Abstract
Extracellular biomineralization proteins such as salivary statherin control the growth of hydroxyapatite (HAP), the principal component of teeth and bones. Despite the important role that statherin plays in the regulation of hard tissue formation in humans, the surface recognition mechanisms involved are poorly understood. The protein-surface interaction likely involves very specific contacts between the surface atoms and key protein side chains. This study demonstrates, for the first time, the power of combining near-edge X-ray absorption fine structure (NEXAFS) spectroscopy with element labeling to quantify the orientation of individual side chains. In this work, the 15 amino acid N-terminal binding domain of statherin, SN15, has been adsorbed onto HAP surfaces and the orientations of both phenylalanine rings F7 and F14 have been determined using NEXAFS analysis and fluorine labels at individual phenylalanine sites. The NEXAFS-derived phenylalanine tilt angles have been verified with sum frequency generation spectroscopy.
The interaction and attachment of proteins to surfaces play a key role in biomaterials, tissue engineering, drug delivery, diagnostics and biomimetics.1-3 True molecular level design in these fields will require high-resolution structure characterization techniques to assess the conformation and orientation of adsorbed proteins along with specific structural motifs used by proteins to interact with surfaces. Traditional X-ray diffraction (XRD) or nuclear magnetic resonance (NMR) structure analysis of protein crystals and solutions are extremely successful methods for obtaining high-resolution structures.4, 5 However, the conventional variants of XRD and NMR methods used for protein crystals or solutions do not have sufficient sensitivity for structure determination of monolayer or submonolayer concentrations of proteins bound on surfaces. As a result, of the tens of thousands of protein structures reported, not a single structure of a protein on an inorganic surface has been solved.
Growing numbers of researchers are exploring possible routes to probe protein structure on surfaces using surface analytical tools such as sum frequency generation (SFG) spectroscopy,6-20 infrared and Raman spectroscopy,21, 22 time-of-flight secondary ion mass spectrometry (ToF-SIMS)6, 23-27 and near-edge X-ray absorption fine structure (NEXAFS) spectroscopy.8,10,28-31 These techniques can provide information about the conformation, binding and orientation of surface bound proteins. However, structural data for individual side chains within surface proteins, a crucial prerequisite for solving protein structures, has thus far only been obtained with SFG spectroscopy. 9 NEXAFS spectroscopy has been of particular interest in this context owing to its inherent sensitivity to chemical bonds and molecular structure. It has been used to probe the global orientation and secondary structure of proteins, where libraries of amino acid and peptide spectra facilitate spectral assignments.29-34 However, individual side chain structures within proteins have not yet been studied with this technique. In this communication we report a first observation of individual amino acid orientations in a surface bound protein using the combination of NEXAFS spectra and side chain element labeling. As a proof of concept, we obtained the phenyl ring orientations for individual phenylalanines (F) in the binding domain of the human biomineralization protein statherin adsorbed onto a hydroxapatite (HAP) surface. Mineralization proteins are excellent model systems for highresolution surface analysis because typically they have high binding affinities and rigid surface structures.3, 35 These proteins act as nature’s crystal engineers and adsorb onto crystal surfaces with precision using specific substrate-surface binding motifs.36
The salivary statherin regulates HAP [Ca10(PO4)6(OH)2] growth in bone and tooth enamel and prevents the buildup of excess HAP by inhibiting spontaneous calcium phosphate growth.37, 38 Statherin also binds calcium ions in solution and inhibits precipitation of calcium ions out of supersaturated salivary solutions of calcium and phosphate ions.39, 40 In addition, statherin has a bacterial binding domain and can act as lubricant.41,42 The amino acid sequence of statherin is DSSEEKFLRRIGRFGYGYGPYQPVPEQPLYPLQPYQPQYQQYTF. 43
Statherin has been investigated in solution using solution NMR and circular dichroism spectroscopy, where it was found that statherin displays some transient helical structure in the N-terminal 15 amino acid peptide HAP binding domain.44 Statherin bound to HAP crystals has been studied with binding isotherms and solid-state NMR (ssNMR). Drobny et al. have published a series of ssNMR studies of the local secondary and tertiary structures of HAP-bound statherin, as well as statherin’s N-terminal 15 amino acid peptide HAP-binding domain, SN15.45-48 In these studies, dipolar recoupling magic angle spinning techniques were used to study the surface proximity and dynamics of key acidic, basic and nonpolar side chains.
