Abstract
Introduction
Exon 20 insertions are the third most common family of EGFR mutations found in non-small cell lung cancer (NSCLC). Little is known about cancers harboring these mutations aside from their lack of response to epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors, impairing the development of effective targeted therapies.
Methods
NSCLC patients with EGFR genotyping were studied using an IRB-approved mechanism. Cancers with exon 20 insertions were indentified, sequences were characterized, and effectiveness of different treatment regimens was reviewed retrospectively. Clinical characteristics and survival were compared with cancers harboring common EGFR mutations and cancers with wild-type EGFR.
Results
1086 patients underwent EGFR genotyping from 2004 to 2012. Twenty seven (2.5%) harbored exon 20 insertions, making up 9.2% of all cancers with documented EGFR mutations. Compared to wild-type cancers, those with exon 20 insertions were more commonly found in never-smokers and Asian patients. Insertion sequences were highly variable, with the most common variant (V769_D770insASV) making up only 22% of cases. Median survival of patients with exon 20 insertions was 16 months, similar to the survival of wild-type cancers and shorter than the survival of cancers with common EGFR mutations.
Conclusions
Patients with EGFR exon 20 insertions have similar clinical characteristics to those with common EGFR mutations but a poorer prognosis. The prevalence of this subset of NSCLC is similar to that of other genotype-defined subsets of lung adenocarcinoma (e.g. those with BRAF mutations, HER2 insertions, ROS1 rearrangements) and is a population of interest for trials of new targeted therapies.
Keywords: Non-small cell lung cancer, EGFR mutations, exon 20 insertions
Introduction
The identification of EGFR mutations in a subset of non-small cell lung cancer (NSCLC) and their association with durable responses to epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors (TKIs) has transformed our approach towards the use of targeted therapies in treating lung cancer.1, 2 This initial discovery has led to a number of parallel discoveries – lung adenocarcinomas harboring ALK and ROS1 rearrangements are both highly sensitive to critozinib,3, 4 while melanomas harboring BRAF V600 mutations are highly sensitivity to BRAF and MEK inhibitors.5, 6 These therapeutic advances have led to numerous efforts to identify genotype-defined subsets of common cancers which can be treated with rationally targeted therapies.
Lung cancers harboring exon 20 insertions EGFR make up one genotype-defined subset of lung cancer that is in need of an effective targeted treatment approach. These were one of the earliest described families of EGFR mutations, identified contemporaneously with the common (and better studied) TKI-sensitive mutations in exon 19 and 21 of EGFR.7, 8 Unlike EGFR exon 19 deletions and exon 21 point mutations, exon 20 insertions are not associated with sensitivity to EGFR TKIs in laboratory studies and in clinical trials,9–11 though these insertion mutations have been shown to be oncogenic when transfected into cell lines.11 Additionally, a family of exon 20 insertions has been described in the HER2 gene,12, 13 which encodes a membrane-bound kinase with significant homology to EGFR, highlighting the potential biological importance of insertion mutations in this part of the kinase domain. Though exon 20 insertions are the third most common family of EGFR mutations and have been identified on routine EGFR genotyping for many years, no prospective studies to our knowledge have specifically targeted this subset of NSCLC, and lung cancers harboring exon 20 insertions have been excluded from many prospective trials of EGFR inhibitors due to the reports of resistance to EGFR TKIs.
To facilitate drug development for this genotype-defined subset of NSCLC, we aimed to characterize our single-center experience of patients with lung cancers carrying EGFR exon 20 insertions. We hypothesized that, despite the presence of EGFR mutations, the clinical outcomes of lung cancers harboring EGFR exon 20 insertions would be similar to EGFR wild-type cancers, supporting the need for effective targeted therapy options for this subset of lung cancer.
Materials and Methods
Patients for this study were identified from two cohorts – a prospective cohort of lung cancer patients who consented to an institutional tissue analysis protocol, and a retrospective cohort of lung cancer patients whose records were reviewed and studied with IRB approval. Of these patients, subjects were deemed eligible for study if they had a diagnosis of NSCLC and had undergone conclusive EGFR genotyping. Patients were grouped into four populations based upon the EGFR genotype of their cancer: (1) EGFR exon 20 insertions, (2) common TKI-sensitive EGFR mutations (exon 19 deletions or L858R point mutations), (3) other rare EGFR mutations, and (4) EGFR wild-type. Those patients with a result of “EGFR mutation positive” but with no additional sequence details available were excluded from the analysis.
