Abstract
One of the paradigms in cancer pathogenesis is the requirement of a cell to undergo transformation from respiration to aerobic glycolysis – the Warburg effect – to become malignant. The demands of a rapidly proliferating cell for carbon metabolites for the synthesis of biomass, energy and redox equivalents, are fundamentally different from the requirements of a differentiated, quiescent cell, but it remains open whether this metabolic switch is a cause or a consequence of malignant transformation. One of the major requirements is the synthesis of lipids for membrane formation to allow for cell proliferation, cell cycle progression and cytokinesis. Enzymes involved in lipid metabolism were indeed found to play a major role in cancer cell proliferation, and most of these enzymes are conserved in the yeast, Saccharomyces cerevisiae. Most notably, cancer cell physiology and metabolic fluxes are very similar to those in the fermenting and rapidly proliferating yeast. Both types of cells display highly active pathways for the synthesis of fatty acids and their incorporation into complex lipids, and imbalances in synthesis or turnover of lipids affect growth and viability of both yeast and cancer cells. Thus, understanding lipid metabolism in S. cerevisiae during cell cycle progression and cell proliferation may complement recent efforts to understand the importance and fundamental regulatory mechanisms of these pathways in cancer.
Keywords: Malignant transformation, Warburg effect, Fatty acids, Membrane lipids, Storage lipids, Saccharomyces cerevisiae
Highlights
► Warburg effect of cancer and Crabtree effect of yeast result in similar physiology. ► Proliferating cells have a high demand for membrane biogenesis. ► Pathways for fatty acid synthesis are highly active in cancer. ► Phospholipid remodeling is important for membrane homeostasis in growing cells.
1. Introduction
In their review “The Hallmarks of Cancer”, Hanahan and Weinberg [1] defined six alterations in cellular processes that are required to turn a normal cell into a cancer cell: response to growth and antigrowth signals, prevention of apoptosis and senescence, induction of angiogenesis, and the ability to colonize other body parts. These milestones in cancer development, however, are exclusively addressing regulatory and signaling processes. In a recent update of their work, the authors expanded their list of required modifications for cancer development by two novel – emerging – hallmarks, namely defense against immune destruction and modifications in energy metabolism [2]. The latter – energy metabolism – is in fact a trait, which has been described in some detail already in the twenties of the last century. Otto Warburg published his observations that the metabolism of cancer cells is shifted from the oxidation of glucose to carbon dioxide and respiration-driven ATP production to fermentative reduction of pyruvate to lactate. Under these conditions, ATP is mainly derived from cytosolic glycolysis, which, however, is a much less efficient pathway to generate energy compared to mitochondrial respiration [3]. Only a few years later, Herbert G. Crabtree showed that mitochondrial respiration in neoplastic tissue is repressed at physiological glucose concentrations [4]. Warburg concluded from his seminal work that cancer cells develop due to defective mitochondrial respiration, forcing the cell to shift to aerobic fermentation, even if oxygen supply is not limiting [5]. Although it has been shown meanwhile that cancer cells are not necessarily compromised in respiratory activity, the switch to high glucose uptake rates and fermentative metabolism is still considered a property of almost all types of tumors [6,7]. The reasons for this physiological switch remain a matter of debate. Reduction of the metabolic flux from pyruvate to lactate results in sensitivity of cancer cells to hypoxic conditions and impaired growth [8]. However, it is unclear whether aerobic glycolysis is a prerequisite for a cell to become neoplastic or if this dramatic switch in metabolism occurs concomitantly to, or after malignant transformation. It has also been suggested that aerobic glycolysis that is accompanied by high glucose uptake rates and acidification of the extracellular environment, is a response to hypoxic conditions and may provide a growth advantage. Similarly, aerobically fermenting yeasts, which convert glucose to ethanol and acetic acid at high rates, prevent competing microorganisms from growth, while being tolerant to high ethanol concentrations and low pH themselves [9]. In addition, reduced mitochondrial activity might contribute to the ability of cancer cells to evade apoptosis. Although aerobic glycolysis generates only 2 mol of ATP per mol of glucose, the overall rate of ATP production might indeed be higher in aerobic glycolysis than in mitochondrial respiration, due to lower costs for enzyme production or higher activities of the fermenting enzymes, compared to tricarboxylic acid (TCA) cycle and respiratory chain enzymes and cofactors [10,11].
2. Cell proliferation: Energy requirements and biomass production
By considering metabolic fluxes it becomes obvious that a rapidly proliferating cell not only relies on sufficient supply of energy in the form of nucleoside triphosphates, but also on a sufficient flux of glucose- or amino acid-derived metabolites into biosynthetic pathways to fuel the duplication of biomass during every cell cycle (Fig. 1). In addition to the consumption of energy and carbon precursors, these biosynthetic activities require numerous anaplerotic reactions, especially in the TCA cycle, and the maintenance of a redox balance for the major redox cofactors, NAD+/NADH and NADP+/NADPH. Since the production of biomass from glucose-derived metabolites is mainly a reductive process that requires NADPH for the synthesis of amino acids and lipids, pathways for reduction of NADP+ have to be activated and the cytosolic NADH has to be rapidly re-oxidized to maintain high glycolytic rates. In contrast, a differentiated non-proliferating cell has no or only minor activity of anaplerotic biosynthetic pathways and can fully oxidize glucose to obtain the maximum possible ATP yield. This demand for a balanced supply of energy, redox equivalents and biomass precursors at high rates may provide an explanation for the switch of cancer cells to aerobic glycolysis, accompanied by high rates of glucose uptake and catabolism.
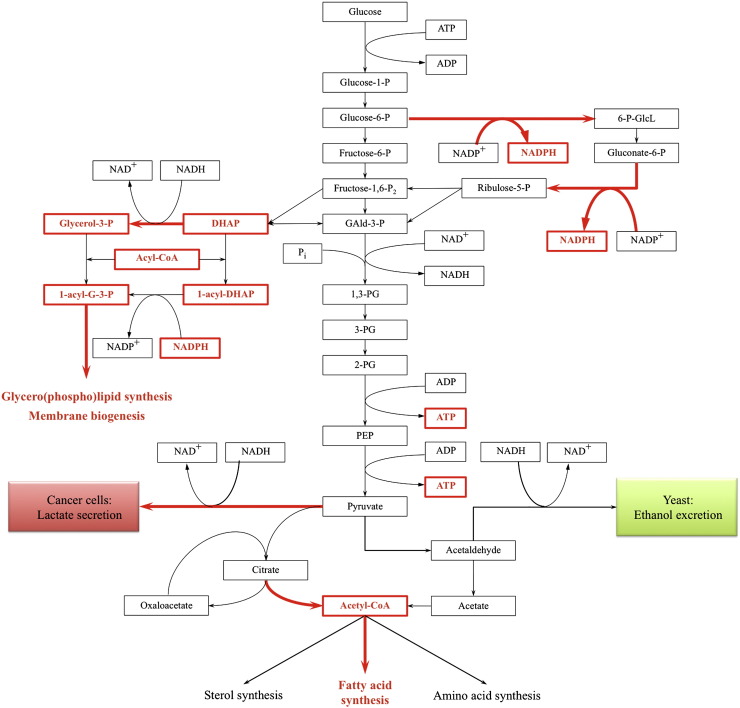
Fig. 1.
Interconnections between central carbon metabolism and lipid synthesis. Yeast and cancer cells rely on increased glucose uptake and high glycolytic activity to fuel the biosynthesis of biomass. For lipid synthesis, the major carbon precursor is acetyl-CoA, derived from the ATP:citrate lyase reaction in humans and from the pyruvate decarboxylase shunt in S. cerevisiae. Essential cofactors in lipid synthesis are ATP and NADPH. In addition, the acyl chain acceptors in phospholipid synthesis, G-3-P and DHAP, are products of glycolysis. Both cancer cells and yeast maintain high glycolytic rates for biomass formation and for ATP production by substrate-level phosphorylation. Most of the NADH derived from glycolysis is reoxidized through cytosolic reactions: in cancer cells, pyruvate is reduced to lactate. In yeast, reduction is preceded by decarboxylation of pyruvate to acetaldehyde, and yields ethanol. Both lactate and ethanol are excreted from the cell. Metabolites and cofactors required for lipid synthesis are printed in red, red arrows indicate reactions known to be up-regulated in cancer cells. DHAP — dihydroxyacetone phosphate, GAld-3-P — glyceraldehyde-3-phosphate, 6-P-GlcL — 6-phosphoglucono-δ-lactone, PEP — phosphoenolpyruvate, 1,3-PG — 1,3-bisphosphoglycerate, 2(3)-PG — 2(3)-phosphoglycerate.
In an attempt to simulate the growth behavior and physiology of a cancer cell with constraint based flux balance analysis (FBA), Shlomi et al. showed that their metabolic model switched from respiration to aerobic glycolysis at high specific growth rates when a constraint for the enzyme solvent capacity of the cellular milieu was introduced [12]. In this simulation, kinetic constants of metabolic enzymes were introduced to allow for the computation of required enzyme concentrations at a given specific growth rate. A shift from respiration with high ATP yield to low yield overflow metabolism occurs when the enzyme concentrations and their respective turnover numbers are in favor of glycolysis, even if this results in a loss of carbon conversion efficiency [12]. Similar results are obtained by FBA with metabolic models of Saccharomyces cerevisiae, although in most of these simulations uptake of oxygen is used as a constraint to shift cells from respiration to fermentation [13], a behavior that is known as the Crabtree effect in yeast.
S. cerevisiae is often considered an organism that is almost entirely committed to fermentative metabolism. Thus, the physiology of a yeast cell and a proliferating cancer cell is very similar with comparable metabolic fluxes in these two cell systems [14]. It has to be noted, however, that the degree to which cancer cells are commited to aerobic glycolysis can vary in a broad range, but not to the same extent as in yeast, where respiratory activity is rather low in the presence of glucose. Aerobic glycolysis in yeast relies on high glucose concentrations: when yeast is cultivated in continuous culture under steady state conditions (chemostat) with glucose as the limiting nutrient, cells gradually switch to respiration when the dilution rate (i.e. the rate of nutrient supply) is reduced: at dilution rates of 0.1 h− 1, i.e. at a specific growth rate μ = 0.1 h− 1, metabolism of yeast is fully respiratory. Indeed, aerobic glycolysis is absent in yeast cultures at glucose concentrations in the range of physiological serum glucose levels in humans (~ 1 g L− 1), but is gradually induced when the glucose concentration is increased above this value. Due to this behavior yeast is a good model to study changes on transcriptional or posttranslational levels or in metabolic fluxes that are associated with the transition from respiration to fermentation [15–17]. The Crabtree effect is genetically determined and reversible in yeast, whereas the Warburg effect in cancer cells seems to be a consequence of spontaneous mutations; it is noteworthy, however, that development of cancer and mortality correlate with blood glucose levels in many cases [18–22]. Hence, increased glucose levels seem to be associated with the switch of neoplastic cells from respiration to aerobic glycolysis, as is also the case in yeast.
Irrespective of the as yet unknown molecular mechanisms that lead to the reprogramming of central carbon metabolism, this transformation apparently provides cancer cells with growth and/or survival advantages that are necessary for their rapid proliferation and invasiveness. Since this metabolic switch has been recognized as an indispensable feature of neoplastic tissue, genes involved in metabolism have become promising targets for cancer therapy; accordingly, the involvement of cellular metabolism in the development and progression of cancer has gained increased attention in recent years. Although mammalian cells are able to take up all major biomass constituents – glucose, amino acids, fatty acids, cholesterol – from the bloodstream, proliferation of cancer cells seems – at least in part – to rely on the endogenous synthesis of these components, as indicated by the metabolic switch to fermentation.
The major metabolic fluxes in a proliferating cell are dedicated to the synthesis of proteins and lipids, whereas only minor fluxes contribute to the synthesis of other components of biomass, such as nucleic acids. Hence, lipid metabolism has an essential function in biomass generation, and is also a major determinant of the cellular redox status. Furthermore, several steps in lipid synthesis have been recognized as being crucial for rapidly growing cancer and also for yeast cells, emphasizing again the metabolic similarities between both types of cells. In the following, we will review in greater detail glycerolipid synthesis and its interconnection with glycolytic fluxes and cellular redox balance, in both yeast and cancer cells.
3. Lipids in yeast and mammals: Functional conservation and structural differences
The basic pathways for the synthesis of membrane glycerolipids are identical in yeast and mammalian cells (Fig. 2), resulting in the same classes of phospholipids (PL), namely phosphatidic acid, phosphatidylinositol, phosphatidylserine, phosphatidylethanolamine, phosphatidylcholine, phosphatidylglycerol, and cardiolipin. Excess fatty acids (FA) that are derived from endogenous de novo synthesis, lipid turnover or nutritional supply are stored as triacylglycerol (TAG) in cytosolic lipid droplets, keeping the total amount of cellular phospholipids within a narrow range. The regulatory mechanisms that control the amount of cellular phospholipids upon varying supplies with FA remain unknown. Major differences between yeast and mammalian lipids exist with respect to their FA composition, which, in yeast, is rather simple, and consists mainly of palmitic and stearic acid, and their monounsaturated derivatives, palmitoleic and oleic acid, respectively [23,24]. In addition to these major FA, polyunsaturated FAs with 18–24 carbon atoms contribute considerably to the total FA pool in mammalian cells, dependent on the type of tissue [25–29]. Yeast membranes contain the inositol-containing sphingolipids, phosphorylceramide, mannosylinositol phosphorylceramide and mannosyldiinositol phosphorylceramide [30,31]. Mammalian sphingolipids are more complex, with sphingomyelin being the major sphingolipid, in addition to various classes of saturated and unsaturated ceramides [32,33]. Cholesterol and ergosterol are essential sterols of cellular membranes in mammals and yeast, respectively. Besides influencing fluidity of membranes they are involved in correct localization and tertiary structure of membrane proteins and in the formation of local asymmetries in the membrane, such as lipid rafts, which are sphingolipid- and sterol-rich microdomains of the plasma membrane [34–40]. Yeast ergosterol and mammalian cholesterol, although structurally similar, are functionally not completely identical [41–44]. Both types of sterols can be esterified with FAs and stored in a biophysically inert form in lipid droplets, together with TAG. Both yeast and mammalian cells store FA as TAG, which might function as a reserve for the production of energy or as a supply of FA for the synthesis of complex lipids, dependent on nutritional state and growth conditions. Especially the regulatory interplay and metabolic inter-conversion between storage lipid (i.e. TAG) and membrane lipid (i.e. PL) have recently been recognized as an important determinant of cellular growth and proliferation in yeast [13,45], whereas the β-oxidation of fatty acids for generation of energy is repressed when glucose is present as the carbon source. Furthermore, peroxisomes are the exclusive site of FA breakdown in yeast, which is in contrast to higher organisms where this pathway is active in both, peroxisomes and mitochondria. In mammals, FA released from TAG are generally assumed to be destined for β-oxidation, although increasing evidence accumulates that indicates additional roles for lipolysis, including the release of metabolic signaling molecules [46]. A role of lipolysis-derived FA in phospholipid synthesis or turnover has not yet been described for mammalian cells.
