Abstract
Inflammation is essential to combat invading microbial pathogens. In this process, the involvement of multiple immune cell populations is critical in mounting an optimum immune response. In the past decade, a new class of non-coding small RNAs, called miRNAs, has emerged as important regulators in biological processes. The important role of miRNAs in inflammation and immune response is highlighted by studies in which deregulation of miRNAs was demonstrated to accompany diseases associated with excessive or uncontrolled inflammation. In this brief review, we summarize the roles of miRNAs that have been characterized in innate and adaptive immune responses. We discuss the role of miRNAs in macrophage polarization, a molecular event that has clear impact on inflammation.
Keywords: microRNA, immune response, macrophage polarization
Introduction
Inflammation plays a critical role in host defense to invading microbial pathogens, and is also essential for the successful repair of tissue damage 1–4. Myeloid derived cell populations, such as monocytes, macrophages, dendritic cells, and granulocytes, that can recognize pathogen associated molecular patterns (PAMPs) and damage associated molecular patterns (DAMPs) present in microbial pathogens or as cellular molecules released by damaged tissues 1. Binding of PAMPs or DAMPs to specific cell surface or intracellular receptors, such as Toll like receptors (TLRs), Nod like receptors (NLRs), and the receptor for advanced glycation endproducts (RAGE), in immune cells initiates a cascade of molecular events that leads to the production of pro-inflammatory cytokines, chemokines, reactive oxygen and nitrogen species, and antimicrobial peptides, as well as enhanced phagocytic activity 1–4. These signaling events are essential for the eradication of infection and rapid clearance of cell debris in damaged tissues 1–4. An appropriately orchestrated inflammatory response is also crucial for the subsequent activation of T lymphocytes and development of specific adaptive immunity 5.
Despite the benefits of inflammation in the protection of host from exogenous and endogenous insults, untimely and/or unnecessarily high degrees of inflammation can cause host tissue damage 6. Therefore, the molecular networks that control the initiation, magnitude, and resolution of inflammation must be properly tuned for maintenance of homeostasis and optimization of host response 6. It is clear that every step of the inflammatory pathway is subject to both positive and negative regulatory events in order to achieve an optimal immune response 3, 7.
In the past decade, a new class of small non-coding RNAs, called microRNAs (miRNAs), has emerged as important regulators in the development of immune and inflammatory responses 6, 8–12. The importance of miRNAs in modulation of normal and pathologic immune function has been shown in multiple studies in which deregulation of miRNAs was demonstrated to accompany diseases associated with excessive or uncontrolled inflammation 6, 8–11. This brief review aims to summarize the role of miRNAs in the regulation of innate immune response and adaptive immunity. It also discusses the role of miRNAs in macrophage polarization, a molecular event that has clear impact on inflammation, wound repair, and tumor progression 13.
Overview of miRNA biogenesis and mode of action
Like protein coding genes, miRNAs are generally transcribed by type II RNA polymerase 14. This feature subjects miRNA expression to positive and negative regulation at the transcriptional level, similar to protein coding genes. Perhaps less well appreciated is the regulation of miRNA biogenesis at the posttranscriptional level. The canonical miRNA biogenesis pathway starts with transcription of the miRNA gene, leading to formation of primary miRNA. The primary miRNA transcript is processed by the endoribonucleases Drosha/DGCR8 in the nucleus before transport into the cytoplasm 14. The resulting pre-miRNA is further processed by the endoribonuclease Dicer that cleaves the looped end of the pre-miRNA to produce mature miRNA that is 20–25 base-pairs in length 14. Mature miRNA is then assembled into the miRNA induced silencing complex (miRISC). Individual miRISC binds to the 3' UTR of target mRNAs using the "seed" sequence in the miRNA through "Watson-Crick" base pairing mechanisms 14. It was previously shown that miRNAs function by inducing target mRNA instability and/or inhibiting mRNA translation 14. However, recent evidence indicates that induction of target mRNA instability is the primary mechanism by which miRNAs suppress target expression 15. Given that miRNAs are suppressors of protein expression, the conventional model of miRNA participation in inflammatory regulation could be simplified into a mechanism in which miRNAs target important regulators in inflammatory pathways.
In addition to their activity in the cytoplasm, miRNAs can be secreted into the extracellular environment by immune cells, with such extracellular miRNA typically being enclosed in exosomes or microvesicles 16. These extracellular miRNAs present in exosomes can be transferred from cell to cell, and regulate gene expression in cells following transfer through canonical binding to their target transcripts. This mechanism of intercellular miRNA transfer is thought to be an important component of communication between cells 16. Recent studies have described an additional mechanism by which miRNAs can regulate inflammation. In these experiments, miR-21 and miR-29a enclosed in exosomes, as well as extracellular synthetic let-7b were found to activate microglia, macrophages and neurons through interactions with TLR7 (TLR8 in humans) in the endosomes of these cells 17, 18. Interestingly, binding of miR-21, miR-29a, and let-7b to TLR7 is not incidental because some other miRNAs, such as miR-16, as well as mutant forms of miR-21, miR-29a or let-7b were unable to bind to TLR7 or stimulate immune response 17, 18. miR-21, miR-29a, and let-7b all have a GU-rich element in their sequence, which has a GU content similar to that of ssRNA40, a known TLR7 ligand 17, 18. The GU-rich element was shown to be critical for binding of these miRNAs to TLR7 17, 18. Conceivably, miRNAs and other small RNAs that have this GU-rich element can also activate immune response by binding to TLR7. These surprising findings demonstrate that extracellular miRNAs are more than biomarkers and can act as paracrine agonists of TLRs.
miRNAs and innate immunity
miRNAs constitute an integrated part of the regulatory networks in innate immunity. The modulation of inflammatory responses by miRNAs is primarily through altered expression of specific miRNAs in stimulated immune cells. More than a dozen of miRNAs have been shown to be upregulated or downregulated in activated immune cells (Table 1). One of the first examples is from Baltimore's group. They found that miR-146a is rapidly upregulated in human monocytic cells stimulated with lipopolysaccharide (LPS), a TLR4 ligand, and acts as a negative feedback regulator of TLR signaling, presumably by targeting tumor necrosis factor receptor-associated factor 6 (TRAF6) and IL-1 receptor-associated kinase 1 (IRAK1) 19. miR-146a targeting of TARF6 and IRAK1 has been confirmed in a more recent study, although done in T cells, from the same group 20. miR-146a upregulation may be an essential component of endotoxin tolerance in innate immune response 21. These findings suggest that an insufficient induction of miR-146a could lead to hyper-activated or prolonged inflammation. Additionally, miR-146a is a negative regulator of vesicular stomatitis virus (VSV) induced type 1 interferon response in macrophages 22. Thus, miR-146a appears to play important roles in the inflammatory response to both bacterial and viral infections. The crucial involvement of miR-146a in inflammation was also highlighted in mice with either global miR-146a deficiency or miR-146a deficiency in regulatory T (Treg) cells 23, 24. These mice develop autoimmune disorders, and their macrophages are hypersensitive to LPS stimulation 23, 24.
