Abstract
Cytosolic double-stranded DNA (dsDNA) triggers type-I interferon production through the endoplasmic reticulum adaptor protein STING (also known as MITA, MPYS and ERIS), which activates the transcription factor IRF3. However, how STING activates IRF3 remains largely unknown. Here we show that STING stimulates IRF3 phosphorylation by the kinase TBK1 in an in vitro reconstitution system. Using this system, we identified a carboxyl terminal region of STING that is both necessary and sufficient to activate TBK1 and stimulate IRF3 phosphorylation. Interestingly, we found that STING interacts with both TBK1 and IRF3, and that mutations in STING that selectively disrupt its binding to IRF3 abrogate IRF3 phosphorylation without impairing TBK1 activation. These results suggest that STING functions as a scaffold to specify and promote IRF3 phosphorylation by TBK1. The scaffolding function of STING and other adaptors may explain why IRF3 is activated only in a subset of signaling pathways that activate TBK1.
INTRODUCTION
Innate immunity is the first line of host defense against invasion by microbial pathogens, including viruses, bacteria, and parasites. The detection of pathogens occurs via the recognition of molecular features of microorganisms known as pathogen-associated molecular patterns (PAMPs) through a set of host pattern recognition receptors (PRRs)(1). Viral RNA is recognized by membrane-bound Toll-like receptors (TLRs) in the endosome in specialized cells or by RIG-I like receptors (RLRs) in the cytosol of most, if not all, cells infected by virus (2). RLRs, including RIG-I, MDA5 and LGP2, are essential for cytosolic viral recognition. Upon viral recognition, RIG-I and MDA5 interact with and activate the adaptor protein MAVS (also known as IPS-1, CARDIF, and VISA) (3–6). Localized mainly on the outer mitochondrial membrane, MAVS triggers a cytosolic signaling cascade leading to the activation of the transcription factors IRF3 and NF-κB, which in turn induce the production of antiviral and proinflammatory cytokines, including type I interferons (IFN-α and IFN-β)(7).
Accumulation of foreign or self DNA in the cytosol can also induce potent innate immune responses (8). Upon infection with DNA viruses or bacteria, the microbial DNA is delivered into the cytosol of mammalian cells, and this cytosolic DNA is detected and triggers an immune response against the pathogen. The sensors for cytosolic DNA have been actively pursued in recent years. While TLR9 is known to detect unmethylated CpG DNA in the endolysosome, the detection of cytosolic DNA is TLR9-independent (9). Recent studies have shown that DNA-dependent RNA polymerase III (Pol-III) converts AT-rich DNA, such as poly(dA-dT), into an RNA species that triggers the RIG-I pathway (10–11). However, in some cell types, such as macrophages and dendritic cells, non-AT-rich DNA can induce IFN-β through a Pol-III independent mechanism (12). Although DNA-dependent activator of IFN (DAI) has been suggested as a sensor of cytosolic DNA (13), DAI-deficient mice still produce interferons in response to B-form DNA and have innate and adaptive immune responses similar to wild-type mice (14). Recent studies suggest that IFI-16, one of the PYHIN family proteins, which contain a pyrin domain and two DNA-binding HIN domains, is an intracellular DNA sensor that induces IFN-β; however, it remains to be determined whether IFI-16 is a cytosolic DNA sensor in vivo (15). Another PYHIN protein, AIM2, is a cytosolic DNA sensor that activates the inflammasome and caspase-1. Current evidence suggests that AIM2 is not involved in the induction of type I interferons by cytosolic DNA (8). More recently, the DNA helicase DDX41 has been shown to function as a DNA sensor that induces type-I interferons in dendritic cells (16).
The endoplasmic reticulum (ER)-localized protein STING (stimulator of interferon genes, also known as MITA, MPYS and ERIS) is critical for regulating the production of IFN in response to cytoplasmic DNA (17–20). STING-deficient cells fail to induce type I IFN production in response to transfection with dsDNA or infection with herpes simplex virus 1 (HSV-1) or Listeria monocytogenes (21). Furthermore, in response to dsDNA stimulation, STING relocalizes from ER to Golgi and assembles into punctate structures together with the kinase TBK1(21–22). It is likely that this recruitment induces TBK1 activation, which results in the phosphorylation of IRF3 and the transcription of type I IFN genes. However, how STING activates TBK1 and IRF3 remains largely unknown.
To dissect the biochemical mechanisms of IRF3 activation by STING, we developed a cell-free system in which STING activates IRF3 in the cytosol. Using this system, we demonstrate that the carboxyl terminus of STING containing just 39 amino acids is necessary and sufficient to activate TBK1. Furthermore, we show that upon DNA stimulation, STING not only activates TBK1 but also recruits IRF3 to TBK1 to activate the IRF3 pathway. Thus, a key role of STING in the DNA signaling pathway is to specify and promote IRF3 phosphorylation by TBK1. These results suggest a new approach to modulate IRF3 functions without affecting other targets of TBK1.
RESULTS
STING Activates IRF3 in an in vitro Reconstitution System
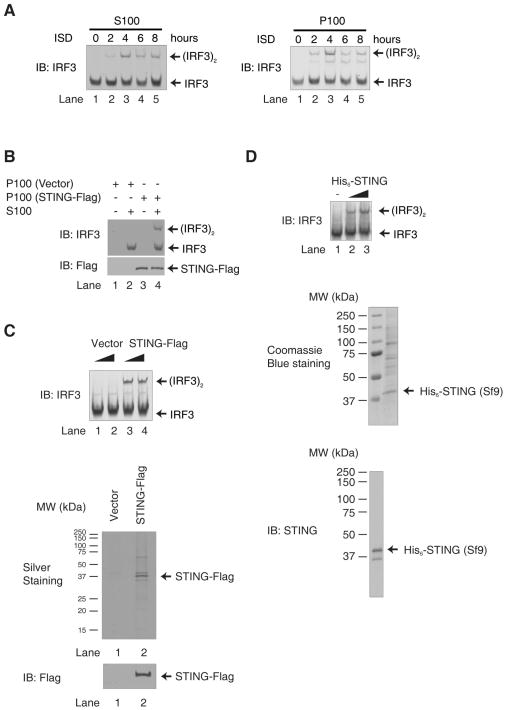
A double-stranded DNA containing 45 nucleotides, known as interferon stimulatory DNA (ISD), induces IFNβ in several cell types through a mechanism dependent on STING, but not RIG-I, MDA5 or MAVS (15, 23). We found that transfection of ISD in the murine fibrosarcoma cell line L929 stimulated IRF3 dimerization, a hallmark of IRF3 activation (Fig. 1A, left panel). To understand how STING, which is localized on the endoplasmic reticulum (ER), activates IRF3 in the cytosol, we established an in vitro assay in which high-speed membrane pellets (P100; containing ER) isolated from ISD-transfected L929 cells were incubated with the cytosolic extracts (S100) from untreated L929 cells in the presence of ATP. P100 from cells transfected with ISD for 2–8 hours were able to cause IRF3 dimerization in the presence of S100, whereas P100 from untransfected cells had no activity (fig. S1A and Fig. 1A, right panel). This activity depends on STING, because P100 from cells depleted of STING by RNAi lost the ability to activate IRF3 (fig. S1, B and C). Consistent with other recent reports (24), we found that STING was dispensable for IRF3 activation in L929 cells by Sendai virus, an RNA virus (fig. S1D).
Figure 1. STING Activates IRF3 in an in vitro Reconstitution System.
