Abstract
Intrinsic antiviral immunity refers to a form of innate immunity that directly restricts viral replication and assembly, thereby rendering a cell non-permissive to a specific class or species of viruses. Intrinsic immunity is conferred by restriction factors that are largely preexisting in certain cell types, although these factors can be further induced by virus infection. Intrinsic viral restriction factors recognize specific viral components, but unlike other pattern recognition receptors that inhibit viral infection indirectly by inducing interferons and other antiviral molecules, intrinsic antiviral factors block viral replication immediately and directly. This review focuses on recent advances in understanding the roles of intrinsic antiviral factors that restrict infection by human immunodeficiency virus (HIV) and influenza virus.
Primordial forms of antiviral immunity - RNAi and CRISPR
One of the earliest forms of antiviral immunity in metazoan evolution is RNA interference (RNAi). RNAi is the predominant mechanism of antiviral defense in plants and invertebrate animals, and it is also a primordial form of immunity to viral infection in vertebrate animals. Infection by RNA viruses leads to the generation of long double-stranded RNA (dsRNA) that are structurally different from host cellular RNAs, which are single stranded with short and often imperfectly matched stem loops1, 2. In plants and invertebrates, Dicer cleaves long viral dsRNA and gives rise to siRNA duplexes. These siRNAs are then loaded into the RNA-induced silencing complex (RISC) to target viral mRNA or genomic RNA for degradation, thereby inhibiting viral replication (Fig. 1a). In plants and nematodes, but not in insects, the antiviral RNAi response is amplified by RNA-dependent RNA polymerases (RdRp) that replicate the incoming viral RNA, which can then be processed by Dicer to generate more siRNAs3–5. Inhibition of RNAi in plants increases susceptibility to many plant viruses6, 7. To counteract antiviral RNAi, many plant and invertebrate viruses have evolved suppressors of RNA silencing (SRS) proteins that are important for establishing infection8, 9.
Figure 1. Evolution of innate immunity.
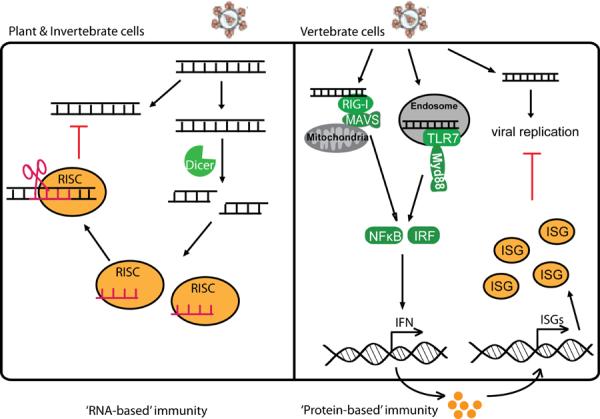
A diagram illustrating the evolutionary progression from `RNA-based' immunity in plant and invertebrate cells (a) to `Protein-based' immunity in vertebrate cells (b). In `RNA-based' immunity, incoming viral RNA is processed by Dicer into small RNAs that directly target the virus through RISC-mediated RNAi. In `Protein-based' immunity, incoming viral RNA is recognized by PRRs that signals to activate IFN expression, which then activates the expression of many interferon-stimulated genes (ISGs) to inhibit viral replication. Some of the ISGs are intrinsic antiviral factors that are constitutively present in certain cell types and can block viral replication immediately and directly.
An even more ancient form of antiviral immunity is the clustered, regularly interspaced short palindromic repeats (CRISPR) system that protects bacteria and archaea from bacteriophages and conjugative plasmids10. In this system, some of the invading DNA sequences from bacteriophages or plasmids are acquired and integrated into the CRISPR loci of the host as repeat elements. The DNA repeats on the CRISPR loci are transcribed and processed into small interfering RNAs (crRNAs) by the bacterial CRISPR-associated (Cas) proteins. crRNAs are incorporated into large Cas protein complexes (e.g, Cascade or Cas6), which then degrade the invading viral DNA in a sequence-specific manner that is guided by crRNAs. This bacterial antiviral mechanism resembles RNAi in that small interfering RNAs are used to guide the destruction of invading nucleic acids with a high degree of sequence specificity. There are also clear differences; for example, the precursors of crRNA are single-stranded RNA and the targets of destruction by crRNA are viral DNA. The CRISPR system has not been found in eukaryotic cells.
Extensive effort has been made to try to demonstrate antiviral RNAi responses in vertebrate animals, especially in mammalian cells11. Most of these efforts failed to recover siRNAs of viral origin in mammalian cells infected with a variety of RNA and DNA viruses12. DNA viruses such as herpesvirus do produce small RNA such as miRNA, but not siRNAs, and the viral miRNAs play an important role in establishing infection13. Compared to the RNA-based immunity of plants and invertebrates, vertebrates have a more versatile interferon (IFN) system, which is an elaborate protein-based antiviral immunity (Fig. 1b). This evolutionary `upgrade' is important for vertebrates to cope with more complex pathogens, the diversity of nucleic acids introduced into the cell, and minimizing off-target effects of RNAi on host mRNAs. Vertebrates do however retain evolutionary `fossils' of the antiviral RNAi machinery. For example, mouse embryonic stem cells (ESCs) express endogenous siRNAs similar to those antiviral siRNA found in plant and invertebrate, although it is unclear whether they target any genes or play any critical roles. Long dsRNA can induce sequence-specific RNAi against target mRNAs in mouse ESCs. These mammalian ESCs lack functional IFN signaling pathways, which might be the reason why they retain remnants of the antiviral RNAi machinery14, 15.
In addition to antiviral RNAi, invertebrates such as Drosophila melanogaster has evolved the Toll signaling pathway that is important for both antimicrobial defense and development of the embryo16. The Drosophila Toll gene is the founding ortholog of mammalian Toll-like receptors (TLRs) that are critical for innate immune responses to pathogens. In Drosophila, gram-positive bacteria and fungi infections activate the Toll pathway that lead to production of antimicrobial peptides, whereas mammalian TLRs trigger proinflammatory and IFN responses16. The remarkable similarity between Drosophila Toll and mammalian TLR signaling pathways and the increased complexity of the latter underscore the evolutionary requirements of more intricate antiviral immunity in mammals. Drosophila also activate the so-called `immune deficiency' (IMD) pathway to induce antimicrobial peptides in response to infection with gram-negative bacteria. The IMD pathway resembles the tumor necrosis factor (TNF) pathway in mammals in that both use similar mechanisms to activate the protein kinases IKK and MAP kinases.
Vertebrate antiviral innate immunity – the IFN response
Vertebrates are constantly challenged by potentially pathogenic microbes that can introduce a variety of proteins and nucleic acids into the cell. To counter this vertebrate cells express many different pattern-recognition receptors (PRRs) that can detect the pathogen-associated molecular patterns (PAMPs) of viruses and other microbes which in turn activate antiviral IFN and proinflammatory responses17, 18. By secretion of IFN, the response can be amplified and spread to surrounding uninfected cells through the JAK-STAT signaling pathway, and thereby activate hundreds of interferon-stimulated genes (ISGs), most of which have profound antiviral effects, such as degradation of viral nucleic acids or inhibition of viral gene expression19 (Fig. 1b).
PRRs are proteins that recognize molecular patterns of microorganisms and trigger innate immune responses to limit microbial infections20, 21. Mammalian PRRs include Toll-like receptors (TLRs), RIG-I-like receptors (RLRs), NOD-like receptors (NLRs) and C-type lectin receptors (CLRs). These receptors activate signaling cascades that lead to the activation of the transcription factors NF-κB and AP-1, which induce proinflammatory cytokines. In the case of viral infection, viral nucleic acids are the major PAMP detected by the host innate immune receptors, which include RLRs in the cytosol and a subfamily of TLRs that localize to the endosomal membrane. TLR7 and TLR9 detect viral RNA and DNA, respectively, in the endosomal lumen of virus-infected cells. These TLRs contain a TIR domain that recruits the adaptor MyD88 from the cytoplasm. MyD88 in turn recruits the protein kinases IRAK1 and IRAK4 and the ubiquitin E3 ligase TRAF622. TRAF6 activates the protein kinase complex IKK, which phosphorylates the NF-κB inhibitor IκB. This phosphorylation targets IκB for degradation by the ubiquitin-proteasome pathway, thereby allowing NF-κB to enter the nucleus to turn on inflammatory genes. MyD88 and TRAF6 on the endosomal membrane also recruit another transcription factor IRF7, which is phosphorylated by IKKα and then enter the nucleus to induce type-I IFNs, especially IFNα (Fig. 1b).