EGFR genotyping for this study was performed at the Laboratory for Molecular Medicine at Harvard University. Specimens deemed to have adequate tumor content were submitted for extraction of somatic DNA, and underwent direct Sanger sequencing of exons 18 through 21 of EGFR as previously described.14 EGFR genotyping performed at a separate CLIA-approved laboratory was also deemed adequate for this analysis, therefore was not repeated at the central laboratory. The amino-acid sequences of the identified exon 20 insertions were characterized and compared.
Patient characteristics at time of lung cancer diagnosis were prospectively collected, including age, gender, race, smoking history (never-smoker was defined as less than 100 cigarettes lifetime), stage (per AJCC VII), and histologic classification. For patients with advanced disease (stage IV or recurrent), survival was calculated from date of diagnosis (for stage IV) or from date of recurrence (for stage I–III) until the date of death from any cause, as in other analyses15. Patients who were alive at the time of analysis were censored at the last known date of follow-up.
The systemic treatment of advanced lung cancers harboring EGFR exon 20 insertions was reviewed to characterize its effectiveness. Because of the small sample size, patients receiving multiple lines of treatment could be considered up to three times, in each of three different treatment settings, but were not studied more than once within a given treatment setting. The first treatment setting included patients receiving erlotinib as a single-agent, regardless of line of therapy. The second treatment setting included patients receiving platinum-based combination chemotherapy, regardless of whether this was before erlotinib (considered first-line) or after erlotinib (considered second-line). The third treatment setting included patients receiving subsequent single-agent cytotoxic chemotherapy, following failure of combination chemotherapy. Combinations of these treatments (e.g. erlotinib plus chemotherapy), investigational treatments, and treatments given in the setting of no evaluable disease (e.g. adjuvant therapies) or without any follow-up imaging were excluded from the analysis. Time to treatment failure (TTF) was determined retrospectively from review of records, and was defined as the time from the start of each therapy until a change of treatment regimen. For those with serial CT imaging available during therapy and measurable disease, maximal tumor shrinkage was determined by blinded radiology review. Objective response rate was calculated using RECIST version 1.1 criteria,16 but response confirmation was not required due to the retrospective nature of the study. Because some patients were studied multiple times in different treatment settings, no statistical analysis was performed comparing each treatment setting.
Categorical data were compared using the Fisher’s exact test, and continuous data were compared using the Wilcoxon rank sum test. The Kaplan-Meier method was used to estimate the overall survival distributions, and the Greenwood formula was used to estimate the variance. Time-to-event distributions were compared using the logrank test. All p-values are two-sided, and no adjustments have been made for multiple comparisons.
Results
A total of 1086 patients had conclusive EGFR genotyping from 2004 to 2012: 29% never smokers, 63% female, 6% Asian, and 99% with non-squamous histology (Table 1). Of the total, 294 (27%) harbored EGFR mutations; 27 of these (2.5%) harbored EGFR exon 20 insertions, constituting 9.2% of all cases with an EGFR mutation. Other genotypes included 128 cancers with exon 19 deletions, 101 cancers with L858R point mutations, 38 cancers harboring other rare EGFR mutations.
Table 1.
Clinical characteristics of NSCLC harboring common TKI-sensitive EGFR mutations (L858R and exon 19 deletions), EGFR exon 20 insertions, and EGFR wild-type cancers.