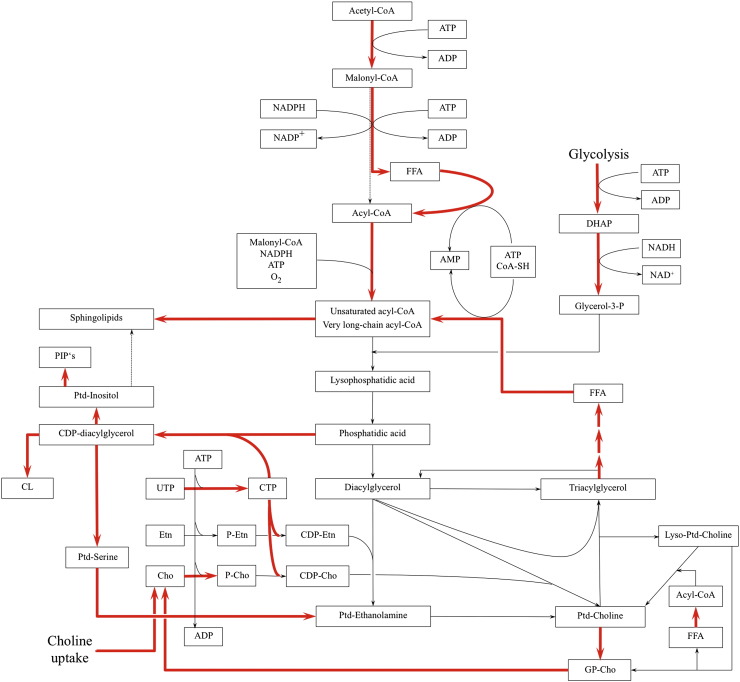
Fig. 2.
Metabolic pathways for the synthesis of glycerolipids. Due to their increased demand for membrane lipids, most cancers show a lipogenic phenotype. Most prominent are the upregulation of acetyl-CoA carboxylase and fatty acid synthase. Additionally, increased remodeling of phosphatidylcholine is observed in many malignous tissues. Pathways for the synthesis of CTP, the energy donor for phospholipid synthesis, of glycerol-3-phosphate, the FA acceptor in PA synthesis, of unsaturated FA and of phosphocholine are activated in some tumors. The storage form of lipids, triacylglycerol, is also involved in the remodeling of membrane lipids in proliferating cells, as suggested by altered regulation of lipases in cancer tissues. Red arrows indicate pathways that were found to play a role in cancer cells. CDP-Cho — CDP-phosphocholine, CDP-Etn — CDP-ethanolamine, Cho — choline, CL — cardiolipin, CTP — cytidine triphosphate, DAG — diacylglycerol, Etn — ethanolamine, FFA — free fatty acids, DHAP — dihydroxyacetone phosphate, G3P — glycerol-3-phosphate, GP-Cho — glycerophosphocholine, PC — phosphatidylcholine, P-Cho — phosphocholine, PE — phosphatidylethanolamine, P-Etn — phosphoethanolamine, PI — phosphatidylinositol, PS — phosphatidylserine, UTP — uridine triphosphate.
4. Central carbon metabolism provides the building blocks for membranes
All pathways for lipid syntheses share one common precursor, acetyl-CoA, and are characterized by a high demand of reductive power in the form of NADPH. For example, synthesis of one molecule of palmitic acid requires 8 acetyl-CoA and 14 NADPH molecules:
| (1) |
According to this equation (for simplification, equations are not balanced for H+, H2O, and inorganic phosphate), synthesis of one glycerophospholipid molecule with one C16 and one C18 FA consumes 17 acetyl-CoA and 30 NADPH molecules. Synthesis of sphingolipids follows a similar stoichiometry. Sterols, although synthesized in a completely different pathway, require comparable amounts of acetyl-CoA and NADPH as the synthesis of glycerophospholipids. Substitution of acetyl-CoA and NADPH in Eq. (1) with glucose as the carbon and energy source and the pentose phosphate pathway as the source for NADPH results in the following equation:
| (2) |
These equations illustrate that proliferating cells have a high demand for carbon precursors and redox equivalents for membrane biogenesis, which makes lipid synthesis one of the major consumers of acetyl-CoA and NADPH in the cell, besides amino acid synthesis. In comparison to these requirements, the consumption of ATP in the pathways for biomass production is moderate and can be met by low respiratory activity or – as in Warburg positive cancers and in fermenting yeast – by substrate level phosphorylation alone, followed by reduction of pyruvate to lactate (ethanol in yeast) for regeneration of NAD+.
Although proliferating cancer cells could satisfy their demand for lipids by the uptake of lipoprotein, the major part of FAs seems to be generated by de novo FA synthesis [47], corroborating the importance of metabolic adaptation to anabolic processes in rapidly proliferating cells.
Notably, acetyl-CoA in S. cerevisiae and mammalian cells is generated by somewhat different pathways (Fig. 1). Crabtree positive yeasts convert most of the pyruvate from glycolysis to acetaldehyde, which is either reduced to ethanol for regeneration of NAD+ or oxidized to acetate. Both ethanol and acetate can be excreted into the medium. Most of the acetate, however, is activated to acetyl-CoA. During fermentation in the presence of high glucose concentration, S. cerevisiae produces approximately 160 molecules of ethanol and only 7 molecules of acetyl-CoA per 100 molecules of glucose taken up from the medium [15], emphasizing the rather wasteful utilization of carbon source. Only minor amounts of pyruvate are converted to mitochondrial acetyl-CoA by pyruvate dehydrogenase, which is further metabolized to 2-ketoglutarate for the synthesis of amino acids. Subsequent reactions of the TCA cycle are transcriptionally repressed and not active at high glycolytic rates.
In mammalian cells, acetyl-CoA is derived from the reaction
| (3) |
which is catalyzed by the cytosolic enzyme ATP:citrate lyase (ACL). This reaction requires high activity of the first steps of the TCA cycle to generate citrate, which has to be translocated to the cytosol. Interestingly, recent data suggest that most of the citrate in cancer cells might be produced by carboxylation of α-ketoglutarate to isocitrate, which is then further converted to citrate. This reductive pathway to citrate requires high anaplerotic activity for α-ketoglutarate and might therefore explain the preference of many tumors for glutamine, which can be easily converted to α-ketoglutarate [48–50].
The requirement for ACL in proliferating tissue is reflected in impaired growth of tumors if the ACL gene is knocked down or if the enzyme activity is inhibited [51,52]. Furthermore, ACL expression and activity are increased in lung cancer cells and increased activity is dependent on the PI3P/Akt pathway [53]. Concomitantly, intracellular neutral lipid pools are filled up when ACL expression is knocked down, which also attenuates the growth rate of lung cancer cells [53]. Increased neutral lipid accumulation as a response to inhibited de novo FA synthesis seems paradoxical. However, S. cerevisiae behaves in a similar way when the growth rate decreases: upon entry into quiescence, cells start to accumulate TAG and steryl esters in lipid droplets. These lipid pools are only rapidly mobilized when growth is re-initiated in the presence of nutrients [54].
In addition to its role as a substrate for lipid and amino acid synthesis, acetyl-CoA is an important regulatory metabolite in the cell, e.g. in histone acetylation. Acetylation of metabolic enzymes has only recently been recognized as a means to regulate fluxes through central carbon metabolic pathways [55–57]. For example, the M2 isoform of pyruvate kinase PKM2, which is the isoform most active in malignant tissue, is acetylated at one of its lysine residues, K305 [58]. This modification results in a reduction of PKM2 activity and protein stability, which is thought to be responsible for attenuating the glycolytic flux and thereby making intermediate metabolites available for biosynthetic reactions. Mutation of K305 to a non-acetylatable glutamine residue increases cellular levels of NADPH, which indicates a higher flux through the pentose phosphate pathway (PPP). If the level of PKM2 acetylation is a function of the cellular concentration of acetyl-CoA, this regulation indeed leads to an increased supply of NADPH for reductive biosynthetic pathways at high acetyl-CoA levels. Whereas PKM1 and PKM2 are splice variants of one gene in mammals, the yeast genome contains two genes encoding for pyruvate kinases, namely CDC19 and PYK2. PYK2 is expressed only at low glucose levels and, like mammalian PKM1, not subject to regulation by fructose-1,6-bisphosphate. Under fermentative conditions, Cdc19 is the active pyruvate kinase that is regulated by fructose-1,6-bisphosphate and through phosphorylation by protein kinase A (PKA). Down-regulation of CDC19 gene expression results in a redirection of carbon fluxes into the TCA cycle, indicating higher respiratory activity. Whether the yeast pyruvate kinase Cdc19 is also acetylated as part of this regulatory loop has not yet been shown [59–62].
Attenuation of glycolysis through PKM2 acetylation might result in increased fluxes through several pathways, namely serine and glycine synthesis from 3-phosphoglycerate, glycerol-3-phosphate from dihydroxyacetone phosphate for glycerolipid synthesis, and the pentose phosphate pathway for reduction of NADP+ and for the synthesis of ribonucleic acids. Flux balance analysis in yeast shows that about 50% of the NADPH is consumed by glutamate dehydrogenase, the major reaction for ammonia assimilation; the remainder is distributed 60:40 between amino acid (AA) and lipid synthesis. In rapidly proliferating mammalian cells, which are relying on import of AA from the environment (at least the essential AA), lipid synthesis is presumably the major consumer of NADPH. Evidence suggests that glucose-6-phosphate dehydrogenase (G6PD), the rate-limiting enzyme in the pentose phosphate pathway, which generates NADPH, indeed plays a role in cancer. It was shown for several tumors that over-expression or increased activity of G6PD induces malignancy or is required for proliferation of cancer cells [63–68]. Increased activity or expression in cancer tissue has also been shown for the subsequent steps of the PPP [69,70]. The tumor suppressor p53 directly binds to G6PD, preventing the enzyme from forming the active homodimer; furthermore mutated forms of p53, such as found in tumors, are unable to inhibit G6PD activity [64]. In yeast, G6PD is encoded by the ZWF1 gene. Zwf1, like human G6PD, is negatively regulated by NADPH levels [71]. Interestingly, an additional inhibition of yeast Zwf1 by palmitoyl-CoA has been reported [72], which indicates a feedback regulation of NADPH production by the end product of fatty acid synthesis, further supporting the assumption that the PPP plays an important role in maintaining a high NADPH/NADP+ ratio for lipid biosynthesis in proliferating cells. These aspects make yeast a promising model for functional studies on the role of G6PD, since human G6PD is able to complement the phenotype of a Zwf1 deficient yeast mutant [73].
5. Fatty acid synthesis: A major sink for acetyl-CoA and NADPH
Tumor cells are characterized by elevated FA synthesis. The first and committed step is the conversion of acetyl-CoA to malonyl-CoA (Fig. 2), catalyzed by acetyl-CoA carboxylase (ACC). Interference with ACC activity, either by use of inhibitors or by silencing gene expression, was shown to be deleterious for several types of cancer cells [74–76]. Treatment of prostate cancer cells with the ACC-specific inhibitor soraphen A leads to growth arrest and, ultimately, cell death. [74]. Mammalian ACC bears at least three phosphorylation sites for AMP activated kinase, AMPK [77], which is one of the major regulators of cellular homeostasis in response to metabolic and energy status of the cell. AMPK phosphorylation of ACC reduces its activity, resulting in reduced fluxes of acetyl-CoA to malonyl-CoA and, hence, attenuates lipogenesis under conditions of low cellular energy load. Other targets of AMPK in lipid metabolism are HMG-CoA reductase, the committed step in sterol synthesis, and hormone sensitive lipase, HSL, which are both inhibited by AMPK phosphorylation. There is little doubt about the connection between reduced activity of AMPK, or its upstream activator LKB1, and cancer development, which has been demonstrated in countless studies (see [78–80] for recent reviews). Thus, reduced activity of AMPK may be one of the major determinants for the metabolic reprogramming of a cancer cell. Recent studies also suggest that AMPK might be linked to the metabolic syndrome, type 2 diabetes or obesity and an increased risk for cancer in patients with one of these disorders. The AMPK homologue in yeast is Snf1; its name – Sucrose Non-Fermenting – stems from the phenotype of mutants that are unable to express invertase, which is required for sucrose breakdown to fructose and glucose [81–83]. Snf1 phosphorylates metabolic enzymes, such as Acc1, as well as transcription factors and histones, and a deletion of the SNF1 gene results in reduced growth rates and increased lipogenesis, due to the inability of the mutant to phosphorylate and inactivate ACC. Interestingly, a SNF1 gene deletion also results in a reduced biomass yield in a glucose-limited chemostat under respiratory conditions, but not in a nitrogen-limited chemostat with fermentative metabolism. These results suggest that yeast AMPK not only plays a role in carbon catabolite repression but also in the switch from respiration to aerobic glycolysis, or vice versa [84].