RLRs include RIG-I, Mda5 and LGP2, all of which share a RNA helicase domain containing a DExD/H (Asp-Glu-X-Asp/His)-box23. RIG-I and Mda5 contain two CARD domains in tandem at the N-termini which are important for their signaling functions. LGP2 lacks the CARD domains that are needed for signaling and likely plays a regulatory role. RIG-I also contains a C-terminal domain (CTD) that binds to viral RNA containing 5'-triphosphate24, 25. The binding of the viral RNA to the CTD of RIG-I induces a conformational change that exposes the N-terminal CARDs, which recruit the ubiquitin E3 ligase TRIM25 to catalyze the synthesis of K63 polyubiquitin chains26–28. These ubiquitin chains bind to and activate RIG-I CARDs26, which then interact with the CARD domain of mitochondrial antiviral signaling protein (MAVS, also known as IPS-1, VISA or CARDIF)29–32. This interaction promotes the aggregation of MAVS into microfibrils through a prion-like mechanism33. MAVS aggregates on the mitochondrial membrane then recruits signaling proteins from the cytoplasm, leading to the activation of IKK and the IKK-like kinase TBK1. TBK1 phosphorylates IRF3, causing IRF3 to dimerize and translocate to the nucleus where it functions together with NF-κB to induce IFNβ and other antiviral molecules (summarized in Fig. 1b). Genetic experiments have demonstrated that RIG-I is essential for immune defense against many RNA viruses, including paramyxoviruses (e.g., Sendai virus and Newcastle disease virus), orthomyxoviruses (e.g., influenza virus), and some positive-strand RNA viruses (e.g, hepatitis C virus and Japanese encephalitis virus). On the other hand, Mda5 is required for interferon induction by picornaviruses (e.g, encephalomyocarditis virus)34. The viral ligands for Mda5 have not been precisely defined, but are thought to consist of long dsRNA containing branched structures35. MAVS is required for interferon induction by both RIG-I and Mda536.
DNA viruses can also induce IFNs through the endoplasmic reticulum membrane protein STING (also known as MITA, MPYS or ERIS)37. A number of proteins have been proposed to detect double-stranded DNA in the cytosol, and these include DAI, IFI16 and DDX4138–40. In addition, RNA polymerase III detects AT-rich DNA in the cytosol and converts the DNA into 5'-ppp RNA which can then activate the RIG-I pathway to induce IFNs41, 42. It remains to be determined whether certain protein functions as a dominant cytosolic DNA sensor in vivo, or whether multiple cytosolic DNA sensors exist and each functions in a distinct cell type to induce IFNs.
Besides inducing IFNs, both DNA and RNA viruses can trigger cell death and induce inflammatory cytokines such as IL-1β through activation of the inflammasome, which belongs to the NLR family43. Whereas viral RNA appears to activate the NLRP3 inflammasome to generate mature IL-1β, viral DNA is instead detected by the AIM2 inflammasome44,45.
`Intrinsic' vs. `innate' immune factors
Numerous other host proteins can also detect viral infections and exert antiviral activities. For example, the deaminase APOBEC3G edits the HIV genome to inhibit HIV replication, and the E3 ubiquitin ligase TRIM5α targets the incoming HIV capsid protein and modulates uncoating of the capsid46. These proteins, herein referred to as intrinsic antiviral factors, can also be classified as PRRs because they directly bind to viral components. However, unlike TLR and RLR, which inhibit viral infections indirectly by activating signaling cascades that result in transcription of new antiviral factors such as IFNs, intrinsic restriction factors inhibit viral replication directly, often before the onset of the IFN response. Thus, intrinsic antiviral factors are pre-existing in certain cell types, although most of these factors can be further induced by IFNs to amplify their antiviral activity. In this review, we will use `intrinsic antiviral factor' exclusively for host factors that can directly recognize viral components and are able to block viral replication immediately. In comparison, `innate immune factor' covers a much broader spectrum, and refers to host factors such as TLR, RLR and NLR, which participate in the recognition, signaling, and orchestration of both innate and adaptive immune responses to viral infection.
Mechanistic studies of intrinsic antiviral factors are important because viruses almost always have to encode proteins or devise strategies that counteract these factors in order to replicate in the host cell. Moreover, expression patterns of intrinsic antiviral factors often determine the permissiveness of a cell type to a virus (or to a mutant virus that lacks the counteracting viral protein). In many cases, this permissiveness vs. non-permissiveness holds the key to discoveries of intrinsic antiviral factors and parallel viral evasion mechanisms (see below). During evolution, many intrinsic antiviral factors are under strong positive selection through co-evolution with the virus. In some cases, they play an important role in limiting cross-species transmission of a virus and thereby determining the viral tropism (see below).
In the following, we will discuss recently discovered intrinsic antiviral factors against HIV-1 and influenza virus. Two other factors, namely RNASE L and Protein kinase R (PKR), are also important intrinsic antiviral factors that are well characterized and extensively reviewed. The 2'-5'-oligoadenylate synthetase (OAS) and RNASE L pathway was one of the first IFN-induced antiviral pathways discovered47. PKR mediates a multifaceted antiviral response by regulating protein translation by the ribosome and by promoting innate immune signaling48. These two factors target a broad spectrum of viruses and will not be included in this review.
Intrinsic immunity against HIV
HIV enters T cells and macrophages by the binding of the viral envelope protein gp120 to CD4, which enables the membrane proximal portion of the envelope gp41 subunit to bind to the co-receptor CCR5 or CXCR4 on the target cell membrane, triggering viral envelope fusion with the plasma membrane. HIV can also bind to cell surface lectins and enter cells by endocytosis, which is the predominant mode of entry into dendritic cells (DCs) and also occurs in macrophages. Once the viral core is released into the cytosol, HIV reverse transcriptase converts RNA into DNA within the reverse transcription complex (RTC). The RTC matures into the preintegration complex (PIC), which delivers reverse-transcribed HIV DNA to the nucleus for chromosomal integration. Few copies of HIV DNA integrate, leaving behind the bulk of HIV DNA in the cytosol to be cleared by host enzymes. Once the viral genomic DNA is integrated into a host chromosome, viral transcription is activated by host pathways with the assistance of HIV protein Tat. HIV mRNAs are all capped and polyadenylated, like host RNAs. The unspliced RNA is both translated to generate Gag-Pol gene products and incorporated as genomic RNA into nascent virions at cell membrane sites where the envelope and capsid proteins assemble before budding (Fig. 2).
Figure 2. Intrinsic antiviral factors against HIV-1.
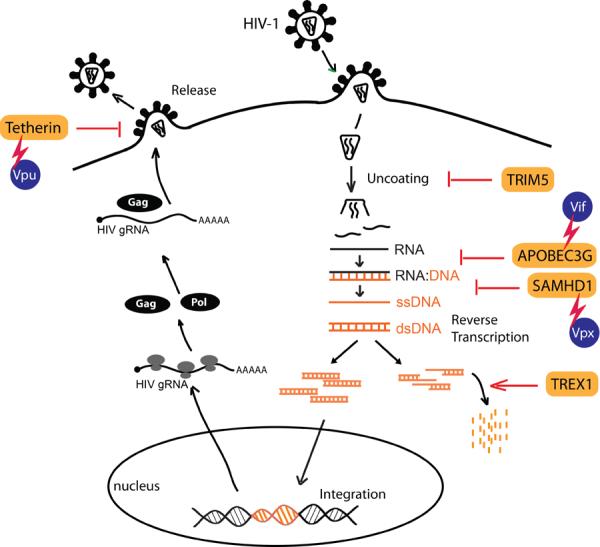
Many steps of the HIV-1 life cycle are targeted by intrinsic antiviral factors. HIV-1 has evolved strategies to counteract these intrinsic antiviral factors, through accessory proteins or other unknown mechanisms that are currently under investigation.