The role of phenylalanine in protein-HAP interactions is of particular interest because it has the largest HAP binding affinity of any amino acids with a non-polar side chain.49 Because two phenylalanines residues are located within the HAP binding domain of statherin, we set out to explore the role that these side chains may perform in HAP-protein recognition. It has been proposed for example that phenylalanine may interact with HAP surfaces by exposing the delocalized electrons of the phenyl ring.49 Phenylalanines do not nucleate ions because they are non-polar, but they may play a role in the recognition of HAP surfaces.
SN15 13C{31P} REDOR NMR experiments showed the phenylalanine residues of F7 and F14 to be oriented quite differently relative to the HAP surface: a small REDOR response indicated the phenyl ring of F7 was located at least 7 Å from the HAP surface while the phenyl ring of F14 elicited a much stronger 13C{31P} REDOR signal indicating it is located within 4 Å of the HAP surface.46
Here we use NEXAFS spectroscopy to study the orientations of these F7 and F14 side chains. NEXAFS has proven to be a powerful tool to probe protein27 structure and binding on surfaces. NEXAFS is extremely sensitive to surface chemical bonds and can provide detailed information about the chemistry and orientation of surface species. One difficulty of protein structure analysis using NEXAFS is the wide variety and abundance of chemical bonds in all proteins and most peptides. While the backbone amide bonds with their different orientations throughout a protein can sometimes be averaged to an apparent amide vector and, thus, provide some general understanding of the orientation of entire proteins,6, 8, 28 this approach can obviously not be applied to understanding details in the side chain structure. To address this problem, we synthesized two SN15 peptides with hydrogens in the phenylalanine rings substituted by fluorine either in F7 (SN15-F7) or F14 (SN15-F14). The unique C–F bonds acted as residue-specific element labels and could then be individually interrogated with NEXAFS spectroscopy (Figure 1).
Figure 1.
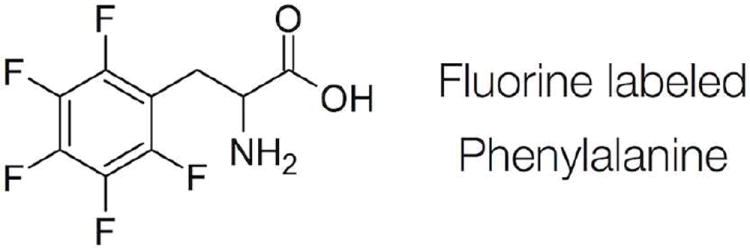
Labeling scheme used for the NEXAFS analysis.
The SN15-F7 and SN15-F14 peptides were adsorbed onto artificial HAP surfaces, prepared by mineral precipitation from a supersaturated simulated body fluid.50 HAP growth and peptide monolayer formation was verified using X-ray photoelectron spectroscopy and ToF-SIMS (see supporting information for details).
