adult RSCs. Abbreviations: CMZ, ciliary marginal zone; GCL, ganglion cell layer; INL/ONL: inner/outer nuclear layer; NR, neural retina; RPE, retinal pigmented epithelium.
Hes4 Maintains Presumptive RSCs in a Proliferative and Undifferentiated State
Our gain of function analysis revealed that Hes4 misexpression prevents retinal precursors from committing toward retinal fates. Of note, Hes4 ability to block neurogenesis is a function shared by many other Hes family members that are known to transcriptionally repress or functionally antagonize several proneural factors [24]. In particular, cHairy1 has proved to be required to prevent CMZ cells from differentiating into neurons and to be sufficient to endow progenitor cells of the CR with CMZ-like cell properties [38]. We found here that the role of Hes4 during Xenopus retinal development may also extend to an inhibition of the RPE differentiation program. Besides, our results indicate that the Hes4-dependent blockade of neuronal and RPE determination is accompanied by maintenance in a proliferative state. Whether the former effect is a consequence of the latter or vice versa remains to be determined. However, both appear tightly coupled since Hes4-induced defective neurogenesis, as observed in lipofection experiments, can be rescued by forcing cells to exit the cell cycle. We thus conclude that the Hes4 phenotype observed in our overexpression experiments reflects a function dedicated to maintain presumptive adult RSCs in an undifferentiated and proliferative state towards adulthood.
Hes4 Regulates Cell Cycle Kinetics
Our study highlights that Hes4 might also constitute a crucial regulator of cell cycle kinetics. Cell cycle speed is known to vary during the time course of vertebrate retinogenesis [44, 56, 57], a phenomenon that has been associated with the transition from proliferative to neurogenic divisions in other parts of the central nervous system [58]. However, comparative analysis of proliferation kinetics between different retinal compartments at a given stage has not been performed so far. We found in this study that Hes4-expressing cells at the NR/RPE border of the optic vesicle exhibit a much longer cell cycle compared to NR precursors. This likely constitutes a hallmark of neural stem cells as observed in adult RSCs of the CMZ ([6] and our own unpublished data). How cell cycle speed is modulated in these cells is poorly documented. We here demonstrated that Hes4 prolongs cell cycle duration, mainly through a lengthening of G1 phase. In line with this result, sustained overexpression of Hes1 in brain neural progenitors was previously shown to downregulate expression of cell cycle regulators such as CyclinD1 and CyclinE2 and to cause G1 phase retardation [26]. Our data thus strongly support the hypothesis that Hes4 regulates some aspects of RSC homeostasis through the maintenance of slow cell cycle kinetics.
Hes4 Is Tightly Regulated by Multiple Inputs
We recently discovered that opposed and counterbalancing functions of Wnt and Hedgehog signaling modulate neural stem/progenitor cell proliferation in the postembryonic CMZ and that the two pathways reciprocally regulate each other's activity [41]. In line with this model, we found that the two pathways exhibit opposite effects on Hes4 expression in the forming CMZ.
The Hes4-dependent maintenance in the cell cycle associated with slow cell cycle kinetics is highly reminiscent of the phenotype resulting from Hedgehog signaling blockade in the retina [12]. Consistent with this, but in sharp contrast with Hes1 regulation in the mouse retina [53], we found that the Hedgehog pathway negatively impacts on Hes4 expression in the forming CMZ. In line with our previously proposed model [12, 59], it would thus be interesting to investigate whether Hedgehog-driven downregulation of Hes4 is required for the transition from a slow cycling RSC phenotype to a fast cycling progenitor state (Fig. 7B).
Our results also highlight that the canonical Wnt pathway (a) is active in the presumptive RSC subpopulation and (b) positively modulates Hes4 levels in the forming CMZ. These results suggest that Wnt signaling may function upstream Hes4 in the maintenance of RSCs in a proliferative and undifferentiated state during retinogenesis (Fig. 7B). This hypothesis is substantiated by the fact that the Wnt pathway is required for proliferation in the postembryonic Xenopus CMZ [10], and that Wnt-dependent prevention of chick CMZ cell differentiation is mediated by cHairy1 [38]. A positive link between the Wnt receptors frizzled 5/8 and Hes1 gene has also recently been observed in the context of cell proliferation in the mouse neuroretina [60]. Besides, activation of the Wnt pathway through overexpression of stabilized β-catenin in mouse [61] or in the zebrafish apc mutant [62] results in lower proliferation rates in the peripheral retina. It is therefore tempting to speculate that Wnt signaling might control as well Hes4-driven slow cell cycle speed of neural stem cells in the Xenopus retina.