| All patients | Common EGFR mutations | Exon 20 insertions | EGFR wild-type | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| n=1086 | % | n=229 | % | P* | n=27 | % | P** | n=792 | % | |
| Age: | ||||||||||
| median | 62 | 60 | 0.65 | 60 | 0.23 | 62 | ||||
| range | 20–95 | 30–95 | 25–80 | 20–88 | ||||||
|
| ||||||||||
| Smoking status: | ||||||||||
| Never | 313 | 29% | 120 | 52% | 0.84 | 15 | 56% | <0.001 | 161 | 20% |
| Ever | 773 | 71% | 109 | 48% | 12 | 44% | 631 | 80% | ||
|
| ||||||||||
| Gender: | ||||||||||
| Female | 679 | 63% | 168 | 73% | 0.82 | 19 | 70% | 0.24 | 462 | 58% |
| Male | 407 | 37% | 61 | 27% | 8 | 30% | 330 | 42% | ||
|
| ||||||||||
| Race: | ||||||||||
| Asian | 62 | 6% | 25 | 11% | 0.52 | 4 | 15% | 0.02 | 30 | 4% |
| Non-Asian | 1024 | 94% | 204 | 89% | 23 | 85% | 762 | 96% | ||
|
| ||||||||||
| Stage: | ||||||||||
| IV | 579 | 53% | 133 | 58% | 0.3 | 19 | 70% | 0.05 | 401 | 51% |
| I–III | 507 | 47% | 96 | 42% | 8 | 30% | 391 | 49% | ||
|
| ||||||||||
| Histology: | ||||||||||
| Non-squamous | 1071 | 99% | 226 | 99% | 1 | 27 | 100% | 1 | 780 | 98% |
| Squamous | 15 | 1% | 3 | 1% | 0 | 0% | 12 | 2% | ||
Comparison of common EGFR mutations and exon 20 insertions
Comparison of EGFR wild-type cancers and exon 20 insertions
The characteristics of cancers with EGFR exon 20 insertions are shown in Table 1, along with the clinical characteristics of cancers which carried common EGFR mutations and those which were EGFR wild-type. No significant differences were identified between the cancers carrying exon 20 insertions and those with the common EGFR mutations. However, cancers with exon 20 insertions were more commonly seen in patients who are never-smokers (p<0.001) and in patients of an Asian race (p=0.02) when compared to EGFR wild-type cancers, similar to the patients with the common EGFR mutations. There was a trend toward an increased proportion of cancers with exon 20 insertions being stage IV at diagnosis (p=0.05); this may in part be due to referral bias, as our cancer center has a research program focusing on treatments for advanced EGFR-mutant lung cancers.
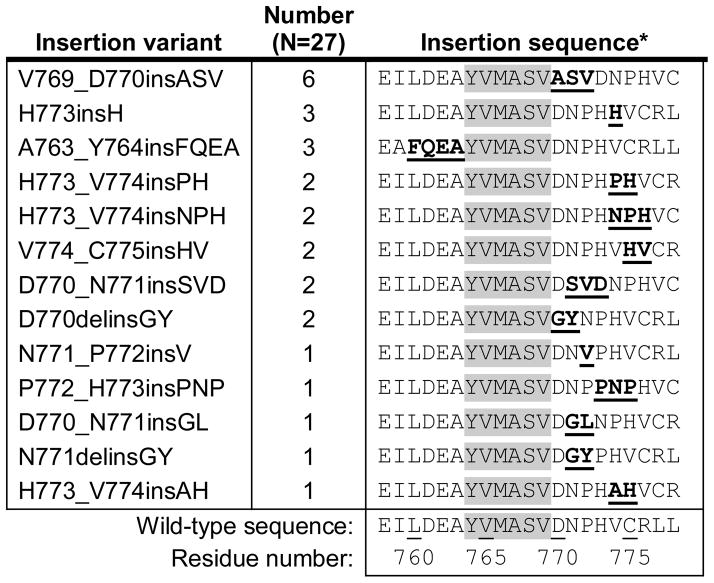
The amino acid sequences of the 27 EGFR exon 20 insertions included 13 different variants occurring at 7 different amino acid positions (Table 2). Review of the amino acid sequences revealed a large degree of variability in insertion length (1, 2, 3, or 4 residues), insertion sequence, and insertion position. Only 8 of the 13 variants occurred more than once. To contextualize the variety of EGFR exon 20 insertions identified, we considered the variety of insertions in EGFR exon 19 and in HER2 exon 20 from the two largest published series on these families of mutations (Figure 1).13, 17 The most common variants of EGFR exon 19 insertions and HER2 exon 20 insertions each make up the majority of cases, but the most common variant of EGFR exon 20 insertions (V769_D770insASV) constitutes only 22% of the total cohort. Table 2 shows the sequence of each variant, registered around residues Y764 through V769, a section that spans the terminus of the C-helix (at M766) and is uninvolved by any insertions. Reviewing these sequences, there are few trends and few recurring substitutions at any specific position. For example, while the most common variant shares the insertion of a serine and valine at positions 771 and 772 with D770_N771insSVD, no other insertion variants include a serine residue.