Table 1.
miRNAs in innate immunity
| miRNA | Targets | Effect |
|---|---|---|
| miR-9 | NF-κB1 | Negative regulator of TLR4 signaling |
| miR-19 | TLR2 | Decreases TLR2 mediated inflammation |
| miR-21 | PDCD4, IL-12 p35 | Negative regulator of TLR4 signaling |
| miR-27b | PPAR-γ | Enhances response to LPS |
| miR-105 | TLR2 | Decreases TLR2 mediated inflammation |
| miR-106a | IL-10 | Decreases IL-10 |
| miR-125b | TNF-α; IRF4 | Diminishes inflammation; enhances macrophage activation |
| miR-145 | TIRAP | Inhibits TLR signaling |
| miR-146a | TRAF6, IRAK1, IRAK2 | Negative regulator of TLR signaling |
| miR-155 | AID; MyD88; TAB2; Pellino-1; IKKε; SHIP-1; SOCS1; C/EBP-β |
Enhances inflammation; negative feedback regulation |
| miR-223 | IKKα; Pknox1 | Proinflammatory activation of macrophages |
| Let-7i, let-7e | TLR4 | Downregulate inflammatory signaling |
miR-21 is induced in monocytic cells by LPS stimulation 25. Upregulated miR-21 was found to dampen LPS induced NF-κB activation and IL-6 expression, but to enhance IL-10 expression 25. The anti-inflammatory effect of miR-21 lies in targeting tumor suppressor PDCD4, a pro-inflammatory protein 25. miR-21 can also trigger inflammatory responses by binding to TLR7 and TLR8 17. Therefore, the functional consequence of miR-21 upregulation in inflammatory conditions may be determined by many factors, such as the local concentration of extracellular miR-21 and the kinetics of mRNA targeting mediated by intracellular miR-21. It is certainly possible that at early time points miR-21 upregulation is pro-inflammatory through binding to TLR7/8. However, downregulation of PDCD4 at later time points by miR-21 may play a role in returning inflammatory responses to baseline levels, thereby maintaining homeostasis of immune responses.
miR-155 is induced in macrophages in response to both bacterial and viral derived antigens that activate TLR4, TLR2, TLR3, or TLR9 19, 26–28. An early study found that miR-155 is a negative regulator of inflammatory response to LPS in dendritic cells. miR-155 was shown to target TGF-β activated kinase 1 binding protein 2 (TAB2) and Pellino-1, adaptors in the signaling complex that activates IκB kinase β (IKKβ). These results suggest that miR-155 is part of a negative feedback loop to dampen inflammatory responses 29. However, most evidence indicates that miR-155 is pro-inflammatory. This proinflammatory effect of miR-155 likely occurs via repression of negative regulators of inflammation, including suppressor of cytokine signaling 1 (SOCS1) and phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase 1 (SHIP1) which are both direct targets of miR-155 9, 26, 30. Through the downregulation of these anti-inflammatory pathways, miR-155 is able to potentiate inflammation. miR-155 can also increase the half-life of TNF-α transcripts, although the mechanism for this effect is presently unknown 28, 31, 32. miR-155 deficient mice have decreased immune responses, whereas miR-155-overexpressing mice develop a myeloproliferative disorder resembling chronic inflammation and also suffer from hematopoietic cancers 6, 33–36. Given the evidence that miR-155 plays both positive and negative regulatory roles in immune responses, it would appear that the effects of this miRNA are dependent on the cellular context.
Different miRNAs seem to show synergies that result in fine tuning of inflammatory responses. An example is the induction of both miR-155 and miR-21 in TLR ligand stimulated immune cells. In these settings, miR-155 may have early effects in promoting inflammatory response by targeting SOCS1 and SHIP1 9, 26, 30. This action is later dampened by miR-21 induced IL-10 because IL-10 can inhibit miR-155, allowing SHIP1 to be de-repressed with resultant inhibition of TLR signaling 37.
Several miRNAs regulate aspects of innate immune responses by targeting signaling components in inflammatory pathways. For example, miR-223, miR-105, miR-19, and let-7 family members directly target TLR2, TLR3 or TLR4 11, 26, 38–40. Thus, these miRNAs can impact the ability of immune cells to engage with TLR specific extracellular ligands. miR-147 is induced following the activation of TLR2, TLR3, or TLR4 in both a Myd88- and TRIF- dependent manner, and acts as a negative regulator to prevent excessive inflammatory responses in macrophages 41. miR-9 is induced by LPS in both neutrophils and monocytes, and negatively regulates NF-κB dependent inflammatory responses by suppressing the expression of NFKB1 transcripts 42. Some miRNAs, such as Let-7i and miR-125b, are downregulated in response to LPS and microbial infections. The reduced expression of these miRNAs was shown to lead to enhanced immune responses 28, 32, 40, 43. Therefore, miRNAs are important components of feed-forward networks in inflammatory amplification. Ectopic expression of miR-125b has also been shown to promote macrophage activation, as reflected by findings that overexpression of miR-125b in macrophages increases costimulatory factor expression and responsiveness to IFN-γ 44. IRF4, a direct target of miR-125b, could mediate the effect of miR-125b to promote macrophage activation because IRF4 knockdown in macrophages mimics the miR-125b overexpression phenotype 44.
miRNA and adaptive immunity
miRNAs regulate T cell development, differentiation and activation
T cells directly carry out specific types of inflammatory responses and their activation is dependent upon presentation of specific antigens in the context of MHC by antigen-presenting cells, a function performed predominately by innate immune cells that are the first to come into contact with infectious agents 6, 9.