In addition to its regulation at the level of gene expression and phosphorylation, ACC activity may also be regulated in a feedback loop by the product of FA synthesis, acyl-CoA: ACC activity in mammals [85–87] and presumably also in yeast [88], is inhibited by palmitoyl-CoA and palmitoleoyl-CoA, respectively. Interestingly, FA profiles are shifted towards longer chain length (C18:0 and C18:1) in yeast snf1 mutants. The resulting lower levels of C16 fatty acids in cells with impaired Snf1 function may result in an ever increased flux from acetyl-CoA to malonyl-CoA, thereby driving FA production due to the observed changes in FA chain length distribution.
The second enzyme in de novo fatty acid synthesis, fatty acid synthase (FASN), utilizes acetyl-CoA as a starter molecule and requires one malonyl-CoA, one ATP and one NADPH molecule for each elongation step. FASN generates free fatty acid (in mammals) or acyl-CoA (in yeast), typically 16 carbon atoms in length. Since its description as an important player in development and growth of malignant tissue [89] many studies have shown that FASN is one of the key metabolic enzymes in uncontrolled proliferation of most cancer cells. Thus, FASN has become one of the major targets in the search for anticancer therapies, because its inhibition results in arrest of tumor growth and induction of apoptosis (for recent reviews, see [90–92]). FASN is also one of the metabolic genes directly regulated by the oncogene HER2. In breast cancer, overexpression of HER2 correlates with increased expression and activity of FASN [93], although FASN expression is not directly controlled by HER2 but rather by sterol regulatory element-binding protein (SREBP), the general transcriptional activator of lipogenic genes, via the PI3K/Akt pathway [94,95]. A recent study also suggests an important role of the endoplasmic reticulum lipid raft-associated protein 2, ERLIN2, in activation of SREBP. ERLIN2 is overexpressed in breast tumors and seems to be required for activation of SREBP1c by a mechanism that involves binding of INSIG1 (insulin-induced gene 1) by ERLIN2. In addition, ERLIN2 is involved in the accumulation of triacylglycerol in cytosolic lipid droplets, indicating a role of this protein in the distribution of fatty acids between membrane and storage lipids [96,97].
Whereas the product of fatty acid synthase in yeast is acyl-CoA, mammalian FASN releases free fatty acid, which has to be activated to acyl-CoA, a reaction catalyzed by acyl-CoA synthetase (ACS). Several ACS isozymes have been shown to be upregulated in various types of cancers and an inhibitor of ACS, Triacsin C, was shown to induce apoptosis in glioma cells [98–100]. Hence, not only the synthesis of FA per se, but also the downstream pathways appear to be activated in cancer cells.
6. Desaturation of fatty acids
Functionality of cellular membranes and activity of membrane bound proteins are dependent on the FA composition of membrane lipids. To establish and maintain the correct composition, C16 and C18 saturated FA (SFA) derived from de novo synthesis are desaturated, yielding mono- or polyunsaturated FA (UFA), and elongated to very long-chain fatty acids, up to C26. Whereas the role of elongating enzymes in cancer cells is only poorly studied [101], the desaturation of palmitic and stearic acid to palmitoleic and oleic acid, respectively, is considered an important factor in the synthesis of membrane lipids for mitogenic cells. For serum-derived FAs, a positive correlation between increased oleic acid levels or a decreased saturation index and breast cancer risk has been suggested [102,103], although conflicting data exist [104,105]. Tumors with the lipogenic phenotype show a higher ratio of saturated to unsaturated FA, and treatment of cancer cells with the ACC inhibitor soraphen A results in increased levels of unsaturated FAs. The authors argue that an increased saturation index serves as a protective mechanism due to a more rigid membrane architecture, and reduces oxidative stress that may result from oxidation of unsaturated FA (UFA) [106].
Desaturation of FA relies on a complex reaction mechanism that involves electron transfers between cytochrome b5, NADH, molecular oxygen and the fatty acid substrate, catalyzed by the enzyme stearoyl-CoA desaturase (SCD). Reports about transcriptional upregulation of SCD1, the major desaturase in humans, in neoplastic tissue are scarce [107,108]. Nevertheless, the activity of the protein is indispensable for cellular proliferation, and a reduction of its activity in tumors results in growth inhibition [109,110]. Intriguingly, reduced activity of SCD1 seems to have an inhibitory effect on de novo fatty acid synthesis, resulting in reduced levels of most lipid classes and increased apoptotic cell death, suggesting a feedback regulatory mechanism to down-regulate ACC and/or FASN. One possibility for such a mechanism would be the inhibition of ACC by palmitoyl-CoA: reduced activity of the desaturase results in an increase in palmitoyl-CoA, thereby increasing the allosteric inhibition of ACC by palmitoyl-CoA. In contrast, increased activity of SCD1 is expected to result in a depletion of saturated FA and, hence, in increased ACC activity. In addition, it was found that AMPK-catalyzed phosphorylation and inactivation of ACC are enhanced in cells with reduced SCD1 activity due to increased phosphorylation/activation of AMPK [110]. On the other hand, higher activity of the desaturase, resulting in a shift of the FA composition to UFA, would also be detrimental for the cell. Over-expression of the yeast ∆9 desaturase Ole1 in cancer cells has shown that these cells suffer from a dramatic increase in their sensitivity to tumor necrosis factor, due to increased membrane fluidity [111]. These results corroborate the importance of an equilibrated supply of saturated and unsaturated fatty acids for membrane lipids. For yeast, we have recently shown that neutral lipid synthesis is involved in the maintenance of this equilibrium. When mutants lacking the ability to buffer excess FA into triacylgycerol are challenged with exogenous oleic acid this results in a rapid increase of oleate-containing membrane lipids and subsequent rapid loss of viability; saturated FA, however, showed no such effect [23]. SFA may or may not be irreversibly converted to UFA in the presence of oxygen to maintain the correct FA composition, and this process is strictly regulated by the activity of the Ole1 protein. Proliferating yeast cells are readily capable of dealing with a significant reduction of the levels of UFA; in contrast, over-expression of the OLE1 gene rather results in reduced growth rates and abnormal budding [112], indicating that increased desaturase activity is in fact detrimental for the cell.
Fatty acid desaturases are subject to tight control at the transcriptional level from yeast to humans [113,114], on the level of mRNA stability in yeast [115], and also with respect to protein stability through the ubiquitin-proteasome-dependent ER-associated degradation pathway [116,117]. Furthermore, evidence suggests that at least the yeast enzyme is also regulated by the degree of saturation of the membrane environment [118]. As a consequence of its putative role in the regulation of upstream enzymes involved in FA synthesis (see above) and the importance of a balanced supply of growing cells with SFA and UFA, SCD1 might become a promising target in anti-cancer drug discovery.
7. Building cellular membranes: Phospholipid synthesis
Fatty acids are incorporated into glycerophospholipids by sequential trans-esterification of two acyl-CoA molecules to glycerol-3-phosphate (G3P), yielding phosphatidic acid (PA). PA is either activated to cytidyldiphosphate-diacylglycerol, the precursor for all phospholipids, or dephosphorylated to diacylglycerol (DAG) by the enzyme lipin (LPIN, Pah1 in yeast); DAG may be utilized for triacylglycerol synthesis (see below), or channeled into the synthesis of phosphatidylethanolamine and phosphatidylcholine (PC) via the CDP-choline (also termed ‘Kennedy’)-pathway (Fig. 2). Several of the key enzymes in PL synthesis are up-regulated in cancer cells. The acceptor in the acyl-transferase reactions, G3P, is generated by the reduction of dihydroxyacetone phosphate (DHAP), an intermediate of glycolysis. The NADH-dependent glycerol-3-phosphate dehydrogenase (G3PD), which catalyzes this step, was shown to be up-regulated in bladder cancer [119]. Up-regulation of this pathway might serve a growing cell in several ways: first, the flux to G3P provides the substrate for PL synthesis and may perhaps be limiting for glycerophospholipid synthesis during growth; second, the reaction yields cytosolic NAD+, which can be used in glycolysis; third, G3PD is part of the glycerol phosphate shuttle: the complete reaction cycle regenerates cytosolic NAD+ by transferring electrons to mitochondrial FADH2 and, therefore, results in higher ATP levels, but also in re-oxidation of G3P to DHAP. The full cycle is operating in mammals, whereas in fermenting yeast it is incomplete due to glucose repression of the mitochondrial G3P dehydrogenase that catalyzes the oxidation of G3P. Hence, the G3P shunt in yeast is only used for recycling of NAD+ and the synthesis of G3P for lipid synthesis and glycerol, which plays an important role in osmoregulation. For the yeast Y. lipolytica it was shown that overexpression of G3PD, deletion of the second enzyme of the shuttle, GUT2, or a combination of both, had no effect on lipid accumulation under standard growth conditions, suggesting that the regeneration of NAD+ may be more important in this pathway than the supply with G3P [120]. It is noteworthy that the acceptor for the first acyl-transferase reaction in S. cerevisiae can be both, G3P and DHAP. Acylation of DHAP yields 1-acyl-DHAP, which has to be reduced to LPA prior to the second acyl transfer, a reaction catalyzed by the NADPH-dependent dehydrogenase Ayr1 (Fig. 1). The regulatory mechanisms controlling the flux distribution between DHAP and G3P acylation are unknown. However, overexpression of Ayr1 in yeast leads to growth arrest, demonstrating that the equilibrium between these two pathways plays a crucial role in cellular proliferation [121]. 1-acyl-DHAP reductase activity has also been detected in mammalian cells but the gene coding for this activity was not identified [122].
Synthesis of all phospholipids, except for the synthesis of PA, requires cytidine triphosphate as an energy donor, either for the conversion of PA to CDP-DAG, or for the activation of phosphocholine (PCho) and phosphoethanolamine to CDP-choline and CDP-ethanolamine, respectively (Fig. 2). In yeast, the phosphorylation of DAG to PA by diglyceride kinase is also CTP-dependent, whereas this reaction is driven by ATP hydrolysis in other organisms [123]. These observations indicate an increased metabolic demand for CTP in proliferating cells. Indeed, increased CTP synthase (CTPS) activity has been found in several cancers [124–126], and negative effects of the CTPS inhibitor cyclopentenyl cytosine on growth of malignant cell lines have been described [125,127]. CTPS apparently is mostly regulated at the post-translational level: in addition to the well-known product inhibition, CTPS activity in S. cerevisiae is regulated through phosphorylation at several sites on the protein. The major yeast CTPS, Ura7, is a substrate of cAMP activated protein kinase (PKA) and protein kinase C (PKC) at different serine residues, in both cases with stimulating effects on enzyme activity [128,129]. Both human CTP synthases were shown to functionally complement yeast mutants lacking CTPS. CTPS1 is phosphorylated by PKA and PKC, like the yeast Ura7 enzyme [130,131], and cyclopentenyl cytosine also acts as an inhibitor of CTPS activity in yeast. Mutations of Ura7 and Ura8 that lead to a reduction of CTP product inhibition decrease cyclopentenyl cytosine sensitivity of the enzymes; these mutations concomitantly stimulate neutral lipid synthesis, and lead to a de-repression of inositol biosynthesis and a shift of PC synthesis from the CDP-DAG pathway to the Kennedy pathway [132].
8. Phospholipid remodeling pathways
The correlation of increased CTPS activity with a metabolic shift to the CDP-choline pathway in yeast shows parallels to the observed changes in PL metabolism of cancer cells. CDS1, the human CDP-DAG synthase 1, was shown to be transcriptionally down-regulated in hepatocellular carcinomas [133]. This decrease in CDS1 transcript levels is an unexpected finding considering the general up-regulation of pathways involved in membrane lipid synthesis, but it might be compensated for by an increased flux through the Kennedy pathway. It has to be noted, however, that down-regulation of CDS1 has not yet been described for other types of cancer. Furthermore, the Kennedy pathway is also the major pathway for PC synthesis in non-cancerous cells [134], and the conclusion that cancer cells rely more strongly on the Kennedy pathway for PC synthesis than non-cancerous cells might be too simplistic. On the other hand, elevated levels of PCho are observed in a wide variety of tumors [135], and choline kinase, which catalyzes the conversion of choline to PCho is known to play a role in many cancers [136]. Similarly, up-regulation of choline transporters was observed in breast cancer cell lines [137,138]. Cki1, the choline kinase in yeast, is phosphorylated by PKA and PKC, both stimulating its activity [139,140]. PKA regulation is also assumed for human choline kinase [141], however, no increased activity was so far reported for other enzymes of the Kennedy pathway. In addition, findings that not only PCho, but also glycerophosphocholine (GPCho) levels are increased in some cancer tissues [135] point to the importance of an increased PC turnover in these cells rather than at a shift from the CDP-DAG to the Kennedy pathway.
PCho may also be derived from PC catabolism through phospholipase C, yielding diacylglycerol and PCho, or from recycling of GPCho, which is the product of PC deacylation by phospholipase B. Both reactions contribute to the remodeling of phospholipids, a process that is required to maintain the correct steady-state composition of PL species and their acyl-chain distribution (Fig. 2). Phospholipase C-catalyzed turnover of PC is required for the Ras oncogene mediated mitogenic signal cascade that is important for cellular proliferation [142]. Hence, both the remodeling of PC to obtain an optimum FA composition and the production of signaling molecules, GPCho, PCho, and DAG, might be important for cellular growth. In S. cerevisiae, two enzymes are known to significantly contribute to PC turnover during exponential growth: the structurally and functionally conserved neuropathy target esterase, Nte1, acts as a phospholipase B and hydrolyzes PC derived from the Kennedy pathway. Notably, this enzyme is also involved in the transcriptional regulation of genes involved in phospholipid metabolism [143–145]. Deletion of the NTE1 gene results in the down-regulation of choline uptake to compensate for the reduced degradation of PC. In HeLa cells, NTE activity was shown to be regulated in a cell cycle dependent manner, with highest activity during G1 [146]. The second enzyme that degrades metabolically relevant amounts of PC is the phospholipid-dependent diacylglycerol acyltransferase Lro1 (LCAT Related Open reading frame), which uses a fatty acid from PC to acylate DAG to TAG. The human homolog of Lro1, lecithin cholesterol acyltransferase (LCAT) catalyzes a similiar reaction. However, the role of LCAT is rather the regulation of cholesterol ester levels in serum than intracellular PC remodeling. Hence, a functional homolog of Lro1 in humans remains to be identified. The byproduct of Lro1, lyso-PC, is either further deacylated to FA and GPCho or reacylated to PC by an acyl-CoA-dependent acyltransferase (Fig. 2). During exponential growth Lro1 is the major pathway for TAG synthesis in yeast, underlining the importance of PL remodeling during cellular growth [147]. Both Nte1 and Lro1 catalyze reactions that, either directly or indirectly, promote synthesis of TAG and contribute to the adjustment of the molecular species composition of membrane phospholipids. Notably, several strains mutated in genes involved in PC metabolism were recently found to have additional defects in neutral lipid homeostasis [148]. These results suggest that the metabolic crosstalk between membrane and storage lipids in proliferating cells is required for maintaining the optimum membrane composition with regard to both FA and PL composition (Fig. 3).