A. ISD activates IRF3 in L929 cells. L929 cells were transfected with ISD for the indicated time. The cytosolic supernatant (S100) was analyzed by native gel electrophoresis to observe the dimerization of endogenous IRF3 (left panel). The high-speed membrane fraction (P100) isolated from ISD stimulated cells was incubated with cytosolic extracts from untreated L929 (S100), and the dimerization of IRF3 was analyzed by native gel electrophoresis (right panel). IB: Immunoblot
B. IRF3 activation by the membrane fraction (P100) of STING-expressing cells. Cytosolic extracts from HeLa cells (S100) were incubated with P100 from HEK293T cells overexpressing STING-Flag. IRF3 dimerization was analyzed by native gel electrophoresis.
C & D. IRF3 activation by STING. STING-Flag (C) and His6-STING (D) were expressed in and purified from HEK293T and Sf9 cells, respectively, and then incubated with HeLa S100 and ATP followed by native gel electrophoresis. Aliquots of the proteins were stained by silver or Coomassie Blue, or by immunoblotting.
Overexpression of STING in cells is known to cause IRF3 dimerization and IFNβ induction (17, 19). Similarly, in our in vitro assay, incubation of P100 from STING-overexpressing HEK293T cells with cytosolic extracts from untreated HeLa cells led to IRF3 dimerization (Fig. 1B, lane 4). In contrast, P100 from the vector-transfected cells had no activity (lane 2). To determine if STING is responsible for this activity, we purified STING-Flag protein from the transfected cells using the Flag antibody affinity resin and incubated it with HeLa cytosolic extracts. The STING protein caused IRF3 dimerization in the extracts (Fig. 1C, top; lanes 3& 4). We also expressed recombinant full-length His6-STING in the insect cells Sf9 and purified the protein using nickel affinity resin. Although it proved difficult to purify the full-length STING protein possibly due to the presence of multiple membrane spanning domains, it was evident that STING expressed in Sf9 cells was capable of activating IRF3 in ATP-supplemented cytosolic extracts (Fig. 1D, top; lanes 2 & 3).
STING C-terminus is Important for IRF3 Activation in vitro
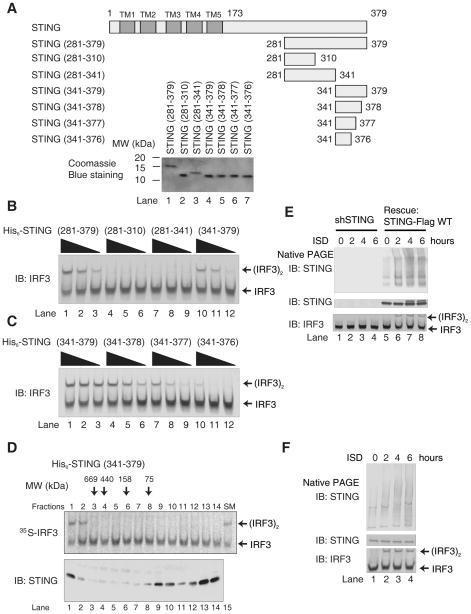
To delineate the domains of STING that are required for IRF3 activation, we first expressed His6-STING (181–379), which lacks all transmembrane (TM) domains, in Sf9 cells (fig. S2A). After purification, His6-STING (181–379) still contained a small amount of contaminating proteins (fig. S2B). Nevertheless, it was evident that His6-STING (181–379) was capable of activating IRF3 in cytosolic extracts (fig. S2C), suggesting that the TM domains of STING were dispensable for IRF3 activation in this in vitro assay. Since deletion of the TM domains is known to prevent IRF3 activation in cells (17), a fraction of the recombinant STING fragment may be ectopically activated by overexpression, bypassing the requirement for the TM domains (see below).
We constructed a series of deletion mutants of STING and expressed them in E. coli (Fig. 2A). After purification, the protein fragments were incubated with ATP-supplemented cytosolic extracts from HeLa cells (Fig. 2, B and C). Immunoblotting for IRF3 showed that the STING fragment spanning residues 281–379 was able to cause IRF3 dimerization (Fig. 2B, lanes 1–3). IRF3 dimerization was confirmed using in vitro translated [35S]-IRF3 as a substrate (fig. S2D) and by detecting phosphorylated IRF3 with an antibody specific for phosphoserine 396 (fig. S2E), a residue known to be important for IRF3 activation(25). Furthermore, mutation of two other serines to alanine at the C terminus of IRF3, Ser385 and Ser386, which have also been shown to be phosphorylated in response to stimulation and are important for IRF3 activation (26), abolished the dimerization of IRF3 in vitro (fig. S2F). Taken together, these results show that recombinant STING (281–379) purified from E. coli induces the phosphorylation and subsequent dimerization of IRF3.
Figure 2. STING C-terminus is Important for IRF3 Activation in vitro.
A. Diagrams of full-length and truncated STING used in this study. TM, transmembrane domain. His6-tagged proteins expressed in and purified from E. coli were analyzed by Coomassie Blue staining (lower panel).
B & C. His6-STING deletion mutants were incubated with ATP-supplemented HeLa S100, and then IRF3 dimerization was analyzed by native gel electrophoresis.
D. His6-STING (341–379) was fractionated by gel filtration on Superdex-200. Each fraction was analyzed by IRF3 dimerization assay (upper panel) and immunoblotting (lower panel). SM: starting material.
E. L929 cells stably expressing mouse STING shRNA with or without the simultaneous expression of wild type human STING-Flag were transfected with ISD for the indicated time and then membrane fractions (P100) were resolved by native-PAGE or SDS-PAGE followed by immunoblotting using a STING antibody. Cytosolic extracts (S100) were also immunoblotted with IRF3 antibody after native PAGE.
F. Similar to E, except that mouse macrophage RAW 264.7 cells were transfected with ISD to detect the aggregation of endogenous STING.
Interestingly, a STING fragment containing only 39 residues (341–379) was sufficient to activate IRF3 in the cytosolic extracts (Fig. 2B, lanes 10–12). Further deletion of just 3 amino acids from the C-terminus abrogated this activity (Fig. 2C, lanes 10–12). When His6-STING (341–379) was fractionated by gel filtration, a small amount of the protein eluted in the high molecular weight fractions (Fig. 2D, lanes 1–2). Strikingly, only the high molecular weight fractions were able to activate IRF3 when incubated with the cytosolic extracts. Furthermore, native gel electrophoresis revealed a smear of high-molecular weight STING aggregates that appeared after 2 hours of ISD stimulation in L929 cells in which endogenous STING was knocked down and replaced with STING-Flag; this smear disappeared in cells depleted of STING by RNAi (Fig. 2E). The endogenous STING aggregates were also observed in mouse macrophage RAW 264.7 cells in response to ISD stimulation (Fig. 2F). These results suggest that STING may form aggregates to activate IRF3.
