Abstract
Summary
The importance of genetic laboratory models such as mice and rats becomes evident when there is poor understanding of the nature of human disease. Many rat models for human disease, created over the years by phenotype-driven strategies, now provide a foundation for the identification of their genetic determinants. These models are especially valuable with the emerging need for validation of genes found in genome-wide association studies for complex diseases. The manipulation of the rat genome using engineered zinc-finger nucleases now introduces a key technology for manipulating the rat genome, which is broadly applicable. The ability to generate knockout rat models using zinc-finger nuclease technology will now enable its full emergence as an exceptional physiological and genetic research model.
The domestication of the rodents for genetic research has its origins in the late 19th to early 20th century [1, 2]. While the mouse became the favorite for geneticists due to its smaller size, breeding fecundity and availability of spontaneous mutant strains, the rat quickly gained favor among physiologists and other researchers in part because their larger size makes them more amenable to experimental manipulation and for drug discovery and development, leading to a significantly greater understanding of rat physiology and how it compares to humans [3–7]. Its utility is facilitated by 150 years of accumulated knowledge about disease pathways, mechanisms, physiology and the development of more than 300 different rat strains to study clinically relevant traits [3, 5]. Many strains were created by phenotype selection for different disease ‘flavors’, like high- or low-renin hypertension models [8], diabetes mellitus types 1 [9] and 2 [10] or renal disease [11], to study the pathways involved in the development of diseases. Strain characterization has been accompanied by the development of tools including genetic markers and genomic resources and databases, leading to the identification of almost 1,700 qualitative trait loci (QTL) [12] in the rat genome corresponding to a diverse range of disease-related phenotypes. These characterized strains and models facilitate the correlation between human candidate disease loci and a particular rat model, while the sequencing and comparative analysis of the rat genome inspire the hunt for new disease alleles.
Narrowing of a QTL region and selection of a candidate disease gene is accomplished mostly by sequence analysis and expression studies in recombinant strains (congenic, recombinant inbred, heterogeneous stocks) and final validation of a gene as causative is traditionally performed by transgenesis [3]. Disease genes can also be pursued using a candidate gene approach where genes and pathways identified in humans are translated to the rat, mostly by sequencing and expression. Both types of studies can further support the involvement of a gene in the development of a disease, however, these approaches have taken many years to find the causative variants and have identified a limited number of disease genes [3].
With the continuing identification of hundreds of candidate SNPs and their correlating genes by Genome-Wide Association Studies (GWAS) in humans, along with a lack of functional information for the majority of genes in the mammalian genome, the need for rodent models whose genome can be efficiently engineered has become more important than ever. Traditionally, this has not been possible in the rat despite its physiological knowledge base and developed models, due to a lack of technologies for manipulating the genome on a broad scale. Luckily, recent advances in producing genetically modified rats, especially the application of zinc-finger nuclease technology to produce knockout rats, are now allowing these barriers to be crossed.
Genetic manipulation of the rat
When gain-of-function studies can be applied to validate the role of a candidate disease gene by overexpression, standard transgenesis approaches that apply to mice also work in rats, evidenced by more than 2000 published studies using transgenic rats [5]. Embryo pronucleus microinjection of naked DNA fragments [13], bacterial or yeast artificial chromosomes [14], as well as lentivirus transduction of embryos by injection into the perivitelline space [15, 16], will each result in germline transgenesis. Conversely, chemical mutagenesis using ethylnitrosourea (ENU) has been used to generate loss-of-function, or ‘knockout’, mutations in valuable disease genes such as Apc, Srt, Brca1 and 2, Nos1 and Tgfbr2 in rats ([17–20]; C. Moreno, unpublished) and random insertional mutagenesis using gene-trap Sleeping Beauty transposons [21, 22] has generated more than 120 mutant rat models of cancer, eye development, immunodeficiency, and others (http://www.knockoutrat.org). Combined, these random mutagenesis efforts have already produced a number of disease models and many are available to the research community through the Rat Resource & Research Center (http://www.nrrrc.missouri.edu).
The establishment of the mouse embryonic stem cell (ESC) [23], and later discovery that these cells permit precise homologous recombination [24, 25], has enabled the targeted disruption of mouse genes on interest and the development of thousands of gene knockout strains. It is now hoped that the first authentic germline-competent ESCs from rats [26, 27] or the induced pluriopotent rat cells (iPSCs) [28–30] will finally allow for homologous recombination in the rat to engineer precise knockouts of genes in some strains.
The fact that the rat ESC is currently available for a limited number of strains is an important point. The rat community faces some important challenges due to the widespread development and characterization of so many unique inbred strains and disease models. It is of particular importance in the case of complex genetic traits, where multiple chromosome loci within a strain interact to produce a phenotype. Ideally, a gene targeting technology would be applicable to many rat strains to capitalize on the known biology of these established models. Luckily, while ESC technology continues to develop for widespread use, an alternative technology has emerged which allows for the rapid production of targeted mutations in any strain.
ZFN-mediated gene disruption
The broad need in the research community for technology to manipulate cellular genomes has propelled the development of a new classification of engineered molecular tools called Zinc-Finger Nucleases (ZFNs). ZFNs are engineered proteins developed for the purpose of introducing site-specific mutations in cellular genomes and their activity is dependent on two highly conserved processes– the DNA binding affinity of a zinc-finger protein motif and cellular processes of DNA damage repair. These proteins are designed with a high degree of specificity to interact with an investigator-specified sequence.