Figure 2 shows fluorine K-edge NEXAFS spectra collected using X-ray incidence angles of 70°, 55° and 20° for SN15-F7 and SN15-F14 monolayer films on HAP. The low surface density of the fluorinated rings lead to a comparatively low signal/noise ratio. Assuming a width of ~15 Å and a length of ~28 Å, we estimate the surface area per peptide and, thus, per fluorophenyl ring to be about 420 Å2. The fluorophenyl surface density is about 15-19 times lower than typical aromatic self-assembled monolayers that typically have a 22 - 27 Å2 footprint per molecule.51 We compensated for the low signal/noise ratio by averaging over 5-10 spectra collected at different positions on the samples for each angle. The spectra exhibit characteristic absorption resonances of the fluorophenyl rings. The feature near 690.9 eV is the most prominent peak for both peptides and it can be clearly assigned to the fluorine 1s → π1*(C–C) transition.52, 53 A weaker peak near 693.8 eV is assigned to the corresponding π2* resonance, overlapping with contributions from an F 1s → σ*(C–F) excitation. Since the intense π1* resonance is representative of the π*-orbital which is delocalized over the entire fluorophenyl molecule, we can use the angular dependence of the resonance intensity to determine the orientation of the rings using well established procedures for the analog hydrogenated phenyl moieties.54, 55 Spectra from both, SN15-F7 and SN15-F14, exhibit a considerable linear dichroism (i.e., a dependence of the absorption resonance intensity on the X-ray’s incidence angle). A significant dichroism is considered a fingerprint of orientational order. The intensity of photo excitation resonances depends on the relative orientations of the electric field vector of the X-rays with respect to the target molecular orbital. The transition dipole moment (TDM) of π* resonances is oriented perpendicular to the phenyl ring plane. A higher intensity at an X-ray incident angle of 70° compared to 20° suggests, that the fluorophenyl moieties in SN15-F7 and SN15-F14 have a predominately upright orientation with respect to the surface. The difference in the angle dependence between SN15-F7 and SN15-F14 (larger for SN15-F7) implies a more tilted orientation for F14 with respect to the surface normal (i.e. the F14 ring plane is closer to parallel to the surface). The lower dichroic ratio for F14 could also result from a lower degree of order or a higher mobility. However, since ssNMR results for SN15 on HAP particles show that F14 is located closer to the surfaces than F7,46 F14 is expected to have a stronger interaction with the surface and, thus, a more ordered configuration than F7. This view is supported by the sum frequency generation (SFG) data discussed below. An additional, detailed ssNMR study of phenylalanine dynamics in HAP-bound SN15 is underway and will be published in the future.
Figure 2.
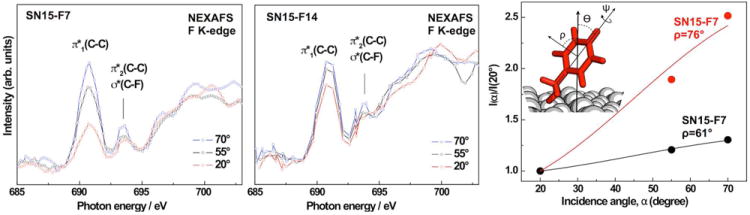
Left & middle panels: F K-edge NEXAFS spectra of SN15-F7 and SN15-F14 adsorbed onto on HAP surfaces acquired at X-ray incidence angles of 70°, 55°, and 20°. Right panel: (inset) Angles used in the characterization of SN15. Angular dependence of the π1* resonance intensity ratio I(α)/I(20°) for SN15-F7 and SN15-F14. The tilt angles ρ of the phenyl ring plane normal with respect to the surface normal are also indicated.
The ring orientation was evaluated by monitoring the intensity of the π1* absorption resonances as a function of the X-ray incidence angle α. The resulting intensity dependence was analyzed according to an established procedure for aromatic rings published in a standard NEXAFS text book by Stöhr.54 The π1* intensities for different incidence angles α are plotted and then fit to the following expression:54
where A is a constant, P is the polarization factor of the synchrotron light, α is the angle of X-ray incidence, and ρ is the average tilt angle of the TDM associated to the molecular orbital (all angles are defined with respect to the surface normal). Intensity normalization problems were minimized by considering intensity ratios, I(α)/I(20°) instead of the absolute values (see Figure 2 for details of the involved angles). The X-ray incidence angle dependence is displayed in Figure 2, along with the respective fits according to eq 1. The phenyl ring plane normal is parallel to the orientation of the π1* orbitals. The derived tilt angles ρ for the normal of the fluorophenyl ring planes are 76° and 61° ±7° for F7 and F14, respectively.