Mucosal innate immunity is the first line of defense against HIV-1 during the early phase of infection, and it also plays a vital role in shaping ensuing adaptive immune responses. The types of innate immunity involved in HIV-1 infection can be divided into two major forms, namely cellular and intracellular innate immunity. Cellular innate immunity includes functions of DCs such as Langerhans cells that are amongst the first group of cells that contact HIV-1 at the site of infection, and that can mediate trans-infection of CD4+ T cells49. γδ+ T cells offer innate responses to HIV through generating antiviral factors, such as RANTES, MIP-1α and MIP-1β50. Natural killer (NK) cells also play important roles in cellular innate immunity against HIV by eliminating infected cells and modulating DC functions51. Intracellular innate immunity includes intrinsic immunity mediated by host factors that play important roles in restricting HIV-1 replication, such as APOBEC3G, TRIM5α, Tetherin/BST-2 and SAMHD152–54 (Table 1). All of these restriction factors are also ISGs. HIV-1 counteracts some of these restriction factors via accessory proteins, and avoid upregulation of other antiviral ISGs in infected target cells55, 56.
Table 1.
Intrinsic antiviral factors
| Name | Target virus | Key roles | References |
|---|---|---|---|
| APOBEC3G | HIV-1, SIV, EIAV, MLV, foamy virus, Hepatitis B virus | Edit C to U in HIV DNA (negative strand); inhibit reverse transcription and integration | 61, 62, 117–120 |
| TRIM5α | HIV-1, MLV | Block uncoating of the incoming virion; promote innate immune signaling by sensing retrovirus capsid | 68, 69, 72, 73 |
| TETHERIN/BST-2/CD317 | HIV-1, MLV, HTLV-1, Ebola virus, KSHV | Block release of enveloped viruses | 74, 75, 77 |
| SAMHD1 | HIV-1 | Inhibit HIV replication in myeloid cells, likely through regulating cellular dNTP supply | 53, 54, 83, 121 |
| TREX1 | HIV-1 | Remove cytosolic nonproductive RT DNA; inhibit innate immune responses to HIV-1 | 55 |
| IFITM family | Influenza virus, Dengue virus, West Nile Virus | Block cytosolic entry | 96, 122 |
| IFIT family | Influenza virus | Recognize 5'-ppp and the lack of 2'-O-methylation in viral RNA and inhibit translation | 106 – 108 |
| MxA | Influenza virus, other RNA viruses | Block transcription | 109 – 111 |
| RNASE L | Many RNA viruses | Cleave ssRNA in U-rich sequences; activate antiviral innate immunity | 47, 123 |
| PKR | Many RNA viruses | Inhibits virus translation by protein phosphorylation; promote innate immune signaling | 48 |
APOBEC3
APOBEC3G (originally named CEM15) was one of the first intrinsic antiviral factors identified for HIV-1. It was discovered by investigating the function of an HIV-1 accessory protein called viral infectivity factor (Vif)57, 58. Vif was known to be essential for HIV replication in certain cell lines (such as CEM-SS, SupT1), but not in others (such as CEM, CD4+ T cells), and the effect of Vif is dependent on virus-producing cells. Heterokaryon fusion of permissive and non-permissive cells yielded cells that display non-permissive phenotypes, suggesting that a dominant antiviral factor exists in non-permissive cells. This antiviral factor was identified as APOBEC3G through a cDNA expression screen for genes specifically expressed in non-permissive cells, and the ability to convert permissive to non-permissive cells upon expression57.
APOBEC3G belongs to a family of cytidine deaminases that contains 7 members in primates (APOBEC3A, B, C, DE, F, G, and H). The mouse homolog of this family contains only one gene, mApobec3, underscoring the remarkable evolutionary diversification that occurred at this locus. Indeed, the APOBEC3 gene locus displays strong evidence of positive selection during evolution of primates59. APOBEC3G and 3F are the predominant restriction factors for HIV-1. They are packaged into HIV-1 virions through interaction with nucleocapsid portion of HIV Gag. Upon infection of target cells and during reverse transcription, APOBEC3G edits C to U in single-stranded HIV DNA (negative strand), which results in G→A mutation in the HIV genome. G→A mutations often lead to premature stop codons that partially contributed to the reduced replication. Such G→A mutations are also frequently found in HIV DNA isolated from AIDS patients60. APOBEC3G also inhibit reverse transcription and chromosomal integration through yet to be defined mechanisms that are independent of its deaminase activity61, 62.
HIV-1 Vif counteracts APOBEC3G by promoting its ubiquitination by an E3 ligase complex consisting of Cul5, Elongin B and C, and Rbx1. This ubiquitination targets APOBEC3G for degradation by the proteasome in virus producing cells58. The interaction between Vif and APOBEC3G is species-specific; e.g. African green monkey APOBEC3G contains a single amino acid change at position 128 and is completely resistant to HIV-1 Vif-mediated degradation63–66. As such, this interaction is currently a major antiretroviral therapeutic target.
TRIM5α
Restriction activity against HIV-1 in some non-permissive cell lines can be saturated by high multiplicity of infection (MOI)67. One such restriction factor was originally named Ref1 that restricts HIV-1 replication in Old World Monkey lung fibroblasts. This restriction factor was later determined to be TRIM5α, which is a member of the tripartite motif family (TRIM) that shares a common organization at the N terminus containing a RING domain, a B-box domain and a coiled-coil domain68. The RING domain is commonly found in E3 ubiquitin ligase and the B-box domain determines substrate specificity. The C terminus of TRIM5α contains a B30.2 domain that binds to the capsid of the incoming virion and is most important for restriction. TRIM5α, specifically the B30.2 domain, is also a major determinant of retrovirus species tropism46. For example, human TRIM5α potently restricts MLV (N-tropic, a strain replicates in Swiss/NIH mice, as opposed to a closely related stain, B-tropic, in Balb/c mice), but does not restrict HIV-1 or SIV from Rhesus macaque (SIVmac). Rhesus macaque TRIM5α restricts HIV-1, but not SIVmac. The importance of TRIM5α and its capsid binding activity was underscored again by the discovery of the TRIM5α-cyclophilin A (TRIMCyp) fusion protein in owl monkey cells69. TRIMCyp occurs naturally by in-frame fusion where CypA replaces the B30.2 domain. CypA also binds to the HIV-1 capsid and TRIMCyp potently restricts HIV-1 through mechanisms similar to TRIM5α.
TRIM5α blocks retrovirus replication early, and prior to reverse transcription, likely during the process of uncoating46. As a ubiquitin E3 ligase, TRIM5α acts in both proteasome-dependent and -independent pathways. For example, inhibiting proteasome during infection or disrupting TRIM5α E3 ligase activity only partially alleviates the restriction70, 71. TRIM5α promotes rapid uncoating of HIV-1 capsid in vitro72. Recently, TRIM5α was also found to promote innate immune signaling and to act as a pattern recognition receptor for capsid of many retroviruses, including MLV, HIV and SIV73. TRIM5α expression in 293T cells activated AP-1 and NF-κB, but not type I IFN, by promoting the synthesis of free K63-linked polyubiquitin chains, which bind and activate the kinase TAK1.
TETHERIN
Tetherin, also known as BST-2 or CD317, was discovered through characterization of HIV-1 accessory protein Vpu74, 75. Vpu enhances the release of HIV and other retroviral virions, thereby promoting replication. Similar to studies on Vif, heterokaryon fusion experiments suggested that a dominant restriction factor exists in non-permissive cells for Vpu-deficient HIV. Such a factor was also found to be interferon inducible, since IFNa treatment could convert permissive cells to cells that are non-permissive to Vpu-deficient HIV76. Tetherin was later identified through comparative microarray analysis74. The topology of Tetherin is unique, including an N terminal cytoplasmic domain, a transmembrane domain, extracellular long coiled-coil domain and a C terminal glycosylphosphatidylinositol (GPI) membrane anchor. The short cytoplasmic domain binds the clathrin adaptors for endocytosis. Tetherin is thought to hold virions at the cell surface, by inserting the GPI membrane anchor into the virion envelope or by dimerization of two Tetherin molecules each anchoring at the host cell membrane and the virion envelope, respectively. Tethered virions will then be internalized by endocytosis and subsequently degraded in the endosomes77. Vpu promotes the degradation of Tetherin, thereby facilitating HIV infection.