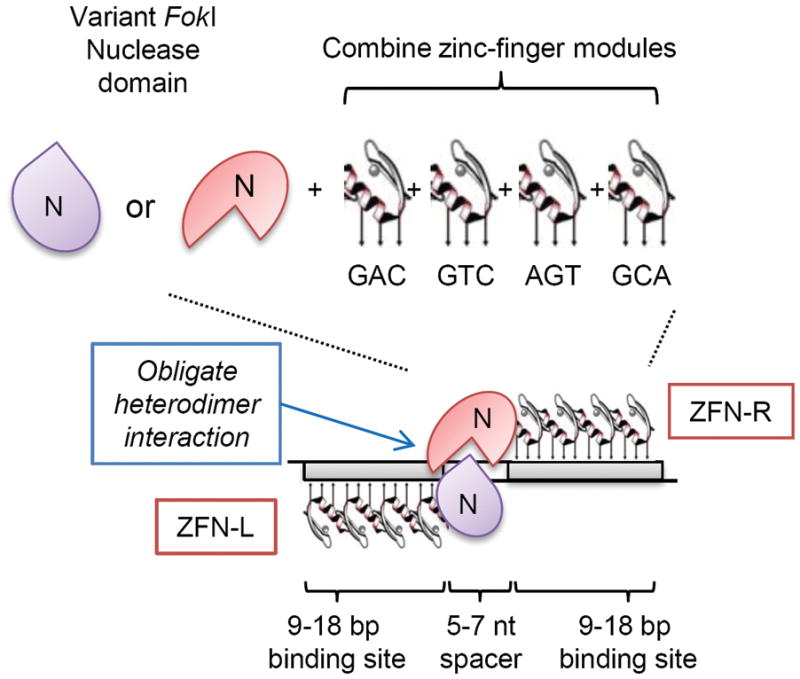
Target sequence recognition and specificity of zinc-finger nucleases are determined by three main factors, the amino acid sequences of the individual fingers, the number of fingers, and the interaction of the nuclease domain (Fig 1). Zinc-finger nucleases (ZFN) consist of two functional domains, a domain consisting of an array of sequence-specific DNA-binding zinc-fingers and the nuclease domain of the type IIs restriction endonuclease FokI. Individual zinc finger ‘modules’ are combined to form zinc-finger array domains where the amino acids within each finger interact with three nucleotides in a sequence-specific manner [31]. It takes a left (ZFN-L) and right (ZFN-R) proteins to cleave a target sequence because the nuclease domain (N) must dimerize to function. Published active reagents contain anywhere from 3–6 zinc fingers for the left or right ZFN [32–44], thus each of the left and right ZFN interact with 9–18 nucleotides and, because they work in pairs, 18–36 nucleotides of target-sequence specificity separated in the middle by a spacer sequence of 5–7 base pairs where the endonuclease domains interact to cleave the DNA. More recently developed ZFNs also use obligate heterodimer variant forms of the FokI endonuclease domain where amino acid residues have been altered to ensure that only a left and right ZFN can pair to cause cleavage, further improving the specificity of the system [39, 45].
Figure 1.
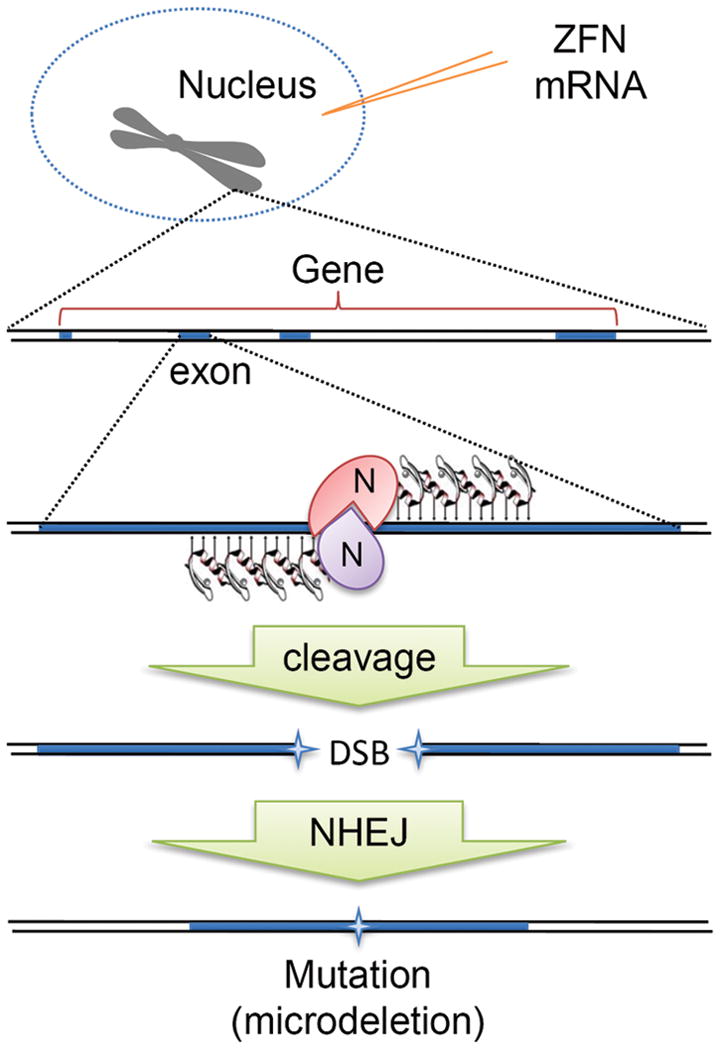
When a pair of ZFNs, referred to as a left and right ZFN, precisely bind to a target sequence such as the exon of a gene, the nuclease domain cleaves the DNA to introduce a double strand break (DSB), like a pair of genomic scissors. The nonhomologous end-joining (NHEJ) repair mechanisms will repair the DSB, often at the expense of small bits of sequence information, disrupting the gene.
The art of developing ZFNs for genome engineering is dependent on the existing libraries of reagents. The complexity of the available libraries of characterized zinc-finger modules ensures that most genes can be targeted, although certain sequences are considered more ideal (sequences where one or more individual finger recognition triplets contain 5′-GNN-3′ [42]). Small genes will, of course, contain fewer of these ideal target sites and therefore fewer active ZFN pairs will be identified. In addition, and for reasons that are not completely understood, even carefully designed ZFNs sometimes lack specificity or lack activity. These current limitations could prevent designing suitable ZFN reagents for some target genes, however both commercial and academic groups are continuously expanding the zinc-finger module libraries to increase the chances of a successful design.
The principal of ZFN-mediated gene disruption is depicted in Figure 2. Two engineered ZFN proteins designed to interact with a gene sequence with high fidelity are introduced into a cell and use their nuclease activity to cause a double-strand breakage (DSB) in the chromosome in a gene’s protein-coding sequence. This stimulates innate cellular DNA repair mechanisms to fix the DSB or the cell will likely die. The most active DNA repair mechanism in mammalian cells for repairing DSBs is the nonhomologous end-joining pathway (NHEJ) although homology-dependent repair (HDR) using an available template containing homologous sequence occurs at a lower frequency [46]. NHEJ is template independent and is therefore an imperfect repair mechanism which frequently results in deletion and/or insertion of sequence information, resulting in a potential mutation (Fig 2) [47]. If the mutation disrupts the production of a normal gene transcript or protein, a knockout mutation has been generated.
Figure 2.