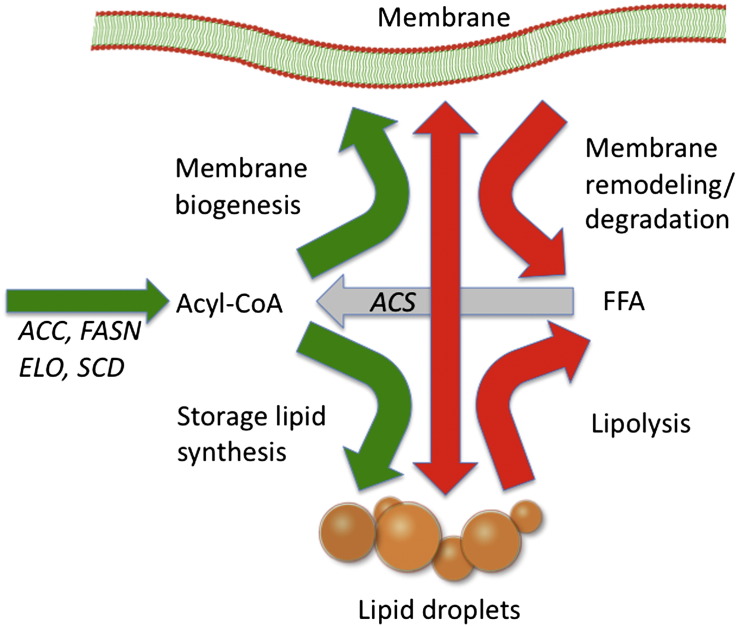
Fig. 3.
Phospholipid remodeling in proliferating cells. The scheme shows the fluxes of fatty acids in glycerolipid synthesis and the assumed involvement of storage lipids in the remodeling of membranes. FA are either incorporated into phospholipids for membrane biogenesis or into triacylglycerol for storage. Both membrane and storage lipids are subject to turnover during growth, resulting in an acyl-CoA pool that is fed by de novo synthesis, lipolysis of TAG, and phospholipase B action on phospholipids. In yeast, FA from phospholipids can also be directly incorporated into storage lipids by PC:diacylglycerol transferase activity and diacylglyerol derived from the first step of TAG lipolysis may directly serve as a substrate for synthesis of PE and PC in the Kennedy pathway or for the DAG kinase to generate phosphatidic acid (see main text for details).
9. Neutral lipid homeostasis: How to deal with excess fatty acids
All eukaryotic cells store fat in intracellular lipid droplets (LD). Dependent on the organism and tissue, LDs are composed of TAG, steryl esters and retinyl esters, with TAG being the predominant lipid class. The classical view of the LD as an inert storage organelle for metabolic energy in the form of TAG has changed during the last years and it is now considered an organelle that is actively involved in lipid turnover, membrane formation and cellular proliferation. In humans, the major site for lipid storage is the adipose tissue but neutral lipid deposits exist in virtually all cells, albeit at lower levels. FA from adipocyte LDs are released and routed to β-oxidation when the body needs more energy than is available from dietary intake. Conversely, when the energy requirement of the cells is lower than the nutritional supply, excess energy and carbon are channeled into one of the storage depots, glycogen or fat. Notably, yeast cells proliferating in the presence of high glucose concentrations accumulate only little TAG, and only at the onset of starvation and entry into quiescence, the remaining carbon source is converted into fat, resulting in high TAG levels in stationary phase. When environmental conditions improve such to support growth, TAG stores are rapidly degraded and the fatty acids are channeled into phospholipid synthesis for fast progression through the first cell division cycle. In contrast to higher organisms, the pathway for β-oxidation of FA is strongly repressed during growth of yeast on glucose [13,54].
The first evidence that neutral lipid homeostasis might play an important role in cancer cells came from studies on the effects of bexarotene, a synthetic ligand for retinoid X receptors (RXR), on mammary tumors. RXR are members of the nuclear hormone receptor family, with 9-cis retionic acid as the binding ligand. As homodimers or as heterodimers with peroxisome proliferator-activated receptors (PPARs) they regulate a plethora of processes in cellular development and metabolism [149–151]. Synthetic RXR ligands, also termed rexinoids, are under investigation as potent agents in therapy for several cancers. Bexarotene induces the accumulation of triacylglycerol in cytosolic lipid droplets and leads to an arrest of cell proliferation, with PPARγ agonists acting synergistically to inhibit tumor growth [152]. In cells treated with bexarotene mRNA expression of acyl-CoA synthetase 1, stearoyl-CoA dehydrogenase and diacylglycerol acyltransferase, key enzymes of FA and TAG metabolism, are induced; another important factor in lipid homeostasis, adipophilin, is elevated at the protein level upon treatment [152]. Furthermore, the authors found a strong inverse correlation between growth attenuation and neutral lipid accumulation in cancer cells, which is also observed in yeast: in a genome-wide approach to identify yeast mutants with aberrant lipid droplet morphology, strains with reduced growth rates were significantly enriched in the group of mutants with elevated lipid content [153].
Growth inhibition by rexinoids may operate by attenuating cell cycle progression. Studies with the rexinoid LG100268 showed that expression of a key cell cycle regulator, cyclin D1, is strongly reduced in the presence of the compound [154]. A growth arrest in cells with active lipid synthesis may trigger the cells to redirect the flux of FA from membrane lipid synthesis to the synthesis of storage lipids, to avoid an accumulation of excess membranes. This hypothesis is supported by the fact that PPARγ agonists act synergistically with rexinoids [152]. However, the detailed mechanisms of rexinoid action remain to be elucidated. Since the PPAR/RXR system can be reconstituted in yeast [155,156], genome-wide drug screens in this model organism might help in this endeavor.
Changes in lipid composition of tumor cells during apoptopic cell death can be seen in this context. Several studies suggest that initiation of apoptosis in cancer cells is accompanied by a remodeling of glycerolipids, resulting in a drop of PC levels and an increase in TAG [157,158]. As in the studies targeting RXR/PPARs, accumulation of storage lipids is the consequence of attenuated growth. It is tempting to speculate whether an increase of TAG synthesis might result in insufficient membrane synthesis and therefore, in a reversal of cause and effect, in growth arrest in proliferating cells. For baker's yeast it has already been shown that an increase in activity of Lro1 results in reduced PC levels and increased TAG content, but an impact on cellular proliferation has not been investigated in that study [159].
Storage of excess fatty acids into TAG or steryl esters is not an essential process in yeast under normal growth conditions, as shown with a mutant lacking all pathways for neutral lipid synthesis. However, challenging this mutant with exogenous fatty acids results in proliferation of intracellular membranes and cell death [23,160]. Hence, the inability to detoxify excess FA by storage into LD can become detrimental for a cell and interventions in the pathways for neutral lipid synthesis might be a means to target proliferation of cancers with a lipogenic phenotype. The assumption that lipid storage might be an important pathway in some types of cancer is supported by evidence for upregulation of ERLIN2 (see Section 5) in breast cancer and its involvement in lipid droplet formation [97].
10. Neutral lipid mobilization: A role for TAG lipases in cell proliferation?
When cells need the fatty acids stored in TAG for the production of energy or for the incorporation into membrane lipids, lipases are activated to hydrolyze the ester bonds and release free FA. One of the four TAG lipases in yeast, Tgl4, is activated by phosphorylation by cyclin-dependent kinase 1, CDK1, a highly conserved key regulator of cell cycle progression in yeast and mammalian cells. These findings link TAG breakdown and the released metabolites directly to the control of cell cycle events [45]. Lipolysis-derived DAG can be phosphorylated by diglyceride kinase to PA and further processed to membrane lipids during the TAG mobilization phase [161], confirming that TAG in fermenting yeast is rather a storage pool for the rapid synthesis of membrane lipids than a reserve for energy production through β-oxidation [13]. In addition, some evidence suggests that TAG-derived DAG may also be channeled directly into the Kennedy pathway for phospholipid synthesis. It might be speculated that FA are stored as TAG during phases of reduced cell growth to support rapid membrane lipid synthesis at later points of the cell division cycle, by utilizing TAG-derived FA or DAG, in addition to de novo synthesis. Such a periodic accumulation and degradation of neutral lipid would enable the cell to finish a cell cycle, even if deprived of nutrients due to changing environmental conditions once the cell is committed to enter a new round of cell division. The assumption that both TAG synthesis and lipolysis are simultaneously active processes in proliferating cells is supported by the observation that lipase-deficient yeast mutants accumulate more TAG than WT cells at any given time point, without significant impact on cell size or growth rate. Hence, FA synthesis is stimulated in these mutants, suggesting a role of lipases or the products of lipolysis in control of FA synthesis. Overall, the reactions for de novo synthesis and degradation of glycerolipids may result in a constant exchange of FA between phospholipids and TAG to maintain the correct FA and PL composition of membranes in proliferating cells (Fig. 3).
It remains to be shown whether these events also occur in mammalian cells, in particular in proliferating cancer cells. The functionally conserved ortholog of Tgl4 in mammals, adipose triglyceride lipase (ATGL), is also regulated by phosphorylation [162]. Regulation of lipolysis by a cell cycle-dependent kinase in humans has not been demonstrated yet, but there is clear evidence that lipolytic activity is controlled by AMPK. Phosphorylation of ATGL by AMPK results in increased enzymatic activity in adipocytes. On the other hand, phosphorylation of hormone-sensitive lipase HSL, the second enzyme in the lipolytic cascade, through AMPK seems to result in inhibition of its translocation to the lipid droplet and impaired lipolysis. Treatment of cells with activators of AMPK, such as AICAR, results in increased activity of ATGL and in repression of HSL [163–165]. AMPK-mediated stimulation of ATGL and inhibition of HSL would result in a halt of lipolysis after the inital step and in the need for an alternative pathway to avoid accumulation of diacylglycerol, for example the conversion of DAG to PC in the Kennedy pathway. However, regulation by AMPK is only one of several mechanisms that control activity of ATGL and HSL. Furthermore, in non-adipose tissue other TAG lipases besides ATGL might contribute considerably to TAG lipolysis. Therefore, the changes in lipolytic activity in cancer might depend on several factors, including transcriptional regulation as well as control of metabolic activity through PKA, AMPK, and cell cycle dependent kinases. For AMPK, it might be expected that defects in the LKB1/AMPK pathway, as observed in many cancers, affect this regulatory network and result in altered TAG metabolism, but such a connection has not been investigated yet. Notably, also expression of monoacylglycerol lipase, which catalyzes the final step in the lipolytic cascade, is increased in colorectal cancer and evidence suggests that its activity is required for rapid cell proliferation, since a gene knockdown resulted in reduced tumor growth and elevated apoptotic cell death [166].
ATGL needs the coactivator Comparative Gene Identification-58, CGI-58, for maximum activity [167]. CGI-58 was shown to possess acyltransferase activity, required for the acylation of lysophosphatidic acid to PA [168]. Interestingly, such an acyltransferase activity has also been shown for the yeast TAG lipases Tgl3 and Tgl5. Deletion of TGL3 or TGL5 resulted in reduced PL levels and overexpression in an increase of PL [169], again suggesting a direct involvement of storage lipid pools in remodeling of membranes during growth (Fig. 3). In mammals, TAG lipolysis is generally regarded as a means to generate energy by β-oxidation of the released FA. Whether these FA can also be used for lipid remodeling in proliferating cells remains to be shown but the activity of CGI-58 as an acyltransferase suggests such a pathway.
Lipolysis of fats seems to play a role not only in intracellular lipid homeostasis of proliferating cells. Cachexia, the detrimental degradation and loss of fat and muscle tissue that is associated with multiple types of cancer, is clearly linked to alterations in neutral lipid turnover [170]. ATGL−/− ko mice are resistant to cachexia and similar results were obtained, although with less significance, for HSL−/− mice. Moreover, the size of tumors in lipase mutant models was reduced in comparison to wild type mice, indicating a role of TAG lipolysis not only in cachexia, but also in the development of the tumor itself. In this study, TAG lipase activity in white adipose tissue of cachexic patients was significantly higher than in control groups [170].
11. Conclusion
Recent findings provide ample evidence that lipid metabolism plays an important role in the development and growth of neoplastic tissue. Alterations in the pathways for synthesis of fatty acids and their incorporation into phospholipids or storage into TAG are reported for a large number of tumors. To identify potential targets for cancer therapy, however, a more detailed understanding of the regulatory network that controls lipid homeostasis is required.
In the past, work with yeast has provided a great deal of mechanistic insight into fundamental cellular processes that are also relevant for mammalian cells, such as TOR signaling, organelle biogenesis, the secretory pathway, and cell cycle progression. Now, is yeast as a single celled organism, also a suitable model to understand cancer development? There is clear evidence that yeast that typically grows in colonies, also bears properties of multicellular complexity, illustrated by features such as the conserved mechanism of programmed cell death [171] and physiological differences within a colony [172]. Most notably, the metabolic reprogramming in cancer cells is similar to the physiology of fermenting yeast cells, and the high level of pathway conservation in lipid metabolism offers great potential for further mechanistic exploration. The convenience of yeast regarding its genetic manipulation and growth conditions, availability of large mutant collections and high-content screens as well as drug susceptibility and detailed knowledge about physiology and metabolism make this eukaryote an excellent experimental model for such studies.