Tetherin targets many other enveloped viruses, such as other retroviruses (MLV, HTLV-1), filoviruses (Ebola virus), and herpesvirus (KSHV)77. Each of these viruses counteracts Tetherin by encoding a viral protein that binds to Tetherin and either promotes its degradation (the K5 protein of KSHV) or inhibits its function through an unknown mechanism (the glycoprotein of Ebola virus).
Like APOBEC3G and TRIM5α, Tetherin also rapidly evolves under positive selection. Tetherin has species specificity that may have contributed to shaping the evolution of primate lentiviruses46. Many primate lentiviruses, mostly SIV isolates, do not encode Vpu. These SIV isolates use Nef to counteract the simian orthologs of Tetherin, which features a five amino acid insertion in the cytoplasmic domain which is not present in human Tetherin. Nef specifically binds to the extra five amino acids in simian Tetherin, and is thus unable to target human Tetherin. HIV-1 originated from chimapanzee SIV (SIVcpz) through a cross-species transmission to humans. SIVcpz uses Nef to counteract chimpanzee Tetherin, whereas HIV-1 had to adapt and use Vpu to antagonize human Tetherin in order to survive the new host environment (because the Nef-targeting motif is missing).
Tetherin also plays a role in immune signaling. Tetherin is a ligand for ILT7 (immunoglobulin-like transcript 7), a membrane receptor selectively expressed in plasmacytoid dendritic cells (pDC). The binding of Tetherin to ILT7 leads to inhibition of TLR-mediated IFN responses in pDCs78. Tetherin was also found to activate NF-κB in a large-scale screening79. The implication of this in viral infection remains to be shown, but one could imagine a simple evasion strategy where enveloped viruses antagonize Tetherin to promote virion release and at the same time downregulate NF-κB signaling to dampen the host immune response.
SAMHD1 and TREX1
HIV-1 replication is very inefficient in cells of the myeloid linage, especially DC. This myeloid-specific restriction can be overcome by treating cells with virus like particles (VLPs) containing accessory protein Vpx from SIVmac or HIV-280. HIV-1 does not encode Vpx, and Vpx-deficient SIVmac or HIV-2 fails to replicate in DC. The host restriction factor targeted by Vpx was recently identified as SAMHD153, 54. SAMHD1 appears to inhibit HIV-1 reverse transcription53 and innate immune responses to HIV (at least in MDDCs)81. SAMHD1 is the only mammalian protein that contains both a SAM domain that is predicted to mediate protein-protein interaction and a HD domain that has nucleotide phosphohydrolase activity82. Vpx binds SAMHD1 and brings it to DCAF1 and DDB-CUL4 E3 ubiquitin ligase complex for ubiquitination and subsequent degradation. Structural and biochemical studies of the HD domain revealed that SAMHD1 is a potent dGTP-stimulated triphosphohydrolase that converts deoxynucleoside triphosphates (dNTPs) to deoxynucleoside (dN) and triphosphate83. Therefore, SAMHD1 is likely to regulate the dNTP pool in myeloid cells, thus blocking HIV reverse transcription by throttling the dNTP supply. It remains to be determined how the dNTP concentration in different cell types impacts HIV reverse transcription, and whether that correlates with SAMHD1 expression (or the lack thereof). SAMHD1 may play an additional role in limiting innate immune signaling to HIV replication, particularly in DCs. Indeed, DCs rendered permissive to HIV-1 infection through the expression of Vpx, which causes SAMHD1 degradation, induces type-I IFNs through cellular cyclophilin A and IRF381. Thus, HIV-1 avoids infecting DCs such that it does not induce IFNs but stealthily passes through DCs to facilitate its infection of helper T cells. Consistent with an inhibitory effect of SAMHD1 on IFN induction, mutations in the SAMHD1 gene are associated with Aicardi-Goutières Syndrome (AGS)84, an autoimmune disorder and a neurological brain disease that is a genetic mimic of congenital viral infection. AGS patients have elevated levels of IFNα.
Interestingly, another AGS-associated gene, TREX1, has been shown to be important for HIV replication, specifically innate immune responses to HIV DNA55. TREX1 is a 3' exonuclease that contains three well-conserved Exo motifs at the N terminus and a hydrophobic region at the C terminus that is important for its localization to the cytoplasm and endoplasmic reticulum (ER). In Trex1−/− mouse cells and human CD4+ T cells and macrophages in which TREX1 was depleted by RNAi, cytosolic HIV DNA accumulated and HIV infection induced IFN that inhibited HIV replication and spreading. TREX1 bound to cytosolic HIV DNA and digested excess non-productive HIV DNA that would otherwise activate interferon expression via a pathway dependent on the kinase TBK1, the adaptor STING and the transcription factor IRF355. TREX1 also prevents autoimmunity induced by DNA derived from endogenous retroelements85, which may explain the AGS disease in patients carrying loss of function mutations of TREX1.
Although both SAMHD1 and TREX1 are associated with the same autoimmune disease, they exhibit opposite effects on HIV-1 replication. SAMHD1 is antiviral whereas TREX1 is proviral for HIV-1, through distinct mechanisms. Interestingly, both proteins appear to target the reverse transcription step, by limiting the dNTP supply (SAMHD1) or by inhibiting immune recognition of non-productive RT products (TREX1). HIV RT DNA can also trigger a proinflammatory response in nonproductively infected CD4+ T cells in tonsil, which promotes T cell killing86. Taken together, HIV reverse transcription is becoming an increasingly important process in the HIV life cycle that is carefully regulated by a concerted effort of viral and host factors. Nucleic acids generated by reverse transcription are also targeted by host immune surveillance. Further investigation is required to provide additional insight into the dynamic interaction between HIV reverse transcription and innate immune signaling and how it influences the establishment of infection and HIV pathogenesis.
Intrinsic immunity against influenza virus
Influenza A virus is a negative sense (−) single-stranded RNA virus of the orthomyxovirus family. Influenza A virus enters host cells through attachment of viral hemagglutinin (HA) to host cell receptor containing α-2,3- or α-2,6-linked sialic acid moieties, followed by endocytosis87 (Fig. 3). Acidification of the endocytosed vesicle promotes fusion of viral envelope and the endosome membrane followed by release of the viral RNA-protein (RNP) complex into the cytoplasm. The viral RNP complex then translocates into the nucleus, where (−) strand viral RNA is converted to complementary (+) RNA and mRNA. Viral mRNAs are exported to the cytoplasm for translation to generate a total of 11 viral proteins. Some of the viral proteins (M1 and NEP) are essential for genome replication and transcription and they shuttle in and out of the nucleus to promote the production of more viral RNP complexes. Other viral proteins are transported through the host protein secretary pathway to the plasma membrane, where new viral particles form. The Non-structure protein NS1 inhibits host IFN-mediated antiviral responses and thus promotes the pathogenesis of the influenza virus88.
Figure 3. Intrinsic antiviral factors against influenza virus.
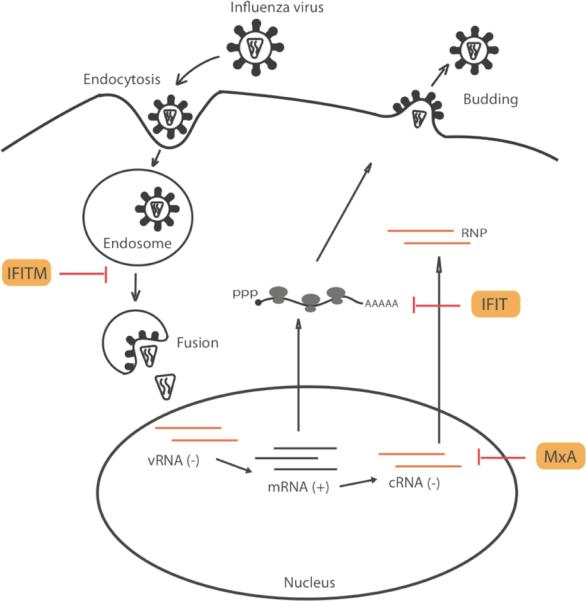
A diagram of influenza virus life cycle and known intrinsic antiviral factors against influenza virus.