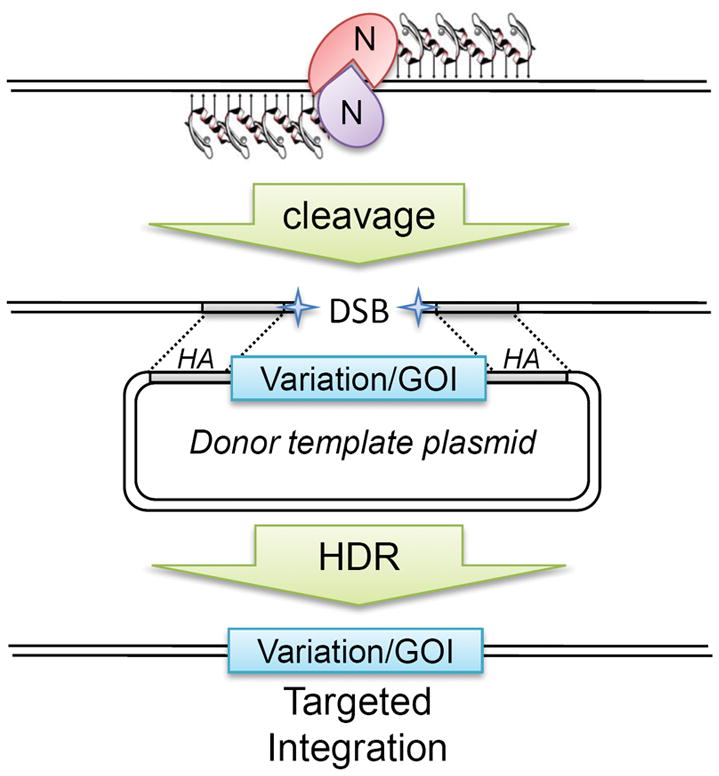
Targeted integration of new sequence variations or new genes of interest (GOI) have been stimulated by ZFN-mediated cleavage and homology-directed repair (HDR) in cultured cells (see Table 1). ZFNs are co-delivered to the cell with a circular double-stranded plasmid containing the desired variation or gene flanked by homology arm (HA) sequences. After cleavage, DNA repair mechanisms can use the plasmid as a template to repair the broken chromosome and introduce the new sequence seamlessly into the genome. Typically, a selectable marker gene is necessary to identify the targeted integration events.
There are mutliple places to obtain ZFN reagents for genome editing in model species - one commercial source and reagents that are available in the public domain. Sigma-Aldrich, Inc. licenses and distributes ZFN technology from Sangamo BioSciences, Inc., the world’s leading commercial developer of zinc-finger technology. Obtaining ZFN reagents to disrupt a target gene is as simple as providing the target sequence and any information about known paralogs, and pseudogenes with high sequence similarity to the gene of interest. Bioinformatics tools are used to identify ideal ZFN target sites within exon sequences. Ideally, suitable sites are found within the first two-thirds of the gene coding sequence to ensure a severe truncation of the gene product. Sigma then combines precharacterized dual zinc-finger ‘modules’ with known specificity for hexamer sequences are combined to make 4-, 5-, or 6- finger ZFNs. Sites are screened for uniqueness in the target genome and as many as 16 ZFN pairs are assembled for testing in a cellular proxy system, in the case of rats, by transfection into cultured rat cells [48]. A functional pair of ZFN reagents, in a ready-to-use format, is then delivered to the investigator. The cost for Sigma’s custom ZFN design and validation is currently $25,000, a price which initially brings about significant pause and exceeds the budgets of many labs. However, the cost includes design and construction of multiple reagents and pre-validation to ensure active reagents are obtained. Coincidently, production of a typical knockout mouse model via gene targeting in mouse ESCs routinely exceeds this cost.
Of important note, the use of Sigma ZFNs also currently comes with a $7,500 ZFN License Agreement fee which contains restrictions on the use of the ZFNs and the modified animals that result from their use, including forbidding the licensing, sale, or other distribution of the animal model and limiting the number of animals that can be bred (for rodents) without further permission. However, the fee itself can typically be waived if the investigator agrees to transfer title of the modified animals to Sigma for potential commercial distribution at the completion of their study.
On the other hand, sources of characterized zinc-finger modules and reagents are available from academic groups, allowing investigators to engineer their own reagents, potentially at a reduced cost and without significant restrictions [37, 49, 50]. Some of these platforms use libraries of pre-characterized single-finger modules to assemble 3-finger ZFN reagents and have yielded functional zinc-finger arrays for use in several systems [51], however, for reasons that are not completely understood, the success rate with some design strategies is low [52, 53]. The Oligomerized Pooled ENgineering (OPEN) platform was developed by academic researchers for developing custom ZFN reagents using combination of a bacterial two-hybrid system and testing in cultured cells [37], is available to researchers at cost, and has dramatically increased the success rate of generating active ZFN reagents for a broad range of targets [54].
To date, engineered ZFNs have been used to generate site-specific mutations at endogenous loci in cultured cells and embryos from several species. The applications have ranged from targeted gene therapy to the production of gene-specific knockout animals. For the purpose of this review, the application of ZFN technology to vertebrate models is outlined in Table 1, although much of the technology and experimental approaches were anticipated by work in flies, worms [35], and plants [55, 56]. Only the commercial reagents have thus far been demonstrated to knock out genes in mammalian embryos as described below, however, both the commercial and OPEN platforms have yielded active pairs of ZFN reagents suitable for genome editing in mammalian cells [37], zebrafish embryos [36, 38, 54] and, presumably, will be applicable to other species.
Table 1.