The overall significance of miRNAs in T cell development and activation was primarily established by using global and specific miRNA deficient mice. Early studies observed that disruption of miRNA biogenesis by conditional removal of Dicer in the early stages of T lymphocyte development results in reduced T cell numbers in the thymus and peripheral lymphoid organs, and causes aberrant T helper (Th) cell differentiation and cytokine production 45, 46. Recent studies have confirmed the crucial involvement of specific miRNAs in T lymphocyte biology and in the pathogenesis of autoimmune diseases associated with T cell perturbation. T cells deficient in miR-155, a miRNA expressed in activated CD4+ T cells, exhibit a Th2 bias under neutral conditions in vitro 47, 48, indicating that miR-155 is normally required for a Th1-type response and for the polarization of T cells towards a pro-inflammatory phenotype. miR-155−/− mice are defective in Th1 and Th17 cell production during autoimmune inflammation 47, 48. These observations may help explain the increased resistance of miR-155−/− mice to experimental autoimmune encephalomyelitis (EAE), colitis, and collagen induced arthritis (CIA) 35, 49–51. The inhibitory effect of miR-155 on Th2 differentiation is thought to be mediated through effects on c-Maf, which is expressed in Th2 cells and promotes the development of this lineage 48.
miR-155 not only regulates effector T cell functions, but also is involved in the development of T regulator (Treg) cells. Although miR-155−/− mice have a reduced number of Tregs in the thymus and peripheral lymphoid tissues, the suppressive activity of miR-155−/− Tregs remains intact 52. This suggests that miR-155 is critical to Treg development, but is not essential to the suppressive functions of Tregs. However, miR-146a, one of the major miRNAs expressed in Treg cells, is critical for their suppressive functions. Deficiency of miR-146a in Treg cells resulted in breakdown of immunological tolerance manifested by fatal IFN-γ-dependent immune-mediated lesions in a variety of organs 23. This was likely due to augmented expression and activation of signal transducer and activator transcription 1 (Stat1), a direct target of miR-146a 23.
miRNAs have been well characterized in the regulation of T cell differentiation and clonal expansion. miR-181a has a significant influence on positive selection by heightening TCR signaling during thymic development 53. These effects are achieved in part by the downregulation of multiple phosphatases, leading to elevated steady-state levels of phosphorylated intermediates and reduction of the T cell receptor signaling threshold 53. Inhibition of miR-181a led to excessive reactions to self-peptides that are normally sufficient only for positive selection 54. Thus, miR-181a contributes to clonal selection and to preventing autoreactive T cell clones from reaching the periphery and potentially causing autoimmune disorders 53. miR-17-92 cluster miRNAs are involved in cell survival by repressing Bim and PTEN, both of which potentiate cell death 55. Overexpression of the miR-17-92 cluster in transgenic mice results in T cell populations that show more proliferation and less activation-induced cell death, and is associated with lymphoproliferative disease 55. miR-326 was shown to target Ets, a transcription factor that negatively regulates Th17 development, and overexpression of miR-326 led to the promotion of Th17 differentiation and IL-17 production 56.
miRNAs regulate B cell development, differentiation, and activation
B cell development and differentiation start with the proliferative expansion of progenitor cells that undergo sequential rearrangements of their antigen receptor genes to produce a diverse, clonally selected receptor repertoire 57. Cells expressing functional, nonself-reactive receptors are positively selected to become the peripheral pool of mature B cells. Major subsets of mature B cells are B2 B cells, including follicular B cells that are responsible for T cell-dependent antibody responses and marginal zone (MZ) B cells that can respond rapidly to infectious agents in the blood, and B1 B cells that home to the peritoneal and pleural cavities and represent an important source of “natural” antibodies in the blood as an evolutionarily selected first line of defense against pathogens 57.
Many cytokines and transcription factors play critical regulatory roles in B lymphopoiesis. In recent years, the role of miRNAs in B cell development and differentiation has been increasingly appreciated. The first report of miRNA regulation of B cell development came in a seminal study showing that miR-181 was highly expressed in B cells and that ectopic expression of miR-181 guided B-cell development 58.
With the establishment of additional miRNA deficient mice, there has been further understanding of the regulatory role of miRNAs at different stages of B cell development. miR-150 was found to be highly expressed in mature and resting B lymphocytes, a pattern that is inversely correlated to the transcriptional factor c-Myb 57. miR-150 deficiency leads to B1 cell expansion and enhanced humoral immune responses, a phenomenon that is opposite to that observed in mice with B cell-specific deletion of c-Myb, which is accompanied by severe block of B cell development at the pro- to pre-B transition and disappearance of B1 B cells 59. Of note, c-Myb was found to be a target of miR-150, and ectopic expression of miR-150 in mice mirrored the observations in c-Myb−/− mice 60. These findings have established a central role for miR-150 in B cell development 57.
miR-155 plays a crucial role in B cell activation. B cells lacking miR-155 generate reduced extrafollicular and germinal center responses and fail to produce high-affinity IgG1 antibodies 48. These phenomena are thought to be mediated by de-repressing of miR-155 targets in B cells, such as Pu.1, SHIP1 and AID (activation-induced cytidine deaminase) 48, 61. However, it should be noted that none of the above targets are solely responsible for the perturbation in B cell activation in miR-155−/− cells. This concept is supported by an elegant study that used transgenic mice carrying a mutation in the putative miR-155 binding site in the 3' UTR of AID (called Aicda155 mice) 62. AID is required for immunoglobulin gene diversification in B lymphocytes, and also promotes chromosomal translocations 62. Aicda155 caused an increase in steady-state Aicda mRNA and protein amounts by increasing the half-life of this mRNA, resulting in a high degree of Myc-Igh translocations 62. Compared to Aicda155 mice, a similar but more pronounced translocation phenotype was also found in miR-155-deficient mice 62.
Specific miRNAs are clearly essential for the development, differentiation, activation, and function of T and B lymphocytes (Table 2). The temporal and spatial expression of miRNAs must be tightly controlled in the development of adaptive immune responses. Dysregulation of specific miRNAs involved in these T and B cell related events is likely to lead to chronic inflammation and autoimmune diseases.
Table 2.
miRNAs in adaptive immunity
| miRNA | Targets | Effect |
|---|---|---|
| miR-10 | Ncor2; Bcl6 | Treg↑ |
| miR-17-92 | Bim; PTEN | CD4 T cell proliferation↑ |
| miR-29b | T-bet; IFN-γ | Th1↓ |
| miR-146a | IRAK1; TRAF6; Stat1 | TCR signaling↓; Treg↑ |
| miR-155 | c-Maf | Th1↑; Th17↑; Treg survival↑; Th2↓ |
| miR-181a | SHP1,2; DUSP5,6 | TCR signaling↑ |
| miR-182 | Foxo1 | CD4 T cell expansion↑ |
| miR-301a | PIAS3 | Th17↑ |
| miR-326 | Ets | Th17↑ |
| miR-150 | c-Myb | B1 cell expansion↓ |
| miR-155 | Pu.1; SHIP1; AID | antibody secretion↑; class-switch recombination↑ |
| miR-181 | B cell expansion↑ |
miRNA in macrophage polarization
Macrophages display remarkable plasticity, with the ability to undergo dynamic transition between different functional phenotypes 63, 64. Macrophages activated by TLR ligands and/or IFN-γ are called M1 macrophages (also referred to as classically activated macrophages) 63–65. Conversely, stimulation of macrophages with Th2 cytokines, such as IL-4 or IL-13, immune complexes plus TLR ligands, IL-10, TGF-β, or glucocorticoids induces the generation of M2-type macrophages (also called alternatively activated macrophages) 63–65. M1 macrophages produce high amounts of proinflammatory cytokines and nitric oxide (NO) by expressing inducible NO synthase (iNOS), and are critical for eradicating bacterial, viral and fungal infections 63–65. M2 macrophages are characterized by their high expression of markers of alternative activation, such as arginase-1 (Arg1), Chitinase 3-like 3 (also called YM-1), and found in inflammatory zone 1 (Fizz1), and regulate responses to parasite infection, tissue remodeling, angiogenesis and tumor progression 63, 64, 66. The classical M2 macrophage markers in mice, such as Arg1, YM-1, and FIZZ-1, are not induced in human macrophages even in the presence of IL 4 67. Therefore, one should be cautious when applying findings obtained with murine M2 macrophages into pathophysiologic states in humans where M2 macrophages play an important role.