Acknowledgments
Work in the authors' laboratories is supported by grants from the Austrian Science Fund FWF, projects SFB 3005 LIPOTOX and W901-B12 to S.D.K. and the Austrian Federal Ministry for Science and Research, project 'GOLD — Genomics of Lipid-Associated Disorders' in the framework of the Austrian Genome Project “GEN-AU Genome research in Austria” to K.N.
References
- 1.Hanahan D., Weinberg R.A. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 2.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 3.Warburg O., Posener K., Negelein E. Ueber den Stoffwechsel der Carcinomzelle. Biochem. Z. 1924;152:309–344. [Google Scholar]
- 4.Crabtree H.G. Observations on the carbohydrate metabolism of tumours. Biochem. J. 1929;23:536–545. doi: 10.1042/bj0230536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 6.Funes J.M., Quintero M., Henderson S., Martinez D., Qureshi U., Westwood C., Clements M.O., Bourboulia D., Pedley R.B., Moncada S., Boshoff C. Transformation of human mesenchymal stem cells increases their dependency on oxidative phosphorylation for energy production. Proc. Natl. Acad. Sci. U. S. A. 2007;104:6223–6228. doi: 10.1073/pnas.0700690104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moreno-Sanchez R., Rodriguez-Enriquez S., Marin-Hernandez A., Saavedra E. Energy metabolism in tumor cells. FEBS J. 2007;274:1393–1418. doi: 10.1111/j.1742-4658.2007.05686.x. [DOI] [PubMed] [Google Scholar]
- 8.Fantin V.R., St-Pierre J., Leder P. Attenuation of LDH-A expression uncovers a link between glycolysis, mitochondrial physiology, and tumor maintenance. Cancer Cell. 2006;9:425–434. doi: 10.1016/j.ccr.2006.04.023. [DOI] [PubMed] [Google Scholar]
- 9.Gatenby R.A., Gillies R.J. Why do cancers have high aerobic glycolysis? Nat. Rev. Cancer. 2004;4:891–899. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- 10.Molenaar D., van Berlo R., de Ridder D., Teusink B. Shifts in growth strategies reflect tradeoffs in cellular economics. Mol. Syst. Biol. 2009;5:323. doi: 10.1038/msb.2009.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pfeiffer T., Schuster S., Bonhoeffer S. Cooperation and competition in the evolution of ATP-producing pathways. Science. 2001;292:504–507. doi: 10.1126/science.1058079. [DOI] [PubMed] [Google Scholar]
- 12.Shlomi T., Benyamini T., Gottlieb E., Sharan R., Ruppin E. Genome-scale metabolic modeling elucidates the role of proliferative adaptation in causing the Warburg effect. PLoS Comput. Biol. 2011;7:e1002018. doi: 10.1371/journal.pcbi.1002018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zanghellini J., Natter K., Jungreuthmayer C., Thalhammer A., Kurat C.F., Gogg-Fassolter G., Kohlwein S.D., von Grunberg H.H. Quantitative modeling of triacylglycerol homeostasis in yeast–metabolic requirement for lipolysis to promote membrane lipid synthesis and cellular growth. FEBS J. 2008;275:5552–5563. doi: 10.1111/j.1742-4658.2008.06681.x. [DOI] [PubMed] [Google Scholar]
- 14.Diaz-Ruiz R., Uribe-Carvajal S., Devin A., Rigoulet M. Tumor cell energy metabolism and its common features with yeast metabolism. Biochim. Biophys. Acta. 2009;1796:252–265. doi: 10.1016/j.bbcan.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 15.Gombert A.K., Moreira dos Santos M., Christensen B., Nielsen J. Network identification and flux quantification in the central metabolism of Saccharomyces cerevisiae under different conditions of glucose repression. J. Bacteriol. 2001;183:1441–1451. doi: 10.1128/JB.183.4.1441-1451.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van den Brink J., Canelas A.B., van Gulik W.M., Pronk J.T., Heijnen J.J., de Winde J.H., Daran-Lapujade P. Dynamics of glycolytic regulation during adaptation of Saccharomyces cerevisiae to fermentative metabolism. Appl. Environ. Microbiol. 2008;74:5710–5723. doi: 10.1128/AEM.01121-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van den Brink J., Daran-Lapujade P., Pronk J.T., de Winde J.H. New insights into the Saccharomyces cerevisiae fermentation switch: dynamic transcriptional response to anaerobicity and glucose-excess. BMC Genomics. 2008;9:100. doi: 10.1186/1471-2164-9-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jee S.H., Ohrr H., Sull J.W., Yun J.E., Ji M., Samet J.M. Fasting serum glucose level and cancer risk in Korean men and women. JAMA. 2005;293:194–202. doi: 10.1001/jama.293.2.194. [DOI] [PubMed] [Google Scholar]
- 19.Kellenberger L.D., Bruin J.E., Greenaway J., Campbell N.E., Moorehead R.A., Holloway A.C., Petrik J. The role of dysregulated glucose metabolism in epithelial ovarian cancer. J. Oncol. 2010;2010:514310. doi: 10.1155/2010/514310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krone C.A., Ely J.T. Controlling hyperglycemia as an adjunct to cancer therapy. Integr. Cancer Ther. 2005;4:25–31. doi: 10.1177/1534735404274167. [DOI] [PubMed] [Google Scholar]
- 21.Liu H., Ma Q., Li J. High glucose promotes cell proliferation and enhances GDNF and RET expression in pancreatic cancer cells. Mol. Cell. Biochem. 2011;347:95–101. doi: 10.1007/s11010-010-0617-0. [DOI] [PubMed] [Google Scholar]
- 22.Sieri S., Muti P., Claudia A., Berrino F., Pala V., Grioni S., Abagnato C.A., Blandino G., Contiero P., Schunemann H.J., Krogh V. Prospective study on the role of glucose metabolism in breast cancer occurrence. Int. J. Cancer. 2012;130:921–929. doi: 10.1002/ijc.26071. [DOI] [PubMed] [Google Scholar]
- 23.Petschnigg J., Wolinski H., Kolb D., Zellnig G., Kurat C.F., Natter K., Kohlwein S.D. Good fat, essential cellular requirements for triacylglycerol synthesis to maintain membrane homeostasis in yeast. J. Biol. Chem. 2009;284:30981–30993. doi: 10.1074/jbc.M109.024752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schneiter R., Brugger B., Sandhoff R., Zellnig G., Leber A., Lampl M., Athenstaedt K., Hrastnik C., Eder S., Daum G., Paltauf F., Wieland F.T., Kohlwein S.D. Electrospray ionization tandem mass spectrometry (ESI-MS/MS) analysis of the lipid molecular species composition of yeast subcellular membranes reveals acyl chain-based sorting/remodeling of distinct molecular species en route to the plasma membrane. J. Cell Biol. 1999;146:741–754. doi: 10.1083/jcb.146.4.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kokatnur M.G., Oalmann M.C., Johnson W.D., Malcom G.T., Strong J.P. Fatty acid composition of human adipose tissue from two anatomical sites in a biracial community. Am. J. Clin. Nutr. 1979;32:2198–2205. doi: 10.1093/ajcn/32.11.2198. [DOI] [PubMed] [Google Scholar]
- 26.Ma J., Folsom A.R., Shahar E., Eckfeldt J.H. Plasma fatty acid composition as an indicator of habitual dietary fat intake in middle-aged adults. The Atherosclerosis Risk in Communities (ARIC) Study Investigators. Am. J. Clin. Nutr. 1995;62:564–571. doi: 10.1093/ajcn/62.3.564. [DOI] [PubMed] [Google Scholar]
- 27.Phinney S.D., Fisler J.S., Tang A.B., Warden C.H. Liver fatty acid composition correlates with body fat and sex in a multigenic mouse model of obesity. Am. J. Clin. Nutr. 1994;60:61–67. doi: 10.1093/ajcn/60.1.61. [DOI] [PubMed] [Google Scholar]
- 28.Ristic V., Tepsic V., De Luka S.R., Vrbaski S.R. Phospholipid content and fatty acid composition in the rat heart after chronic diazepam treatment. Physiol. Res. 1998;47:115–118. [PubMed] [Google Scholar]
- 29.Svennerholm L. Distribution and fatty acid composition of phosphoglycerides in normal human brain. J. Lipid Res. 1968;9:570–579. [PubMed] [Google Scholar]
- 30.Cowart L.A., Obeid L.M. Yeast sphingolipids: recent developments in understanding biosynthesis, regulation, and function. Biochim. Biophys. Acta. 2007;1771:421–431. doi: 10.1016/j.bbalip.2006.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dickson R.C., Sumanasekera C., Lester R.L. Functions and metabolism of sphingolipids in Saccharomyces cerevisiae. Prog. Lipid Res. 2006;45:447–465. doi: 10.1016/j.plipres.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 32.Merrill A.H., Jr., Wang M.D., Park M., Sullards M.C. (Glyco)sphingolipidology: an amazing challenge and opportunity for systems biology. Trends Biochem. Sci. 2007;32:457–468. doi: 10.1016/j.tibs.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 33.Pruett S.T., Bushnev A., Hagedorn K., Adiga M., Haynes C.A., Sullards M.C., Liotta D.C., Merrill A.H., Jr. Biodiversity of sphingoid bases (“sphingosines”) and related amino alcohols. J. Lipid Res. 2008;49:1621–1639. doi: 10.1194/jlr.R800012-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chattopadhyay A., Paila Y.D., Jafurulla M., Chaudhuri A., Singh P., Murty M.R., Vairamani M. Differential effects of cholesterol and 7-dehydrocholesterol on ligand binding of solubilized hippocampal serotonin1A receptors: implications in SLOS. Biochem. Biophys. Res. Commun. 2007;363:800–805. doi: 10.1016/j.bbrc.2007.09.040. [DOI] [PubMed] [Google Scholar]
- 35.Gimpl G., Burger K., Fahrenholz F. Cholesterol as modulator of receptor function. Biochemistry. 1997;36:10959–10974. doi: 10.1021/bi963138w. [DOI] [PubMed] [Google Scholar]
- 36.Hanzal-Bayer M.F., Hancock J.F. Lipid rafts and membrane traffic. FEBS Lett. 2007;581:2098–2104. doi: 10.1016/j.febslet.2007.03.019. [DOI] [PubMed] [Google Scholar]
- 37.Pang L., Graziano M., Wang S. Membrane cholesterol modulates galanin-GalR2 interaction. Biochemistry. 1999;38:12003–12011. doi: 10.1021/bi990227a. [DOI] [PubMed] [Google Scholar]
- 38.Rietveld A., Simons K. The differential miscibility of lipids as the basis for the formation of functional membrane rafts. Biochim. Biophys. Acta. 1998;1376:467–479. doi: 10.1016/s0304-4157(98)00019-7. [DOI] [PubMed] [Google Scholar]
- 39.Shouffani A., Kanner B.I. Cholesterol is required for the reconstruction of the sodium- and chloride-coupled, gamma-aminobutyric acid transporter from rat brain. J. Biol. Chem. 1990;265:6002–6008. [PubMed] [Google Scholar]
- 40.Telbisz A., Muller M., Ozvegy-Laczka C., Homolya L., Szente L., Varadi A., Sarkadi B. Membrane cholesterol selectively modulates the activity of the human ABCG2 multidrug transporter. Biochim. Biophys. Acta. 2007;1768:2698–2713. doi: 10.1016/j.bbamem.2007.06.026. [DOI] [PubMed] [Google Scholar]
- 41.Kitson S.M., Mullen W., Cogdell R.J., Bill R.M., Fraser N.J. GPCR production in a novel yeast strain that makes cholesterol-like sterols. Methods. 2011;55:287–292. doi: 10.1016/j.ymeth.2011.09.023. [DOI] [PubMed] [Google Scholar]
- 42.Lagane B., Gaibelet G., Meilhoc E., Masson J.M., Cezanne L., Lopez A. Role of sterols in modulating the human mu-opioid receptor function in Saccharomyces cerevisiae. J. Biol. Chem. 2000;275:33197–33200. doi: 10.1074/jbc.C000576200. [DOI] [PubMed] [Google Scholar]
- 43.Opekarova M., Tanner W. Specific lipid requirements of membrane proteins–a putative bottleneck in heterologous expression. Biochim. Biophys. Acta. 2003;1610:11–22. doi: 10.1016/s0005-2736(02)00708-3. [DOI] [PubMed] [Google Scholar]
- 44.Xu X., Bittman R., Duportail G., Heissler D., Vilcheze C., London E. Effect of the structure of natural sterols and sphingolipids on the formation of ordered sphingolipid/sterol domains (rafts). Comparison of cholesterol to plant, fungal, and disease-associated sterols and comparison of sphingomyelin, cerebrosides, and ceramide. J. Biol. Chem. 2001;276:33540–33546. doi: 10.1074/jbc.M104776200. [DOI] [PubMed] [Google Scholar]
- 45.Kurat C.F., Wolinski H., Petschnigg J., Kaluarachchi S., Andrews B., Natter K., Kohlwein S.D. Cdk1/Cdc28-dependent activation of the major triacylglycerol lipase Tgl4 in yeast links lipolysis to cell-cycle progression. Mol. Cell. 2009;33:53–63. doi: 10.1016/j.molcel.2008.12.019. [DOI] [PubMed] [Google Scholar]
- 46.Zechner R., Zimmermann R., Eichmann T.O., Kohlwein S.D., Haemmerle G., Lass A., Madeo F. FAT SIGNALS–lipases and lipolysis in lipid metabolism and signaling. Cell Metab. 2012;15:279–291. doi: 10.1016/j.cmet.2011.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ookhtens M., Kannan R., Lyon I., Baker N. Liver and adipose tissue contributions to newly formed fatty acids in an ascites tumor. Am. J. Physiol. 1984;247:R146–R153. doi: 10.1152/ajpregu.1984.247.1.R146. [DOI] [PubMed] [Google Scholar]