The primary target of influenza virus is epithelial cells in the respiratory tract. Macrophages and DC in the airway can also be infected by influenza virus, and these cells play an important role in host innate and adaptive immune responses to the virus. Influenza virus can be recognized by multiple PRRs, including RLR, TLR and NLR. In infected fibroblasts, the cytosolic RNA sensor RIG-I recognizes 5'-triphosphate of influenza virus genomic RNA and triggers IFN through MAVS and IRF3. In fact, studies of influenza virus played an important role in understanding molecular mechanisms of the RIG-I signaling pathway25. As a counterstrike, viral protein NS1 helps the virus to evade innate immune detection by sequestering viral RNA or by binding to RIG-I and other protein required for RIG-I signaling25. In pDC, the single-stranded RNA genome of influenza virus can be recognized by TLR7 in the endosomes, and induce production of proinflammatory cytokines and IFN89. The TLR3-dependent inflammatory response has also been implicated in influenza virus infected lung epithelial cells and induction of acute pneumonia90, 91. Influenza virus infection also activates NLRP3 inflammasomes as an innate immune response that contributes to the adaptive immune response89. Initial evidence for NLRP3 activation came from an observation that influenza virus-infected human macrophages produce IL-1β and IL-18 through a caspase-1-dependent pathway92. It is not until recently that mechanistic details started to emerge. Influenza virus induces IL-1β production by enhancing the transcription of pro-IL-1β and NLRP3 (signal 1) and by activating NLRP3 inflammasomes (signal 2)93–95. Signal 1 is triggered by the detection of viral RNA by TLR7, which activates NF-κB. Signal 2 is contributed by multiple sources that all depend on viral M2 protein, including ionic imbalance of trans-Golgi pH, potassium efflux through the P2X7 receptor ion channel, and elevation of cellular reactive oxygen species (ROS). Studies using inflammasome-deficient mice found that the inflammasome complex is dispensable for early viral clearance, but is essential for late stage viral clearance93–95. Further investigation is needed to elucidate the role of inflammasome in mediating host innate and adaptive responses to influenza virus.
The IFITM family
The interferon induced transmembrane (IFITM) genes belong to a family of small ISGs containing IFITM1, 2 and 3. IFITM3 was identified in two genome-wide screens (RNAi96 and yeast-two-hybrid97) as a host restriction factor for influenza A virus. Expression of IFITM genes is induced by influenza virus infection. RNAi knockdown or genetic knockout of IFITM genes increased influenza virus replication and overexpression of IFITM proteins potently inhibited viral replication. IFITM proteins likely block infection early during entry96, although mechanistic details remain unclear. Interestingly, avian cells do not appear to encode a homologue to IFITM3, raising the possibility that IFITMs might influence viral tropism. Moreover, IFITM proteins also inhibit the replication of some flaviviruses, including dengue virus and West Nile virus96. Infection of DNA viruses such as cytomegalovirus and herpes simplex virus also induce IFITM gene expression98, 99, although there is no clear evidence for an antiviral activity of IFITM proteins against DNA viruses.
IFITM proteins have also been implicated in cancer100. IFITM1 and 3 associate with cell surface antigen CD81 that is expressed in a variety of cancers. All IFITM proteins contain a conserved CD225 domain that has antiproliferative activity; such activity can be enhanced by IFN treatment101. Overexpression of IFITM3 inhibited proliferation of IFN-sensitive melanoma cells, whereas IFITM1 knockdown hampered IFNγ-mediated antiproliferative effect102. Comparison of IFITM genes from different organisms revealed higher than normal sequence variations, suggesting that these genes are under positive selection during evolution100. Taken together, IFITM proteins represent attractive targets for therapeutics against virus and cancer.
The IFIT family
IFIT (IFN-induced protein with tetratricopeptide repeats [TPRs]) family contains four members in humans, namely IFIT1 (ISG56), IFIT2 (ISG54), IFIT3 (ISG60) and IFIT5 (ISG58). All IFIT proteins are cytoplasmic proteins that contain multiple TPRs. TPRs are helix-turn-helix structures that mediate protein-protein interactions and assembly of protein complexes103. IFIT1 was initially found to inhibit cellular translation by binding the eIF3 initiation factor104. This activity is part of the nonspecific antiviral response triggered by IFN. IFIT1 also inhibits cytoplasmic sensing of viral RNA by binding adaptor protein STING thereby disrupting the formation of signaling scaffolds105. This negative feedback of IFN signaling regulated by IFIT1 is considered to be important for preventing IFN overdrive in infected cells that may be toxic for the host.
More recently, IFIT proteins were found to recognize viral RNA that contains a 5'-triphosphate (5'-ppp) moiety or lacks 2'-O-methylation106–108. Cellular mRNA usually contains a 5'-guanosine cap that stabilizes the mRNA for translation and differentiates self from non-self viral genomic RNAs that often contain 5'-ppp. Cellular mRNAs are also methylated at the 2'-O position, the purpose of which was unclear because it does not contribute to translation or stability. Many RNA viruses also encode a methyltransferase that methylates the 2'-O position of viral RNA to mimic host mRNA. It is now clear that this modification is important for these viruses to evade host restriction by IFIT proteins. Viruses defective in methyltransferase exhibited enhanced sensitivity to IFN treatment in an IFIT-dependent manner106, 107. Thus, 2'-O-methylation of host mRNAs is critical for self vs. non-self distinction for at least some RNA viruses. Interestingly, mRNAs from plants or plant viruses do not contain 2'-O-methylation, and they don't have IFN responses or orthologs of IFIT genes. This represents yet another evolutionary sophistication that is acquired by vertebrates. IFIT1 also binds viral genomic RNA containing 5'-ppp, similar to RIG-I, and exerts a direct antiviral activity 108. Instead of activating IFN, the binding of IFIT proteins to 5'-ppp viral RNA inhibits viral translation and replication. Consistent with having a direct antiviral activity, knockdown of IFIT1 results in more viral replication without affecting the IFN response. Moreover, this antiviral activity is orchestrated by at least three members of the IFIT family as a protein complex. The structural basis for IFIT recognition of 5'-ppp and the absence of 2'-O-methylation in RNA molecules is unclear. Similarly, the mechanisms of inhibition following recognition by IFIT proteins remain to be determined.
MxA
The human myxovirus resistant protein 1 (MxA) is a GTPase with broad antiviral activities. MxA shares similar domain structures with the dynamin family of large GTPases, including an N-terminal GTPase domain, a self-oligomerization domain, and a C-terminal GTPase effector domain. Both human MxA protein and the mouse Mx1 protein have antiviral activity against influenza virus infection. Mx1−/− mice succumb to influenza virus infection whereas wild type mice are highly resistant and survive high dose challenges109, 110. Murine Mx1 localizes to the nucleus and blocks primary transcription of influenza vRNA. Human MxA is cytoplasmic and blocks later in the influenza virus life cycle such as secondary transcription and viral replication111. Different strains of influenza virus vary in their sensitivity to Mx proteins, which is influenced by viral NP protein112, 113. The structure of MxA revealed that it might form an oligomeric ring structure around viral nucleocapsid, thereby inhibiting viral replication114. It is unclear how human MxA can inhibit a broad spectrum of RNA viruses, some of which replicates in the nucleus, and whether MxA acts by recognizing a common viral component or structure.
Conclusions and perspectives
The rapid progress in innate immunity research in the past decade is breathtaking. Several families of innate immune sensors have been discovered and the signaling pathways that they trigger are being rigorously investigated. By comparison, less is understood of the intrinsic antiviral factors and their mechanisms of action. The intense medical interest in HIV, influenza and other viruses are beginning to reveal some of the host restriction factors that limit infection by these viruses. These studies also provide striking examples of the arms race between the host and virus during their co-evolution. Further studies of individual viral proteins will continue to shed light on the host immunity that attacks different viruses. Conversely, investigation of host antiviral factors will illuminate how viruses evade the host immune surveillance. The studies of viral tropism in certain cell types have had an excellent track record of discovering important antiviral factors such as APOBEC3G, TRIM5α, TETHERIN and SAMHD1. There is no reason to think that such a successful track record will end anytime soon.