Endogenous gene targeting in vertebrate animal models and cultured cells
| Organism | Gene a | cell type | Ref |
|---|---|---|---|
| Gene Disruption | |||
| Zebrafish | golden | embryo b | [36] |
| Zebrafish | notail | embryo b | [36] |
| Zebrafish | dopamine transporter | embryo | [54] |
| Zebrafish | gridlock | embryo | [54] |
| Zebrafish | hypoxia inducible factor 1a | embryo | [54] |
| Zebrafish | telomerase | embryo | [54] |
| Zebrafish | transferrin receptor protein 2 | embryo | [54] |
| Zebrafish | vascular endothelial growth factor- 2 receptor | embryo | [38] |
| Rat | immunoglobulin M | embryo b | [44] |
| Rat | ras-related protein Rab38 | embryo b | [44] |
| Rat | interleukin 2 receptor, gamma | embryo b | [57] |
| Human | cystic fibrosis transmembrane conductance regulator | HEK293 | [37] |
| Human | homeobox protein B13 | HEK293 | [37] |
| Human | vascular endothelial growth factor A | HEK293 | [37] |
| Human | C-C chemokine receptor 5 | various human cells b,c | [61, 65, 66] |
| Hamster | bak | CHO b | [67] |
| Hamster | bax | CHO b | [67] |
| Hamster | glutamine synthase | CHO b | [68] |
| Hamster | alpha-1,6-fucosyltransferase | CHO b | [68] |
| Hamster | dihydrofolate reductase | CHO b | [68, 69] |
| Targeted Integration | |||
| mouse | tyrosinase | Melan-C | [61] |
| human | octamer-4 | hESC b | [62] |
| human | adeno-associated virus integration site 1 | hESC, hiPSC b | [62] |
| human | pituitary homeobox 3 | hESC, hiPSC b | [62] |
| human | interleukin 2 receptor, gamma | various human cells b | [39, 40, 60, 70] |
only endogenous targeted loci are listed
commercially designed ZFN reagents used
open-source ZFNs were designed for some studies
Rapid gene knockout in the rat
Targeted genome modification in the rat begins with the careful selection of the target sequence and acquiring the ZFN reagents. The existing rat gene and transcript annotations are frequently generated by comparative species analysis combined with gene prediction algorithms run on the assembly. Because the rat genome sequence still contains many gaps, these gene models are not always accurate and therefore expressed sequence tag (EST) and direct sequence comparison with gene models from other species are used to generate the best target sequence information. We also recommend direct sequencing of the target region in the target strain of interest since a single nucleotide difference (due to the presence of a SNP or error in the genome sequence) can interfere with designing effective ZFN reagents. The validated sequence is then scanned for ideal target sites to design ZFN reagents.
The process of delivering the ZFNs to the rat embryo is essentially identical to techniques used to produce transgenic mice and rats, where ZFN-encoding nucleic acids are microinjected into newly fertilized embryos [48]. Although we previously reported cytoplasmic delivery by microinjection [44], typically, ZFN mRNA is injected into the male pronucleus after which it is transported to the cytoplasm for translation into ZFN proteins which re-enter the nucleus to cleave the target sequence. Occasionally, a mutation occurs which is recovered after transferring the embryos to surrogate females and birth. The animals are screened by a simple PCR-based assays followed by sequencing [48]. Animals carrying frameshift alleles in the gene coding sequence are then considered as potential knockouts of the gene of interest.
The initial description of ZFN genome editing at three distinct loci in the rat genome revealed several important and beneficial attributes of this technology [44]. First, the availability of the green fluorescent protein (GFP) lentivirus-transgenic rat [16] revealed that delivery of mRNA-encoded ZFNs to the rat embryo results in site specific mutagenesis at the 1-cell stage. Thus, the mutation occurs before the cell has a chance to divide, leading to whole-animal mono- or, at lower frequency, bi-allelic knockout of the target gene with very little mosaicism [44]. This is unlike the zebrafish reports, where ZFN activity occurs during a period of rapid cell division, leading to mosaic animals [36, 38]. Second, mutations in rat embryos typically arise as microdeletions of a single to a few hundred nucleotides, although microdeletions can sometimes be accompanied by insertion of new sequence at the target site ([48] and PhysGen Knockout team, unpublished observations). When these microdeletions and insertions are targeted to a coding exon of a gene, the probability of a frameshift or other mutation that will disrupt gene function is high. Third, a beneficial attribute of the technology that cannot be overemphasized for the reasons mentioned above, is that it is proven to be applicable to multiple inbred and outbred rat strains [44]. While there will be some variability in the health and ease of manipulating embryos from some rat strains, ZFN technology will allow researchers to knock out their favorite gene in their favorite strain. Finally, the fact that ZFN proteins are highly active during the early hours of embryo development, leading to founder animals with high percentages of mutagenized chromosomes means that the production of gene knockout rats is very rapid. From selection of a target gene to obtaining the ZFN reagents to microinjection and production of knockout founder animals can be as little as 3–4 months. Recently, new report of gene knockout for the interleukin 2 receptor gamma (Il2rg) gene by Mashimo, et al. demonstrated similar findings [57].
Despite the best efforts and care in the design of ZFN reagents, they do have the potential to cause off-target cleavage and mutations and this remains a key criticism of the technology. These events can be observed by first predicting where these effects might occur by scanning for similar sequences to the target site in the genome [36, 38] and screening them for ZFN modification. Typically, if great care is put into selecting the target site and reagents which have high specificity are used, these events are minimized. For example, no evidence of off-target cleavage by the engineered reagents has been observed in the two rat studies using 5- and 6-finger ZFNs [44, 57]. However, other reagents utilizing 3-finger ZFNs have sometimes demonstrated off-target effects [36, 38, 39, 45]. Obviously, for gene therapeutic applications, these effects must be minimized to the greatest extent possible, but in animal models, the occasional off-target event may be less important if they can be bred away. Nevertheless, highly related sequences among members of clustered gene families may be particularly susceptible to off-target cleavage and these events would be difficult to separate by recombination. In these cases, care should be taken to screen mutant founders for these secondary events.
The advent of ZFN technology opens new doors to rapidly knock out candidate human disease genes and open new avenues to understanding disease mechanisms. In some cases, however, more precise engineering will be required to be able to assign function to some genes which are essential at early stages of development (are lethal when knocked out) or to introduce a new sequence into the rat genome. These types of strategies will require a more complex engineering of the rat genome and ZFNs can play a role.
ZFNs are diverse tools for manipulating genomes
While NHEJ is the most active DNA repair mechanism, homology-dependent repair can be dramatically stimulated by double strand breakages in chromosomes [58] and several groups are now using ZFNs to facilitate targeted integration of new sequences into genomes by homologous recombination (Table 1). In this application, the ZFNs are introduced with a donor template harboring homologous sequence to regions flanking the ZFN target site (Fig. 3). ZFN cleavage prompts the cell to use the donor template to repair the chromosome, leading to precise integration of the donor sequence. ZFNs can stimulate the targeted integration of new sequences from a few to more than a thousand-fold increase over spontaneous homologous recombination. To date, ZFNs have been used for targeted integration of a few base pairs or even 9-kilobase expression constructs into Drosophila embryos and cultured human cells [33, 39, 40, 59–61] and, recently, ZFNs have been used to target expression cassettes into human ESCs and iPSCs [62, 63]. It will be of great interest to combine the emerging technologies for rat genome engineering, such as the rat ESC and iPSC with ZFN technology to produce more complex manipulation of the rat genome such as conditional knockout alleles, humanized rat models where a human gene is inserted precisely into the rat homologous gene locus, or even more imaginative alterations to the rat genome.