In addition to these opposite influences of Wnt and Hedgehog signaling, we found that Hes4 is able to repress its own transcription. Such a negative feedback loop has previously been reported for Hes1 and is known to contribute to oscillations of its expression in brain neural precursors [26, 63]. However, Hes4-expressing cells in the retina exhibit features that are strikingly reminiscent of boundary cell properties (constitutive repression of proneural genes and reduced proliferation rates). In these boundary cells Hes1, like other Hes-related genes, does not oscillate and is rather persistently expressed at high levels [20, 27]. This sustained upregulation is due to the action of Id factors that prevent Hes1 from binding to its own promoter [64]. As shown here, Id2 and Hes4 exhibit a similar expression pattern in the developing retina. It would thus be interesting to determine whether a similar interaction leading to sustained Hes4 expression might be at work in RSCs.
CONCLUSION
Altogether, our study opens new avenue onto the cell-of-origin of adult RSCs and the involvement of the transcriptional repressor Hes4 in their maintenance during embryonic development. Our results indeed suggest that Hes4 expression at the NR/RPE border of the optic vesicle foreshadows the adult RSC pool of the CMZ. Besides, our functional analysis converges toward a model whereby Hes4 acts as a safeguard of neural stemness features in this cell subpopulation by preventing its differentiation and maintaining it in a slowly proliferative state.
Acknowledgments
We thank E. Bellefroid and M. Nichane for providing the Hes4 expressing constructs and F. Cremisi for the pCS2-Xgadd-45γ plasmid. We are very grateful to Pr. R. Nowakowski for providing the Excel sheet and for helpful discussions on EdU cumulative labeling. We thank S. Ohnuma and L. Bally-Cuif for critical reading of the manuscript and G. Luque for statistical advises. We are also grateful to C. de Meideros for animal care. W.E.Y. was granted by MNERT and Retina France. C.B. is an ANR and Region Ile de France fellow (DIM Stem Pole). This research was supported by grants to M.P. from the ANR (06-JCJC-0108-01 and 056054), CNRS, Univ. Paris-Sud, Retina France, ARC (1108), IFR144 and to K.V. from the Research Foundation-Flanders, Interuniversity Attraction Poles IAP7, the Belgian Association against Cancer and the Concerted Research Actions (GOA) of Ghent University.
DISCLOSURE OF POTENTIAL C ONFLICTS OF INTEREST
The authors indicate no potential conflicts of interest.
Supplementary material
Additional Supporting Information may be found in the online version of this article.
Figure S1. Hes4 expression on whole mount embryos. Whole mount in situ hybridization analysis of Hes4 expression (lateral views of tailbud heads, anterior to the right). In addition to the eye, Hes4 expression is detected in several parts of the brain and spinal cord, in the branchial arches (white arrow in B-E) and in the pronephros (black arrowhead in B-D). For more details on Hes4 expression pattern in whole mount embryos, see [65]. Scale Bar: 300 μm.
Figure S2. Expression pattern of the retinal stem cell markers Id2 and Wnt8b during retinogenesis. In situ hybridization analysis of Id2 and Wnt8b expression during retinal development. Following expression in the presumptive RPE at the optic vesicle stage (stage 24), both genes get restricted to the peripheral RPE and forming CMZ in the optic cup (stage 32). Their expression in the CMZ of the mature retina (stage 41) is confined to the most peripheral region where neural stem cells reside (see magnifications of the dorsal CMZ on the right panels). Note that both Id2 and Wnt8b exhibit a more intense staining dorsally than ventrally. Scale Bar: 50 μm.