Table 2.
Amino acid sequences of the exon 20 insertion variants identified in this series, and their relative position in the exon (underlined).
|
Insertions sequences are registered around the uninvolved residues Y764-V769 (highlighted), an area including the final residue of the C-helix (M766).
Figure 1.
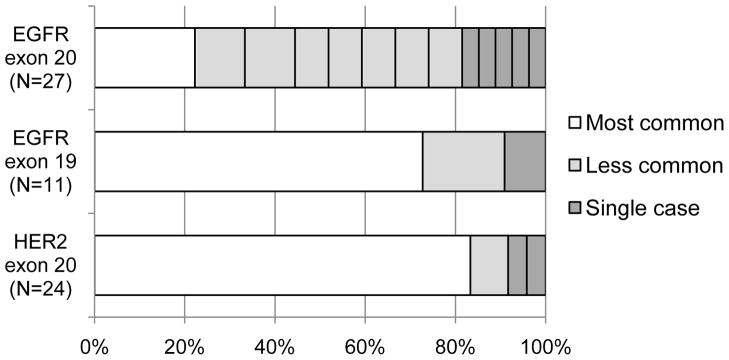
Relative prevalence of different insertions variants found in NSCLC. The most common insertions variant in EGFR exon 20 (V769_D770insASV) constitutes only 22% of the total cohort. This contrasts with the relative prevalence of insertions variants in EGFR exon 19 and HER2 exon 20 from the largest published series of each 13, 17, where the most common variant makes up a large majority of the cases.
Nineteen patients with EGFR exon 20 insertions received treatment for advanced disease, with a total of 34 different treatment courses studied (Figure 2). Eight patients received erlotinib, with a median TTF of 2.4 months (range 1.0–4.5 months); of 5 patients with imaging available, none demonstrated an objective response. Seventeen patients received combination chemotherapy with a median TTF of 5.9 months (range 1.3–17.7 months); of 12 patients with imaging available, 7 demonstrated an objective response. Nine patients received subsequent cytotoxic chemotherapy with a median TTF of 2.3 months (range 1.2–7.0 months); of 6 patients with imaging available, 1 demonstrated an objective response.
Figure 2.
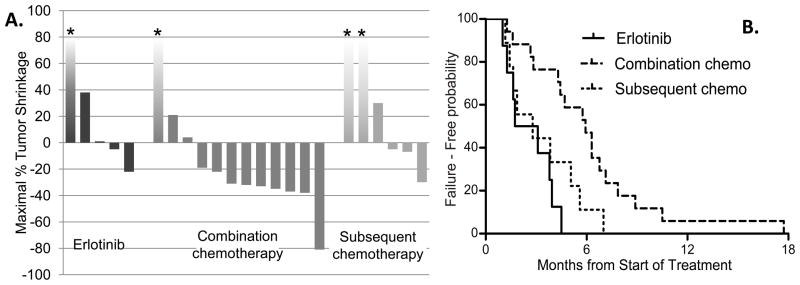
Activity of different systemic treatment approaches against advanced NSCLC harboring an EGFR exon 20 insertion. Treatment settings studied are treatment with erlotinib, treatment with combination chemotherapy, and treatment with subsequent cytotoxic chemotherapy. Patients treated in multiple settings are included in each setting studied, therefore no statistical comparison is performed. (A) Waterfall plots show maximal tumor shrinkage. Bars marked with asterisks represent non-quantifiable progression (i.e. non-target disease or new lesions). (B) Time to treatment failure.