The polarization of macrophages has been the focus of recent studies, particularly with regard to transcriptional regulation 63. Transcriptional factors NF-κB, AP-1, CCAAT/enhancer-binding protein α (C/EBP-α), PU.1, and IFN-regulatory factor 5 (IRF5) participate in TLR ligand induced M1 activation, whereas STAT6, peroxisome proliferator-activated receptor (PPAR)-γ, IRF4, C/EBP-β, and Kruppel-like factor 4 (KLF4) are involved in the polarization of M2 macrophages 63. Enzymes involved in epigenetic regulation, such as Jumonji domain containing 3 (JMJD3) and histone deacetylase 3 (HDAC3), also play important roles in M2 macrophage polarization 68.
As discussed above, many miRNAs are involved in inflammation by either diminishing or potentiating inflammatory response through targeting important regulators in signaling networks. Given that TLR stimulation induces M1 macrophage polarization, one could arbitrarily consider the inflammation modulating miRNAs as those that can regulate M1 polarization. However, it remains unclear if these miRNAs also participate in M2 macrophage polarization and, if so, what are the potential regulatory targets for such an action. There was an effort to systemically identify miRNAs that have altered expression in differentially polarized human monocyte derived macrophages 69. However, only a few miRNAs were found to have more than 2-fold change of expression in M1 macrophages or one of the three M2 subtypes 69. Among these miRNAs, all but two are low abundant passenger strands 69. Although there are examples that passenger strands do have regulatory roles 70, they are generally excluded from miRISC 71. Thus, the functional significance of these miRNAs in polarized macrophages remains to be determined. Furthermore, previously identified miRNAs with altered expression in inflamed immune cells, such as miR-21 and miR-146a, were not picked up in this study, possibly due to a great abundance of these miRNAs under basal conditions in human monocyte-derived macrophages 69. Nevertheless, this was the first systemic attempt to uncover M1 and M2 related miRNAs in a model with definitive induction of macrophage polarization.
Efforts to delineate the role of miRNAs in macrophage activation in inflammatory diseases have been fruitful. One of the best examples is miR-124, a brain-specific miRNA 72. miR-124 is highly expressed in microglia, but not in other tissue macrophages. miR-124 is downregulated in activated microglia from the central nerve system (CNS) of mice with experimental autoimmune encephalomyelitis (EAE), a mouse model of the human disease multiple sclerosis 72. EAE is characterized by inflammation of the CNS, and is associated with microglia activation and infiltration of T cells and leukocytes. Overexpression of miR-124 diminished M1 polarization and enhanced M2 polarization in BMDMs, as reflected by reduced expression of the surface markers CD45, CD11b, F4/80, MHC class II, and CD86, but increased expression of the M2 phenotypic markers FIZZ1 and Arg1 72. In contrast, knockdown of miR-124 enhanced the expression of the surface markers CD45 and MHC class II in BMDMs co-cultured with neural and astroglial cells 72. Furthermore, systemic administration of miR-124 in vivo inhibited the development of EAE and reduced CNS inflammation 72. This study identified C/EBP-α as the mediator of the miR-124 effect on macrophage polarization 72. Although it remains unclear how C/EBP-α suppresses M2 polarization, it is one of the few cases in which a specific miRNA has been shown to regulate macrophage plasticity.
Another elegant example of miRNA participation in macrophage polarization is the discovery that miR-223 regulates adipose tissue inflammation and insulin resistance 73. In that study, miR-223 was found to be differentially expressed during macrophage polarization, with upregulation in LPS treated macrophages, but downregulation in IL-4 treated BMDMs 73. miR-223-deficient macrophages were hypersensitive to LPS stimulation, whereas such macrophages exhibited delayed responses to IL-4 compared with controls 73. Furthermore, miR-223-deficient mice exhibited an increase in adipose tissue inflammatory responses and decreased adipose tissue insulin signaling 73. The importance of macrophage miR-223 was also confirmed by increased adipose tissue inflammation and insulin resistance in mice with transplantation of bone marrow from miR-223-deficient mice 73. Pknox1 was identified in these studies as a genuine target of miR-223. The expression of Pknox1 is inversely correlated with miR-223 levels in activated BMDMs and adipose tissue. The function of Pknox1 as a target of miR-223 in regulating macrophage polarization was validated in gain-of-function and loss-of-function analyses in BMDMs 73. However, a key question how Pknox1 regulates macrophage polarization has not been answered.
Akt kinase has been shown to be both pro- and anti-inflammatory in immune cells, including in macrophages 74, 75. Recent studies addressed the role of Akt isoforms in differentially contributing to macrophage polarization 76. Akt1 ablation in macrophages gave rise to a M1 macrophage phenotype, and Akt2 ablation resulted in a M2 phenotype 76. Akt2−/− mice were more resistant to LPS-induced endotoxin shock and to dextran sulfate sodium (DSS)-induced colitis than were wild-type mice, whereas Akt1−/− mice were more sensitive 76. These phenomena converge on miR-155, whose expression was repressed in naive and in LPS-stimulated Akt2−/− macrophages. The miR-155 target, C/EBP-β, appears to play a key role in this process 76. C/EBP-β, a hallmark of M2 macrophages that regulates Arg1, was up-regulated upon Akt2 ablation or silencing. Overexpression or silencing of miR-155 confirmed its central role in Akt isoform-dependent M1/M2 polarization of macrophages 76. Furthermore, knocking down miR-155 had a direct impact on IL-13 induced expression of M2 phenotypic markers by derepressing IL-13Rα1, leading to increased Stat6 phosphorylation 77. These studies support the hypothesis that miR-155 has a key role in determining macrophage plasticity, with the ability to skew macrophages toward the M1 phenotype.