Figure 3.
The laboratory rat has a vast knowledge base and genomic tools which demonstrate its utility as a model system for studying human physiology and disease. Zinc-finger nuclease (ZFN) and, hopefully, stem cell technologies now reduce the hurdles which previously existed to the investigator wishing to study the function of a particular gene in a physiological or disease process in rats.
Clinical Relevance: Disease Gene Validation
The application of ZFN-mediated genome editing to zebrafish and rats is changing the views of these species as genetic research models, an attitude which will impact our understanding of human disease and enhance care. Valuable models are already beginning to emerge. For instance, Mashimo, et al. recently developed a Knockout rat for Interleukin 2 receptor gamma with ZFN, inducing X-Linked Severe Combined Immunodeficiency (X-SCID), one of the most common forms of human SCID. This model can be used as a tool for pre-clinical testing as well as a base for xenotransplantation and cancer research [57]. In addition the Sigma Advanced Genetic Engineering (SAGE) lab is developing several valuable models for studies of neurobiology, toxicology, cardiology, cancer, and immunology (http://www.sageresearchmodels.com).
Besides developing models such as these for known diseases and validation of new candidate genes causing monogenic disease, ZFNs will enable identification and validation of genes involved in complex diseases. Numerous linkage and association studies have identified candidate genes for disease susceptibility. Validation of these genes by gene manipulation is confounded by the ‘complexity’ of the disease, that is, there needs to be several susceptibility genes for a gene to have an effect on a phenotype - a susceptible background. This has been shown by the different effects of gene knockout in different strain backgrounds in mouse [64]. However, in established disease models which have been selected by phenotyping over generations and have accumulated susceptibility genes for complex diseases, ZFN technology provides a very powerful tool for gene validation. For example, the PhysGen Knockout team is currently using ZFN technology to knockout 100 genes for hypertension and renal disease in the sensitized Dahl S hypertensive rat as a validation tool for different published GWAS candidate loci and providing these initially characterized models to the research community (for more information, see http://rgd.mcw.edu/wg/physgenknockouts). We expect that the susceptible hypertensive background of the Dahl S and consomic strains will better allow us to see an effect of these genes on blood pressure, since these strains lack the compensatory mechanisms for returning blood pressure to normal values.
Summary and Future Directions
Rat has been classically used to study physiology and disease related to humans and, in recent years, for the search of genes involved in complex diseases. New technologies such as ZFN-mediated engineering and stem cells complete the genetic toolbox now available to the researcher who wishes to use the rat (Fig. 4). Linkage analysis in rats and humans will continue to identify many candidate genes related to different diseases and combining the knockout approach with established rat disease models will enable the study of the effects of gene function in the context of well characterized phenotypes and pathways, increasing our understanding of the mechanism of action of genes in disease. This accelerated knowledge, capitalizing on the wealth of information obtained after more than 100 years of research, will lead to therapeutic strategies in humans in a shorter timeframe.
Figure 4. The rat model is fully enabled by new technologies.
The laboratory rat provides a vast knowledge base and genomic tools which demonstrate its utility as a model system for studying human physiology and disease. ZFN and, hopefully, stem cell technologies now reduce the hurdles which previously existed to the investigator wishing to study the function of a particular gene in a physiological or disease process in rats.
Much is left to be developed for routine ZFN use in the rat and several applications of ZFNs to editing the rat genome in stem cells and embryos, including targeted integration, must be explored. Its application enables new studies which have previously been impossible or cost prohibitive in other mammalian model systems other than the mouse. This technology platform is amenable to both small and large laboratories alike with the only requirements being the ZFN reagents, access to an embryo microinjection facility, and routine molecular biology skills to PCR-amplify and sequence the target site. Moving forward, the pursuit of gene targets by different laboratories will develop an understanding of the reproducibility of the technology. The PhysGen Knockout team has now knocked out more than 40 genes using the commercial reagents (unpublished). It will be interesting to see how rat researchers choose to implement gene targeting strategies in their studies. More importantly, in addition to zebrafish and rats, additional species will likely soon be shown to be amenable to ZFN-mediated genome engineering and researchers will finally be afforded the valuable opportunity to choose which model system and strain background best fits the disease or process they are studying.
References
- 1.Baker HJ, Lindsey JR, Weisbroth SH. The laboratory rat: volume I -biology and diseases. Academic Press; New York: 1979. [Google Scholar]
- 2.Nishioka Y. The origin of common laboratory mice. Genome. 1995;38:1–7. doi: 10.1139/g95-001. [DOI] [PubMed] [Google Scholar]