Survival analysis was performed on the 839 patients with advanced disease (553 stage IV at diagnosis and 286 recurrent), with a median follow-up of 30 months. The 24 patients with advanced lung cancers harboring EGFR exon 20 insertions had a median survival of 16.5 months (95% CI: 10.4 - NA) from date of advanced disease. Their survival was shorter than the 183 patients with common EGFR mutations (median 33.0 months, 95% CI 28.7–40.6, p=0.06, Figure 3A), but was not dissimilar from the survival of the 632 patients with EGFR wild-type lung cancers (median 20.0 months, 17.4–22.9, p=0.60, Figure 3B).
Figure 3.
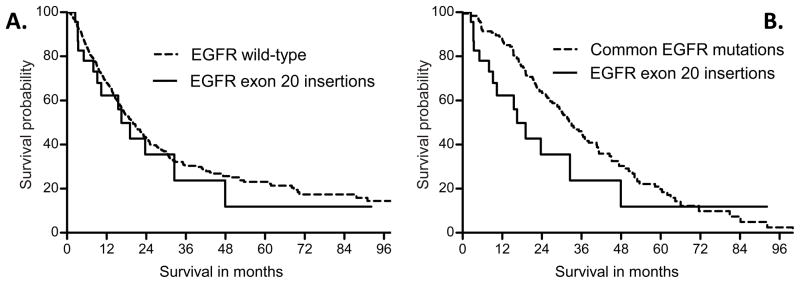
Survival from date of advanced disease by EGFR genotype. Lung cancers with exon 20 insertions have a median survival of 16.5 months, shorter than the median survival of 33.0 months in those with common EGFR mutations (A, p=0.06) but similar to the median survival of 20.0 months in EGFR wild-type cancers (B, p=0.60).
Discussion
We have identified that lung cancers harboring EGFR exon 20 insertions exhibit clinical characteristics similar to lung cancers carrying common EGFR mutations, with a tendency to occur in never-smokers and Asian patients. This is consistent with these insertions being driver mutations, and with the potential for durable benefit from an effective targeted therapy. However, patients with this family of mutations have a survival similar to that seen in EGFR wild-type lung cancers. While patients with advanced lung cancers carrying TKI-sensitive EGFR mutations had a median survival of 33.0 months, similar to reports from other academic centers,18 patients with cancers carrying exon 20 insertions had a median survival of only 16.5 months. It should be noted that, while these medians are longer than those seen in first-line treatment studies, they are similar to those seen in other analyses measuring survival from date of advanced disease.15
Our finding that 9.2% of EGFR-mutant lung cancers harbor exon 20 insertions is somewhat higher than the 4–5% prevalence found by some prior analyses.10, 19, 20 While this higher prevalence may reflect referral bias, it is also expected that the prevalence of these mutations will vary depending upon which genotyping technique is used. Sanger sequencing, used in this analysis and in many commercial labs, is able to detect all varieties of insertion mutations. In contrast, mutation-specific assays such as the EGFR PCR kit by Qiagen (Valencia, CA) includes only three exon 20 insertion variants among the specific EGFR mutations that it tests for, such that it would have detected fewer than half of the cases with exon 20 insertions in our series. Going forward, efforts to study this subset of lung cancer will need to use genotyping assays that are not mutation-specific, or else a portion of these cancers might be missed.
Given our prevalence findings, we estimate that lung cancers with EGFR exon 20 have an annual incidence of approximately 2000–4000 patients in the United States, not dissimilar from the expected incidence of other uncommon genotype-defined subsets of NSCLC such as those with ROS1 rearrangements, BRAF mutations, or HER2 insertions.4, 13, 21 While detection of ROS1 rearrangements currently requires an independent break-apart FISH assay, exon 20 insertions can be routinely detected during standard-of-care EGFR genotyping. Since patients with this genotype will continue to emerge during standard-of-care EGFR genotyping, we encourage other investigators to develop trials for lung cancers with EGFR exon 20 insertions, towards the goal of potentially developing an orphan drug for this genotype-defined rare disease.22 One treatment strategy that has shown preliminary efficacy in EGFR-mutant lung cancers that are resistant to TKI is HSP90 inhibition, with responses seen in preclinical models as well as in some patients.23–25 A clinical trial is in development studying the HSP90 inhibitor AUY922 in lung cancers harboring EGFR exon 20 insertions, which would be first prospective trial specifically targeting this genotype-defined subset of NSCLC. Alternatively, new kinase inhibitors may need to be developed against this specific family of kinase mutations, or developed jointly for HER2 insertion mutations. To facilitate these efforts, a transgenic mouse model of NSCLC carrying a HER2 exon 20 insertion has been generated by our colleagues,26 and a model harboring an EGFR exon 20 insertion is in development.