- 48.Metallo C.M., Gameiro P.A., Bell E.L., Mattaini K.R., Yang J., Hiller K., Jewell C.M., Johnson Z.R., Irvine D.J., Guarente L., Kelleher J.K., Vander Heiden M.G., Iliopoulos O., Stephanopoulos G. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2011;481:380–384. doi: 10.1038/nature10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mullen A.R., Wheaton W.W., Jin E.S., Chen P.H., Sullivan L.B., Cheng T., Yang Y., Linehan W.M., Chandel N.S., DeBerardinis R.J. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature. 2011;481:385–388. doi: 10.1038/nature10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wise D.R., Ward P.S., Shay J.E., Cross J.R., Gruber J.J., Sachdeva U.M., Platt J.M., DeMatteo R.G., Simon M.C., Thompson C.B. Hypoxia promotes isocitrate dehydrogenase-dependent carboxylation of alpha-ketoglutarate to citrate to support cell growth and viability. Proc. Natl. Acad. Sci. U. S. A. 2011;108:19611–19616. doi: 10.1073/pnas.1117773108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hatzivassiliou G., Zhao F., Bauer D.E., Andreadis C., Shaw A.N., Dhanak D., Hingorani S.R., Tuveson D.A., Thompson C.B. ATP citrate lyase inhibition can suppress tumor cell growth. Cancer Cell. 2005;8:311–321. doi: 10.1016/j.ccr.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 52.Bauer D.E., Hatzivassiliou G., Zhao F., Andreadis C., Thompson C.B. ATP citrate lyase is an important component of cell growth and transformation. Oncogene. 2005;24:6314–6322. doi: 10.1038/sj.onc.1208773. [DOI] [PubMed] [Google Scholar]
- 53.Migita T., Narita T., Nomura K., Miyagi E., Inazuka F., Matsuura M., Ushijima M., Mashima T., Seimiya H., Satoh Y., Okumura S., Nakagawa K., Ishikawa Y. ATP citrate lyase: activation and therapeutic implications in non-small cell lung cancer. Cancer Res. 2008;68:8547–8554. doi: 10.1158/0008-5472.CAN-08-1235. [DOI] [PubMed] [Google Scholar]
- 54.Kurat C.F., Natter K., Petschnigg J., Wolinski H., Scheuringer K., Scholz H., Zimmermann R., Leber R., Zechner R., Kohlwein S.D. Obese yeast: triglyceride lipolysis is functionally conserved from mammals to yeast. J. Biol. Chem. 2006;281:491–500. doi: 10.1074/jbc.M508414200. [DOI] [PubMed] [Google Scholar]
- 55.Choudhary C., Kumar C., Gnad F., Nielsen M.L., Rehman M., Walther T.C., Olsen J.V., Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 56.Kim S.C., Sprung R., Chen Y., Xu Y., Ball H., Pei J., Cheng T., Kho Y., Xiao H., Xiao L., Grishin N.V., White M., Yang X.J., Zhao Y. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol. Cell. 2006;23:607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 57.Zhao S., Xu W., Jiang W., Yu W., Lin Y., Zhang T., Yao J., Zhou L., Zeng Y., Li H., Li Y., Shi J., An W., Hancock S.M., He F., Qin L., Chin J., Yang P., Chen X., Lei Q., Xiong Y., Guan K.L. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327:1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lv L., Li D., Zhao D., Lin R., Chu Y., Zhang H., Zha Z., Liu Y., Li Z., Xu Y., Wang G., Huang Y., Xiong Y., Guan K.L., Lei Q.Y. Acetylation targets the M2 isoform of pyruvate kinase for degradation through chaperone-mediated autophagy and promotes tumor growth. Mol. Cell. 2011;42:719–730. doi: 10.1016/j.molcel.2011.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Boles E., Schulte F., Miosga T., Freidel K., Schluter E., Zimmermann F.K., Hollenberg C.P., Heinisch J.J. Characterization of a glucose-repressed pyruvate kinase (Pyk2p) in Saccharomyces cerevisiae that is catalytically insensitive to fructose-1,6-bisphosphate. J. Bacteriol. 1997;179:2987–2993. doi: 10.1128/jb.179.9.2987-2993.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bond C.J., Jurica M.S., Mesecar A., Stoddard B.L. Determinants of allosteric activation of yeast pyruvate kinase and identification of novel effectors using computational screening. Biochemistry. 2000;39:15333–15343. doi: 10.1021/bi001443i. [DOI] [PubMed] [Google Scholar]
- 61.Portela P., Moreno S., Rossi S. Characterization of yeast pyruvate kinase 1 as a protein kinase A substrate, and specificity of the phosphorylation site sequence in the whole protein. Biochem. J. 2006;396:117–126. doi: 10.1042/BJ20051642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pearce A.K., Crimmins K., Groussac E., Hewlins M.J., Dickinson J.R., Francois J., Booth I.R., Brown A.J. Pyruvate kinase (Pyk1) levels influence both the rate and direction of carbon flux in yeast under fermentative conditions. Microbiology. 2001;147:391–401. doi: 10.1099/00221287-147-2-391. [DOI] [PubMed] [Google Scholar]
- 63.Frederiks W.M., Vizan P., Bosch K.S., Vreeling-Sindelarova H., Boren J., Cascante M. Elevated activity of the oxidative and non-oxidative pentose phosphate pathway in (pre)neoplastic lesions in rat liver. Int. J. Exp. Pathol. 2008;89:232–240. doi: 10.1111/j.1365-2613.2008.00582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jiang P., Du W., Wang X., Mancuso A., Gao X., Wu M., Yang X. p53 regulates biosynthesis through direct inactivation of glucose-6-phosphate dehydrogenase. Nat. Cell Biol. 2011;13:310–316. doi: 10.1038/ncb2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kuo W., Lin J., Tang T.K. Human glucose-6-phosphate dehydrogenase (G6PD) gene transforms NIH 3T3 cells and induces tumors in nude mice. Int. J. Cancer. 2000;85:857–864. doi: 10.1002/(sici)1097-0215(20000315)85:6<857::aid-ijc20>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 66.Li D., Zhu Y., Tang Q., Lu H., Li H., Yang Y., Li Z., Tong S. A new G6PD knockdown tumor-cell line with reduced proliferation and increased susceptibility to oxidative stress. Cancer Biother. Radiopharm. 2009;24:81–90. doi: 10.1089/cbr.2008.0494. [DOI] [PubMed] [Google Scholar]
- 67.Vizan P., Alcarraz-Vizan G., Diaz-Moralli S., Solovjeva O.N., Frederiks W.M., Cascante M. Modulation of pentose phosphate pathway during cell cycle progression in human colon adenocarcinoma cell line HT29. Int. J. Cancer. 2009;124:2789–2796. doi: 10.1002/ijc.24262. [DOI] [PubMed] [Google Scholar]
- 68.Wang J., Yuan W., Chen Z., Wu S., Chen J., Ge J., Hou F. Overexpression of G6PD is associated with poor clinical outcome in gastric cancer. Tumour Biol. 2012;33:95–101. doi: 10.1007/s13277-011-0251-9. [DOI] [PubMed] [Google Scholar]
- 69.Nerurkar V.R., Ishwad C.S., Seshadri R., Naik S.N., Lalitha V.S. Glucose-6-phosphate dehydrogenase and 6-phosphogluconate dehydrogenase activities in normal canine mammary gland and in mammary tumours and their correlation with oestrogen receptors. J. Comp. Pathol. 1990;102:191–195. doi: 10.1016/s0021-9975(08)80124-7. [DOI] [PubMed] [Google Scholar]
- 70.Ou K., Yu K., Kesuma D., Hooi M., Huang N., Chen W., Lee S.Y., Goh X.P., Tan L.K., Liu J., Soon S.Y., Bin Abdul Rashid S., Putti T.C., Jikuya H., Ichikawa T., Nishimura O., Salto-Tellez M., Tan P. Novel breast cancer biomarkers identified by integrative proteomic and gene expression mapping. J. Proteome Res. 2008;7:1518–1528. doi: 10.1021/pr700820g. [DOI] [PubMed] [Google Scholar]
- 71.Llobell A., Lopez-Ruiz A., Peinado J., Lopez-Barea J. Glutathione reductase directly mediates the stimulation of yeast glucose-6-phosphate dehydrogenase by GSSG. Biochem. J. 1988;249:293–296. doi: 10.1042/bj2490293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kawaguchi A., Bloch K. Inhibition of glucose 6-phosphate dehydrogenase by palmitoyl coenzyme A. J. Biol. Chem. 1974;249:5793–5800. [PubMed] [Google Scholar]
- 73.Grabowska D., Jablonska-Skwiecinska E., Plochocka D., Chelstowska A., Lewandowska I., Witos I., Majewska Z., Rokicka-Milewska R., Burzynska B. A novel mutation in the glucose-6-phosphate dehydrogenase gene in a subject with chronic nonspherocytic hemolytic anemia–characterization of enzyme using yeast expression system and molecular modeling. Blood Cells Mol. Dis. 2004;32:124–130. doi: 10.1016/j.bcmd.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 74.Beckers A., Organe S., Timmermans L., Scheys K., Peeters A., Brusselmans K., Verhoeven G., Swinnen J.V. Chemical inhibition of acetyl-CoA carboxylase induces growth arrest and cytotoxicity selectively in cancer cells. Cancer Res. 2007;67:8180–8187. doi: 10.1158/0008-5472.CAN-07-0389. [DOI] [PubMed] [Google Scholar]
- 75.Brusselmans K., De Schrijver E., Verhoeven G., Swinnen J.V. RNA interference-mediated silencing of the acetyl-CoA-carboxylase-alpha gene induces growth inhibition and apoptosis of prostate cancer cells. Cancer Res. 2005;65:6719–6725. doi: 10.1158/0008-5472.CAN-05-0571. [DOI] [PubMed] [Google Scholar]
- 76.Chajes V., Cambot M., Moreau K., Lenoir G.M., Joulin V. Acetyl-CoA carboxylase alpha is essential to breast cancer cell survival. Cancer Res. 2006;66:5287–5294. doi: 10.1158/0008-5472.CAN-05-1489. [DOI] [PubMed] [Google Scholar]
- 77.Davies S.P., Sim A.T., Hardie D.G. Location and function of three sites phosphorylated on rat acetyl-CoA carboxylase by the AMP-activated protein kinase. Eur. J. Biochem. 1990;187:183–190. doi: 10.1111/j.1432-1033.1990.tb15293.x. [DOI] [PubMed] [Google Scholar]
- 78.Cabarcas S.M., Hurt E.M., Farrar W.L. Defining the molecular nexus of cancer, type 2 diabetes and cardiovascular disease. Curr. Mol. Med. 2010;10:744–755. doi: 10.2174/156652410793384187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Luo Z., Zang M., Guo W. AMPK as a metabolic tumor suppressor: control of metabolism and cell growth. Future Oncol. 2010;6:457–470. doi: 10.2217/fon.09.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.van Veelen W., Korsse S.E., van de Laar L., Peppelenbosch M.P. The long and winding road to rational treatment of cancer associated with LKB1/AMPK/TSC/mTORC1 signaling. Oncogene. 2011;30:2289–2303. doi: 10.1038/onc.2010.630. [DOI] [PubMed] [Google Scholar]
- 81.Carlson M., Osmond B.C., Botstein D. Mutants of yeast defective in sucrose utilization. Genetics. 1981;98:25–40. doi: 10.1093/genetics/98.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hedbacker K., Carlson M. SNF1/AMPK pathways in yeast. Front. Biosci. 2008;13:2408–2420. doi: 10.2741/2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zimmermann F.K., Kaufmann I., Rasenberger H., Haubetamann P. Genetics of carbon catabolite repression in Saccharomycess cerevisiae: genes involved in the derepression process. Mol. Gen. Genet. 1977;151:95–103. doi: 10.1007/BF00446918. [DOI] [PubMed] [Google Scholar]
- 84.Zhang J., Vaga S., Chumnanpuen P., Kumar R., Vemuri G.N., Aebersold R., Nielsen J. Mapping the interaction of Snf1 with TORC1 in Saccharomyces cerevisiae. Mol. Syst. Biol. 2011;7:545. doi: 10.1038/msb.2011.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Goodridge A.G. Regulation of the activity of acetyl coenzyme A carboxylase by palmitoyl coenzyme A and citrate. J. Biol. Chem. 1972;247:6946–6952. [PubMed] [Google Scholar]
- 86.Rubink D.S., Winder W.W. Effect of phosphorylation by AMP-activated protein kinase on palmitoyl-CoA inhibition of skeletal muscle acetyl-CoA carboxylase. J. Appl. Physiol. 2005;98:1221–1227. doi: 10.1152/japplphysiol.00621.2004. [DOI] [PubMed] [Google Scholar]
- 87.Trumble G.E., Smith M.A., Winder W.W. Purification and characterization of rat skeletal muscle acetyl-CoA carboxylase. Eur. J. Biochem. 1995;231:192–198. doi: 10.1111/j.1432-1033.1995.tb20686.x. [DOI] [PubMed] [Google Scholar]