Recent technological advances such as genome-wide RNAi screens provide additional avenues to discovering new host antiviral factors, as exemplified by the recent success in identifying many host proteins and non-coding RNAs (e.g, microRNA) that either permit or impede the replication of HIV and influenza virus. As we gain experience and develop new tools to differentiate true positive hits from false-negative or false-positive hits, the large scale screening approaches will be applied to more viruses with better success rates. The next challenge will be to investigate the function of novel antiviral factors and elucidate their mechanisms of action. Knowledge gained from these studies will be very powerful for designing the future generation of antiviral therapies.
The repeated failure in producing an effective HIV vaccine is a humbling reminder that we still lack a full understanding of immune responses against HIV. In fact, we know very little about innate immune responses against retroviruses in general. Even the existence of intracellular innate immunity against retrovirus is a subject of debate, because retroviral infection normally does not trigger the production of cytokines or interferons. However, retroviruses, including HIV, clearly activate T cells and B cells in vivo. A recent study shows that TLR7 detects the entry of the murine retrovirus MMTV into host cells and activates humoral immune responses115. Emerging literature also reveals the importance of NK cells as the cytotoxic arm of innate immunity during HIV transmission51, 116. The recent findings that HIV ceases replication in professional IFN producing cells, such as DCs due to the presence of the restriction factor SAMHD1 and that HIV co-opts the host protein TREX1 to suppress IFN production in target cells reinvigorate research on innate and intrinsic immunity against HIV. This line of research may be key to developing effective treatments for HIV and other devastating pathogens.
References
- 1.Hamilton AJ, Baulcombe DC. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- 2.Zamore PD, Tuschl T, Sharp PA, Bartel DP. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- 3.Aoki K, Moriguchi H, Yoshioka T, Okawa K, Tabara H. In vitro analyses of the production and activity of secondary small interfering RNAs in C. elegans. EMBO J. 2007;26:5007–5019. doi: 10.1038/sj.emboj.7601910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Diaz-Pendon JA, Li F, Li WX, Ding SW. Suppression of antiviral silencing by cucumber mosaic virus 2b protein in Arabidopsis is associated with drastically reduced accumulation of three classes of viral small interfering RNAs. Plant Cell. 2007;19:2053–2063. doi: 10.1105/tpc.106.047449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Umbach JL, Cullen BR. The role of RNAi and microRNAs in animal virus replication and antiviral immunity. Genes & development. 2009;23:1151–1164. doi: 10.1101/gad.1793309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mourrain P, et al. Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell. 2000;101:533–542. doi: 10.1016/s0092-8674(00)80863-6. [DOI] [PubMed] [Google Scholar]
- 7.Dalmay T, Horsefield R, Braunstein TH, Baulcombe DC. SDE3 encodes an RNA helicase required for post-transcriptional gene silencing in Arabidopsis. EMBO J. 2001;20:2069–2078. doi: 10.1093/emboj/20.8.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Voinnet O, Pinto YM, Baulcombe DC. Suppression of gene silencing: a general strategy used by diverse DNA and RNA viruses of plants. Proc Natl Acad Sci U S A. 1999;96:14147–14152. doi: 10.1073/pnas.96.24.14147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li HW, Ding SW. Antiviral silencing in animals. FEBS Lett. 2005;579:5965–5973. doi: 10.1016/j.febslet.2005.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marraffini LA, Sontheimer EJ. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat Rev Genet. 2010;11:181–190. doi: 10.1038/nrg2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cullen BR. Is RNA interference involved in intrinsic antiviral immunity in mammals? Nat Immunol. 2006;7:563–567. doi: 10.1038/ni1352. [DOI] [PubMed] [Google Scholar]
- 12.Pfeffer S, et al. Identification of microRNAs of the herpesvirus family. Nat Methods. 2005;2:269–276. doi: 10.1038/nmeth746. [DOI] [PubMed] [Google Scholar]
- 13.Plaisance-Bonstaff K, Renne R. Viral miRNAs. Methods Mol Biol. 2011;721:43–66. doi: 10.1007/978-1-61779-037-9_3. [DOI] [PubMed] [Google Scholar]
- 14.Wianny F, Zernicka-Goetz M. Specific interference with gene function by double-stranded RNA in early mouse development. Nat Cell Biol. 2000;2:70–75. doi: 10.1038/35000016. [DOI] [PubMed] [Google Scholar]
- 15.Yang S, Tutton S, Pierce E, Yoon K. Specific double-stranded RNA interference in undifferentiated mouse embryonic stem cells. Mol Cell Biol. 2001;21:7807–7816. doi: 10.1128/MCB.21.22.7807-7816.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferrandon D, Imler J-L, Hetru C, Hoffmann JA. The Drosophila systemic immune response: sensing and signalling during bacterial and fungal infections. Nature reviews Immunology. 2007;7:862–874. doi: 10.1038/nri2194. [DOI] [PubMed] [Google Scholar]
- 17.Janeway CA., Jr. Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb Symp Quant Biol. 1989;54(Pt 1):1–13. doi: 10.1101/sqb.1989.054.01.003. [DOI] [PubMed] [Google Scholar]
- 18.Iwasaki A, Medzhitov R. Regulation of adaptive immunity by the innate immune system. Science. 2010;327:291–295. doi: 10.1126/science.1183021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bowie AG, Unterholzner L. Viral evasion and subversion of pattern-recognition receptor signalling. Nat Rev Immunol. 2008;8:911–922. doi: 10.1038/nri2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity. 2011;34:637–650. doi: 10.1016/j.immuni.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 21.Ronald PC, Beutler B. Plant and animal sensors of conserved microbial signatures. Science. 2010;330:1061–1064. doi: 10.1126/science.1189468. [DOI] [PubMed] [Google Scholar]
- 22.Skaug B, Jiang X, Chen ZJ. The role of ubiquitin in NF-kappaB regulatory pathways. Annu Rev Biochem. 2009;78:769–796. doi: 10.1146/annurev.biochem.78.070907.102750. [DOI] [PubMed] [Google Scholar]
- 23.Yoneyama M, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 24.Hornung V, et al. 5'-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 25.Pichlmair A, et al. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5'-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 26.Zeng W, et al. Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell. 2010;141:315–330. doi: 10.1016/j.cell.2010.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gack MU, et al. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446:916–920. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- 28.Kowalinski E, et al. Structural Basis for the Activation of Innate Immune Pattern-Recognition Receptor RIG-I by Viral RNA. Cell. 2011;147:423–435. doi: 10.1016/j.cell.2011.09.039. [DOI] [PubMed] [Google Scholar]
- 29.Kawai T, et al. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 30.Meylan E, et al. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 31.Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 32.Xu LG, et al. VISA Is an Adapter Protein Required for Virus-Triggered IFN-beta Signaling. Mol Cell. 2005;19:727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 33.Hou F, et al. MAVS Forms Functional Prion-like Aggregates to Activate and Propagate Antiviral Innate Immune Response. Cell. 2011;146:448–461. doi: 10.1016/j.cell.2011.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kato H, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 35.Pichlmair A, et al. Activation of MDA5 requires higher-order RNA structures generated during virus infection. J Virol. 2009;83:10761–10769. doi: 10.1128/JVI.00770-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun Q, et al. The specific and essential role of MAVS in antiviral innate immune responses. Immunity. 2006;24:633–642. doi: 10.1016/j.immuni.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 37.Barber GN. Innate immune DNA sensing pathways: STING, AIMII and the regulation of interferon production and inflammatory responses. Current opinion in immunology. 2011 doi: 10.1016/j.coi.2010.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takaoka A, et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448:501–505. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- 39.Unterholzner L, et al. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Z, et al. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat Immunol. 2011;12:959–965. doi: 10.1038/ni.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chiu Y-H, Macmillan JB, Chen ZJ. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138:576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ablasser A, et al. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat Immunol. 2009;10:1065–1072. doi: 10.1038/ni.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Davis BK, Wen H, Ting JP. The inflammasome NLRs in immunity, inflammation, and associated diseases. Annu Rev Immunol. 2011;29:707–735. doi: 10.1146/annurev-immunol-031210-101405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schroder K, Tschopp J. The inflammasomes. Cell. 2010;140:821–832. doi: 10.1016/j.cell.2010.01.040. [DOI] [PubMed] [Google Scholar]