- 3.Aitman TJ, Critser JK, Cuppen E, Dominiczak A, Fernandez-Suarez XM, Flint J, Gauguier D, Geurts AM, Gould M, Harris PC, Holmdahl R, Hubner N, Izsvak Z, Jacob HJ, Kuramoto T, Kwitek AE, Marrone A, Mashimo T, Moreno C, Mullins J, Mullins L, Olsson T, Pravenec M, Riley L, Saar K, Serikawa T, Shull JD, Szpirer C, Twigger SN, Voigt B, Worley K. Progress and prospects in rat genetics: a community view. Nat Genet. 2008;40:516–522. doi: 10.1038/ng.147. [DOI] [PubMed] [Google Scholar]
- 4.Gill TJ, 3rd, Smith GJ, Wissler RW, Kunz HW. The rat as an experimental animal. Science. 1989;245:269–276. doi: 10.1126/science.2665079. [DOI] [PubMed] [Google Scholar]
- 5.Jacob HJ, Kwitek AE. Rat genetics: attaching physiology and pharmacology to the genome. Nat Rev Genet. 2002;3:33–42. doi: 10.1038/nrg702. [DOI] [PubMed] [Google Scholar]
- 6.James MR, Lindpaintner K. Why map the rat? Trends Genet. 1997;13:171–173. doi: 10.1016/s0168-9525(97)01130-x. [DOI] [PubMed] [Google Scholar]
- 7.Kola I. Putting the rat on the map. Nat Biotechnol. 2004;22:529–531. doi: 10.1038/nbt0504-529. [DOI] [PubMed] [Google Scholar]
- 8.Dupont J, Dupont JC, Froment A, Milon H, Vincent M. Selection of three strains of rats with spontaneously different levels of blood pressure. Biomedicine. 1973;19:36–41. [PubMed] [Google Scholar]
- 9.Like AA, Butler L, Williams RM, Appel MC, Weringer EJ, Rossini AA. Spontaneous autoimmune diabetes mellitus in the BB rat. Diabetes. 1982;31:7–13. doi: 10.2337/diab.31.1.s7. [DOI] [PubMed] [Google Scholar]
- 10.Kimura K, Toyota T, Kakizaki M, Kudo M, Takebe K, Goto Y. Impaired insulin secretion in the spontaneous diabetes rats. Tohoku J Exp Med. 1982;137:453–459. doi: 10.1620/tjem.137.453. [DOI] [PubMed] [Google Scholar]
- 11.Kreisberg JI, Karnovsky MJ. Focal glomerular sclerosis in the fawn-hooded rat. Am J Pathol. 1978;92:637–652. [PMC free article] [PubMed] [Google Scholar]
- 12.Shimoyama M, Hayman GT, Laulederkind SJF, Nigam R, Lowry TF, Petri V, Smith JR, Wang S-J, Munzenmaier DH, Dwinell MR, Twigger SN, Jacob HJ the RGDT. The Rat Genome Database Curators: Who, What, Where, Why. PLoS Comput Biol. 2009;5:e1000582. doi: 10.1371/journal.pcbi.1000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mullins JJ, Peters J, Ganten D. Fulminant hypertension in transgenic rats harbouring the mouse Ren-2 gene. Nature. 1990;344:541–544. doi: 10.1038/344541a0. [DOI] [PubMed] [Google Scholar]
- 14.Takahashi R, Ito K, Fujiwara Y, Kodaira K, Hirabayashi M, Ueda M. Generation of transgenic rats with YACs and BACs: preparation procedures and integrity of microinjected DNA. Exp Anim. 2000;49:229–233. doi: 10.1538/expanim.49.229. [DOI] [PubMed] [Google Scholar]
- 15.Dann CT, Alvarado AL, Hammer RE, Garbers DL. Heritable and stable gene knockdown in rats. PNAS. 2006;103:11246–11251. doi: 10.1073/pnas.0604657103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Michalkiewicz M, Michalkiewicz T, Geurts AM, Roman RJ, Slocum GR, Singer O, Weihrauch D, Greene AS, Kaldunski M, Verma IM, Jacob HJ, Cowley AW., Jr Efficient transgenic rat production by a lentiviral vector. Am J Physiol Heart Circ Physiol. 2007;293:H881–894. doi: 10.1152/ajpheart.00060.2007. [DOI] [PubMed] [Google Scholar]
- 17.Amos-Landgraf JM, Kwong LN, Kendziorski CM, Reichelderfer M, Torrealba J, Weichert J, Haag JD, Chen KS, Waller JL, Gould MN. A target-selected Apc-mutant rat kindred enhances the modeling of familial human colon cancer. Proc Natl Acad Sci USA. 2007;104:4036– 4041. doi: 10.1073/pnas.0611690104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smits BMG, Mudde J, Plasterk RHA, Cuppen E. Target-selected mutagenesis of the rat. Genomics. 2004;83:332–334. doi: 10.1016/j.ygeno.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 19.Zan Y, Haag JD, Chen KS, Shepel LA, Wigington D, Wang YR, Hu R, Lopez-Guajardo CC, Brose HL, Porter KI. Production of knockout rats using ENU mutagenesis and a yeast-based screening assay. Nat Biotechnol. 2003;21:645– 651. doi: 10.1038/nbt830. [DOI] [PubMed] [Google Scholar]
- 20.Homberg JR, Olivier JD, Smits BM, Mul JD, Mudde J, Verheul M, Nieuwenhuizen OF, Cools AR, Ronken E, Cremers T, Schoffelmeer AN, Ellenbroek BA, Cuppen E. Characterization of the serotonin transporter knockout rat: a selective change in the functioning of the serotonergic system. Neuroscience. 2007;146:1662–1676. doi: 10.1016/j.neuroscience.2007.03.030. [DOI] [PubMed] [Google Scholar]
- 21.Kitada K, Ishishita S, Tosaka K, Takahashi R, Ueda M, Keng VW, Horie K, Takeda J. Transposon-tagged mutagenesis in the rat. Nat Methods. 2007;4:131–133. doi: 10.1038/nmeth1002. [DOI] [PubMed] [Google Scholar]
- 22.Lu B, Geurts AM, Poirier C, Petit DC, Harrison W, Overbeek PA, Bishop CE. Generation of rat mutants using a coat color-tagged Sleeping Beauty transposon system. Mamm Genome. 2007;18:338–346. doi: 10.1007/s00335-007-9025-5. [DOI] [PubMed] [Google Scholar]
- 23.Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 24.Doetschman T, Maeda N, Smithies O. Targeted mutation of the Hprt gene in mouse embryonic stem cells. Proc Natl Acad Sci U S A. 1988;85:8583–8587. doi: 10.1073/pnas.85.22.8583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thomas KR, Folger KR, Capecchi MR. High frequency targeting of genes to specific sites in the mammalian genome. Cell. 1986;44:419–428. doi: 10.1016/0092-8674(86)90463-0. [DOI] [PubMed] [Google Scholar]
- 26.Buehr M, Meek S, Blair K, Yang J, Ure J, Silva J, McLay R, Hall J, Ying QL, Smith A. Capture of authentic embryonic stem cells from rat blastocysts. Cell. 2008;135:1287–1298. doi: 10.1016/j.cell.2008.12.007. [DOI] [PubMed] [Google Scholar]