STING Directly Activates TBK1 in vitro
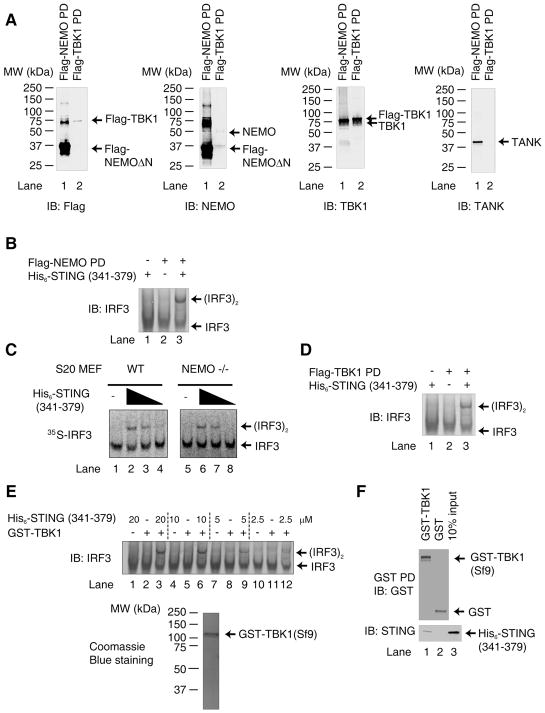
We next attempted to isolate the components of the cytosolic extract (S100) involved in STING-dependent IRF3 activation. Recent studies have shown that NEMO-deficient cells fail to activate IRF3 in response to infection by certain RNA viruses (27–28), suggesting that NEMO, which forms a complex with TANK and TBK1, is important for IRF3 activation in the RIG-I pathway. Because the majority of endogenous NEMO is incorporated into the IKK complex, which contains the catalytic subunits IKKα and IKKβ in addition to NEMO, we reconstituted NEMO deficient MEFs with Flag-NEMO that lacks the N-terminal 85 residues (Flag-NEMO ΔN) (28). Flag-NEMO ΔN cannot form a complex with IKKα or IKKβ but can still interact with TBK1 and TANK, thereby allowing us to pull down the NEMO/TANK/TBK1 complex (Fig. 3A). This complex, herein referred to as Flag-NEMO PD, supported IRF3 dimerization in the presence of His6-STING(341–379) and ATP (Fig. 3B; lane 3).
Figure 3. STING Directly Activates TBK1 in vitro.
A. MEF cells stably expressing Flag-NEMO-ΔN were used to isolate endogenous TBK1 by virtue of its association with Flag-NEMO-ΔN. The composition of this complex, denoted as Flag-NEMO PD, was analyzed by immunoblotting with the indicated antibodies. HEK293T cells stably overexpressing Flag-TBK1 were used to isolate recombinant TBK1 protein, denoted here as Flag-TBK1 PD.
B. Flag-NEMO PD, which contained endogenous TBK1 as shown in A, was incubated with His6-STING (341–379, ~2 μM) and His6-IRF3 (~40 nM) in the presence of ATP. IRF3 dimerization was analyzed by native gel electrophoresis.
C. S20 from wild type or NEMO deficient MEFs was incubated with varying concentrations of His6-STING (341–379) (0.375, 0.75 and 1.5 μM) together with 35S-IRF3. IRF3 dimerization was analyzed by native gel electrophoresis followed by autoradiography.
D. Similar to B, except that Flag-TBK1 pull-down (PD) complex was used.
E. GST-TBK1 was purified from Sf9 cells and analyzed by Coomassie blue staining (lower panel). The purified protein (200 nM) was incubated with His6-IRF3 (200 nM) and varying concentrations of His6-STING (341–379) in the presence of ATP, then IRF3 dimerization was analyzed by native PAGE.
F. GST-TBK1 or GST (1 μg) was incubated with 2 μg of His6-STING (341–379) and then pulled down with glutathione Sepharose, followed by immunoblotting. The input represents 10% of the amount of STING used in the pull-down experiments.
To determine whether NEMO is required for IRF3 activation by STING, S20 from wild type or NEMO-deficient MEFs was incubated with His6-STING (341–379) together with 35S-IRF3. As shown in Fig. 3C, the cytosolic extracts from both wild type (lanes 2–4) and NEMO-deficient (lanes 6–8) MEFs supported IRF3 activation by STING, suggesting that NEMO is dispensable for IRF3 activation by STING. Furthermore, overexpression of STING activated a luciferase reporter driven by interferon stimulation regulatory element (ISRE) even in NEMO-deficient MEFs, whereas MAVS failed to activate the reporter in the absence of NEMO but was able to activate the reporter when NEMO was restored (fig. S3). Thus, unlike the MAVS pathway (28), NEMO is not required for IRF3 activation by STING.
To determine the role of TBK1 in IRF3 activation by STING, lysates from HEK293T cells stably expressing Flag-TBK1 were subject to immunopurification using anti-Flag-M2 agarose. This TBK1 complex, herein referred to as Flag-TBK1 PD, which contained a small amount of NEMO (Fig. 3A), supported IRF3 dimerization in the presence of His6-STING (341–379) and ATP (Fig. 3D, lane 3). To examine whether TBK1 alone is required for this assay, GST-tagged TBK1 was expressed in and purified from Sf9 cells (Fig. 3E, lower panel). It has been shown previously that TBK1 purified from Sf9 cells is capable of phosphorylating IRF3 if used in excess in vitro (29); therefore, we used a limiting amount of TBK1 (200 nM) and titrated the amounts of His6-STING (341–379) used in this assay to observe STING-dependent TBK1 activation (Fig. 3E, top panel). GST-TBK1 alone did not induce IRF3 activation; however, in the presence of increasing amounts of His6-STING (341–379), GST-TBK1 caused IRF3 dimerization (lanes 3, 6 and 9). We also found that GST-TBK1, but not GST, bound to His6-STING (341–379) in the GST pull-down assay (Fig. 3F). Taken together, these results suggest that the C-terminus of STING binds to and activates TBK1 to phosphorylate IRF3.
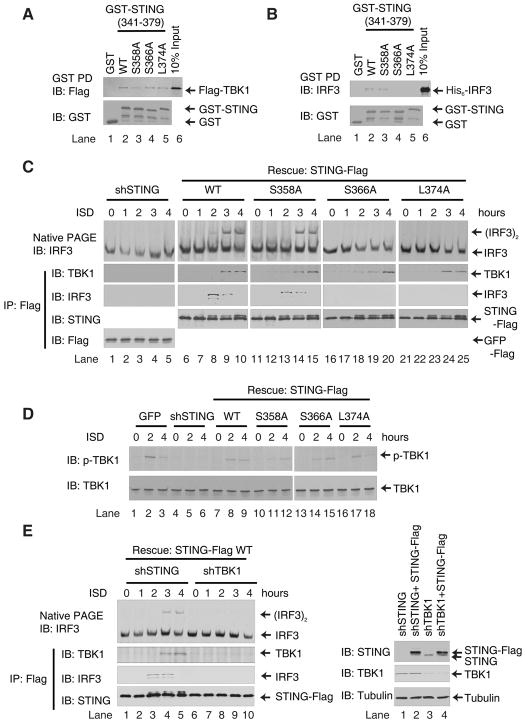
Ser366 and Leu374 of STING are Important for IRF3 Activation
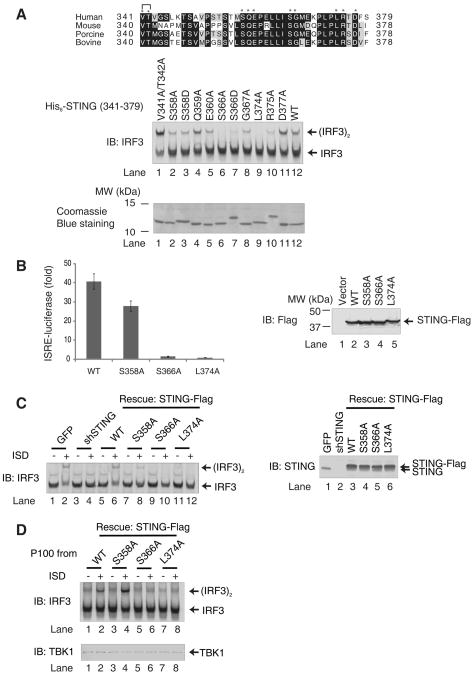
Sequence analyses of the STING protein revealed that the carboxyl-terminal amino acids are highly conserved across species, suggesting that this region has biological importance (Fig. 4A). To determine the residues of STING (341–379) that are important for IRF3 activation, we constructed a series of STING (341–379) mutants in which the conserved residues were replaced by alanine (V341A and T342A, S358A, Q359A, E360A, S366A, G367A, L374A, R375A, or D377A). We also constructed STING mutants that mimic phosphorylation of serine residues (S358D and S366D). These mutants were expressed in and purified from E. coli and tested for their ability to activate IRF3 in vitro (Fig. 4A). Two mutations, S366A and L374A, completely abolished IRF3 dimerization (lanes 6 and 9), whereas other mutations were largely tolerated. Notably, the S358A mutant (lane 2), which has been previously reported to be functionally defective (19), was partially active in causing IRF3 dimerization in our in vitro assay. The S366D mutant was also largely inactive (lane 7), suggesting that the functional defect of the Ser366 mutation may not be due to a phosphorylation defect.