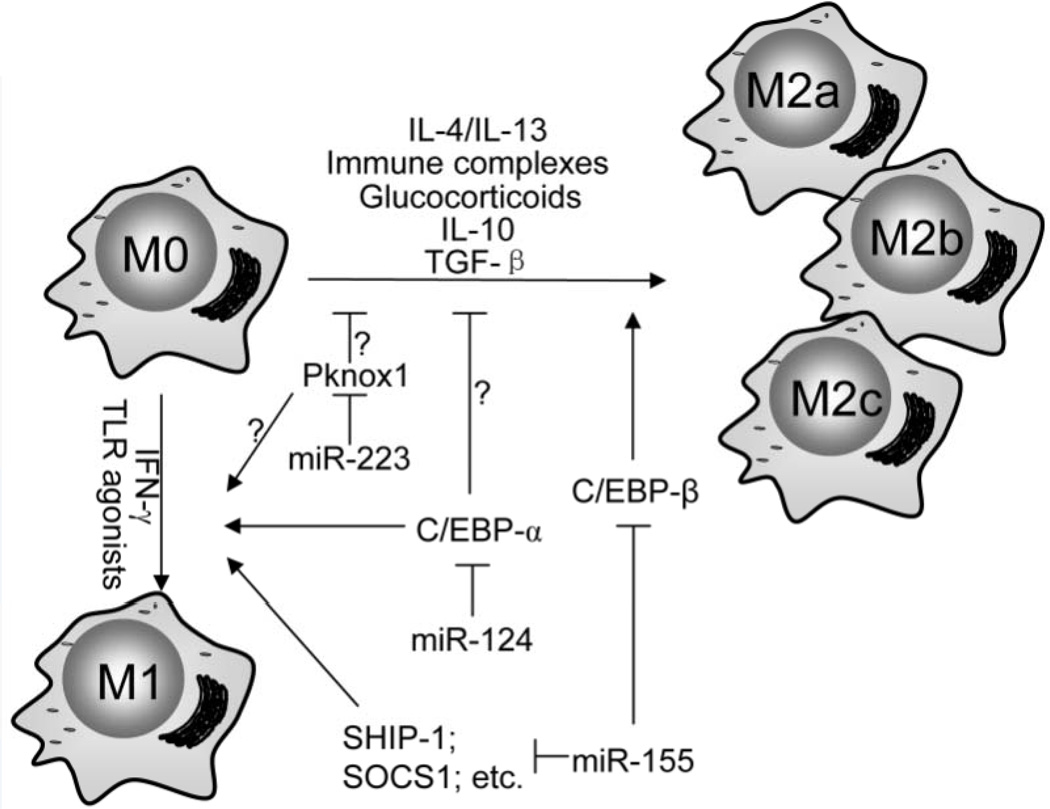
In summary, although many miRNAs can regulate inflammatory response in macrophages, only a few miRNAs are shown to participate in both M1 and M2 macrophage polarization. This certainly reflects the fact that only a limited number of protein regulators are known to be involved in both processes (Figure 1). Although additional miRNAs that can modulate macrophage plasticity await identification, there is even more effort required to reveal the mechanism by which the targets of those miRNAs regulate this important event.
Figure 1.
A few miRNAs, such as miR-124, miR-155 and miR-223, have been identified to regulate both M1 and M2 macrophage polarization. However, it is unknown if the large number of miRNAs that regulate inflammatory response also modulate macrophage plasticity.
Concluding remarks
In just a few short years, miRNA research in the immune system has produced a wealth of knowledge firmly supporting the concept that miRNAs have a central role in modulating inflammatory responses. With the identification of specific miRNAs that play key roles in negative or positive regulatory pathways in innate and adaptive immunity and with the demonstration that dysregulation of specific miRNAs is associated with inflammatory diseases, the field has opened new avenues for uncovering novel targets in treating pathological inflammatory conditions.
There is no doubt that additional miRNAs will be shown to participate in a variety of inflammatory responses. However, there are a number of intriguing questions that remain inadequately answered. First, it is clear that every stage of the inflammatory response involves more than one regulatory miRNA. However, an improved understanding of how multiple miRNAs can collaborate to properly balance inflammatory responses is needed. This will provide a better understanding of how miRNA networks can work together to regulate inflammation and other immune responses. Second, since even one miRNA is capable of targeting a large number of mRNAs, it remains challenging to identify the specific targets of any single miRNA. Most studies primarily rely on computational algorithms to predict target transcripts and often select to study one or two targets known to be involved in inflammatory response. This approach can certainly introduce bias and runs the risk of missing key targets with presently unknown but important regulatory functions. To overcome these limitations, a number of techniques, such as RIP-ChIP, RIP-Seq, and PAR-Clip, have been developed to capture bona fide miRNA targets. These advanced techniques to identify miRNA targets have been thoroughly reviewed elsewhere 78. Thirdly, better understanding of how modest suppression or fine tuning of multiple targets achieves optimal immune responses will provide more complete insights into how miRNAs modulate central events in inflammation, including immune cell development and activation.
The development of atherosclerotic lesions is shaped by immune responses and their regulation 79. Given that miRNAs are centrally involved in innate and adaptive immune responses, miRNAs that participate in such processes have been shown to participate in the regulation of atherosclerosis 79. Thus, delineation of the mechanisms by which miRNAs modulate immune response is likely to lead to therapeutic targeting of specific miRNAs that may be efficacious in preventing or treating atherosclerosis.
Acknowledgments
We apologize to colleagues whose work could not be cited due to space limitations. We also regret that we cited some review papers instead of the original articles due to the concise nature of this review.
Sources of funding - G.L's laboratory is supported by NIH grants HL105473, HL097218, HL076206, and an AHA award 10SDG4210009.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure - None.