- 45.Mehle A, Doudna JA. A host of factors regulating influenza virus replication. Viruses. 2010;2:566–573. doi: 10.3390/v2020566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wolf D, Goff SP. Host restriction factors blocking retroviral replication. Annu Rev Genet. 2008;42:143–163. doi: 10.1146/annurev.genet.42.110807.091704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chakrabarti A, Jha BK, Silverman RH. New insights into the role of RNase L in innate immunity. J Interferon Cytokine Res. 2011;31:49–57. doi: 10.1089/jir.2010.0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pindel A, Sadler A. The role of protein kinase R in the interferon response. Journal of interferon & cytokine research : the official journal of the International Society for Interferon and Cytokine Research. 2011;31:59–70. doi: 10.1089/jir.2010.0099. [DOI] [PubMed] [Google Scholar]
- 49.Geijtenbeek TB, et al. DC-SIGN-ICAM-2 interaction mediates dendritic cell trafficking. Nat Immunol. 2000;1:353–357. doi: 10.1038/79815. [DOI] [PubMed] [Google Scholar]
- 50.Lehner T, et al. The emerging role of innate immunity in protection against HIV-1 infection. Vaccine. 2008;26:2997–3001. doi: 10.1016/j.vaccine.2007.11.060. [DOI] [PubMed] [Google Scholar]
- 51.Alter G, et al. HIV-1 adaptation to NK-cell-mediated immune pressure. Nature. 2011;476:96–100. doi: 10.1038/nature10237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kirchhoff F. Immune evasion and counteraction of restriction factors by HIV-1 and other primate lentiviruses. Cell Host Microbe. 2010;8:55–67. doi: 10.1016/j.chom.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 53.Hrecka K, et al. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature. 2011;474:658–661. doi: 10.1038/nature10195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Laguette N, et al. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature. 2011;474:654–657. doi: 10.1038/nature10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yan N, Regalado-Magdos AD, Stiggelbout B, Lee-Kirsch MA, Lieberman J. The cytosolic exonuclease TREX1 inhibits the innate immune response to human immunodeficiency virus type 1. Nat Immunol. 2010;11:1005–1013. doi: 10.1038/ni.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yan N, Lieberman J. Gaining a foothold: how HIV avoids innate immune recognition. Curr Opin Immunol. 2010;23:1–8. doi: 10.1016/j.coi.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sheehy A, Gaddis N, Choi J, Malim MH. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- 58.Sheehy A, Gaddis N, Malim MH. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat Med. 2003;9:1404–1407. doi: 10.1038/nm945. [DOI] [PubMed] [Google Scholar]
- 59.Sawyer SL, Emerman M, Malik HS. Ancient adaptive evolution of the primate antiviral DNA-editing enzyme APOBEC3G. PLoS Biol. 2004;2:E275. doi: 10.1371/journal.pbio.0020275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vartanian JP, Meyerhans A, Asjo B, Wain-Hobson S. Selection, recombination, and G----A hypermutation of human immunodeficiency virus type 1 genomes. J Virol. 1991;65:1779–1788. doi: 10.1128/jvi.65.4.1779-1788.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bishop KN, Verma M, Kim E-Y, Wolinsky SM, Malim MH. APOBEC3G inhibits elongation of HIV-1 reverse transcripts. PLoS Pathog. 2008;4:e1000231. doi: 10.1371/journal.ppat.1000231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mbisa JL, et al. Human immunodeficiency virus type 1 cDNAs produced in the presence of APOBEC3G exhibit defects in plus-strand DNA transfer and integration. J Virol. 2007;81:7099–7110. doi: 10.1128/JVI.00272-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bogerd HP, Doehle BP, Wiegand HL, Cullen BR. A single amino acid difference in the host APOBEC3G protein controls the primate species specificity of HIV type 1 virion infectivity factor. Proc Natl Acad Sci U S A. 2004;101:3770–3774. doi: 10.1073/pnas.0307713101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mangeat B, Turelli P, Liao S, Trono D. A single amino acid determinant governs the species-specific sensitivity of APOBEC3G to Vif action. J Biol Chem. 2004;279:14481–14483. doi: 10.1074/jbc.C400060200. [DOI] [PubMed] [Google Scholar]
- 65.Schrofelbauer B, Chen D, Landau NR. A single amino acid of APOBEC3G controls its species-specific interaction with virion infectivity factor (Vif) Proc Natl Acad Sci U S A. 2004;101:3927–3932. doi: 10.1073/pnas.0307132101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xu H, et al. A single amino acid substitution in human APOBEC3G antiretroviral enzyme confers resistance to HIV-1 virion infectivity factor-induced depletion. Proc Natl Acad Sci U S A. 2004;101:5652–5657. doi: 10.1073/pnas.0400830101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Towers G, Collins M, Takeuchi Y. Abrogation of Ref1 retrovirus restriction in human cells. J Virol. 2002;76:2548–2550. doi: 10.1128/jvi.76.5.2548-2550.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stremlau M, et al. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 2004;427:848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- 69.Sayah DM, Sokolskaja E, Berthoux L, Luban J. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature. 2004;430:569–573. doi: 10.1038/nature02777. [DOI] [PubMed] [Google Scholar]
- 70.Diaz-Griffero F, et al. Rapid turnover and polyubiquitylation of the retroviral restriction factor TRIM5. Virology. 2006;349:300–315. doi: 10.1016/j.virol.2005.12.040. [DOI] [PubMed] [Google Scholar]
- 71.Anderson JL, et al. Proteasome inhibition reveals that a functional preintegration complex intermediate can be generated during restriction by diverse TRIM5 proteins. J Virol. 2006;80:9754–9760. doi: 10.1128/JVI.01052-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stremlau M, et al. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5alpha restriction factor. Proc Natl Acad Sci U S A. 2006;103:5514–5519. doi: 10.1073/pnas.0509996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pertel T, et al. TRIM5 is an innate immune sensor for the retrovirus capsid lattice. Nature. 2011;472:361–365. doi: 10.1038/nature09976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Neil S, Zang T, Bieniasz P. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature. 2008;451:425–430. doi: 10.1038/nature06553. [DOI] [PubMed] [Google Scholar]
- 75.Van Damme N, et al. The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe. 2008;3:245–252. doi: 10.1016/j.chom.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Neil S, Eastman S, Jouvenet N, Bieniasz P. HIV-1 Vpu promotes release and prevents endocytosis of nascent retrovirus particles from the plasma membrane. PLoS Pathog. 2006;2:e39. doi: 10.1371/journal.ppat.0020039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Evans DT, Serra-Moreno R, Singh RK, Guatelli JC. BST-2/tetherin: a new component of the innate immune response to enveloped viruses. Trends in microbiology. 2010;18:388–396. doi: 10.1016/j.tim.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cao W, et al. Regulation of TLR7/9 responses in plasmacytoid dendritic cells by BST2 and ILT7 receptor interaction. J Exp Med. 2009;206:1603–1614. doi: 10.1084/jem.20090547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Matsuda A, et al. Large-scale identification and characterization of human genes that activate NF-kappaB and MAPK signaling pathways. Oncogene. 2003;22:3307–3318. doi: 10.1038/sj.onc.1206406. [DOI] [PubMed] [Google Scholar]
- 80.Goujon C, et al. With a little help from a friend: increasing HIV transduction of monocyte-derived dendritic cells with virion-like particles of SIV(MAC) Gene Ther. 2006;13:991–994. doi: 10.1038/sj.gt.3302753. [DOI] [PubMed] [Google Scholar]
- 81.Manel N, et al. A cryptic sensor for HIV-1 activates antiviral innate immunity in dendritic cells. Nature. 2010;467:214–217. doi: 10.1038/nature09337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Aravind L, Koonin EV. The HD domain defines a new superfamily of metal-dependent phosphohydrolases. Trends Biochem Sci. 1998;23:469–472. doi: 10.1016/s0968-0004(98)01293-6. [DOI] [PubMed] [Google Scholar]