Figure 4. Ser366 and Leu374 of STING are Important for IRF3 Activation.
A. Sequence alignment of the carboxy-termini of human, mouse, porcine and bovine STING using Clustal W2. Asterisks indicate residues that were mutated in this study. WT and mutated STING fragments were expressed in and purified from E. coli as His6-tagged proteins and analyzed by Coomassie blue staining (lower panel). Each protein (2 μM) was tested in IRF3 dimerization assay (upper panel).
B. HEK293T cells were transiently transfected with full-length STING-Flag (WT) and its mutants (S368A, S366A and L374A) together with an ISRE-luciferase reporter (left panel). The error bars represent the variation ranges of duplicate experiments. The expression of STING proteins was confirmed by immunoblotting (right panel).
C. L929 cells stably expressing mouse STING shRNA with or without the simultaneous expression of wild type or mutant (S358A, S366A and L374A) human STING-Flag were transfected with ISD for 4 hours. The dimerization of endogenous IRF3 was analyzed by native gel electrophoresis (left panel). The expression endogenous and rescued STING proteins were confirmed by immunoblotting (right panel).
D. L929 cells in which endogenous STING was replaced with wild type or mutant STING-Flag as shown in (C) were transfected with ISD or mock treated. Membrane pellets (P100) from these cells were incubated with His6-IRF3 in the presence of ATP, and IRF3 dimerization was analyzed by native PAGE (upper panel). Each P100 fraction used in this assay was also blotted with a TBK1 antibody (lower panel).
To determine whether Ser358, Ser366 and Leu 374 of STING are important for its function in cells, we transfected HEK293T cells with full-length STING containing the point mutations (S358A, S366A and L374A) together with the ISRE-luciferase reporter plasmid (Fig. 4B). Whereas wild-type STING activated the ISRE-luciferase reporter, the STING S366A and L374A mutants were completely defective. The S358A mutant exhibited 75% of the activity of the WT protein. Collectively, these results indicate that residues Ser366 and Leu374 of STING are important for IRF3 activation.
We further investigated the role of STING C-terminal residues in ISD-induced IRF3 activation by complementation experiments in which endogenous STING in L929 cells was replaced by WT and mutant STING. L929 cells stably expressing shRNA against STING or expressing a GFP control were transfected with ISD for 4 hours, and then cytosolic extracts were prepared to measure IRF3 dimerization (Fig. 4C). RNAi of STING abolished ISD-induced IRF3 dimerization (lane 4), which was rescued by restoring the expression of WT STING (lane 6), but not the S366A or L374A mutant (lanes 10 and 12). S358A STING partially rescued IRF3 dimerization in these cells (lane 8). Consistent with these results, membrane pellets (P100) isolated from ISD-stimulated L929 cells expressing WT or S358A STING were capable of causing IRF3 dimerization, whereas those from cells expressing S366A or L374A were inactive (Fig. 4D). These results demonstrate that Ser366 and Leu374 of STING are essential for ISD-induced IRF3 activation in cells.
STING Recruits IRF3 to TBK1 to Induce IRF3 Activation
To explore the mechanism by which IRF3 is activated by wild type and S358A STING, but not S366A or L374A STING, we examined the interaction between STING and TBK1 or IRF3 in vitro. Wild type and mutant GST-STING (341–379) as well as GST were incubated with Flag-TBK1 or His6-IRF3 and then pulled down with glutathione-Sepharose and analyzed by immunoblotting. Wild type STING and all of the mutant STING proteins associated with Flag-TBK1 in vitro (Fig. 5A). Interestingly, wild type and S358A STING, but not S366A or L374A STING, associated with His6-IRF3 (Fig. 5B). Thus, defective binding to IRF3 may underlie the inability of S366A and L374A STING to promote IRF3 phosphorylation by TBK1.
Figure 5. STING Recruits IRF3 to TBK1 to Induce IRF3 Activation.
A & B. Wild type or mutant GST-STING (341–379) or GST was incubated with Flag-TBK1 (A) or His6-IRF3 (B) and then pulled down with glutathione Sepharose followed by immunoblotting. The input represents 10% of TBK1 or IRF3 used in the pull-down experiments.
C. L929 cells stably expressing STING shRNA and those in which endogenous STING was replaced with WT or mutated STING-Flag were stimulated with ISD for the indicated lengths of time. Cell lysates were immunoprecipitated with anti-Flag agarose and then immunoblotted with the indicated antibodies. Aliquots of the cell lysates were immunoblotted for IRF3 following native PAGE (top).
D. Similar to C, except that cell lysates were resolved by SDS-PAGE, followed by immunoblotting with antibodies against TBK1 or TBK1 phosphorylated at Ser172.
E. L929 cells, in which STING or TBK1 was depleted by shRNA, were engineered to express STING-Flag using a lentiviral vector. The cells were transfected with ISD for the indicated lengths of time, and then the STING complex was immunoprecipitated using anti-Flag agarose and analyzed by immunoblotting with the indicated antibodies. Aliquots of the cell lysates were immunoblotted with an IRF3 antibody following native PAGE (top).
Finally, we examined the interactions between STING, IRF3 and TBK1 in L929 cells in which endogenous STING was replaced with full-length STING harboring different mutations. Following ISD stimulation, WT and S358A STING were able to interact with both TBK1 and IRF3 (Fig. 5C, lanes 6–15). On the other hand, S366A and L374A STING interacted with TBK1 but not IRF3 (lanes 16–25). These observations were confirmed by immunostaining in cells. Confocal fluorescence microscopy revealed that two hours after ISD stimulation, IRF3 had translocated into the nuclei in L929 cells in which endogenous STING was replaced with WT and S358A STING, but not S366A and L374A STING. At this time, WT and all mutant STING proteins formed punctuate-like aggregates at perinuclear regions and co-localized with TBK1 (fig. S4, A and C). These results suggest that IRF3 translocates into the nucleus promptly after phosphorylation by TBK1. To trap the complex of STING and IRF3, we expressed HA-tagged IRF3 mutated in S385A and S386A (2A) by retroviral transduction. This mutant did not translocate into the nucleus after ISD stimulation and appeared to co-localize with WT andS358A STING, but not S366A and L374A STING (fig. S4B). In contrast, WT and all mutant STING proteins appeared to co-localize with TBK1 after ISD stimulation (fig. S4C).