References
- 1.Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 2.Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol. 2010;11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 3.Medzhitov R, Horng T. Transcriptional control of the inflammatory response. Nat Rev Immunol. 2009;9:692–703. doi: 10.1038/nri2634. [DOI] [PubMed] [Google Scholar]
- 4.Jeannin P, Jaillon S, Delneste Y. Pattern recognition receptors in the immune response against dying cells. Curr Opin Immunol. 2008;20:530–537. doi: 10.1016/j.coi.2008.04.013. [DOI] [PubMed] [Google Scholar]
- 5.Mills KH. TLR-dependent T cell activation in autoimmunity. Nat Rev Immunol. 2011;11:807–822. doi: 10.1038/nri3095. [DOI] [PubMed] [Google Scholar]
- 6.O'Connell RM, Rao DS, Baltimore D. microRNA regulation of inflammatory responses. Annu Rev Immunol. 2012;30:295–312. doi: 10.1146/annurev-immunol-020711-075013. [DOI] [PubMed] [Google Scholar]
- 7.Ruland J. Return to homeostasis: downregulation of NF-kappaB responses. Nat Immunol. 2011;12:709–714. doi: 10.1038/ni.2055. [DOI] [PubMed] [Google Scholar]
- 8.Alam MM, O'Neill LA. MicroRNAs and the resolution phase of inflammation in macrophages. Eur J Immunol. 2011;41:2482–2485. doi: 10.1002/eji.201141740. [DOI] [PubMed] [Google Scholar]
- 9.Contreras J, Rao DS. MicroRNAs in inflammation and immune responses. Leukemia. 2012;26:404–413. doi: 10.1038/leu.2011.356. [DOI] [PubMed] [Google Scholar]
- 10.Dai R, Ahmed SA. MicroRNA, a new paradigm for understanding immunoregulation, inflammation, and autoimmune diseases. Transl Res. 2011;157:163–179. doi: 10.1016/j.trsl.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O'Neill LA, Sheedy FJ, McCoy CE. MicroRNAs: the fine-tuners of Toll-like receptor signalling. Nat Rev Immunol. 2011;11:163–175. doi: 10.1038/nri2957. [DOI] [PubMed] [Google Scholar]
- 12.Xiao C, Rajewsky K. MicroRNA control in the immune system: basic principles. Cell. 2009;136:26–36. doi: 10.1016/j.cell.2008.12.027. [DOI] [PubMed] [Google Scholar]
- 13.Murray PJ, Wynn TA. Protective and pathogenic functions of macrophage subsets. Nat Rev Immunol. 2011;11:723–737. doi: 10.1038/nri3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vickers KC, Remaley AT. Lipid-based carriers of microRNAs and intercellular communication. Curr Opin Lipidol. 2012;23:91–97. doi: 10.1097/MOL.0b013e328350a425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fabbri M, Paone A, Calore F, Galli R, Gaudio E, Santhanam R, Lovat F, Fadda P, Mao C, Nuovo GJ, Zanesi N, Crawford M, Ozer GH, Wernicke D, Alder H, Caligiuri MA, Nana-Sinkam P, Perrotti D, Croce CM. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci U S A. 2012;109:E2110–E2116. doi: 10.1073/pnas.1209414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lehmann SM, Kruger C, Park B, Derkow K, Rosenberger K, Baumgart J, Trimbuch T, Eom G, Hinz M, Kaul D, Habbel P, Kalin R, Franzoni E, Rybak A, Nguyen D, Veh R, Ninnemann O, Peters O, Nitsch R, Heppner FL, Golenbock D, Schott E, Ploegh HL, Wulczyn FG, Lehnardt S. An unconventional role for miRNA: let-7 activates Toll-like receptor 7 and causes neurodegeneration. Nat Neurosci. 2012;15:827–835. doi: 10.1038/nn.3113. [DOI] [PubMed] [Google Scholar]
- 19.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang L, Boldin MP, Yu Y, Liu CS, Ea CK, Ramakrishnan P, Taganov KD, Zhao JL, Baltimore D. miR-146a controls the resolution of T cell responses in mice. J Exp Med. 2012;209:1655–1670. doi: 10.1084/jem.20112218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nahid MA, Pauley KM, Satoh M, Chan EK. miR-146a is critical for endotoxin-induced tolerance: IMPLICATION IN INNATE IMMUNITY. J Biol Chem. 2009;284:34590–34599. doi: 10.1074/jbc.M109.056317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hou J, Wang P, Lin L, Liu X, Ma F, An H, Wang Z, Cao X. MicroRNA-146a feedback inhibits RIG-I-dependent Type I IFN production in macrophages by targeting TRAF6, IRAK1, and IRAK2. J Immunol. 2009;183:2150–2158. doi: 10.4049/jimmunol.0900707. [DOI] [PubMed] [Google Scholar]
- 23.Lu LF, Boldin MP, Chaudhry A, Lin LL, Taganov KD, Hanada T, Yoshimura A, Baltimore D, Rudensky AY. Function of miR-146a in controlling Treg cell-mediated regulation of Th1 responses. Cell. 2010;142:914–929. doi: 10.1016/j.cell.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boldin MP, Taganov KD, Rao DS, Yang L, Zhao JL, Kalwani M, Garcia-Flores Y, Luong M, Devrekanli A, Xu J, Sun G, Tay J, Linsley PS, Baltimore D. miR-146a is a significant brake on autoimmunity, myeloproliferation, and cancer in mice. J Exp Med. 2011;208:1189–1201. doi: 10.1084/jem.20101823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sheedy FJ, Palsson-McDermott E, Hennessy EJ, Martin C, O'Leary J, Ruan Q, Johnson DP, Chen Y, O'Neill LA. Negative regulation of TLR4 via targeting of the proinflammatory tumor suppressor PDCD4 by the microRNA miR-21. Nat Immunol. 2010;11:141–147. doi: 10.1038/ni.1828. [DOI] [PubMed] [Google Scholar]
- 26.Androulidaki A, Iliopoulos D, Arranz A, Doxaki C, Schworer S, Zacharioudaki V, Margioris AN, Tsichlis PN, Tsatsanis C. The kinase Akt1 controls macrophage response to lipopolysaccharide by regulating microRNAs. Immunity. 2009;31:220–231. doi: 10.1016/j.immuni.2009.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O'Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci U S A. 2007;104:1604–1609. doi: 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tili E, Michaille JJ, Cimino A, Costinean S, Dumitru CD, Adair B, Fabbri M, Alder H, Liu CG, Calin GA, Croce CM. Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-alpha stimulation and their possible roles in regulating the response to endotoxin shock. J Immunol. 2007;179:5082–5089. doi: 10.4049/jimmunol.179.8.5082. [DOI] [PubMed] [Google Scholar]