A model was developed for SN-15 adsorbed onto HAP indicating F7 and F14 ring orientations that are consistent with the NEXAFS data and published constraints based on ssNMR data. The NEXAFS-determined tilt angles were tested with independent measurements of the phenyl ring orientations using SFG spectroscopy. We have shown previously that SFG can probe the orientations of individual side chains on surfaces if combined with isotope labels.9 Here we take advantage of the fact that the fluorine labeled SN15 peptides only have one hydrogenated aromatic side chain. Details of the experimental setup and the data analysis can be found in the supporting information. Since the aromatic C-H resonances are well separated from the background of aliphatic modes below 3000 cm-1, the phenylalanine rings can be probed individually with SFG. Figure 3 shows SFG spectra recorded using ssp (spolarized SFG, s-polarized visible and p-polarized IR), ppp and sps polarization combinations for SN15-F14 and SN15-F7. The main spectral features are resonances near 3022 cm-1 and 3060 cm-1 related to the ν20A and ν2 modes of the phenyl rings, respectively.56, 57 The ring tilt and twist angles (Θ and ψ, see Figure 2 for details) were determined utilizing ν2 peak intensity ratios determined from the ssp, ppp and sps spectra following published procedures for polystyrene phenyl rings.56 This analysis yielded tilt and twist angles of Θ=11° and ψ=15° for F7 and Θ=34° and ψ=35° for F14. The general accuracy of these values is ±6° which reflects experimental factors and the analysis procedure. The orientation of the ring plane normal ρ is related to the tilt and twist angles according to:
Figure 3.

SFG spectra for SN15-F7 and SN15-F14 recorded in situ in PBS buffer using different polarization combinations. The spectra of SN15-F7 are representative of ring F14 and the spectra of SN15-F14 are representative of ring F7.
The resulting SFG-determined ring orientations ρ are 80° for F7 and 63° for F14. These values compare well with the NEXAFS-determined angles (76° and 61° for F7 and F14, respectively).
For additional insight into the phenylalanine side chain structure, the NEXAFS- and SFG-determined ring orientations and positions can be combined with published structural data obtained from ssNMR. Using distance measurements and side chain dynamics, Gibson at al. have determined, that F14 is close to the HAP (≈4 Å) with its 2‵–4‵ and 5‵–6‵ axes at equal distances to the surface.46 F7 was found to be at a larger distance from the surface (>6.5 Å). NEXAFS is sensitive to ring orientation but cannot directly distinguish between different pointing directions. Comparing NMR and NEXAFS results (distance, ring rotation), we conclude that the phenyl ring of F14 is pointing toward the surface with a C1-C4 axis angle of 33°. F7 was found to be more dynamic than F14 according to ssNMR and most likely undergoes two-site jumps between different orientations with high frequency. The NEXAFS and SFG-based orientation is, thus, a timeaverage of the two conformers and a study combining ssNMR line shape simulations and NEXAFS analysis is underway to address this complex question in more detail. Based on the NMR data, we can conclude that on average F7 is pointing away from the surface with a ring orientation of 11°. Based on this information and steric constraints from published ssNMR data about the orientation of neighboring side chains,42,58,59 we can estimate the F7 and F14 side chain orientations. Figure 4 displays a possible structure for SN15 on HAP that takes into account the NEXAFS and SFG angles, surface distance data as well as structural parameters from previous ssNMR studies. Admittedly, this model is only one of a number of possible structures. However, this study has shown that NEXAFS spectroscopy can provide detailed information about side chain structure if combined with element labels. A promising approach to further refine our picture of SN15 on HAP will be to combine our findings with additional experimental data (e.g. from SFG amide I measurements) and use the determined structural parameters to guide and constrain computational methods such as molecular dynamics simulations.58,60
Figure 4.
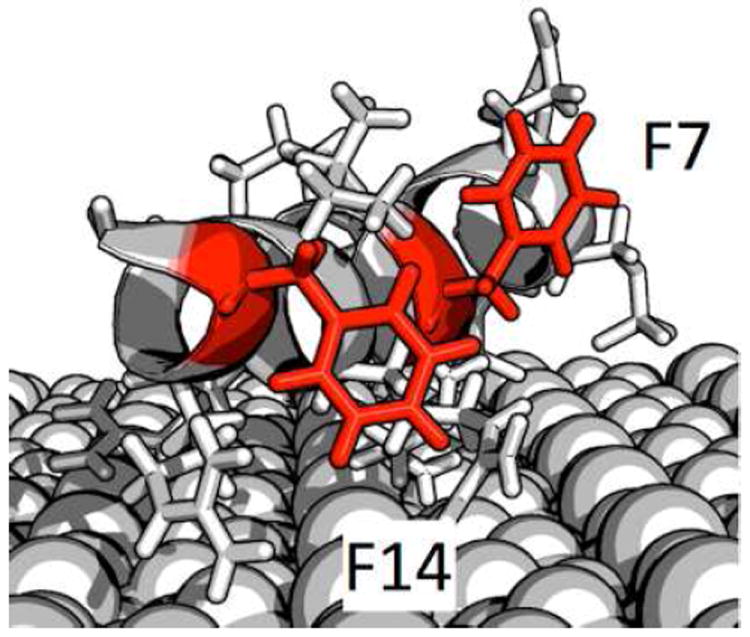
A model of SN-15 adsorbed onto HAP indicating F7 and F14 ring orientations that are consistent with the NEXAFS and SFG data as well as published constrains based on ssNMR data.