Figure S3. Hes4 is expressed in domains exhibiting high Wnt and low Hedgehog activity. Comparative in situ hybridization analysis of Hes4 expression profile with that of eGFP (in Wnt-responsive transgenic embryos) and Gli1 (as a readout of hedgehog signalling activity). Black and red dotted lines respectively delineate the RPE/NR border and the stem cell zone of the CMZ. Red and black arrows respectively point to the differentiating central RPE and the retinal margin. Drawings on the right schematize Hes4 expression and domains with Wnt or Hedgehog activity at stage 25 and 35. Scale Bar: 50 μm. Hes4 and retinal stemness
Figure S4. Hes4 represses its own transcription. (A) Whole mount in situ hybridization analysis of N-Tubulin and Hes4 expression on stage 23 embryos injected with Hes4VP16-GR mRNA. Arrows indicate ectopic expression in the epidermis (anterior on the right, dorsal side up). Of note, a similar result was obtained with a probe designed in the 3′UTR of the Hes4 gene, confirming that it was not due to crosshybridization with the Hes4VP16-GR mRNA. (B) Whole mount in situ hybridization analysis of Rx and Hes4 expression on stage 20 embryos, following injection of control or Hes4 morpholinos (Mo). Note the reduction of Rx staining in the eye field (black arrow) and the increase of Hes4 expression (white arrows) on the injected side (Inj). N indicates the proportion of embryos exhibiting the corresponding phenotype. Scale bar: 500 μm (A) or 300 μm (B).
Figure S5. Hes4 expression is insensitive to a 24-hour Notch signalling blockade. (A-E) In situ hybridization analysis following a 24- (from stage 25 to stage 35; A, B) or a 48- hour (from stage 25 to stage 39; C, D) treatment with the Notch inhibitor DAPT. (A, C) Expression of the Notch target gene HRT1 (lateral views of the head) serves as a control of DAPT efficiency [42]. Arrows point to the brain labelling, which is clearly decreased upon DAPT treatment. (B, D) Typical Hes4 staining on retinal sections from control or treated embryos as indicated. Shown on the right are higher magnifications of the dorsal CMZ, delineated with dotted lines. (E) Quantification of Hes4 staining area in the dorsal CMZ in each condition. (F) qPCR analysis of retinal Hes4 expression at stage 35 following a 24-hour DAPT treatment. N-Tubulin serves as a control of drug efficiency. Scale bar: 50 μm.
Figure S6. Hes4 overexpression prolongs proliferation of retinal progenitors. (A) Typical sections of stage 32 retinas lipofected with GFP and immunostained for both GFP and BrdU. White and yellow arrows point to a GFP+/BrdU- and to a GFP+/BrdU+ cell, Hes4 and retinal stemness respectively. (B) Quantification of BrdU incorporation (3-hour pulse) in retinal clones at stage 32 or 35, following Hes4 or Hes2 lipofection. Scale Bar: 50 μm.
Figure S7. Hes4-dependent reduction of neurogenesis can be rescued by forcing cell cycle exit. (A, B) Analysis of cell type distribution in stage 41 retinas, following Hes4, Xgadd-45γ or Hes4 plus Xgadd-45γ lipofection. (A) Typical sections of retinas transfected with the indicated construct. (B) Quantification of neurons and glia among transfected cells in each condition. (C) Birthdating experiments in retinas lipofected with Hes4, Xgadd-45γ or Hes4 plus Xgadd-45γ. GCL: ganglion cell layer; INL/ONL: inner/outer nuclear layer. Scale Bar: 25 μm.