Consistent with the confocal staining data, in cells expressing S366A and L374A STING, ISD induced phosphorylation of TBK1 at Ser172, a residue known to be important for TBK1 activation (Fig. 5D, lanes 13–18) (30). In contrast, IRF3 dimerization was defective in these cells (Fig. 5C, lanes 16–25). This uncoupling of TBK1 activation and IRF3 phosphorylation was recapitulated in our in vitro assay, in which WT or mutated His6-STING (341–379) proteins were incubated with ATP supplemented cytosolic extracts from HeLa cells (fig. S5). The WT and S358A STING fragments, but not those containing S366A or L374A, were able to support IRF3 phosphorylation. On the other hand, all of these proteins stimulated TBK1 phosphorylation. The inability of STING S366A and L374A mutants to support IRF3 phosphorylation was not due to a defect in STING aggregation, as a fraction of these proteins still eluted from the gel filtration column as high molecular weight species (fig. S6A). Moreover, full-length STING harboring the S366A or L374A mutation still formed punctate-like aggregates in L929 cells stimulated with ISD (fig. S4). Native gel electrophoresis showed that WT and all mutant STING proteins formed high molecular weight aggregates after stimulation of L929 cells with ISD (fig. S6B). Taken together, these results suggest that S366A and L374A mutations in STING selectively disrupt its interaction with IRF3 but not TBK1, thereby blocking IRF3 phosphorylation by TBK1.
Interestingly, all STING proteins underwent a gel mobility shift after 2 hours of ISD stimulation (e.g., Fig. 5C, lanes 8–10; fig. S7A). This shift was abolished by treatment with calf intestinal phosphatase (CIP), indicating STING phosphorylation (fig. S7B). In contrast to ISD stimulation, Sendai virus infection did not cause a detectable mobility shift of STING (fig. S7C). These results suggest that STING is specifically phosphorylated in cells stimulated with ISD. The phosphorylation of STING appeared to precede that of IRF3 (Fig. 5C). Interestingly, depletion of TBK1 by RNAi prevents the association between STING and IRF3 as well as the phosphorylation of STING (Fig. 5E and fig. S7D). These results suggest that TBK1 phosphorylates STING before IRF3, and that phosphorylation of STING may strengthen its interaction with IRF3, which further promotes IRF3 phosphorylation by TBK1.
To map the sites of STING phosphorylation induced by ISD, L929 cells in which endogenous STING was replaced with human STING-Flag were stimulated with ISD for 4 hours, and then STING protein was affinity purified and analyzed by tandem mass spectrometry. In addition to Ser358, we detected phosphorylation of Ser353 and Ser379 of STING in ISD stimulated cells (fig. S7E). However, we found no evidence of phosphorylation of Ser366 in vitro or in vivo. Consistent with this result, S366A STING was phosphorylated in cells stimulated with ISD (Fig. 5C, lanes 18–20). Further, mimicking the phosphorylation of Ser366 with aspartic acid (S366D) did not rescue the activity of STING in vitro (Fig. 4A, lane 7). Therefore, currently there is no evidence of phosphorylation of STING at Ser366. It is possible that, like L374A, the S366A mutation impairs the interaction between STING and IRF3, but not TBK1. These mutations uncouple the roles of STING in activating TBK1 and specifying IRF3 phosphorylation, thereby revealing the mechanism by which IRF3 is selectively activated in some but not all pathways that stimulate TBK1 (see Discussion and Fig. 6).
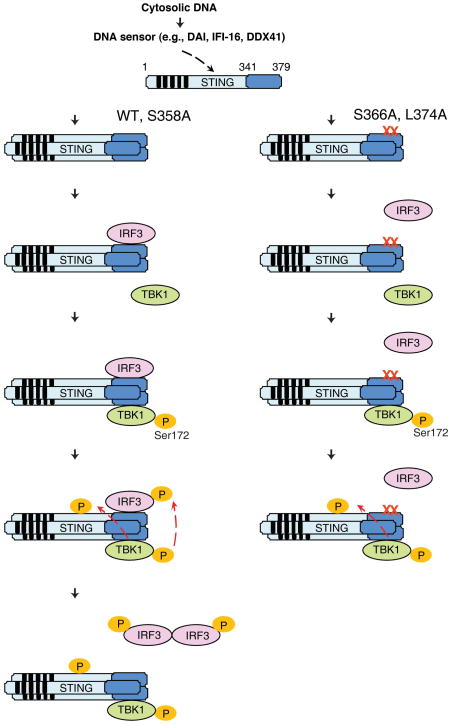
Figure 6. A model of IRF3 Activation by STING.
Following detection of cytosolic DNA with a DNA sensor, STING forms oligomers on ER or other intracellular membranes. The C-terminus of STING then recruits IRF3 and TBK1, facilitating IRF3 phosphorylation by TBK1 (left). The mutation of STING at Ser366 or Leu374 does not impair its ability to recruit and activate TBK1, as evidenced by the phosphorylation of TBK1 and STING in ISD-stimulated cells harboring these mutations (right). However, S366A or L374A mutation abolishes the interaction between STING and IRF3, thereby preventing IRF3 phosphorylation by TBK1.
DISCUSSION
It is commonly assumed that activation of a protein kinase is synonymous to phosphorylation of its substrates. This assumption has been applied to many kinase assays that rely on detection of activated kinases with phospho-specific antibodies or with surrogate substrates rather than physiological substrates. However, accumulating evidence suggests that activation of a kinase does not equate to phosphorylation of a physiological substrate. Recent studies of TBK1 provide a striking example of the uncoupling of protein kinase activation and substrate phosphorylation (30–31). It is now well established that TBK1 is responsible for IRF3 phosphorylation in response to stimulation of several intracellular receptors that induce type-I interferons, including TLR3, TLR4, RIG-I, MDA5 as well cytosolic DNA sensors that signal through STING. IRF3 is not activated in inflammatory pathways triggered by other TLRs, such as TLR2 and TLR5, or by receptors for TNF or IL-1, all of which are known to activate NF-κB. However, TLR ligands and IL-1β could activate TBK1, raising the question of why activated TBK1 does not lead to IRF3 phosphorylation (30–31). The answer to this question may lie in the fact that signaling pathways that lead to IRF3 activation engage specific adaptor proteins, such as TRIF for TLR3 and TLR4, MAVS for RIG-I and MDA5, and STING for the DNA sensing pathways.
Through in vitro reconstitution of STING-dependent IRF3 activation, we obtained evidence that may explain why IRF3 is activated only in a subset of pathways that stimulate TBK1. We found that STING binds to both TBK1 and IRF3 and that this binding is mediated through a short C-terminal fragment of STING (Fig. 6). Importantly, we identified two residues, Ser366 and Leu374, at the C-terminus of STING that are dispensable for TBK1 binding and activation, but required for IRF3 binding and phosphorylation. These results indicate that STING not only mediates TBK1 activation, but also specifies IRF3 for phosphorylation by TBK1. Although IL-1 and TNF can also stimulate TBK1, these ligands do not activate STING, hence no IRF3 phosphorylation. It will be very interesting to determine whether other adaptors such as MAVS and TRIF also activate IRF3 through the dual mechanism of stimulating TBK1’s catalytic activity and specifying IRF3 for phosphorylation by TBK1.
Burdette et al recently showed that STING binds directly to cyclic-di-GMP, which is known to induce type-I interferons (32). A mouse STING mutant containing three substitutions at the C-terminus, S357A, E359A and S365A (equivalent to human STING S358A, E360A, S366A), retained less than 10% of IFNβ inducing activity of the wild type protein when it is overexpressed. Further, ectopic expression of this mutant in HEK293 cells did not confer IFNβ induction in response to cyclic-di-GMP. By knocking down endogenous STING in L929 cells and replacing it with human STING, we demonstrated that a single S366A mutation abolished its ability to bind and activate IRF3 following ISD stimulation (Figure 5C).