- 88.Shirra M.K., Patton-Vogt J., Ulrich A., Liuta-Tehlivets O., Kohlwein S.D., Henry S.A., Arndt K.M. Inhibition of acetyl coenzyme A carboxylase activity restores expression of the INO1 gene in a snf1 mutant strain of Saccharomyces cerevisiae. Mol. Cell. Biol. 2001;21:5710–5722. doi: 10.1128/MCB.21.17.5710-5722.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kuhajda F.P., Jenner K., Wood F.D., Hennigar R.A., Jacobs L.B., Dick J.D., Pasternack G.R. Fatty acid synthesis: a potential selective target for antineoplastic therapy. Proc. Natl. Acad. Sci. U. S. A. 1994;91:6379–6383. doi: 10.1073/pnas.91.14.6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Flavin R., Peluso S., Nguyen P.L., Loda M. Fatty acid synthase as a potential therapeutic target in cancer. Future Oncol. 2010;6:551–562. doi: 10.2217/fon.10.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Menendez J.A., Lupu R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat. Rev. Cancer. 2007;7:763–777. doi: 10.1038/nrc2222. [DOI] [PubMed] [Google Scholar]
- 92.Menendez J.A., Vazquez-Martin A., Ortega F.J., Fernandez-Real J.M. Fatty acid synthase: association with insulin resistance, type 2 diabetes, and cancer. Clin. Chem. 2009;55:425–438. doi: 10.1373/clinchem.2008.115352. [DOI] [PubMed] [Google Scholar]
- 93.Menendez J.A., Ropero S., Mehmi I., Atlas E., Colomer R., Lupu R. Overexpression and hyperactivity of breast cancer-associated fatty acid synthase (oncogenic antigen-519) is insensitive to normal arachidonic fatty acid-induced suppression in lipogenic tissues but it is selectively inhibited by tumoricidal alpha-linolenic and gamma-linolenic fatty acids: a novel mechanism by which dietary fat can alter mammary tumorigenesis. Int. J. Oncol. 2004;24:1369–1383. [PubMed] [Google Scholar]
- 94.Yang Y.A., Han W.F., Morin P.J., Chrest F.J., Pizer E.S. Activation of fatty acid synthesis during neoplastic transformation: role of mitogen-activated protein kinase and phosphatidylinositol 3-kinase. Exp. Cell Res. 2002;279:80–90. doi: 10.1006/excr.2002.5600. [DOI] [PubMed] [Google Scholar]
- 95.Yang Y.A., Morin P.J., Han W.F., Chen T., Bornman D.M., Gabrielson E.W., Pizer E.S. Regulation of fatty acid synthase expression in breast cancer by sterol regulatory element binding protein-1c. Exp. Cell Res. 2003;282:132–137. doi: 10.1016/s0014-4827(02)00023-x. [DOI] [PubMed] [Google Scholar]
- 96.Garcia M.J., Pole J.C., Chin S.F., Teschendorff A., Naderi A., Ozdag H., Vias M., Kranjac T., Subkhankulova T., Paish C., Ellis I., Brenton J.D., Edwards P.A., Caldas C. A 1 Mb minimal amplicon at 8p11-12 in breast cancer identifies new candidate oncogenes. Oncogene. 2005;24:5235–5245. doi: 10.1038/sj.onc.1208741. [DOI] [PubMed] [Google Scholar]
- 97.Wang G., Zhang X., Lee J.S., Wang X., Yang Z., Zhang K. Endoplasmic reticulum factor ERLIN2 regulates cytosolic lipid content in cancer cells. Biochem. J. 2012;446:415–425. doi: 10.1042/BJ20112050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cao Y., Dave K.B., Doan T.P., Prescott S.M. Fatty acid CoA ligase 4 is up-regulated in colon adenocarcinoma. Cancer Res. 2001;61:8429–8434. [PubMed] [Google Scholar]
- 99.Mashima T., Sato S., Okabe S., Miyata S., Matsuura M., Sugimoto Y., Tsuruo T., Seimiya H. Acyl-CoA synthetase as a cancer survival factor: its inhibition enhances the efficacy of etoposide. Cancer Sci. 2009;100:1556–1562. doi: 10.1111/j.1349-7006.2009.01203.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yamashita Y., Kumabe T., Cho Y.Y., Watanabe M., Kawagishi J., Yoshimoto T., Fujino T., Kang M.J., Yamamoto T.T. Fatty acid induced glioma cell growth is mediated by the acyl-CoA synthetase 5 gene located on chromosome 10q25.1-q25.2, a region frequently deleted in malignant gliomas. Oncogene. 2000;19:5919–5925. doi: 10.1038/sj.onc.1203981. [DOI] [PubMed] [Google Scholar]
- 101.Tamura K., Makino A., Hullin-Matsuda F., Kobayashi T., Furihata M., Chung S., Ashida S., Miki T., Fujioka T., Shuin T., Nakamura Y., Nakagawa H. Novel lipogenic enzyme ELOVL7 is involved in prostate cancer growth through saturated long-chain fatty acid metabolism. Cancer Res. 2009;69:8133–8140. doi: 10.1158/0008-5472.CAN-09-0775. [DOI] [PubMed] [Google Scholar]
- 102.Pala V., Krogh V., Muti P., Chajes V., Riboli E., Micheli A., Saadatian M., Sieri S., Berrino F. Erythrocyte membrane fatty acids and subsequent breast cancer: a prospective Italian study. J. Natl. Cancer Inst. 2001;93:1088–1095. doi: 10.1093/jnci/93.14.1088. [DOI] [PubMed] [Google Scholar]
- 103.Saadatian-Elahi M., Toniolo P., Ferrari P., Goudable J., Akhmedkhanov A., Zeleniuch-Jacquotte A., Riboli E. Serum fatty acids and risk of breast cancer in a nested case–control study of the New York University Women's Health Study. Cancer Epidemiol Biomarkers Prev. 2002;11:1353–1360. [PubMed] [Google Scholar]
- 104.Crowe F.L., Allen N.E., Appleby P.N., Overvad K., Aardestrup I.V., Johnsen N.F., Tjonneland A., Linseisen J., Kaaks R., Boeing H., Kroger J., Trichopoulou A., Zavitsanou A., Trichopoulos D., Sacerdote C., Palli D., Tumino R., Agnoli C., Kiemeney L.A., Bueno-de-Mesquita H.B., Chirlaque M.D., Ardanaz E., Larranaga N., Quiros J.R., Sanchez M.J., Gonzalez C.A., Stattin P., Hallmans G., Bingham S., Khaw K.T., Rinaldi S., Slimani N., Jenab M., Riboli E., Key T.J. Fatty acid composition of plasma phospholipids and risk of prostate cancer in a case–control analysis nested within the European Prospective Investigation into Cancer and Nutrition. Am. J. Clin. Nutr. 2008;88:1353–1363. doi: 10.3945/ajcn.2008.26369. [DOI] [PubMed] [Google Scholar]
- 105.Brasky T.M., Till C., White E., Neuhouser M.L., Song X., Goodman P., Thompson I.M., King I.B., Albanes D., Kristal A.R. Serum phospholipid fatty acids and prostate cancer risk: results from the prostate cancer prevention trial. Am. J. Epidemiol. 2011;173:1429–1439. doi: 10.1093/aje/kwr027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rysman E., Brusselmans K., Scheys K., Timmermans L., Derua R., Munck S., Van Veldhoven P.P., Waltregny D., Daniels V.W., Machiels J., Vanderhoydonc F., Smans K., Waelkens E., Verhoeven G., Swinnen J.V. De novo lipogenesis protects cancer cells from free radicals and chemotherapeutics by promoting membrane lipid saturation. Cancer Res. 2010;70:8117–8126. doi: 10.1158/0008-5472.CAN-09-3871. [DOI] [PubMed] [Google Scholar]
- 107.Li J., Ding S.F., Habib N.A., Fermor B.F., Wood C.B., Gilmour R.S. Partial characterization of a cDNA for human stearoyl-CoA desaturase and changes in its mRNA expression in some normal and malignant tissues. Int. J. Cancer. 1994;57:348–352. doi: 10.1002/ijc.2910570310. [DOI] [PubMed] [Google Scholar]
- 108.Lu J., Pei H., Kaeck M., Thompson H.J. Gene expression changes associated with chemically induced rat mammary carcinogenesis. Mol. Carcinog. 1997;20:204–215. doi: 10.1002/(sici)1098-2744(199710)20:2<204::aid-mc7>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 109.Scaglia N., Chisholm J.W., Igal R.A. Inhibition of stearoylCoA desaturase-1 inactivates acetyl-CoA carboxylase and impairs proliferation in cancer cells: role of AMPK. PLoS One. 2009;4:e6812. doi: 10.1371/journal.pone.0006812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Scaglia N., Igal R.A. Inhibition of Stearoyl-CoA Desaturase 1 expression in human lung adenocarcinoma cells impairs tumorigenesis. Int. J. Oncol. 2008;33:839–850. [PubMed] [Google Scholar]
- 111.Gyorfy Z., Benko S., Kusz E., Maresca B., Vigh L., Duda E. Highly increased TNF sensitivity of tumor cells expressing the yeast delta 9-desaturase gene. Biochem. Biophys. Res. Commun. 1997;241:465–470. doi: 10.1006/bbrc.1997.7835. [DOI] [PubMed] [Google Scholar]
- 112.Stukey J.E., McDonough V.M., Martin C.E. Isolation and characterization of OLE1, a gene affecting fatty acid desaturation from Saccharomyces cerevisiae. J. Biol. Chem. 1989;264:16537–16544. [PubMed] [Google Scholar]
- 113.Ntambi J.M., Miyazaki M. Regulation of stearoyl-CoA desaturases and role in metabolism. Prog. Lipid Res. 2004;43:91–104. doi: 10.1016/s0163-7827(03)00039-0. [DOI] [PubMed] [Google Scholar]
- 114.Zhang S., Skalsky Y., Garfinkel D.J. MGA2 or SPT23 is required for transcription of the delta9 fatty acid desaturase gene, OLE1, and nuclear membrane integrity in Saccharomyces cerevisiae. Genetics. 1999;151:473–483. doi: 10.1093/genetics/151.2.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gonzalez C.I., Martin C.E. Fatty acid-responsive control of mRNA stability. Unsaturated fatty acid-induced degradation of the Saccharomyces OLE1 transcript. J. Biol. Chem. 1996;271:25801–25809. doi: 10.1074/jbc.271.42.25801. [DOI] [PubMed] [Google Scholar]
- 116.Braun S., Matuschewski K., Rape M., Thoms S., Jentsch S. Role of the ubiquitin-selective CDC48(UFD1/NPL4)chaperone (segregase) in ERAD of OLE1 and other substrates. EMBO J. 2002;21:615–621. doi: 10.1093/emboj/21.4.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kato H., Sakaki K., Mihara K. Ubiquitin-proteasome-dependent degradation of mammalian ER stearoyl-CoA desaturase. J. Cell Sci. 2006;119:2342–2353. doi: 10.1242/jcs.02951. [DOI] [PubMed] [Google Scholar]
- 118.Tatzer V., Zellnig G., Kohlwein S.D., Schneiter R. Lipid-dependent subcellular relocalization of the acyl chain desaturase in yeast. Mol. Biol. Cell. 2002;13:4429–4442. doi: 10.1091/mbc.E02-04-0196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Turyn J., Schlichtholz B., Dettlaff-Pokora A., Presler M., Goyke E., Matuszewski M., Kmiec Z., Krajka K., Swierczynski J. Increased activity of glycerol 3-phosphate dehydrogenase and other lipogenic enzymes in human bladder cancer. Horm. Metab. Res. 2003;35:565–569. doi: 10.1055/s-2003-43500. [DOI] [PubMed] [Google Scholar]
- 120.Dulermo T., Nicaud J.M. Involvement of the G3P shuttle and beta-oxidation pathway in the control of TAG synthesis and lipid accumulation in Yarrowia lipolytica. Metab. Eng. 2011;13:482–491. doi: 10.1016/j.ymben.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 121.Athenstaedt K., Daum G. 1-Acyldihydroxyacetone-phosphate reductase (Ayr1p) of the yeast Saccharomyces cerevisiae encoded by the open reading frame YIL124w is a major component of lipid particles. J. Biol. Chem. 2000;275:235–240. doi: 10.1074/jbc.275.1.235. [DOI] [PubMed] [Google Scholar]
- 122.Datta S.C., Ghosh M.K., Hajra A.K. Purification and properties of acyl/alkyl dihydroxyacetone-phosphate reductase from guinea pig liver peroxisomes. J. Biol. Chem. 1990;265:8268–8274. [PubMed] [Google Scholar]
- 123.Han G.S., O'Hara L., Siniossoglou S., Carman G.M. Characterization of the yeast DGK1-encoded CTP-dependent diacylglycerol kinase. J. Biol. Chem. 2008;283:20443–20453. doi: 10.1074/jbc.M802866200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ellims P.H., Gan T.E., Medley G. Cytidine triphosphate synthetase activity in lymphoproliferative disorders. Cancer Res. 1983;43:1432–1435. [PubMed] [Google Scholar]
- 125.Verschuur A.C., Van Gennip A.H., Leen R., Meinsma R., Voute P.A., van Kuilenburg A.B. In vitro inhibition of cytidine triphosphate synthetase activity by cyclopentenyl cytosine in paediatric acute lymphocytic leukaemia. Br. J. Haematol. 2000;110:161–169. doi: 10.1046/j.1365-2141.2000.02136.x. [DOI] [PubMed] [Google Scholar]
- 126.Williams J.C., Kizaki H., Weber G., Morris H.P. Increased CTP synthetase activity in cancer cells. Nature. 1978;271:71–73. doi: 10.1038/271071a0. [DOI] [PubMed] [Google Scholar]
- 127.Schimmel K.J., Gelderblom H., Guchelaar H.J. Cyclopentenyl cytosine (CPEC): an overview of its in vitro and in vivo activity. Curr. Cancer Drug Targets. 2007;7:504–509. doi: 10.2174/156800907781386579. [DOI] [PubMed] [Google Scholar]
- 128.Yang W.L., Bruno M.E., Carman G.M. Regulation of yeast CTP synthetase activity by protein kinase C. J. Biol. Chem. 1996;271:11113–11119. doi: 10.1074/jbc.271.19.11113. [DOI] [PubMed] [Google Scholar]
- 129.Yang W.L., Carman G.M. Phosphorylation and regulation of CTP synthetase from Saccharomyces cerevisiae by protein kinase A. J. Biol. Chem. 1996;271:28777–28783. doi: 10.1074/jbc.271.46.28777. [DOI] [PubMed] [Google Scholar]