REFERENCES
- 1.Yoshii C, Ueda Y, Okamoto M, et al. Neural retinal regeneration in the anuran amphibian Xenopus laevis post-metamorphosis: Transdifferentiation of retinal pigmented epithelium regenerates the neural retina. Dev Biol. 2007;303:45–56. doi: 10.1016/j.ydbio.2006.11.024. [DOI] [PubMed] [Google Scholar]
- 2.Wetts R, Serbedzija GN, Fraser SE. Cell lineage analysis reveals multipotent precursors in the ciliary margin of the frog retina. Dev Biol. 1989;136:254–263. doi: 10.1016/0012-1606(89)90146-2. [DOI] [PubMed] [Google Scholar]
- 3.Casarosa S, Leone P, Cannata S, et al. Genetic analysis of metamorphic and premetamorphic Xenopus ciliary marginal zone. Dev Dyn. 2005;233:646–651. doi: 10.1002/dvdy.20369. [DOI] [PubMed] [Google Scholar]
- 4.Perron M, Kanekar S, Vetter ML, et al. The genetic sequence of retinal development in the ciliary margin of the Xenopus eye. Dev Biol. 1998;199:185–200. doi: 10.1006/dbio.1998.8939. [DOI] [PubMed] [Google Scholar]
- 5.Centanin L, Hoeckendorf B, Wittbrodt J. Fate restriction and multipotency in retinal stem cells. Cell Stem Cell. 2011;9:553–562. doi: 10.1016/j.stem.2011.11.004. [DOI] [PubMed] [Google Scholar]
- 6.Xue X, Harris W. Using myc genes to search for stem cells in the ciliary margin of the Xenopus Retina. Dev Neurobiol. 2012;72:475–490. doi: 10.1002/dneu.20887. [DOI] [PubMed] [Google Scholar]
- 7.Locker M, Borday C, Perron M. Stemness or not stemness? Current status and perspectives of adult retinal stem cells. Curr Stem Cell Res Ther. 2009;4:118–130. doi: 10.2174/157488809788167382. [DOI] [PubMed] [Google Scholar]
- 8.Raymond PA, Barthel LK, Bernardos RL, et al. Molecular characterization of retinal stem cells and their niches in adult zebrafish. BMC Dev Biol. 2006;6:36. doi: 10.1186/1471-213X-6-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cerveny KL, Cavodeassi F, Turner KJ, et al. The zebrafish flotte lotte mutant reveals that the local retinal environment promotes the differentiation of proliferating precursors emerging from their stem cell niche. Development. 2010;137:2107–2115. doi: 10.1242/dev.047753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Denayer T, Locker M, Borday C, et al. Canonical Wnt signaling controls proliferation of retinal stem/progenitor cells in postembryonic Xenopus eyes. Stem Cells. 2008;26:2063–2074. doi: 10.1634/stemcells.2007-0900. [DOI] [PubMed] [Google Scholar]
- 11.Kubo F, Takeichi M, Nakagawa S. Wnt2b controls retinal cell differentiation at the ciliary marginal zone. Development. 2003;130:587–598. doi: 10.1242/dev.00244. [DOI] [PubMed] [Google Scholar]
- 12.Locker M, Agathocleous M, Amato MA, et al. Hedgehog signaling and the retina: Insights into the mechanisms controlling the proliferative properties of neural precursors. Genes Dev. 2006;20:3036–3048. doi: 10.1101/gad.391106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moshiri A, McGuire CR, Reh TA. Sonic hedgehog regulates proliferation of the retinal ciliary marginal zone in posthatch chicks. Dev Dyn. 2005;233:66–75. doi: 10.1002/dvdy.20299. [DOI] [PubMed] [Google Scholar]
- 14.Parain K, Mazurier N, Bronchain O, et al. A large scale screen for neural stem cell markers in Xenopus retina. Dev Neurobiol. 2012;72:491–506. doi: 10.1002/dneu.20973. [DOI] [PubMed] [Google Scholar]
- 15.Wehman AM, Staub W, Meyers JR, et al. Genetic dissection of the zebrafish retinal stem-cell compartment. Dev Biol. 2005;281:53–65. doi: 10.1016/j.ydbio.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 16.Cerveny KL, Varga M, Wilson SW. Continued growth and circuit building in the anamniote visual system. Dev Neurobiol. 2012;72:328–345. doi: 10.1002/dneu.20917. [DOI] [PubMed] [Google Scholar]