- 83.Goldstone DC, et al. HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature. 2011 doi: 10.1038/nature10623. [DOI] [PubMed] [Google Scholar]
- 84.Rice G, et al. Mutations involved in Aicardi-Goutières syndrome implicate SAMHD1 as regulator of the innate immune response. Nat Genet. 2009 doi: 10.1038/ng.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Stetson DB, Ko JS, Heidmann T, Medzhitov R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 2008;134:587–598. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Doitsh G, et al. Abortive HIV Infection Mediates CD4 T Cell Depletion and Inflammation in Human Lymphoid Tissue. Cell. 2010;143:789–801. doi: 10.1016/j.cell.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Medina RA, García-Sastre A. Influenza A viruses: new research developments. Nature reviews Microbiology. 2011;9:590–603. doi: 10.1038/nrmicro2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fernandez-Sesma A. The influenza virus NS1 protein: inhibitor of innate and adaptive immunity. Infect Disord Drug Targets. 2007;7:336–343. doi: 10.2174/187152607783018754. [DOI] [PubMed] [Google Scholar]
- 89.Pang IK, Iwasaki A. Inflammasomes as mediators of immunity against influenza virus. Trends in immunology. 2011;32:34–41. doi: 10.1016/j.it.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Le Goffic R, et al. Detrimental contribution of the Toll-like receptor (TLR)3 to influenza A virus-induced acute pneumonia. PLoS Pathog. 2006;2:e53. doi: 10.1371/journal.ppat.0020053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Guillot L, et al. Involvement of toll-like receptor 3 in the immune response of lung epithelial cells to double-stranded RNA and influenza A virus. J Biol Chem. 2005;280:5571–5580. doi: 10.1074/jbc.M410592200. [DOI] [PubMed] [Google Scholar]
- 92.Pirhonen J, Sareneva T, Kurimoto M, Julkunen I, Matikainen S. Virus infection activates IL-1 beta and IL-18 production in human macrophages by a caspase-1-dependent pathway. J Immunol. 1999;162:7322–7329. [PubMed] [Google Scholar]
- 93.Ichinohe T, Lee HK, Ogura Y, Flavell R, Iwasaki A. Inflammasome recognition of influenza virus is essential for adaptive immune responses. J Exp Med. 2009;206:79–87. doi: 10.1084/jem.20081667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Thomas PG, et al. The intracellular sensor NLRP3 mediates key innate and healing responses to influenza A virus via the regulation of caspase-1. Immunity. 2009;30:566–575. doi: 10.1016/j.immuni.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Allen IC, et al. The NLRP3 inflammasome mediates in vivo innate immunity to influenza A virus through recognition of viral RNA. Immunity. 2009;30:556–565. doi: 10.1016/j.immuni.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Brass AL, et al. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell. 2009;139:1243–1254. doi: 10.1016/j.cell.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Shapira SD, et al. A physical and regulatory map of host-influenza interactions reveals pathways in H1N1 infection. Cell. 2009;139:1255–1267. doi: 10.1016/j.cell.2009.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhu H, Cong JP, Shenk T. Use of differential display analysis to assess the effect of human cytomegalovirus infection on the accumulation of cellular RNAs: induction of interferon-responsive RNAs. Proc Natl Acad Sci U S A. 1997;94:13985–13990. doi: 10.1073/pnas.94.25.13985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nicholl MJ, Robinson LH, Preston CM. Activation of cellular interferon-responsive genes after infection of human cells with herpes simplex virus type 1. J Gen Virol. 2000;81:2215–2218. doi: 10.1099/0022-1317-81-9-2215. [DOI] [PubMed] [Google Scholar]
- 100.Siegrist F, Ebeling M, Certa U. The small interferon-induced transmembrane genes and proteins. Journal of interferon & cytokine research : the official journal of the International Society for Interferon and Cytokine Research. 2011;31:183–197. doi: 10.1089/jir.2010.0112. [DOI] [PubMed] [Google Scholar]
- 101.Gutterman JU. Cytokine therapeutics: lessons from interferon alpha. Proc Natl Acad Sci U S A. 1994;91:1198–1205. doi: 10.1073/pnas.91.4.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yang G, Xu Y, Chen X, Hu G. IFITM1 plays an essential role in the antiproliferative action of interferon-gamma. Oncogene. 2007;26:594–603. doi: 10.1038/sj.onc.1209807. [DOI] [PubMed] [Google Scholar]
- 103.D'Andrea LD, Regan L. TPR proteins: the versatile helix. Trends Biochem Sci. 2003;28:655–662. doi: 10.1016/j.tibs.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 104.Guo J, Hui DJ, Merrick WC, Sen GC. A new pathway of translational regulation mediated by eukaryotic initiation factor 3. EMBO J. 2000;19:6891–6899. doi: 10.1093/emboj/19.24.6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Li Y, et al. ISG56 is a negative-feedback regulator of virus-triggered signaling and cellular antiviral response. Proc Natl Acad Sci U S A. 2009;106:7945–7950. doi: 10.1073/pnas.0900818106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Daffis S, et al. 2'-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature. 2010;468:452–456. doi: 10.1038/nature09489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zust R, et al. Ribose 2'-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat Immunol. 2011;12:137–143. doi: 10.1038/ni.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pichlmair A, et al. IFIT1 is an antiviral protein that recognizes 5'-triphosphate RNA. Nat Immunol. 2011;12:624–630. doi: 10.1038/ni.2048. [DOI] [PubMed] [Google Scholar]
- 109.Salomon R, et al. Mx1 gene protects mice against the highly lethal human H5N1 influenza virus. Cell Cycle. 2007;6:2417–2421. doi: 10.4161/cc.6.19.4779. [DOI] [PubMed] [Google Scholar]
- 110.Tumpey TM, et al. The Mx1 gene protects mice against the pandemic 1918 and highly lethal human H5N1 influenza viruses. J Virol. 2007;81:10818–10821. doi: 10.1128/JVI.01116-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Pavlovic J, Haller O, Staeheli P. Human and mouse Mx proteins inhibit different steps of the influenza virus multiplication cycle. J Virol. 1992;66:2564–2569. doi: 10.1128/jvi.66.4.2564-2569.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Dittmann J, et al. Influenza A virus strains differ in sensitivity to the antiviral action of Mx-GTPase. J Virol. 2008;82:3624–3631. doi: 10.1128/JVI.01753-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zimmermann P, Manz B, Haller O, Schwemmle M, Kochs G. The viral nucleoprotein determines Mx sensitivity of influenza A viruses. J Virol. 2011;85:8133–8140. doi: 10.1128/JVI.00712-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gao S, et al. Structural basis of oligomerization in the stalk region of dynamin-like MxA. Nature. 2010;465:502–506. doi: 10.1038/nature08972. [DOI] [PubMed] [Google Scholar]
- 115.Kane M, et al. Innate immune sensing of retroviral infection via Toll-like receptor 7 occurs upon viral entry. Immunity. 2011;35:135–145. doi: 10.1016/j.immuni.2011.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Martin MP, et al. Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat Genet. 2007;39:733–740. doi: 10.1038/ng2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Mangeat B, et al. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature. 2003;424:99–103. doi: 10.1038/nature01709. [DOI] [PubMed] [Google Scholar]
- 118.Löchelt M, et al. The antiretroviral activity of APOBEC3 is inhibited by the foamy virus accessory Bet protein. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:7982–7987. doi: 10.1073/pnas.0501445102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Turelli P, Mangeat B, Jost S, Vianin S, Trono D. Inhibition of hepatitis B virus replication by APOBEC3G. Science (New York, NY) 2004;303:1829. doi: 10.1126/science.1092066. [DOI] [PubMed] [Google Scholar]
- 120.Harris RS, et al. DNA deamination mediates innate immunity to retroviral infection. Cell. 2003;113:803–809. doi: 10.1016/s0092-8674(03)00423-9. [DOI] [PubMed] [Google Scholar]
- 121.Powell RD, Holland PJ, Hollis T, Perrino FW. The Aicardi-Goutieres syndrome gene and HIV-1 restriction factor SAMHD1 is a dGTP-regulated deoxynucleotide triphosphohydrolase. The Journal of biological chemistry. 2011 doi: 10.1074/jbc.C111.317628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Feeley EM, et al. IFITM3 Inhibits Influenza A Virus Infection by Preventing Cytosolic Entry. PLoS pathogens. 2011;7:e1002337. doi: 10.1371/journal.ppat.1002337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Malathi K, Dong B, Gale M, Silverman RH. Small self-RNA generated by RNase L amplifies antiviral innate immunity. Nature. 2007;448:816–819. doi: 10.1038/nature06042. [DOI] [PMC free article] [PubMed] [Google Scholar]