The highly variable length and position of EGFR exon 20 insertions contrasts with the more consistent insertions seen in exon 19 of EGFR and in exon 20 of HER2.13, 17 Looking at the sequences of the insertions (Table 2), there is no consistent substitution seen that is shared by the entire spectrum. This contrasts with deletions and insertions in exon 19 of EGFR, which commonly involve a substitution at L747 to either a proline or a serine, a substitution that may underlie the oncogenicity of these types of mutations by disfavoring the inactive conformation of the mutant EGFR kinase.17 Supporting the importance of this residue, exon 19 deletions that do not include a substitution at 747 have been found to have poorer outcomes when treated with TKI.27 The lack of structural consistency among the exon 20 insertions may indicate a particularly varied biology among this family of mutations. Indeed, cell line studies have recently indicated that one variant of exon 20 insertions, A763_Y764insFQEA, is actually sensitive to treatment with erlotinib.28 In our cohort, only one patient harboring this insertion variant was treated with erlotinib, and objective response assessment identified stable disease lasting 4 months. Given that our understanding of the biology of these mutations is still evolving, it would be valuable if, when radiographic responses are seen to EGFR TKI in lung cancers with EGFR exon 20 insertions, they be reported with mention of the specific sequence of the insertion variant.29 For example, the one published report of response to an irreversible TKI also involved a rare exon 20 insertion variant, D770delinsGY, which may suggest distinct biologic characteristics.30
Our retrospective analysis of treatment effectiveness was limited in its ability to evaluate different treatment approaches because of the small number of lung cancers harboring exon 20 insertions. However, by considering each patient multiple times when treated in different treatment settings, we were able to generate an initial sense of how some therapies work in this subset of cancers. Combination chemotherapy resulted in a sizeable proportion of patients having objective responses with a median TTF of 5.9 months; this is consistent with the results seen when treating advanced lung adenocarcinoma in general. Subsequent chemotherapy was relatively less effective, as was erlotinib, suggesting that previously-treated NSCLC carrying EGFR exon 20 insertions may be an appropriate setting for the development of new therapies. Though such analyses are merely descriptive and based upon retrospective data, we hope that presenting them in the medical literature may facilitate later efforts for drug development.
In conclusion, we have found that lung cancers harboring EGFR exon 20 insertions have the clinical characteristics of other lung cancers carrying targetable oncogenes and are just as prevalent, but exon 20 insertions are associated with a poorer survival than common TKI-sensitive EGFR mutations. Previously treated NSCLC carrying EGFR exon 20 insertions represent a population of oncogene-addicted cancers in need of targeted therapy development.
Acknowledgments
Source of Funding and Conflicts of Interest: This work was supported in part by the National Cancer Institute at the National Institutes of Health (R01-CA135257, P50-CA090578, K23-CA157631). G.R. Oxnard is a consultant/advisory board member for Genentech and Boehringer Ingelheim. D.M. Jackman is a consultant/advisory board member for Genentech and Foundation Medicine. B.E. Johnson is a consultant/advisory board member for Genentech, Pfizer, Chugai, Astra-Zeneca, Kew and Transgenomics. P.A. Janne is a consultant/advisory board member for Boehringer Ingelheim, Roche, Genentech, Abbot, Teva, Astra-Zeneca, Pfizer, and Sanofi. B.E. Johnson and P.A. Janne are coinventors on a patent held by the Dana-Farber Cancer Institute for the use of EGFR genotyping, and receive a share of post-market licensing revenue distributed by DFCI.
Footnotes
Other authors have no potential conflicts of interest to disclose.