- 17.Locker M, El Yakoubi W, Mazurier N, et al. A decade of mammalian retinal stem cell research. Arch Ital Biol. 2010;148:59–72. [PubMed] [Google Scholar]
- 18.Davis RL, Turner DL. Vertebrate hairy and enhancer of split related proteins: Transcriptional repressors regulating cellular differentiation and embryonic patterning. Oncogene. 2001;20:8342–8357. doi: 10.1038/sj.onc.1205094. [DOI] [PubMed] [Google Scholar]
- 19.Kageyama R, Ohtsuka T, Kobayashi T. The Hes gene family: Repressors and oscillators that orchestrate embryogenesis. Development. 2007;134:1243–1251. doi: 10.1242/dev.000786. [DOI] [PubMed] [Google Scholar]
- 20.Baek JH, Hatakeyama J, Sakamoto S, et al. Persistent and high levels of Hes1 expression regulate boundary formation in the developing central nervous system. Development. 2006;133:2467–2476. doi: 10.1242/dev.02403. [DOI] [PubMed] [Google Scholar]
- 21.Bonev B, Pisco A, Papalopulu N. MicroRNA-9 reveals regional diversity of neural progenitors along the anterior–posterior axis. Dev Cell. 2011;20:19–32. doi: 10.1016/j.devcel.2010.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hatakeyama J, Bessho Y, Katoh K, et al. Hes genes regulate size, shape and histogenesis of the nervous system by control of the timing of neural stem cell differentiation. Development. 2004;131:5539–5550. doi: 10.1242/dev.01436. [DOI] [PubMed] [Google Scholar]
- 23.Ishibashi M, Moriyoshi K, Sasai Y, et al. Persistent expression of helix-loop-helix factor HES-1 prevents mammalian neural differentiation in the central nervous system. EMBO J. 1994;13:1799–1805. doi: 10.1002/j.1460-2075.1994.tb06448.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kageyama R, Ohtsuka T, Kobayashi T. Roles of Hes genes in neural development. Dev Growth Differ. 2008;50(suppl 1):S97–S103. doi: 10.1111/j.1440-169X.2008.00993.x. [DOI] [PubMed] [Google Scholar]
- 25.Ohtsuka T, Sakamoto M, Guillemot F, et al. Roles of the basic helix-loop-helix genes Hes1 and Hes5 in expansion of neural stem cells of the developing brain. J Biol Chem. 2001;276:30467–30474. doi: 10.1074/jbc.M102420200. [DOI] [PubMed] [Google Scholar]
- 26.Shimojo H, Ohtsuka T, Kageyama R. Oscillations in notch signaling regulate maintenance of neural progenitors. Neuron. 2008;58:52–64. doi: 10.1016/j.neuron.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 27.Stigloher C, Chapouton P, Adolf B, et al. Identification of neural progenitor pools by E(Spl) factors in the embryonic and adult brain. Brain Res Bull. 2008;75:266–273. doi: 10.1016/j.brainresbull.2007.10.032. [DOI] [PubMed] [Google Scholar]
- 28.Lee HY, Wroblewski E, Philips GT, et al. Multiple requirements for Hes1 during early eye formation. Dev Biol. 2005;284:464–478. doi: 10.1016/j.ydbio.2005.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takatsuka K, Hatakeyama J, Bessho Y, et al. Roles of the bHLH gene Hes1 in retinal morphogenesis. Brain Res. 2004;1004:148–155. doi: 10.1016/j.brainres.2004.01.045. [DOI] [PubMed] [Google Scholar]
- 30.Tomita K, Ishibashi M, Nakahara K, et al. Mammalian hairy and enhancer of split homolog 1 regulates differentiation of retinal neurons and is essential for eye morphogenesis. Neuron. 1996;16:723–734. doi: 10.1016/s0896-6273(00)80093-8. [DOI] [PubMed] [Google Scholar]
- 31.Sang L, Coller HA. Fear of commitment: Hes1 protects quiescent fibroblasts from irreversible cellular fates. Cell Cycle. 2009;8:2161–2167. doi: 10.4161/cc.8.14.9104. [DOI] [PubMed] [Google Scholar]
- 32.Sang L, Coller HA, Roberts JM. Control of the reversibility of cellular quiescence by the transcriptional repressor HES1. Science. 2008;321:1095–1100. doi: 10.1126/science.1155998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nagatomo K, Hashimoto C. Xenopus hairy2 functions in neural crest formation by maintaining cells in a mitotic and undifferentiated state. Dev Dyn. 2007;236:1475–1483. doi: 10.1002/dvdy.21152. [DOI] [PubMed] [Google Scholar]