In our in vitro reconstitution system, only three proteins - TBK1, IRF3 and a C-terminal fragment of STING - are necessary and sufficient for STING-dependent IRF3 phosphorylation. This simple system is in contrast to MAVS-dependent phosphorylation of IRF3, which requires K63 polyubiquitination as well as detection of polyubiquitin chains by NEMO (28). Consistent with this disparity, NEMO is required for IRF3 activation by MAVS, but not STING, in cells (fig. S3). Unlike MAVS, which contains several binding sites for TRAF proteins, including TRAF2, TRAF3 and TRAF6, STING contains multiple transmembrane domains but no apparent TRAF binding motifs. Thus, STING activates TBK1 and promotes IRF3 phosphorylation through a rather simple mechanism, namely by direct binding to TBK1 and IRF3. However, our results do not mean that ubiquitination is not involved in the cytosolic DNA signaling pathway. In fact, it has been reported that TRIM56 ubiquitinates STING to modulate its ability to induce immune responses against intracellular DNA(33). Therefore, it is possible that ubiquitination may control a step upstream of or at the level of STING activation.
The mechanism of STING activation by cytosolic DNA remains to be elucidated. Several DNA sensors have been proposed, including DAI, RNA polymerase III and IFI-16 and DDX41(16). Further genetic studies are required to further establish the role of these and other potential DNA sensors in type-I interferon induction (12). In any case, it has been demonstrated that STING is indispensable for interferon induction by cytosolic DNA in many cell types including macrophages (21). Interestingly, microscopic studies have shown that STING forms punctate-like cytoplasmic structures. Previous studies have demonstrated that the TM domains of STING are essential for it to activate IRF3 and induce IFN (17, 19). Indeed, it has been shown that DDX41 interacts with STING in the region spanning the second to fourth transmembrane domains of STING (16). Surprisingly, we found that the C-terminal fragment of STING containing just 39 amino acids is sufficient to support IRF3 phosphorylation by TBK1 in vitro. Interestingly, a fraction of the recombinant STING fragment eluted from the gel filtration column as high molecular weight species, and only these species were capable of promoting IRF3 phosphorylation by TBK1. Native gel electrophoresis also suggests that endogenous STING forms aggregates following ISD stimulation. Thus, it is possible that the detection of cytosolic DNA leads to aggregation and activation of STING on ER or other membranes, and that the TM domains of STING are required for its aggregation. Since a fraction of the recombinant STING fragment already forms active aggregates, the TM domains are no longer required. This mechanism is analogous to that of the activation of MAVS, which forms very large aggregates in response to RNA virus infection (34). The mitochondrial TM domain of MAVS is required for it to form these functional aggregates in cells in response to viral infection. In the in vitro experiments, however, MAVS lacking the TM domain is active because a fraction of the recombinant protein already forms high molecular weight aggregates. The MAVS aggregates are highly potent in activating the IRF3 signaling cascade through a prion-like mechanism (34). Thus far, we have not obtained any evidence that STING forms similar prion-like aggregates in the cytosolic DNA signaling pathway.
Another potential mechanism of STING regulation is suggested by our observation that STING is phosphorylated in response to stimulation by cytosolic DNA. This phosphorylation of STING depends on TBK1. Previous studies have shown that STING is phosphorylated at Ser358 by TBK1 in response to Sendai virus infection (19). Our mass spectrometry analysis confirmed the phosphorylation of STING at Ser358 following ISD stimulation. However, the mutation of Ser358 to alanine only partially impaired the ability of STING to rescue IRF3 activation in STING-deficient cells. In contrast, the mutation at Ser366 or Leu374 of STING completely abolished IRF3 activation by ISD.
Although we have not detected STING phosphorylation at Ser366 and cannot rule out a role for the phosphorylation of this residue, STING mutants bearing the S366A or L374A mutation are still phosphorylated in response to ISD stimulation. The phosphorylation of STING and TBK1 in cells expressing the S366A or L374A mutant suggest that TBK1 is activated in these cells. Therefore, S366A and L374A mutations selectively inhibit IRF3 phosphorylation by activated TBK1.
Our results not only provide a mechanism of specific phosphorylation of IRF3 in the cytosolic DNA sensing pathway, but also suggest that it is possible to identify inhibitors that selectively inhibit IRF3 without affecting the phosphorylation of other TBK1 substrates. Such inhibitors may prove useful for treating certain human diseases. For example, mice lacking IRF3 or interferon receptors are resistant to the lethal effects of infection by the bacterium Listeria monocytogenes or by the parasite Plasmodium falciparum, which causes malaria, the world’s most common infectious disease (12, 35).
MATERIALS AND METHODS
Reagents and Standard Methods
The antibody against STING was generated by immunizing rabbits with the recombinant His6-STING 281–379 protein produced in E. coli. This antibody was affinity purified using a STING antigen column. The other antibodies used were obtained from the following vendors: Santa Cruz Biotech (anti-human IRF3 and anti-NEMO); Sigma (anti-Flag M2, M2-conjugated agarose, anti-HA and anti-tubulin); Cell Signaling (anti-p-IRF3 Ser396 and anti-p-TBK1 Ser172); Invitrogen (anti-mouse IRF3; anti-GST); IMGENEX (anti-TBK1); BioVision (anti-TANK); and Jackson ImmunoResearch Laboratories (anti-mouse Texas Red and anti-rabbit FITC).
ISD was prepared from equimolar amounts of sense DNA oligo, TAC AGA TCT ACT AGT GAT CTA TGA CTG ATC TGT ACA TGA TCT ACA, and the corresponding antisense oligo. The oligos were annealed at 75°C for 30 min before being cooled to room temperature (23). Calf intestine alkaline phosphatase (CIP) was purchased from New England Biolabs. Other chemicals and reagents were from Sigma unless otherwise specified. Sendai virus (Cantell strain, Charles River Laboratories) was used at a final concentration of 100 hemagglutinating units per ml. The procedures for native gel electrophoresis for the detection of IRF3 dimerization, SDS-PAGE, and immunoblotting have been described previously (5).
Expression Constructs and Recombinant Proteins
The human STING plasmid, pcDNA3.1-hMPYS-HA, was kindly provided by Dr. John Cambier (National Jewish Medical and Research Center) (18). The cDNA encoding STING was subcloned into pcDNA3 (Invitrogen) in-frame with a C-terminal Flag tag for expression in mammalian cells. Various mutants were generated using the QuikChange Site-Directed Mutagenesis Kit (Stratagene).
For baculovirus-mediated expression in insect cells, cDNA encoding human STING was inserted into pDEST10 (Invitrogen) in-frame with an N-terminal His6-tag using the gateway system. Human TBK1 was cloned into pDEST20 (Invitrogen) with an N-terminal GST-tag. For expression in E. coli, cDNA encoding the various mutants of human STING was inserted into pDEST17 (Invitrogen) in-frame with an N-terminal His6-tag or into pDEST15 (Invitrogen) in-frame with an N-terminal GST-tag using the gateway system. His6-tagged proteins were purified by nickel affinity chromatography, and GST-tagged proteins were purified by glutathione affinity chromatography. His6- STING (341–379) was further fractionated on Superdex-200-pc equilibrated with Buffer E (20 mM Tris-HCl [pH 7.5], 100 mM NaCl, 10% glycerol, 1 mM EDTA, 0.1 mM PMSF, and 0.5 mM DTT) using the ETTAN system (GE Healthcare) as previously described (34).