Supplementary Material
Acknowledgments
This work was funded in part by NIH grants DE-012554 and EB-002027 (NESAC/BIO). T.W. thanks the German Research Foundation (DFG) for a research fellowship (We 4478/1-1). NEXAFS studies were performed at the NSLS, Brookhaven National Laboratory, which is supported by the U.S. Department of Energy, Division of Materials Science and Division of Chemical Sciences.
Footnotes
Supporting Information. Details of the substrate preparation, peptide synthesis and surface analysis are provided. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Horbett TA, Brash JL. Proteins at Interfaces II: Fundamentals and Applications. American Chemical Society; Washington D. C: 1995. [Google Scholar]
- 2.Castner DG, Ratner BD. Surf Sci. 2002;500:28. [Google Scholar]
- 3.Hildebrand M. Chem Rev. 2008;108:4855. doi: 10.1021/cr078253z. [DOI] [PubMed] [Google Scholar]
- 4.Cavanagh J, Fairbrother WJ, Palmer AG, III, Rance M, Skelton NJ. Protein NMR spectroscopy: principles and practice. 2. Academic Press; Boston: 2007. [Google Scholar]
- 5.Rupp B. Biomolecular Crystallography: Principles, Practice and Application to Structural Biology. Garland Science; New York: 2009. [Google Scholar]
- 6.Baugh L, Weidner T, Baio JE, Nguyen PCT, Gamble LJ, Slayton PS, Castner DG. Langmuir. 2010;26:16434. doi: 10.1021/la1007389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Breen NF, Weidner T, Li K, Castner DG, Drobny GP. J Amer Chem Soc. 2009;131:14148. doi: 10.1021/ja905382m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weidner T, Apte JS, Gamble LJ, Castner DG. Langmuir. 2010;26:3433. doi: 10.1021/la903267x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weidner T, Breen NF, Li K, Drobny GP, Castner DG. P Natl Acad Sci USA. 2010;107:13288. doi: 10.1073/pnas.1003832107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weidner T, Samuel NT, McCrea K, Gamble LJ, Ward RS, Castner DG. Biointerphases. 2010;5:9. doi: 10.1116/1.3317116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Z, Ward R, Tian Y, Malizia F, Gracias DH, Shen YR, Somorjai GA. J Biomed Mater Res. 2002;62:254. doi: 10.1002/jbm.10075. [DOI] [PubMed] [Google Scholar]
- 12.Mermut O, Phillips DC, York RL, McCrea KR, Ward RS, Somorjai GA. J Amer Chem Soc. 2006;128:3598. doi: 10.1021/ja056031h. [DOI] [PubMed] [Google Scholar]
- 13.Phillips DC, York RL, Mermut O, McCrea KR, Ward RS, Somorjai GA. J Phys Chem B. 2007;111:255. [Google Scholar]
- 14.York RL, K BW, Geissler PL, Somorjai GA. Isr J Chem. 2007;47:51. [Google Scholar]
- 15.York RL, Mermut O, Phillips DC, McCrea KR, Ward RS, Somorjai GA. J Phys Chem B. 2007;111:8866. [Google Scholar]
- 16.Wang J, Buck SM, Chen Z. J Phys Chem B. 2002;106:11666. [Google Scholar]