- 34.Nichane M, de Croze N, Ren X, et al. Hairy2–Id3 interactions play an essential role in Xenopus neural crest progenitor specification. Dev Biol. 2008;322:355–367. doi: 10.1016/j.ydbio.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 35.Nichane M, Ren X, Souopgui J, et al. Hairy2 functions through both DNA-binding and non DNA-binding mechanisms at the neural plate border in Xenopus. Dev Biol. 2008;322:368–380. doi: 10.1016/j.ydbio.2008.07.026. [DOI] [PubMed] [Google Scholar]
- 36.Radosevic M, Robert-Moreno A, Coolen M, et al. Her9 represses neurogenic fate downstream of Tbx1 and retinoic acid signaling in the inner ear. Development. 2011;138:397–408. doi: 10.1242/dev.056093. [DOI] [PubMed] [Google Scholar]
- 37.Jouve C, Palmeirim I, Henrique D, et al. Notch signalling is required for cyclic expression of the hairy-like gene HES1 in the presomitic mesoderm. Development. 2000;127:1421–1429. doi: 10.1242/dev.127.7.1421. [DOI] [PubMed] [Google Scholar]
- 38.Kubo F, Nakagawa S. Hairy1 acts as a node downstream of Wnt signaling to maintain retinal stem cell-like progenitor cells in the chick ciliary marginal zone. Development. 2009;136:1823–1833. doi: 10.1242/dev.029272. [DOI] [PubMed] [Google Scholar]
- 39.Matter-Sadzinski L, Puzianowska-Kuznicka M, Hernandez J, et al. A bHLH transcriptional network regulating the specification of retinal ganglion cells. Development. 2005;132:3907–3921. doi: 10.1242/dev.01960. [DOI] [PubMed] [Google Scholar]
- 40.Tran HT, Sekkali B, Van Imschoot G, et al. Wnt/beta-catenin signaling is involved in the induction and maintenance of primitive hematopoiesis in the vertebrate embryo. Proc Natl Acad Sci USA. 2010;107:16160–16165. doi: 10.1073/pnas.1007725107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Borday C, Cabochette P, Parain K, et al. Antagonistic cross-regulation between Wnt and Hedgehog signalling pathways controls post-embryonic retinal proliferation. Development. 2012;139:3499–3509. doi: 10.1242/dev.079582. [DOI] [PubMed] [Google Scholar]
- 42.Taelman V, Van Campenhout C, Solter M, et al. The Notch-effector HRT1 gene plays a role in glomerular development and patterning of the Xenopus pronephros anlagen. Development. 2006;133:2961–2971. doi: 10.1242/dev.02458. [DOI] [PubMed] [Google Scholar]
- 43.Solter M, Locker M, Boy S, et al. Characterization and function of the bHLH-O protein XHes2: Insight into the mechanisms controlling retinal cell fate decision. Development. 2006;133:4097–4108. doi: 10.1242/dev.02567. [DOI] [PubMed] [Google Scholar]
- 44.Decembrini S, Andreazzoli M, Vignali R, et al. Timing the generation of distinct retinal cells by homeobox proteins. Plos Biol. 2006;4:e272. doi: 10.1371/journal.pbio.0040272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ohnuma S, Mann F, Boy S, et al. Lipofection strategy for the study of Xenopus retinal development. Methods. 2002;28:411–419. doi: 10.1016/s1046-2023(02)00260-8. [DOI] [PubMed] [Google Scholar]
- 46.Nowakowski RS, Lewin SB, Miller MW. Bromodeoxyuridine immunohistochemical determination of the lengths of the cell cycle and the DNA-synthetic phase for an anatomically defined population. J Neurocytol. 1989;18:311–318. doi: 10.1007/BF01190834. [DOI] [PubMed] [Google Scholar]
- 47.Arai Y, Pulvers JN, Haffner C, et al. Neural stem and progenitor cells shorten S-phase on commitment to neuron production. Nat Commun. 2011;2:154. doi: 10.1038/ncomms1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ohnuma S, Hopper S, Wang KC, et al. Co-ordinating retinal histogenesis: Early cell cycle exit enhances early cell fate determination in the Xenopus retina. Development. 2002;129:2435–2446. doi: 10.1242/dev.129.10.2435. [DOI] [PubMed] [Google Scholar]