References
- 1.Mok TS, Wu Y-L, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 2.Rosell R, Moran T, Queralt C, et al. Screening for epidermal growth factor receptor mutations in lung cancer. N Engl J Med. 2009;361:958–967. doi: 10.1056/NEJMoa0904554. [DOI] [PubMed] [Google Scholar]
- 3.Kwak EL, Bang Y-J, Camidge DR, et al. Anaplastic Lymphoma Kinase Inhibition in Non Small-Cell Lung Cancer. N Engl J Med. 2010;363:1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bergethon K, Shaw AT, Ignatius Ou S-H, et al. ROS1 Rearrangements Define a Unique Molecular Class of Lung Cancers. J Clin Oncol. 2012;30:863–870. doi: 10.1200/JCO.2011.35.6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flaherty KT, Puzanov I, Kim KB, et al. Inhibition of Mutated, Activated BRAF in Metastatic Melanoma. N Engl J Med. 2010;363:809–819. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flaherty KT, Robert C, Hersey P, et al. Improved Survival with MEK Inhibition in BRAF-Mutated Melanoma. N Engl J Med. 2012;367:107–114. doi: 10.1056/NEJMoa1203421. [DOI] [PubMed] [Google Scholar]
- 7.Kosaka T, Yatabe Y, Endoh H, et al. Mutations of the epidermal growth factor receptor gene in lung cancer: biological and clinical implications. Cancer Res. 2004;64:8919–8923. doi: 10.1158/0008-5472.CAN-04-2818. [DOI] [PubMed] [Google Scholar]
- 8.Shigematsu H, Lin L, Takahashi T, et al. Clinical and Biological Features Associated With Epidermal Growth Factor Receptor Gene Mutations in Lung Cancers. J Natl Cancer Inst. 2005;97:339–346. doi: 10.1093/jnci/dji055. [DOI] [PubMed] [Google Scholar]
- 9.Sasaki H, Endo K, Takada M, et al. EGFR exon 20 insertion mutation in Japanese lung cancer. Lung Cancer. 2007;58:324–328. doi: 10.1016/j.lungcan.2007.06.024. [DOI] [PubMed] [Google Scholar]
- 10.Wu JY, Wu SG, Yang CH, et al. Lung cancer with epidermal growth factor receptor exon 20 mutations is associated with poor gefitinib treatment response. Clin Cancer Res. 2008;14:4877–4882. doi: 10.1158/1078-0432.CCR-07-5123. [DOI] [PubMed] [Google Scholar]
- 11.Greulich H, Chen TH, Feng W, et al. Oncogenic transformation by inhibitor-sensitive and -resistant EGFR mutants. PLoS Med. 2005;2:e313. doi: 10.1371/journal.pmed.0020313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shigematsu H, Takahashi T, Nomura M, et al. Somatic mutations of the HER2 kinase domain in lung adenocarcinomas. Cancer Res. 2005;65:1642–1646. doi: 10.1158/0008-5472.CAN-04-4235. [DOI] [PubMed] [Google Scholar]
- 13.Arcila ME, Chaft JE, Nafa K, et al. Prevalence, clinicopathologic associations and molecular spectrum of ERBB2 (HER2) tyrosine kinase mutations in lung adenocarcinomas. Clin Cancer Res. 2012 doi: 10.1158/1078-0432.CCR-12-0912. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jackman DM, Yeap BY, Sequist LV, et al. Exon 19 Deletion Mutations of Epidermal Growth Factor Receptor Are Associated with Prolonged Survival in Non–Small Cell Lung Cancer Patients Treated with Gefitinib or Erlotinib. Clin Cancer Res. 2006;12:3908–3914. doi: 10.1158/1078-0432.CCR-06-0462. [DOI] [PubMed] [Google Scholar]