- 27.Li P, Tong C, Mehrian-Shai R, Jia L, Wu N, Yan Y, Maxson RE, Schulze EN, Song H, Hsieh CL, Pera MF, Ying QL. Germline competent embryonic stem cells derived from rat blastocysts. Cell. 2008;135:1299–1310. doi: 10.1016/j.cell.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chang M-Y, Kim D, Kim C-H, Kang H-C, Yang E, Moon J-I, Ko S, Park J, Park K-S, Lee K-A, Hwang D-Y, Chung Y, Lanza R, Kim K-S. Direct Reprogramming of Rat Neural Precursor Cells and Fibroblasts into Pluripotent Stem Cells. PLoS One. 5:e9838. doi: 10.1371/journal.pone.0009838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li W, Wei W, Zhu S, Zhu J, Shi Y, Lin T, Hao E, Hayek A, Deng H, Ding S. Generation of Rat and Human Induced Pluripotent Stem Cells by Combining Genetic Reprogramming and Chemical Inhibitors. Cell Stem Cell. 2008 doi: 10.1016/j.stem.2008.11.014. [DOI] [PubMed] [Google Scholar]
- 30.Liao J, Cui C, Chen S, Ren J, Chen J, Gao Y, Li H, Jia N, Cheng L, Xiao H, Xiao L. Generation of Induced Pluripotent Stem Cell Lines from Adult Rat Cells. Cell Stem Cell. 2008 doi: 10.1016/j.stem.2008.11.013. [DOI] [PubMed] [Google Scholar]
- 31.Pavletich NP, Pabo CO. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science. 1991;252:809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- 32.Beumer KJ, Trautman JK, Bozas A, Liu JL, Rutter J, Gall JG, Carroll D. Efficient gene targeting in Drosophila by direct embryo injection with zinc-finger nucleases. Proc Natl Acad Sci U S A. 2008;105:19821–19826. doi: 10.1073/pnas.0810475105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bibikova M, Golic M, Golic KG, Carroll D. Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. Genetics. 2002;161:1169–1175. doi: 10.1093/genetics/161.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carroll D. Progress and prospects: zinc-finger nucleases as gene therapy agents. Gene Ther. 2008;15:1463–1468. doi: 10.1038/gt.2008.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carroll D, Beumer KJ, Morton JJ, Bozas A, Trautman JK. Gene targeting in Drosophila and Caenorhabditis elegans with zinc-finger nucleases. Methods Mol Biol. 2008;435:63–77. doi: 10.1007/978-1-59745-232-8_5. [DOI] [PubMed] [Google Scholar]
- 36.Doyon Y, McCammon JM, Miller JC, Faraji F, Ngo C, Katibah GE, Amora R, Hocking TD, Zhang L, Rebar EJ, Gregory PD, Urnov FD, Amacher SL. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat Biotechnol. 2008;26:702–708. doi: 10.1038/nbt1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maeder ML, Thibodeau-Beganny S, Osiak A, Wright DA, Anthony RM, Eichtinger M, Jiang T, Foley JE, Winfrey RJ, Townsend JA, Unger-Wallace E, Sander JD, Muller-Lerch F, Fu F, Pearlberg J, Gobel C, Dassie JP, Pruett-Miller SM, Porteus MH, Sgroi DC, Iafrate AJ, Dobbs D, McCray PB, Jr, Cathomen T, Voytas DF, Joung JK. Rapid “open-source” engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol Cell. 2008;31:294–301. doi: 10.1016/j.molcel.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meng X, Noyes MB, Zhu LJ, Lawson ND, Wolfe SA. Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Nat Biotechnol. 2008;26:695–701. doi: 10.1038/nbt1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller JC, Holmes MC, Wang J, Guschin DY, Lee YL, Rupniewski I, Beausejour CM, Waite AJ, Wang NS, Kim KA, Gregory PD, Pabo CO, Rebar EJ. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat Biotechnol. 2007;25:778–785. doi: 10.1038/nbt1319. [DOI] [PubMed] [Google Scholar]
- 40.Moehle EA, Rock JM, Lee YL, Jouvenot Y, DeKelver RC, Gregory PD, Urnov FD, Holmes MC. Targeted gene addition into a specified location in the human genome using designed zinc finger nucleases. Proc Natl Acad Sci U S A. 2007;104:3055–3060. doi: 10.1073/pnas.0611478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Porteus MH. Mammalian gene targeting with designed zinc finger nucleases. Mol Ther. 2006;13:438–446. doi: 10.1016/j.ymthe.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 42.Porteus MH, Carroll D. Gene targeting using zinc finger nucleases. Nat Biotechnol. 2005;23:967–973. doi: 10.1038/nbt1125. [DOI] [PubMed] [Google Scholar]
- 43.Santiago Y, Chan E, Liu PQ, Orlando S, Zhang L, Urnov FD, Holmes MC, Guschin D, Waite A, Miller JC, Rebar EJ, Gregory PD, Klug A, Collingwood TN. Targeted gene knockout in mammalian cells using engineered zinc finger nucleases. Proc Natl Acad Sci U S A. 2008;105:5809–5814. doi: 10.1073/pnas.0800940105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Geurts AM, Cost GJ, Freyvert Y, Zeitler B, Miller JC, Choi VM, Jenkins SS, Wood A, Cui X, Meng X, Vincent A, Lam S, Michalkiewicz M, Schilling R, Foeckler J, Kalloway S, Weiler H, Menoret S, Anegon I, Davis GD, Zhang L, Rebar EJ, Gregory PD, Urnov FD, Jacob HJ, Buelow R. Knockout Rats via Embryo Microinjection of Zinc-Finger Nucleases. Science. 2009;325:433. doi: 10.1126/science.1172447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Szczepek M, Brondani V, Buchel J, Serrano L, Segal DJ, Cathomen T. Structure-based redesign of the dimerization interface reduces the toxicity of zinc-finger nucleases. Nat Biotech. 2007;25:786–793. doi: 10.1038/nbt1317. [DOI] [PubMed] [Google Scholar]
- 46.Mao Z, Bozzella M, Seluanov A, Gorbunova V. DNA repair by nonhomologous end joining and homologous recombination during cell cycle in human cells. Cell Cycle. 2008;7:2902–2906. doi: 10.4161/cc.7.18.6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lieber MR. The mechanism of human nonhomologous DNA end joining. J Biol Chem. 2008;283:1–5. doi: 10.1074/jbc.R700039200. [DOI] [PubMed] [Google Scholar]
- 48.Geurts AM, Cost GJ, Remy S, Cui X, Tesson L, Usal C, Menoret S, Jacob HJ, Anegon I, Buelow R. Generation of gene-specific mutated rats using zinc-finger nucleases. Methods Mol Biol. 2010;597:211–225. doi: 10.1007/978-1-60327-389-3_15. [DOI] [PubMed] [Google Scholar]
- 49.Wright DA, Thibodeau-Beganny S, Sander JD, Winfrey RJ, Hirsh AS, Eichtinger M, Fu F, Porteus MH, Dobbs D, Voytas DF, Joung JK. Standardized reagents and protocols for engineering zinc finger nucleases by modular assembly. Nat Protocols. 2006;1:1637–1652. doi: 10.1038/nprot.2006.259. [DOI] [PubMed] [Google Scholar]