- 49.Perron M, Boy S, Amato MA, et al. A novel function for Hedgehog signalling in retinal pigment epithelium differentiation. Development. 2003;130:1565–1577. doi: 10.1242/dev.00391. [DOI] [PubMed] [Google Scholar]
- 50.Kubo F, Nakagawa S. Wnt signaling in retinal stem cells and regeneration. Dev Growth Differ. 2008;50:245–251. doi: 10.1111/j.1440-169X.2008.01033.x. [DOI] [PubMed] [Google Scholar]
- 51.Van Raay TJ, Vetter ML. Wnt/frizzled signaling during vertebrate retinal development. Dev Neurosci. 2004;26:352–358. doi: 10.1159/000082277. [DOI] [PubMed] [Google Scholar]
- 52.Hirata H, Yoshiura S, Ohtsuka T, et al. Oscillatory expression of the bHLH factor Hes1 regulated by a negative feedback loop. Science. 2002;298:840–843. doi: 10.1126/science.1074560. [DOI] [PubMed] [Google Scholar]
- 53.Wall DS, Mears AJ, McNeill B, et al. Progenitor cell proliferation in the retina is dependent on Notch-independent Sonic hedgehog/Hes1 activity. J Cell Biol. 2009;184:101–112. doi: 10.1083/jcb.200805155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nelson BR, Gumuscu B, Hartman BH, et al. Notch activity is downregulated just prior to retinal ganglion cell differentiation. Dev Neurosci. 2006;28:128–141. doi: 10.1159/000090759. [DOI] [PubMed] [Google Scholar]
- 55.Kwan KM, Otsuna H, Kidokoro H, et al. A complex choreography of cell movements shapes the vertebrate eye. Development. 2012;139:359–372. doi: 10.1242/dev.071407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Alexiades MR, Cepko C. Quantitative analysis of proliferation and cell cycle length during development of the rat retina. Dev Dyn. 1996;205:293–307. doi: 10.1002/(SICI)1097-0177(199603)205:3<293::AID-AJA9>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 57.Morris VB, Cowan R. An analysis of the growth of the retinal cell population in embryonic chicks yielding proliferative ratios, numbers of proliferative and non-proliferative cells and cell-cycle times for successive generations of cell cycles. Cell Prolif. 1995;28:373–391. doi: 10.1111/j.1365-2184.1995.tb00079.x. [DOI] [PubMed] [Google Scholar]
- 58.Gotz M, Huttner WB. The cell biology of neurogenesis. Nat Rev Mol Cell Biol. 2005;6:777–788. doi: 10.1038/nrm1739. [DOI] [PubMed] [Google Scholar]
- 59.Agathocleous M, Locker M, Harris WA, et al. A general role of hedgehog in the regulation of proliferation. Cell Cycle. 2007;6:156–159. doi: 10.4161/cc.6.2.3745. [DOI] [PubMed] [Google Scholar]
- 60.Liu C, Bakeri H, Li T, et al. Regulation of retinal progenitor expansion by Frizzled receptors: Implications for microphthalmia and retinal coloboma. Hum Mol Genet. 2012;21:1848–1860. doi: 10.1093/hmg/ddr616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu H, Xu S, Wang Y, et al. Ciliary margin transdifferentiation from neural retina is controlled by canonical Wnt signaling. Dev Biol. 2007;308:54–67. doi: 10.1016/j.ydbio.2007.04.052. [DOI] [PubMed] [Google Scholar]
- 62.Stephens WZ, Senecal M, Nguyen M, et al. Loss of adenomatous polyposis coli (apc) results in an expanded ciliary marginal zone in the zebrafish eye. Dev Dyn. 2010;239:2066–2077. doi: 10.1002/dvdy.22325. [DOI] [PubMed] [Google Scholar]
- 63.Kageyama R, Ohtsuka T, Shimojo H, et al. Dynamic Notch signaling in neural progenitor cells and a revised view of lateral inhibition. Nat Neurosci. 2008;11:1247–1251. doi: 10.1038/nn.2208. [DOI] [PubMed] [Google Scholar]
- 64.Bai G, Sheng N, Xie Z, et al. Id sustains Hes1 expression to inhibit precocious neurogenesis by releasing negative autoregulation of Hes1. Dev Cell. 2007;13:283–297. doi: 10.1016/j.devcel.2007.05.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.