- 15.Johnson ML, Sima CS, Chaft J, et al. Association of KRAS and EGFR mutations with survival in patients with advanced lung adenocarcinomas. Cancer. 2012 doi: 10.1002/cncr.27730. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1. 1) Eur J Cancer. 2009;45:228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 17.He M, Capelletti M, Nafa K, et al. EGFR Exon 19 Insertions: A New Family of Sensitizing EGFR Mutations in Lung Adenocarcinoma. Clin Cancer Res. 2011;18:1790–1797. doi: 10.1158/1078-0432.CCR-11-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oxnard GR, Arcila ME, Sima CS, et al. Acquired resistance to EGFR tyrosine kinase inhibitors in EGFR mutant lung cancer: Distinct natural history of patients with tumors harboring the T790M mutation. Clin Cancer Res. 2011;17:1616–1622. doi: 10.1158/1078-0432.CCR-10-2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yasuda H, Kobayashi S, Costa DB. EGFR exon 20 insertion mutations in non-small-cell lung cancer: preclinical data and clinical implications. The Lancet Oncology. 2012;13:e23–e31. doi: 10.1016/S1470-2045(11)70129-2. [DOI] [PubMed] [Google Scholar]
- 20.Mitsudomi T, Yatabe Y. Mutations of the epidermal growth factor receptor gene and related genes as determinants of epidermal growth factor receptor tyrosine kinase inhibitors sensitivity in lung cancer. Cancer science. 2007;98:1817–1824. doi: 10.1111/j.1349-7006.2007.00607.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paik PK, Arcila ME, Fara M, et al. Clinical Characteristics of Patients With Lung Adenocarcinomas Harboring BRAF Mutations. J Clin Oncol. 2011;29:2046–2051. doi: 10.1200/JCO.2010.33.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gaddipati H, Liu K, Pariser A, et al. Rare Cancer Trial Design: Lessons from FDA Approvals. Clin Cancer Res. 2012 doi: 10.1158/1078-0432.CCR-12-1135. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 23.Garon EB, Moran T, Barlesi F, et al. Phase II study of the HSP90 inhibitor AUY922 in patients with previously treated, advanced non-small cell lung cancer (NSCLC) ASCO Meeting Abstracts. 2012;30:7543. [Google Scholar]
- 24.Shimamura T, Perera SA, Foley KP, et al. Ganetespib (STA-9090), a Non-Geldanamycin HSP90 Inhibitor, has Potent Antitumor Activity in In Vitro and In Vivo Models of Non-Small Cell Lung Cancer. Clin Cancer Res. 2012 doi: 10.1158/1078-0432.CCR-11-2967. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu W, Soga S, Beebe K, et al. Sensitivity of epidermal growth factor receptor and ErbB2 exon 20 insertion mutants to Hsp90 inhibition. Br J Cancer. 2007;97:741–744. doi: 10.1038/sj.bjc.6603950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perera SA, Li D, Shimamura T, et al. HER2YVMA drives rapid development of adenosquamous lung tumors in mice that are sensitive to BIBW2992 and rapamycin combination therapy. Proc Natl Acad Sci U S A. 2009;106:474–479. doi: 10.1073/pnas.0808930106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chung K-P, Wu S-G, Wu J-Y, et al. Clinical Outcomes in Non-Small Cell Lung Cancers Harboring Different Exon 19 Deletions in EGFR. Clin Cancer Res. 2012;18:3470–3477. doi: 10.1158/1078-0432.CCR-11-2353. [DOI] [PubMed] [Google Scholar]
- 28.Costa DB, Yasuda H, Sng NJ, et al. Sensitivity to EGFR inhibitors based on location of EGFR exon 20 insertion mutations within the tyrosine kinase domain of EGFR. ASCO Meeting Abstracts. 2012;30:7523. [Google Scholar]
- 29.Yatabe Y, Pao W, Jett JR. Encouragement to Submit Data of Clinical Response to EGFR-TKIs in Patients With Uncommon EGFR Mutations. Journal of Thoracic Oncology. 2012;7:775–776. doi: 10.1097/JTO.0b013e318251980b. [DOI] [PubMed] [Google Scholar]
- 30.Janne P, Boss DS, Camidge DR, et al. Phase I dose-escalation study of the pan-HER Inhibitor, PF299804, in patients with advanced malignant solid tumors. Clin Cancer Res. 2011;17:1131–1139. doi: 10.1158/1078-0432.CCR-10-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]