- 50.Carroll D, Morton JJ, Beumer KJ, Segal DJ. Design, construction and in vitro testing of zinc finger nucleases. Nat Protoc. 2006;1:1329–1341. doi: 10.1038/nprot.2006.231. [DOI] [PubMed] [Google Scholar]
- 51.Kim J-S, Lee HJ, Carroll D. Genome editing with modularly assembled zinc-finger nucleases. Nat Meth. 7:91–91. doi: 10.1038/nmeth0210-91b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Joung JK, Voytas DF, Cathomen T. Reply to “Genome editing with modularly assembled zinc-finger nucleases”. Nat Meth. 7:91–92. doi: 10.1038/nmeth0210-91b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ramirez CL, Foley JE, Wright DA, Muller-Lerch F, Rahman SH, Cornu TI, Winfrey RJ, Sander JD, Fu F, Townsend JA, Cathomen T, Voytas DF, Joung JK. Unexpected failure rates for modular assembly of engineered zinc fingers. Nat Meth. 2008;5:374–375. doi: 10.1038/nmeth0508-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Foley JE, Yeh J-RJ, Maeder ML, Reyon D, Sander JD, Peterson RT, Joung JK. Rapid Mutation of Endogenous Zebrafish Genes Using Zinc Finger Nucleases Made by Oligomerized Pool ENgineering (OPEN) PLoS ONE. 2009;4:e4348. doi: 10.1371/journal.pone.0004348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wright DA, Townsend JA, Winfrey RJ, Jr, Irwin PA, Rajagopal J, Lonosky PM, Hall BD, Jondle MD, Voytas DF. High-frequency homologous recombination in plants mediated by zinc-finger nucleases. Plant J. 2005;44:693–705. doi: 10.1111/j.1365-313X.2005.02551.x. [DOI] [PubMed] [Google Scholar]
- 56.Lloyd A, Plaisier CL, Carroll D, Drews GN. Targeted mutagenesis using zinc-finger nucleases in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:2232–2237. doi: 10.1073/pnas.0409339102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mashimo T, Takizawa A, Voigt B, Yoshimi K, Hiai H, Kuramoto T, Serikawa T. Generation of knockout rats with X-linked severe combined immunodeficiency (X-SCID) using zinc-finger nucleases. PLoS One. 5:e8870. doi: 10.1371/journal.pone.0008870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jasin M. Genetic manipulation of genomes with rare-cutting endonucleases. Trends Genet. 1996;12:224–228. doi: 10.1016/0168-9525(96)10019-6. [DOI] [PubMed] [Google Scholar]
- 59.Porteus MH, Baltimore D. Chimeric Nucleases Stimulate Gene Targeting in Human Cells. Science. 2003;300:763. doi: 10.1126/science.1078395. [DOI] [PubMed] [Google Scholar]
- 60.Urnov FD, Miller JC, Lee Y-L, Beausejour CM, Rock JM, Augustus S, Jamieson AC, Porteus MH, Gregory PD, Holmes MC. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- 61.Kandavelou K, Ramalingam S, London V, Mani M, Wu J, Alexeev V, Civin CI, Chandrasegaran S. Targeted manipulation of mammalian genomes using designed zinc finger nucleases. Biochemical and Biophysical Research Communications. 2009;388:56–61. doi: 10.1016/j.bbrc.2009.07.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hockemeyer D, Soldner F, Beard C, Gao Q, Mitalipova M, DeKelver RC, Katibah GE, Amora R, Boydston EA, Zeitler B, Meng X, Miller JC, Zhang L, Rebar EJ, Gregory PD, Urnov FD, Jaenisch R. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat Biotech. 2009 doi: 10.1038/nbt.1562. advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zou J, Maeder ML, Mali P, Pruett-Miller SM, Thibodeau-Beganny S, Chou BK, Chen G, Ye Z, Park IH, Daley GQ, Porteus MH, Joung JK, Cheng L. Gene targeting of a disease-related gene in human induced pluripotent stem and embryonic stem cells. Cell Stem Cell. 2009;5:97–110. doi: 10.1016/j.stem.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Crusio WE. Flanking gene and genetic background problems in genetically manipulated mice. Biological Psychiatry. 2004;56:381–385. doi: 10.1016/j.biopsych.2003.12.026. [DOI] [PubMed] [Google Scholar]
- 65.Kim HJ, Lee HJ, Kim H, Cho SW, Kim JS. Targeted genome editing in human cells with zinc finger nucleases constructed via modular assembly. Genome Res. 2009;19:1279–1288. doi: 10.1101/gr.089417.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Perez EE, Wang J, Miller JC, Jouvenot Y, Kim KA, Liu O, Wang N, Lee G, Bartsevich VV, Lee YL, Guschin DY, Rupniewski I, Waite AJ, Carpenito C, Carroll RG, JSO, Urnov FD, Rebar EJ, Ando D, Gregory PD, Riley JL, Holmes MC, June CH. Establishment of HIV-1 resistance in CD4(+) T cells by genome editing using zinc-finger nucleases. Nat Biotechnol. 2008;26:808–816. doi: 10.1038/nbt1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cost GJ, Freyvert Y, Vafiadis A, Santiago Y, Miller JC, Rebar E, Collingwood TN, Snowden A, Gregory PD. BAK and BAX deletion using zinc-finger nucleases yields apoptosis-resistant CHO cells. Biotechnology and Bioengineering. 105:330–340. doi: 10.1002/bit.22541. [DOI] [PubMed] [Google Scholar]
- 68.Liu P-Q, Chan EM, Cost GJ, Zhang L, Wang J, Miller JC, Guschin DY, Reik A, Holmes MC, Mott JE, Collingwood TN, Gregory PD. Generation of a triple-gene knockout mammalian cell line using engineered zinc-finger nucleases. Biotechnology and Bioengineering. 106:97–105. doi: 10.1002/bit.22654. [DOI] [PubMed] [Google Scholar]
- 69.Santiago Y, Chan E, Liu PQ, Orlando S, Zhang L, Urnov FD, Holmes MC, Guschin D, Waite A, Miller JC, Rebar EJ, Gregory PD, Klug A, Collingwood TN. Targeted gene knockout in mammalian cells by using engineered zinc-finger nucleases. Proc Natl Acad Sci U S A. 2008;105:5809–5814. doi: 10.1073/pnas.0800940105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lombardo A, Genovese P, Beausejour CM, Colleoni S, Lee Y-L, Kim KA, Ando D, Urnov FD, Galli C, Gregory PD, Holmes MC, Naldini L. Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat Biotech. 2007;25:1298–1306. doi: 10.1038/nbt1353. [DOI] [PubMed] [Google Scholar]