- 29.Ceppi M, Pereira PM, Dunand-Sauthier I, Barras E, Reith W, Santos MA, Pierre P. MicroRNA-155 modulates the interleukin-1 signaling pathway in activated human monocyte-derived dendritic cells. Proc Natl Acad Sci U S A. 2009;106:2735–2740. doi: 10.1073/pnas.0811073106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O'Connell RM, Chaudhuri AA, Rao DS, Baltimore D. Inositol phosphatase SHIP1 is a primary target of miR-155. Proc Natl Acad Sci U S A. 2009;106:7113–7118. doi: 10.1073/pnas.0902636106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bala S, Marcos M, Kodys K, Csak T, Catalano D, Mandrekar P, Szabo G. Up-regulation of microRNA-155 in macrophages contributes to increased tumor necrosis factor {alpha} (TNF{alpha}) production via increased mRNA half-life in alcoholic liver disease. J Biol Chem. 2011;286:1436–1444. doi: 10.1074/jbc.M110.145870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rajaram MV, Ni B, Morris JD, Brooks MN, Carlson TK, Bakthavachalu B, Schoenberg DR, Torrelles JB, Schlesinger LS. Mycobacterium tuberculosis lipomannan blocks TNF biosynthesis by regulating macrophage MAPK-activated protein kinase 2 (MK2) and microRNA miR-125b. Proc Natl Acad Sci U S A. 2011;108:17408–17413. doi: 10.1073/pnas.1112660108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Costinean S, Sandhu SK, Pedersen IM, Tili E, Trotta R, Perrotti D, Ciarlariello D, Neviani P, Harb J, Kauffman LR, Shidham A, Croce CM. Src homology 2 domain-containing inositol-5-phosphatase and CCAAT enhancer-binding protein beta are targeted by miR-155 in B cells of Emicro-MiR-155 transgenic mice. Blood. 2009;114:1374–1382. doi: 10.1182/blood-2009-05-220814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Costinean S, Zanesi N, Pekarsky Y, Tili E, Volinia S, Heerema N, Croce CM. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc Natl Acad Sci U S A. 2006;103:7024–7029. doi: 10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O'Connell RM, Kahn D, Gibson WS, Round JL, Scholz RL, Chaudhuri AA, Kahn ME, Rao DS, Baltimore D. MicroRNA-155 promotes autoimmune inflammation by enhancing inflammatory T cell development. Immunity. 2010;33:607–619. doi: 10.1016/j.immuni.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O'Connell RM, Rao DS, Chaudhuri AA, Boldin MP, Taganov KD, Nicoll J, Paquette RL, Baltimore D. Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder. J Exp Med. 2008;205:585–594. doi: 10.1084/jem.20072108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McCoy CE, Sheedy FJ, Qualls JE, Doyle SL, Quinn SR, Murray PJ, O'Neill LA. IL-10 inhibits miR-155 induction by toll-like receptors. J Biol Chem. 2010;285:20492–20498. doi: 10.1074/jbc.M110.102111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Philippe L, Alsaleh G, Suffert G, Meyer A, Georgel P, Sibilia J, Wachsmann D, Pfeffer S. TLR2 expression is regulated by microRNA miR-19 in rheumatoid fibroblast-like synoviocytes. J Immunol. 2012;188:454–461. doi: 10.4049/jimmunol.1102348. [DOI] [PubMed] [Google Scholar]
- 39.Benakanakere MR, Li Q, Eskan MA, Singh AV, Zhao J, Galicia JC, Stathopoulou P, Knudsen TB, Kinane DF. Modulation of TLR2 protein expression by miR-105 in human oral keratinocytes. J Biol Chem. 2009;284:23107–23115. doi: 10.1074/jbc.M109.013862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen XM, Splinter PL, O'Hara SP, LaRusso NF. A cellular micro-RNA, let-7i, regulates Toll-like receptor 4 expression and contributes to cholangiocyte immune responses against Cryptosporidium parvum infection. J Biol Chem. 2007;282:28929–28938. doi: 10.1074/jbc.M702633200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu G, Friggeri A, Yang Y, Park YJ, Tsuruta Y, Abraham E. miR-147, a microRNA that is induced upon Toll-like receptor stimulation, regulates murine macrophage inflammatory responses. Proc Natl Acad Sci U S A. 2009;106:15819–15824. doi: 10.1073/pnas.0901216106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bazzoni F, Rossato M, Fabbri M, Gaudiosi D, Mirolo M, Mori L, Tamassia N, Mantovani A, Cassatella MA, Locati M. Induction and regulatory function of miR-9 in human monocytes and neutrophils exposed to proinflammatory signals. Proc Natl Acad Sci U S A. 2009;106:5282–5287. doi: 10.1073/pnas.0810909106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murphy AJ, Guyre PM, Pioli PA. Estradiol suppresses NF-kappa B activation through coordinated regulation of let-7a and miR-125b in primary human macrophages. J Immunol. 2010;184:5029–5037. doi: 10.4049/jimmunol.0903463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chaudhuri AA, So AY, Sinha N, Gibson WS, Taganov KD, O'Connell RM, Baltimore D. MicroRNA-125b potentiates macrophage activation. J Immunol. 2011;187:5062–5068. doi: 10.4049/jimmunol.1102001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cobb BS, Nesterova TB, Thompson E, Hertweck A, O'Connor E, Godwin J, Wilson CB, Brockdorff N, Fisher AG, Smale ST, Merkenschlager M. T cell lineage choice and differentiation in the absence of the RNase III enzyme Dicer. J Exp Med. 2005;201:1367–1373. doi: 10.1084/jem.20050572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Muljo SA, Ansel KM, Kanellopoulou C, Livingston DM, Rao A, Rajewsky K. Aberrant T cell differentiation in the absence of Dicer. J Exp Med. 2005;202:261–269. doi: 10.1084/jem.20050678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, van Dongen S, Grocock RJ, Das PP, Miska EA, Vetrie D, Okkenhaug K, Enright AJ, Dougan G, Turner M, Bradley A. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, Murphy A, Frendewey D, Valenzuela D, Kutok JL, Schmidt-Supprian M, Rajewsky N, Yancopoulos G, Rao A, Rajewsky K. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- 49.Kurowska-Stolarska M, Alivernini S, Ballantine LE, Asquith DL, Millar NL, Gilchrist DS, Reilly J, Ierna M, Fraser AR, Stolarski B, McSharry C, Hueber AJ, Baxter D, Hunter J, Gay S, Liew FY, McInnes IB. MicroRNA-155 as a proinflammatory regulator in clinical and experimental arthritis. Proc Natl Acad Sci U S A. 2011;108:11193–11198. doi: 10.1073/pnas.1019536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oertli M, Engler DB, Kohler E, Koch M, Meyer TF, Muller A. MicroRNA-155 is essential for the T cell-mediated control of Helicobacter pylori infection and for the induction of chronic Gastritis and Colitis. J Immunol. 2011;187:3578–3586. doi: 10.4049/jimmunol.1101772. [DOI] [PubMed] [Google Scholar]
- 51.Murugaiyan G, Beynon V, Mittal A, Joller N, Weiner HL. Silencing microRNA-155 ameliorates experimental autoimmune encephalomyelitis. J Immunol. 2011;187:2213–2221. doi: 10.4049/jimmunol.1003952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kohlhaas S, Garden OA, Scudamore C, Turner M, Okkenhaug K, Vigorito E. Cutting edge: the Foxp3 target miR-155 contributes to the development of regulatory T cells. J Immunol. 2009;182:2578–2582. doi: 10.4049/jimmunol.0803162. [DOI] [PubMed] [Google Scholar]