Cell Culture, DNA Transfection, and Luciferase Reporter Assay
The cells were cultured at 37°C in an atmosphere of 5% (v/v) CO2. HEK293T, HEK293T-Flag-TBK1, and L929 cells were cultured in DMEM supplemented with 10% (v/v) cosmic calf serum (Hyclone) and antibiotics (100 U/ml penicillin and 100 mg/ml streptomycin). RAW 264.7 cells, NEMO-deficient MEFs and MEFs expressing Flag-NEMO that lack the N-terminal 85 residues (Flag-NEMO-ΔN) were cultured in DMEM supplemented with 10% (v/v) FBS (Atlanta) and antibiotics.
For ISD stimulation, L929 cells were transfected in 10-cm plates with 25 μg of ISD using Lipofectamine 2000 (Invitrogen). To measure the activity of WT or mutant STING, HEK293T cells were transfected with 1.8 μg of STING cDNA together with 100 ng ISRE-luciferase reporter and 100 ng pCMV-LacZ as an internal control using calcium phosphate precipitation. The luciferase activity was measured in duplicate on day 3 as previously described (5).
Purification of TBK1 Complexes
For purification of endogenous TBK1 complex, MEFs lacking NEMO or those reconstituted with Flag-NEMO-ΔN were lysed in Buffer A (10 mM Tris-HCl [pH 7.5], 10 mM KCl, 1.5 mM MgCl2, and a protease inhibitor cocktail [Roche]). Following centrifugation at 20,000 g for 30 min, the supernatants from these two types of cells were mixed at a ratio of 8:1, and the mixture was subjected to immunoprecipitation with anti-Flag M2 agarose at 4°C for 4 hours. The agarose beads were washed three times with Buffer A, and the proteins were eluted with 0.2 mg/ml Flag peptide in Buffer B (50 mM Tris-HCl [pH 7.5] and 0.1% CHAPS). The eluted proteins, which contain endogenous TBK1 from MEFs, were stored in Buffer C (20 mM Tris-HCl [pH 7.5] and 10% glycerol) after buffer exchange by repeated dilution and concentration. To purify recombinant Flag-TBK1 complex, HEK293T cells stably expressing human Flag-TBK1 were used to prepare cell lysates and Flag antibody affinity purification was carried out as above.
In vitro Assay for STING-Dependent IRF3 Activation
Biochemical assays for IRF3 activation using cytosolic extracts (S5, S20 and S100) and crude mitochondrial (P5) or ER membrane (P100) fractions of cultured cells were performed as described previously (28). Briefly, cell homogenates in Buffer A were centrifuged at 1,000 g for 5 min to pellet the nuclei. The post nuclear supernatant was further centrifuged at 5,000 g for 10 min to separate the mitochondria (P5) from the cytosolic supernatant (S5). The cytosolic supernatant (S5) was further centrifuged at 100,000 g or 20,000 g for 30 min to separate the membrane (P100 or P20) from the cytosolic supernatant (S100 or S20).
Each 10-μl assay for IRF3 activation contained P5 (or P100), 40 μg of S5 (or S20 or S100), and the indicated recombinant proteins with an ATP Buffer (20 mM HEPES-KOH [pH 7.0], 2 mM ATP, 5 mM MgCl2, and 0.25 M D-mannitol). After incubation at 30°C for 1 hour, the samples were subjected to native gel electrophoresis, and the dimerization of IRF3 was visualized by immunoblotting using the IRF3 antibody. For autoradiography, 35S-IRF3 or the radiolabeled mutant (S385A and S386A; denoted as 2A) was synthesized using the TNT Coupled Reticulocyte Lysate Kit (Promega) supplemented with 35S-methionine. 35S-IRF3 was mixed with the reaction, and IRF3 was visualized by autoradiography using a PhosphorImager (GE Healthcare).
Lentivirus-Mediated RNAi and Rescue with Transgene
The lentiviral knockdown vector, pTY-shRNA-EF1a-puroR-2a-Flag, was kindly provided by Dr. Yi Zhang (University of North Carolina at Chapel Hill) (36). The short-hairpin RNA (shRNA) sequences were cloned into this vector under the U6 promoter. To rescue the knockdown of mouse STING with human STING, STING cDNA and that of the STING mutants were placed downstream of the puromycin-resistance gene and the foot-and-mouth disease virus 2A segment, which enables the multicistronic expression of transgenes using a single promoter. After infection with lentivirus, the cells were selected with puromycin (2 μg/ml) to establish cells that stably express shRNA. The shRNA sequences are as follows (only the sense strand is shown): STING, 5′-GAGCTTGACTCCAGCGGAA-3′; and TBK1, 5′-TCAAGAACTTATCTACGAA-3′.
GST Pull-down Assay
For the binding among STING, TBK1 and IRF3, a GST-tagged protein was incubated with the His6-tagged or Flag-tagged protein in 100 μl buffer containing 10 mM Tris-HCl (pH 7.5), 150 mM NaCl, and 0.1% CHAPS at 4°C for 1 hour. Then, the GST-tagged proteins were pulled down with glutathione Sepharose, and the bound proteins were analyzed by immunoblotting.
Confocal Imaging
L929 cells in which endogenous STING was replaced with WT or mutated STING-Flag were plated onto cover slips in 24-well plates. On the next day, the cells were transfected with ISD for indicated times, and then washed with PBS and fixed in 3.7% formaldehyde in PBS for 15 min. Cells were permeabilized and blocked for 30 min at room temperature in a staining buffer containing Triton X-100 (0.2%) and BSA (3%), and then incubated with an antibody against Flag, IRF3, TBK1 or HA in the staining buffer for 1 hour. After washing three times in the staining buffer, cells were incubated with anti-mouse Texas Red or anti-rabbit FITC for 1 hour. The cover slips, which were washed extensively, were dipped once in water and mounted onto slides using mounting media (VectaShield with DAPI; Vector Laboratories). Imaging of the cells was carried out using Zeiss LSM510 META laser scanning confocal microscopy (Carl Zeiss MicroImaging, Inc).
Supplementary Material
Acknowledgments
We thank Jiaxi Wu, Xiang Chen and Chengzu Long for mapping STING phosphorylation sites by mass spectrometry. We also thank Dr. Inder Verma (Salk Institute) for providing the NEMO-deficient MEF cells, Dr. John Cambier (National Jewish Medical and Research Center) for pcDNA3.1-hMPYS-HA, and Dr. Yi Zhang (University of North Carolina at Chapel Hill) for the pTY lentiviral vector.
Funding: This work was supported by grants from NIH (RO1-GM63692 and RO1-AI93967), Cancer Prevention and Research Institute of Texas (RP110430), and the Welch Foundation (I-1389). Y.T. was supported by a fellowship from the Sankyo Foundation of Life Science. Z.J.C is an Investigator of Howard Hughes Medical Institute.
Footnotes
Author contributions: Y.T. performed the experiments; Y.T. and Z.J.C. analyzed the data and wrote the paper.
Competing interests: The authors declare that they have no competing interests.