- 130.Chang Y.F., Martin S.S., Baldwin E.P., Carman G.M. Phosphorylation of human CTP synthetase 1 by protein kinase C: identification of Ser(462) and Thr(455) as major sites of phosphorylation. J. Biol. Chem. 2007;282:17613–17622. doi: 10.1074/jbc.M702799200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Han G.S., Sreenivas A., Choi M.G., Chang Y.F., Martin S.S., Baldwin E.P., Carman G.M. Expression of Human CTP synthetase in Saccharomyces cerevisiae reveals phosphorylation by protein kinase A. J. Biol. Chem. 2005;280:38328–38336. doi: 10.1074/jbc.M509622200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ostrander D.B., O'Brien D.J., Gorman J.A., Carman G.M. Effect of CTP synthetase regulation by CTP on phospholipid synthesis in Saccharomyces cerevisiae. J. Biol. Chem. 1998;273:18992–19001. doi: 10.1074/jbc.273.30.18992. [DOI] [PubMed] [Google Scholar]
- 133.Yeh K.T., Tang K.P., Chen Y.L., Su W.W., Wang Y.F., Chang J.G. Methylation inactivates expression of CDP-diacylglycerol synthase 1 (CDS1) in hepatocellular carcinoma. Cancer Genomics Proteomics. 2006;3:231–238. [PubMed] [Google Scholar]
- 134.Li Z., Vance D.E. Phosphatidylcholine and choline homeostasis. J. Lipid Res. 2008;49:1187–1194. doi: 10.1194/jlr.R700019-JLR200. [DOI] [PubMed] [Google Scholar]
- 135.Ackerstaff E., Glunde K., Bhujwalla Z.M. Choline phospholipid metabolism: a target in cancer cells? J. Cell. Biochem. 2003;90:525–533. doi: 10.1002/jcb.10659. [DOI] [PubMed] [Google Scholar]
- 136.Ramirez de Molina A., Gallego-Ortega D., Sarmentero-Estrada J., Lagares D., Gomez Del Pulgar T., Bandres E., Garcia-Foncillas J., Lacal J.C. Choline kinase as a link connecting phospholipid metabolism and cell cycle regulation: implications in cancer therapy. Int. J. Biochem. Cell Biol. 2008;40:1753–1763. doi: 10.1016/j.biocel.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 137.Eliyahu G., Kreizman T., Degani H. Phosphocholine as a biomarker of breast cancer: molecular and biochemical studies. Int. J. Cancer. 2007;120:1721–1730. doi: 10.1002/ijc.22293. [DOI] [PubMed] [Google Scholar]
- 138.Katz-Brull R., Degani H. Kinetics of choline transport and phosphorylation in human breast cancer cells; NMR application of the zero trans method. Anticancer. Res. 1996;16:1375–1380. [PubMed] [Google Scholar]
- 139.Choi M.G., Kurnov V., Kersting M.C., Sreenivas A., Carman G.M. Phosphorylation of the yeast choline kinase by protein kinase C. Identification of Ser25 and Ser30 as major sites of phosphorylation. J. Biol. Chem. 2005;280:26105–26112. doi: 10.1074/jbc.M503551200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kim K.H., Carman G.M. Phosphorylation and regulation of choline kinase from Saccharomyces cerevisiae by protein kinase A. J. Biol. Chem. 1999;274:9531–9538. doi: 10.1074/jbc.274.14.9531. [DOI] [PubMed] [Google Scholar]
- 141.Wieprecht M., Wieder T., Geilen C.C. N-[2-bromocinnamyl(amino)ethyl]-5-isoquinolinesulphonamide (H-89) inhibits incorporation of choline into phosphatidylcholine via inhibition of choline kinase and has no effect on the phosphorylation of CTP: phosphocholine cytidylyltransferase. Biochem. J. 1994;297(Pt 1):241–247. doi: 10.1042/bj2970241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Cai H., Erhardt P., Troppmair J., Diaz-Meco M.T., Sithanandam G., Rapp U.R., Moscat J., Cooper G.M. Hydrolysis of phosphatidylcholine couples Ras to activation of Raf protein kinase during mitogenic signal transduction. Mol. Cell. Biol. 1993;13:7645–7651. doi: 10.1128/mcb.13.12.7645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Fernandez-Murray J.P., Gaspard G.J., Jesch S.A., McMaster C.R. NTE1-encoded phosphatidylcholine phospholipase b regulates transcription of phospholipid biosynthetic genes. J. Biol. Chem. 2009;284:36034–36046. doi: 10.1074/jbc.M109.063958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Murray J.P., McMaster C.R. Nte1p-mediated deacylation of phosphatidylcholine functionally interacts with Sec14p. J. Biol. Chem. 2005;280:8544–8552. doi: 10.1074/jbc.M413999200. [DOI] [PubMed] [Google Scholar]
- 145.Zaccheo O., Dinsdale D., Meacock P.A., Glynn P. Neuropathy target esterase and its yeast homologue degrade phosphatidylcholine to glycerophosphocholine in living cells. J. Biol. Chem. 2004;279:24024–24033. doi: 10.1074/jbc.M400830200. [DOI] [PubMed] [Google Scholar]
- 146.Chang P.A., Chen Y.Y., Qin W.Z., Long D.X., Wu Y.J. The role of cell cycle-dependent neuropathy target esterase in cell proliferation. Mol. Biol. Rep. 2011;38:123–130. doi: 10.1007/s11033-010-0085-3. [DOI] [PubMed] [Google Scholar]
- 147.Oelkers P., Cromley D., Padamsee M., Billheimer J.T., Sturley S.L. The DGA1 gene determines a second triglyceride synthetic pathway in yeast. J. Biol. Chem. 2002;277:8877–8881. doi: 10.1074/jbc.M111646200. [DOI] [PubMed] [Google Scholar]
- 148.Fei W., Shui G., Zhang Y., Krahmer N., Ferguson C., Kapterian T.S., Lin R.C., Dawes I.W., Brown A.J., Li P., Huang X., Parton R.G., Wenk M.R., Walther T.C., Yang H. A role for phosphatidic acid in the formation of “supersized” lipid droplets. PLoS Genet. 2011;7:e1002201. doi: 10.1371/journal.pgen.1002201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Berger J., Moller D.E. The mechanisms of action of PPARs. Annu. Rev. Med. 2002;53:409–435. doi: 10.1146/annurev.med.53.082901.104018. [DOI] [PubMed] [Google Scholar]
- 150.Chambon P. A decade of molecular biology of retinoic acid receptors. FASEB J. 1996;10:940–954. [PubMed] [Google Scholar]
- 151.Kliewer S.A., Umesono K., Noonan D.J., Heyman R.A., Evans R.M. Convergence of 9-cis retinoic acid and peroxisome proliferator signalling pathways through heterodimer formation of their receptors. Nature. 1992;358:771–774. doi: 10.1038/358771a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Uray I.P., Rodenberg J.M., Bissonnette R.P., Brown P.H., Mancini M.A. Cancer preventive rexinoid modulates neutral lipid content of mammary epithelial cells through a PPARgamma-dependent mechanism. Mol. Pharmacol. 2012;81:228–238. doi: 10.1124/mol.111.072967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Fei W., Shui G., Gaeta B., Du X., Kuerschner L., Li P., Brown A.J., Wenk M.R., Parton R.G., Yang H. Fld1p, a functional homologue of human seipin, regulates the size of lipid droplets in yeast. J. Cell Biol. 2008;180:473–482. doi: 10.1083/jcb.200711136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Li Y., Zhang Y., Hill J., Shen Q., Kim H.T., Xu X., Hilsenbeck S.G., Bissonnette R.P., Lamph W.W., Brown P.H. The Rexinoid LG100268 prevents the development of preinvasive and invasive estrogen receptor negative tumors in MMTV-erbB2 mice. Clin. Cancer Res. 2007;13:6224–6231. doi: 10.1158/1078-0432.CCR-06-2681. [DOI] [PubMed] [Google Scholar]
- 155.Kassam A., Hunter J., Rachubinski R.A., Capone J.P. Subtype- and response element-dependent differences in transactivation by peroxisome proliferator-activated receptors alpha and gamma. Mol. Cell. Endocrinol. 1998;141:153–162. doi: 10.1016/s0303-7207(98)00085-9. [DOI] [PubMed] [Google Scholar]
- 156.Marcus S.L., Miyata K.S., Rachubinski R.A., Capone J.P. Transactivation by PPAR/RXR heterodimers in yeast is potentiated by exogenous fatty acid via a pathway requiring intact peroxisomes. Gene Expr. 1995;4:227–239. [PMC free article] [PubMed] [Google Scholar]
- 157.Al-Saffar N.M., Titley J.C., Robertson D., Clarke P.A., Jackson L.E., Leach M.O., Ronen S.M. Apoptosis is associated with triacylglycerol accumulation in Jurkat T-cells. Br. J. Cancer. 2002;86:963–970. doi: 10.1038/sj.bjc.6600188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Schmitz J.E., Kettunen M.I., Hu D.E., Brindle K.M. 1H MRS-visible lipids accumulate during apoptosis of lymphoma cells in vitro and in vivo. Magn. Reson. Med. 2005;54:43–50. doi: 10.1002/mrm.20529. [DOI] [PubMed] [Google Scholar]
- 159.Oelkers P., Tinkelenberg A., Erdeniz N., Cromley D., Billheimer J.T., Sturley S.L. A lecithin cholesterol acyltransferase-like gene mediates diacylglycerol esterification in yeast. J. Biol. Chem. 2000;275:15609–15612. doi: 10.1074/jbc.C000144200. [DOI] [PubMed] [Google Scholar]
- 160.Sandager L., Gustavsson M.H., Stahl U., Dahlqvist A., Wiberg E., Banas A., Lenman M., Ronne H., Stymne S. Storage lipid synthesis is non-essential in yeast. J. Biol. Chem. 2002;277:6478–6482. doi: 10.1074/jbc.M109109200. [DOI] [PubMed] [Google Scholar]
- 161.Fakas S., Konstantinou C., Carman G.M. DGK1-encoded diacylglycerol kinase activity is required for phospholipid synthesis during growth resumption from stationary phase in Saccharomyces cerevisiae. J. Biol. Chem. 2011;286:1464–1474. doi: 10.1074/jbc.M110.194308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Zimmermann R., Strauss J.G., Haemmerle G., Schoiswohl G., Birner-Gruenberger R., Riederer M., Lass A., Neuberger G., Eisenhaber F., Hermetter A., Zechner R. Fat mobilization in adipose tissue is promoted by adipose triglyceride lipase. Science. 2004;306:1383–1386. doi: 10.1126/science.1100747. [DOI] [PubMed] [Google Scholar]
- 163.Ahmadian M., Abbott M.J., Tang T., Hudak C.S., Kim Y., Bruss M., Hellerstein M.K., Lee H.Y., Samuel V.T., Shulman G.I., Wang Y., Duncan R.E., Kang C., Sul H.S. Desnutrin/ATGL is regulated by AMPK and is required for a brown adipose phenotype. Cell Metab. 2011;13:739–748. doi: 10.1016/j.cmet.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Daval M., Diot-Dupuy F., Bazin R., Hainault I., Viollet B., Vaulont S., Hajduch E., Ferre P., Foufelle F. Anti-lipolytic action of AMP-activated protein kinase in rodent adipocytes. J. Biol. Chem. 2005;280:25250–25257. doi: 10.1074/jbc.M414222200. [DOI] [PubMed] [Google Scholar]
- 165.Sullivan J.E., Brocklehurst K.J., Marley A.E., Carey F., Carling D., Beri R.K. Inhibition of lipolysis and lipogenesis in isolated rat adipocytes with AICAR, a cell-permeable activator of AMP-activated protein kinase. FEBS Lett. 1994;353:33–36. doi: 10.1016/0014-5793(94)01006-4. [DOI] [PubMed] [Google Scholar]
- 166.Ye L., Zhang B., Seviour E.G., Tao K.X., Liu X.H., Ling Y., Chen J.Y., Wang G.B. Monoacylglycerol lipase (MAGL) knockdown inhibits tumor cells growth in colorectal cancer. Cancer Lett. 2011;307:6–17. doi: 10.1016/j.canlet.2011.03.007. [DOI] [PubMed] [Google Scholar]
- 167.Lass A., Zimmermann R., Haemmerle G., Riederer M., Schoiswohl G., Schweiger M., Kienesberger P., Strauss J.G., Gorkiewicz G., Zechner R. Adipose triglyceride lipase-mediated lipolysis of cellular fat stores is activated by CGI-58 and defective in Chanarin-Dorfman Syndrome. Cell Metab. 2006;3:309–319. doi: 10.1016/j.cmet.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 168.Montero-Moran G., Caviglia J.M., McMahon D., Rothenberg A., Subramanian V., Xu Z., Lara-Gonzalez S., Storch J., Carman G.M., Brasaemle D.L. CGI-58/ABHD5 is a coenzyme A-dependent lysophosphatidic acid acyltransferase. J. Lipid Res. 2010;51:709–719. doi: 10.1194/jlr.M001917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Rajakumari S., Daum G. Janus-faced enzymes yeast Tgl3p and Tgl5p catalyze lipase and acyltransferase reactions. Mol. Biol. Cell. 2010;21:501–510. doi: 10.1091/mbc.E09-09-0775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Das S.K., Eder S., Schauer S., Diwoky C., Temmel H., Guertl B., Gorkiewicz G., Tamilarasan K.P., Kumari P., Trauner M., Zimmermann R., Vesely P., Haemmerle G., Zechner R., Hoefler G. Adipose triglyceride lipase contributes to cancer-associated cachexia. Science. 2011;333:233–238. doi: 10.1126/science.1198973. [DOI] [PubMed] [Google Scholar]
- 171.Carmona-Gutierrez D., Eisenberg T., Buttner S., Meisinger C., Kroemer G., Madeo F. Apoptosis in yeast: triggers, pathways, subroutines. Cell Death Differ. 2010;17:763–773. doi: 10.1038/cdd.2009.219. [DOI] [PubMed] [Google Scholar]
- 172.Cap M., Stepanek L., Harant K., Vachova L., Palkova Z. Cell differentiation within a yeast colony: metabolic and regulatory parallels with a tumor-affected organism. Mol. Cell. 2012;46:436–448. doi: 10.1016/j.molcel.2012.04.001. [DOI] [PubMed] [Google Scholar]