- 53.Li QJ, Chau J, Ebert PJ, Sylvester G, Min H, Liu G, Braich R, Manoharan M, Soutschek J, Skare P, Klein LO, Davis MM, Chen CZ. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell. 2007;129:147–161. doi: 10.1016/j.cell.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 54.Ebert PJ, Jiang S, Xie J, Li QJ, Davis MM. An endogenous positively selecting peptide enhances mature T cell responses and becomes an autoantigen in the absence of microRNA miR-181a. Nat Immunol. 2009;10:1162–1169. doi: 10.1038/ni.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xiao C, Srinivasan L, Calado DP, Patterson HC, Zhang B, Wang J, Henderson JM, Kutok JL, Rajewsky K. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol. 2008;9:405–414. doi: 10.1038/ni1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Du C, Liu C, Kang J, Zhao G, Ye Z, Huang S, Li Z, Wu Z, Pei G. MicroRNA miR-326 regulates TH-17 differentiation and is associated with the pathogenesis of multiple sclerosis. Nat Immunol. 2009;10:1252–1259. doi: 10.1038/ni.1798. [DOI] [PubMed] [Google Scholar]
- 57.Xiao C, Calado DP, Galler G, Thai TH, Patterson HC, Wang J, Rajewsky N, Bender TP, Rajewsky K. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell. 2007;131:146–159. doi: 10.1016/j.cell.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 58.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 59.Thomas MD, Kremer CS, Ravichandran KS, Rajewsky K, Bender TP. c-Myb is critical for B cell development and maintenance of follicular B cells. Immunity. 2005;23:275–286. doi: 10.1016/j.immuni.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 60.Zhou B, Wang S, Mayr C, Bartel DP, Lodish HF. miR-150, a microRNA expressed in mature B and T cells, blocks early B cell development when expressed prematurely. Proc Natl Acad Sci U S A. 2007;104:7080–7085. doi: 10.1073/pnas.0702409104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vigorito E, Perks KL, Abreu-Goodger C, Bunting S, Xiang Z, Kohlhaas S, Das PP, Miska EA, Rodriguez A, Bradley A, Smith KG, Rada C, Enright AJ, Toellner KM, Maclennan IC, Turner M. microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity. 2007;27:847–859. doi: 10.1016/j.immuni.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dorsett Y, McBride KM, Jankovic M, Gazumyan A, Thai TH, Robbiani DF, Di Virgilio M, Reina San-Martin B, Heidkamp G, Schwickert TA, Eisenreich T, Rajewsky K, Nussenzweig MC. MicroRNA-155 suppresses activation-induced cytidine deaminase-mediated Myc-Igh translocation. Immunity. 2008;28:630–638. doi: 10.1016/j.immuni.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lawrence T, Natoli G. Transcriptional regulation of macrophage polarization: enabling diversity with identity. Nat Rev Immunol. 2011;11:750–761. doi: 10.1038/nri3088. [DOI] [PubMed] [Google Scholar]
- 64.Murray PJ, Wynn TA. Obstacles and opportunities for understanding macrophage polarization. J Leukoc Biol. 2011;89:557–563. doi: 10.1189/jlb.0710409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol. 2008;8:958–969. doi: 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ruffell B, Affara NI, Coussens LM. Differential macrophage programming in the tumor microenvironment. Trends Immunol. 2012;33:119–126. doi: 10.1016/j.it.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Martinez FO, Gordon S, Locati M, Mantovani A. Transcriptional profiling of the human monocyte-to-macrophage differentiation and polarization: new molecules and patterns of gene expression. J Immunol. 2006;177:7303–7311. doi: 10.4049/jimmunol.177.10.7303. [DOI] [PubMed] [Google Scholar]
- 68.Takeuch O, Akira S. Epigenetic control of macrophage polarization. Eur J Immunol. 2011;41:2490–2493. doi: 10.1002/eji.201141792. + [DOI] [PubMed] [Google Scholar]
- 69.Graff JW, Dickson AM, Clay G, McCaffrey AP, Wilson ME. Identifying Functional MicroRNAs in Macrophages with Polarized Phenotypes. J Biol Chem. 2012;287:21816–21825. doi: 10.1074/jbc.M111.327031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jazdzewski K, Liyanarachchi S, Swierniak M, Pachucki J, Ringel MD, Jarzab B, de la Chapelle A. Polymorphic mature microRNAs from passenger strand of pre-miR-146a contribute to thyroid cancer. Proc Natl Acad Sci U S A. 2009;106:1502–1505. doi: 10.1073/pnas.0812591106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Diederichs S, Haber DA. Dual role for argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell. 2007;131:1097–1108. doi: 10.1016/j.cell.2007.10.032. [DOI] [PubMed] [Google Scholar]
- 72.Ponomarev ED, Veremeyko T, Barteneva N, Krichevsky AM, Weiner HL. MicroRNA-124 promotes microglia quiescence and suppresses EAE by deactivating macrophages via the C/EBP-alpha-PU.1 pathway. Nat Med. 2011;17:64–70. doi: 10.1038/nm.2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhuang G, Meng C, Guo X, Cheruku PS, Shi L, Xu H, Li H, Wang G, Evans AR, Safe S, Wu C, Zhou B. A Novel Regulator of Macrophage Activation: miR-223 in Obesity-Associated Adipose Tissue Inflammation. Circulation. 2012;125:2892–2903. doi: 10.1161/CIRCULATIONAHA.111.087817. [DOI] [PubMed] [Google Scholar]
- 74.Strassheim D, Asehnoune K, Park JS, Kim JY, He Q, Richter D, Kuhn K, Mitra S, Abraham E. Phosphoinositide 3-kinase and Akt occupy central roles in inflammatory responses of Toll-like receptor 2-stimulated neutrophils. J Immunol. 2004;172:5727–5733. doi: 10.4049/jimmunol.172.9.5727. [DOI] [PubMed] [Google Scholar]
- 75.Luyendyk JP, Schabbauer GA, Tencati M, Holscher T, Pawlinski R, Mackman N. Genetic analysis of the role of the PI3K-Akt pathway in lipopolysaccharide-induced cytokine and tissue factor gene expression in monocytes/macrophages. J Immunol. 2008;180:4218–4226. doi: 10.4049/jimmunol.180.6.4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Arranz A, Doxaki C, Vergadi E, Martinez de la Torre Y, Vaporidi K, Lagoudaki ED, Ieronymaki E, Androulidaki A, Venihaki M, Margioris AN, Stathopoulos EN, Tsichlis PN, Tsatsanis C. Akt1 and Akt2 protein kinases differentially contribute to macrophage polarization. Proc Natl Acad Sci U S A. 2011;109:9517–9522. doi: 10.1073/pnas.1119038109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Martinez-Nunez RT, Louafi F, Sanchez-Elsner T. The interleukin 13 (IL-13) pathway in human macrophages is modulated by microRNA-155 via direct targeting of interleukin 13 receptor alpha1 (IL13Ralpha1) J Biol Chem. 2011;286:1786–1794. doi: 10.1074/jbc.M110.169367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Thomas M, Lieberman J, Lal A. Desperately seeking microRNA targets. Nat Struct Mol Biol. 2010;17:1169–1174. doi: 10.1038/nsmb.1921. [DOI] [PubMed] [Google Scholar]
- 79.Zernecke A. MicroRNAs in the regulation of immune cell functions--implications for atherosclerotic vascular disease. Thromb Haemost. 2012;107:626–633. doi: 10.1160/TH11-08-0603. [DOI] [PubMed] [Google Scholar]