REFERENCES AND NOTES
- 1.Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 2.Yoneyama M, Fujita T. RNA recognition and signal transduction by RIG-I-like receptors. Immunol Rev. 2009;227:54–65. doi: 10.1111/j.1600-065X.2008.00727.x. [DOI] [PubMed] [Google Scholar]
- 3.Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 4.Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 5.Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 6.Xu LG, Wang YY, Han KJ, Li LY, Zhai Z, Shu HB. VISA Is an Adapter Protein Required for Virus-Triggered IFN-beta Signaling. Mol Cell. 2005;19:727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 7.Rehwinkel J, Reise Sousa C. RIGorous detection: exposing virus through RNA sensing. Science. 2010;327:284–286. doi: 10.1126/science.1185068. [DOI] [PubMed] [Google Scholar]
- 8.Barber GN. Innate immune DNA sensing pathways: STING, AIMII and the regulation of interferon production and inflammatory responses. Curr Opin Immunol. 2011;23:10–20. doi: 10.1016/j.coi.2010.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ishii KJ, Coban C, Kato H, Takahashi K, Torii Y, Takeshita F, Ludwig H, Sutter G, Suzuki K, Hemmi H, Sato S, Yamamoto M, Uematsu S, Kawai T, Takeuchi O, Akira S. A Toll-like receptor-independent antiviral response induced by double-stranded B-form DNA. Nat Immunol. 2006;7:40–48. doi: 10.1038/ni1282. [DOI] [PubMed] [Google Scholar]
- 10.Ablasser A, Bauernfeind F, Hartmann G, Latz E, Fitzgerald KA, Hornung V. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat Immunol. 2009;10:1065–1072. doi: 10.1038/ni.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chiu YH, Macmillan JB, Chen ZJ. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138:576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharma S, Deoliveira RB, Kalantari P, Parroche P, Goutagny N, Jiang Z, Chan J, Bartholomeu DC, Lauw F, Hall JP, Barber GN, Gazzinelli RT, Fitzgerald KA, Golenbock DT. Innate Immune Recognition of an AT-Rich Stem-Loop DNA Motif in the Plasmodium falciparum Genome. Immunity. 2011;35:194–207. doi: 10.1016/j.immuni.2011.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takaoka A, Wang Z, Choi MK, Yanai H, Negishi H, Ban T, Lu Y, Miyagishi M, Kodama T, Honda K, Ohba Y, Taniguchi T. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448:501–505. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- 14.Ishii KJ, Kawagoe T, Koyama S, Matsui K, Kumar H, Kawai T, Uematsu S, Takeuchi O, Takeshita F, Coban C, Akira S. TANK-binding kinase-1 delineates innate and adaptive immune responses to DNA vaccines. Nature. 2008;451:725–729. doi: 10.1038/nature06537. [DOI] [PubMed] [Google Scholar]
- 15.Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois CM, Jin T, Latz E, Xiao TS, Fitzgerald KA, Paludan SR, Bowie AG. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Z, Yuan B, Bao M, Lu N, Kim T, Liu YJ. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat Immunol. 2011;12:959–965. doi: 10.1038/ni.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin L, Waterman PM, Jonscher KR, Short CM, Reisdorph NA, Cambier JC. MPYS, a novel membrane tetraspanner, is associated with major histocompatibility complex class II and mediates transduction of apoptotic signals. Mol Cell Biol. 2008;28:5014–5026. doi: 10.1128/MCB.00640-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhong B, Yang Y, Li S, Wang YY, Li Y, Diao F, Lei C, He X, Zhang L, Tien P, Shu HB. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity. 2008;29:538–550. doi: 10.1016/j.immuni.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 20.Sun W, Li Y, Chen L, Chen H, You F, Zhou X, Zhou Y, Zhai Z, Chen D, Jiang Z. ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc Natl Acad Sci U S A. 2009;106:8653–8658. doi: 10.1073/pnas.0900850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saitoh T, Fujita N, Hayashi T, Takahara K, Satoh T, Lee H, Matsunaga K, Kageyama S, Omori H, Noda T, Yamamoto N, Kawai T, Ishii K, Takeuchi O, Yoshimori T, Akira S. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc Natl Acad Sci U S A. 2009;106:20842–20846. doi: 10.1073/pnas.0911267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stetson DB, Medzhitov R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 2006;24:93–103. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 24.Chen H, Sun H, You F, Sun W, Zhou X, Chen L, Yang J, Wang Y, Tang H, Guan Y, Xia W, Gu J, Ishikawa H, Gutman D, Barber G, Qin Z, Jiang Z. Activation of STAT6 by STING Is Critical for Antiviral Innate Immunity. Cell. 2011;147:436–446. doi: 10.1016/j.cell.2011.09.022. [DOI] [PubMed] [Google Scholar]
- 25.Lin R, Heylbroeck C, Pitha PM, Hiscott J. Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol Cell Biol. 1998;18:2986–2996. doi: 10.1128/mcb.18.5.2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoneyama M, Suhara W, Fujita T. Control of IRF-3 activation by phosphorylation. J Interferon Cytokine Res. 2002;22:73–76. doi: 10.1089/107999002753452674. [DOI] [PubMed] [Google Scholar]
- 27.Zhao T, Yang L, Sun Q, Arguello M, Ballard DW, Hiscott J, Lin R. The NEMO adaptor bridges the nuclear factor-kappaB and interferon regulatory factor signaling pathways. Nat Immunol. 2007;8:592–600. doi: 10.1038/ni1465. [DOI] [PubMed] [Google Scholar]
- 28.Zeng W, Xu M, Liu S, Sun L, Chen ZJ. Key role of Ubc5 and lysine-63 polyubiquitination in viral activation of IRF3. Mol Cell. 2009;36:315–325. doi: 10.1016/j.molcel.2009.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Panne D, McWhirter SM, Maniatis T, Harrison SC. Interferon regulatory factor 3 is regulated by a dual phosphorylation-dependent switch. J Biol Chem. 2007;282:22816–22822. doi: 10.1074/jbc.M703019200. [DOI] [PubMed] [Google Scholar]
- 30.Clark K, Peggie M, Plater L, Sorcek RJ, Young ER, Madwed JB, Hough J, McIver EG, Cohen P. Novel cross-talk within the IKK family controls innate immunity. Biochem J. 2011;434:93–104. doi: 10.1042/BJ20101701. [DOI] [PubMed] [Google Scholar]
- 31.Clark K, Takeuchi O, Akira S, Cohen P. The TRAF-associated protein TANK facilitates cross-talk within the I{kappa}B kinase family during Toll-like receptor signaling. Proc Natl Acad Sci U S A. 2011;108:17093–17098. doi: 10.1073/pnas.1114194108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burdette DL, Monroe KM, Sotelo-Troha K, Iwig JS, Eckert B, Hyodo M, Hayakawa Y, Vance RE. STING is a direct innate immune sensor of cyclic di-GMP. Nature. 2011;478:515–518. doi: 10.1038/nature10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsuchida T, Zou J, Saitoh T, Kumar H, Abe T, Matsuura Y, Kawai T, Akira S. The ubiquitin ligase TRIM56 regulates innate immune responses to intracellular double-stranded DNA. Immunity. 2010;33:765–776. doi: 10.1016/j.immuni.2010.10.013. [DOI] [PubMed] [Google Scholar]
- 34.Hou F, Sun L, Zheng H, Skaug B, Jiang QX, Chen ZJ. MAVS Forms Functional Prion-like Aggregates to Activate and Propagate Antiviral Innate Immune Response. Cell. 2011;146:448–461. doi: 10.1016/j.cell.2011.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O’Connell RM, Saha SK, Vaidya SA, Bruhn KW, Miranda GA, Zarnegar B, Perry AK, Nguyen BO, Lane TF, Taniguchi T, Miller JF, Cheng G. Type I interferon production enhances susceptibility to Listeria monocytogenes infection. J Exp Med. 2004;200:437–445. doi: 10.1084/jem.20040712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He J, Nguyen AT, Zhang Y. KDM2b/JHDM1b, an H3K36me2-specific demethylase, is required for initiation and maintenance of acute myeloid leukemia. Blood. 2011;117:3869–3880. doi: 10.1182/blood-2010-10-312736. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.