- 17.Wang J, Chen XY, Clarke ML, Chen Z. P Natl Acad Sci USA. 2005;102:4978. doi: 10.1073/pnas.0501206102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang P, Ramamoorthy A, Chen Z. Langmuir. 2011;27:7760. doi: 10.1021/la201048t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ye S, Nguyen KT, Le Clair SV, Z C. J Struct Biol. 2009;168:61. doi: 10.1016/j.jsb.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fu L, Liu J, Yan ECY. J Amer Chem Soc. 2011;133:8094. doi: 10.1021/ja201575e. [DOI] [PubMed] [Google Scholar]
- 21.Ataka K, Heberle J. Anal Bioanal Chem. 2007;388:47. doi: 10.1007/s00216-006-1071-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pettinger B. Mol Phys. 2010;108:2039. [Google Scholar]
- 23.Baio JE, Weidner T, Interlandi G, Mendoza-Barreraa C, Canavan HE, Michel R, Castner DG. J Vac Sci Technol B. 2011;29:04D113. doi: 10.1116/1.3613919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lhoest JB, Detrait E, van den Bosch de Aguilar P, Bertrand P. J Biomed Mater Res A. 1998;41:95. doi: 10.1002/(sici)1097-4636(199807)41:1<95::aid-jbm12>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 25.Xia N, May CJ, McArthur SL, Castner DG. Langmuir. 2002;18:4090. [Google Scholar]
- 26.Wang H, Castner DG, Ratner BD, Jiang SY. Langmuir. 2004;20:1877. doi: 10.1021/la035376f. [DOI] [PubMed] [Google Scholar]
- 27.Baio JE, Weidner T, Samuel NT, McCrea K, Baugh L, Stayton PS, Castner DG. J Vac Sci Technol B. 2010;28:C5D1. doi: 10.1116/1.3456176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu X, Jang C-H, Zheng F, Jürgensen A, Denlinger JD, Dickson KA, Raines RT, Abbott NL, Himpsel FJ. Langmuir. 2006;22:7719. doi: 10.1021/la060988w. [DOI] [PubMed] [Google Scholar]
- 29.Zubavichus Y, Shaporenko A, Grunze M, Zharnikov M. J Phys Chem B. 2007;111:11866. doi: 10.1021/jp073922y. [DOI] [PubMed] [Google Scholar]
- 30.Zubavichus Y, Zharnikov M, Schaporenko A, Grunze M. J Electron Spectrosc Relat Phenom. 2004;134:25. [Google Scholar]
- 31.Gordon ML, Cooper G, Morin C, Araki T, Turci CC, Kaznatcheev K, Hitchcock AP. J Phys Chem A. 2003;107:6144. [Google Scholar]
- 32.Zubavichus Y, Shaporenko A, Grunze M, Zharnikov M. J Phys Chem A. 2005;109:6998. doi: 10.1021/jp0535846. [DOI] [PubMed] [Google Scholar]
- 33.Zubavichus Y, Shaporenko A, Grunze M, Zharnikov M. J Phys Chem B. 2006;110:3420. doi: 10.1021/jp056118p. [DOI] [PubMed] [Google Scholar]
- 34.Cooper G, Gordon M, Tulumello D, Turci C, Kaznatcheev K, Hitchcock AP. J Electron Spectrosc Relat Phenom. 2004;137:795. [Google Scholar]
- 35.Mann S. Nature. 1988;332:119. [Google Scholar]
- 36.Behrens P, Bäuerlein E, editors. Handbook of Biomineralization: Biomimetic and Bioinspired Chemistry. Vol. 2. Wiley-VCH; Weinheim: 2009. [Google Scholar]
- 37.Proctor GB, Hamdan S, Carpenter GH, Wilde P. Biochem J. 2005;389:111. doi: 10.1042/BJ20042012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hay DI, Moreno EC. Human Saliva: Clinical Chemistry and Microbiology. Vol. 1 CRC; Boca Raton, FL: 1989. [Google Scholar]
- 39.Moreno E, Varughese K, Hay D. Calcified Tissue Int. 1979;28:7. doi: 10.1007/BF02441212. [DOI] [PubMed] [Google Scholar]
- 40.Goobes R, Goobes G, Shaw WJ, Drobny GP, Campbell CT, Stayton PS. Biochemistry. 2007;46:4725. doi: 10.1021/bi602345a. [DOI] [PubMed] [Google Scholar]
- 41.Sekine S, Kataoka K, Tanaka M, Nagata H, Kawakami T, Akaji K, Aimoto S, Shizukuishi S. Microbiology. 2004;150:2373. doi: 10.1099/mic.0.27107-0. [DOI] [PubMed] [Google Scholar]
- 42.Goobes G, Goobes R, Schueler-Furman O, Baker D, Drobny GP. P Natl Acad Sci USA. 2006;103:16083. doi: 10.1073/pnas.0607193103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ulrich EL, Akutsu H, Doreleijers JF, Harano Y, Ioannidis YE, Lin J, Livny M, Mading S, Maziuk D, Miller Z, Nakatani E, Schulte CF, Tolmie DE, Kent Wenger R, Yao H, Markley JL. Nucleic Acids Res. 2008;36:D402. doi: 10.1093/nar/gkm957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Naganagowda GA, Gururaja TL, MJ L. J Biomol Struct Dyn. 1998;16:91. doi: 10.1080/07391102.1998.10508230. [DOI] [PubMed] [Google Scholar]
- 45.Ndao M, Ash JT, Breen NF, Goobes G, Stayton PS, Drobny GP. Langmuir. 2009;25:12136. doi: 10.1021/la901647n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gibson JM, Popham JM, Raghunathan V, Stayton PS, Drobny GP. J Amer Chem Soc. 2006;128:5364. doi: 10.1021/ja056731m. [DOI] [PubMed] [Google Scholar]
- 47.Gibson JM, Raghunathan V, Popham JM, Stayton PS, Drobny GP. J Amer Chem Soc. 2006;127:9350. doi: 10.1021/ja050910m. [DOI] [PubMed] [Google Scholar]
- 48.Shaw WJ, Long JR, Dindot JL, Campbell AA, Stayton PS, Drobny GP. J Amer Chem Soc. 2000;122:1709. [Google Scholar]
- 49.Koutsopoulos S, Dalas E. Langmuir. 2000;16:6739. [Google Scholar]
- 50.Toworfe GK, Composto RJ, Shapiro IM, Ducheyne P. Biomaterials. 2006;27:631. doi: 10.1016/j.biomaterials.2005.06.017. [DOI] [PubMed] [Google Scholar]
- 51.Cyganik P, Buck M. J Amer Chem Soc. 2004;126:5960. doi: 10.1021/ja049146e. [DOI] [PubMed] [Google Scholar]
- 52.Hitchcock AP, Fischer P, Gedanken A, Robin MB. J Phys Chem. 1987;91:531. [Google Scholar]
- 53.Plashkevych O, Yang L, Vahtras O, Ågren H, Pettersson LGM. Chem Phys. 1997;222:125. [Google Scholar]
- 54.Stöhr J. NEXAFS Spectroscopy. Vol. 25 Springer-Verlag; Berlin: 1992. [Google Scholar]
- 55.Chesneau F, Schupbach B, Szelagowska-Kunstman K, Ballav N, Cyganik P, Terfort A, Zharnikov M. Phys Chem Chem Phys. 2010;12:12123. doi: 10.1039/c0cp00317d. [DOI] [PubMed] [Google Scholar]
- 56.Briggman KA, Stephenson JC, Wallace WE, Richter LJ. J Phys Chem B. 2001;105:2785. [Google Scholar]
- 57.Varsanyi G. Assignments for Vibrational Spectra of Seven Hundred Benzene Derivatives. Vol. 1.1. Adam Hilger; London: 1974. pp. 1.2–2. [Google Scholar]
- 58.Masica DL, Ash JT, Ndao M, Drobny GP, Gray JJ. Structure. 2010;18:1678. doi: 10.1016/j.str.2010.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ndao M, Ash JT, Stayton PS, Drobny GP. Surf Sci. 2010;604:L39. doi: 10.1016/j.susc.2010.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Makrodimitris K, Masica DL, Kim ET, Gray JJ. J Amer Chem Soc. 2007;129:13713. doi: 10.1021/ja074602v. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


