Abstract
Membrane fusion is a fundamental requirement in numerous developmental, physiological, and pathological processes in eukaryotes. So far, only a limited number of viral and cellular fusogens, proteins that fuse membranes, have been isolated and characterized. Despite the diversity in structures and functions of known fusogens, some common principles of action apply to all fusion reactions. These can serve as guidelines in the search for new fusogens, and may allow the formulation of a cross-species, unified theory to explain divergent and convergent evolutionary principles of membrane fusion.
Main Text
Introduction
The basic building blocks of all organisms are cells, whose outer surfaces are lipid bilayer-based plasma membranes. The plasma membrane normally functions as a barrier between the inner contents of individual cells and between the cells and the extracellular space. This compartmentalization by membranes allows cells to function as single units with discrete genetic and biochemical programs. However, in various biological processes, these compartments must be extended as cells unite to form syncytial tissues where internal contents are mixed. Although such cell union might in principle proceed through phagocytosis followed by membrane breakdown, many examples of syncytiogenesis involve fusion between plasma membranes of individual cells. In prokaryotes, it is not known whether membrane fusion occurs in processes where two bacteria are united; for example, during conjugation. The fusion of mitochondria from prokaryotic origin, however, raises the possibility that cell fusion does occur in some prokaryotes. In contrast, cell fusion has been identified as an important stage of development in most eukaryotes from yeast to humans. Characterized examples include gamete fusion in protists, fungi, plants, and animals; hypodermis (epidermis) formation in Caenorhabditis elegans; muscle organogenesis in Drosophila melanogaster and vertebrates; and bone and placenta organogenesis in mammals. In these processes, cell fusion is used to unite gametes and other cells, to alter cell differentiation states, and to resculpt organs. In addition to its occurrence in normal physiology, membrane fusion is essential for the progression of different pathological events such as viral entry into host cells and, possibly, tumor progression.
Despite the diversity in the organisms and cell types that utilize cell fusion in normal physiology and pathology, most fusion reactions occur via a common stereotypic sequence of events starting with cell-cell recognition, alignment, and adhesion. This is followed by an actual fusion event that merges the two cells together by the tethering of plasma-membranes, formation of aqueous pores between the membranes, and expansion of these pores to clear the membranes from the fusing cells' junction. The diversity of cell fusion stages suggests the sequential activity of various proteins executing different biological processes that in sum account for cell fusion. Although much is known about several fusion stages within a specific model organism, the field of cell fusion lacks a cross-species, unified picture of the molecular mechanisms of this multistep process (Chen et al., 2007, Chen and Olson, 2005, Oren-Suissa and Podbilewicz, 2007, Shemer and Podbilewicz, 2003).
The Fusogen Concept versus Spontaneous Fusion
In vitro experiments have revealed that in the absence of proteins, fusion between lipid bilayers mimicking the typical composition of biological membranes requires energy input. As a result, even close and long-lived contacts between membranes under physiological conditions do not yield fusion (reviewed in Chernomordik et al., 2006). Thus, membrane fusion is widely assumed to be controlled by, and to rely on, the activity of fusogenic proteins that lower the energy barrier of the process and drive membrane bilayer rearrangements that result in membrane and cell merging (Chernomordik et al., 2006, Jahn et al., 2003). Theoretically, any of the biological fusion reactions for which no specialized fusogen has been identified may represent a hypothetical “spontaneous” fusion (Kerbel et al., 1983) dependent on destabilization of membrane bilayers caused by changes in their composition, and by proteins that normally carry out fusion-unrelated functions (Byrne et al., 2007, Lemaire et al., 2006, Palovuori and Eskelinen, 2000). However, all well-characterized biological fusion events studied so far have been found to rely on the activity of specialized fusogens that directly fuse lipid bilayers (Chernomordik et al., 2006, Jahn et al., 2003).
Because enveloped viruses contain only a small number of proteins and have developed rather simple fusion machineries relying, in many cases, on the activity of one fusogenic protein, identification of viral fusogens is relatively easy. In contrast, validation of a candidate eukaryotic cellular protein as a “true” developmental fusogen in vivo is not a trivial task. Many candidate proteins have been proposed to act as developmental fusogens, yet only a fraction of these proteins have proven to be actual fusogens (reviewed in Oren-Suissa and Podbilewicz, 2007). A bona fide fusogen has to satisfy several criteria, such as being necessary for cell fusion and localizing to the fusion site. Final validation stems from fusogen sufficiency to directly mediate fusion in heterologous cells or liposomes. Verifying these standards for a candidate fusion protein is a major challenge in identifying the missing developmental fusogens, which is a crucial step in deciphering the molecular basis of cell fusion during development (Oren-Suissa and Podbilewicz, 2007).
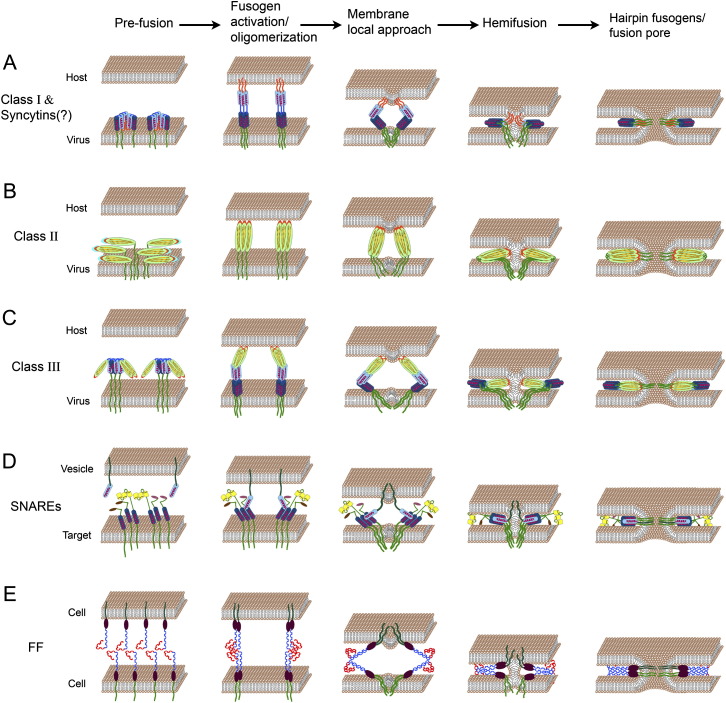
In this review, we discuss the pathway and molecular mechanisms of membrane fusion, beginning from fusion of viruses with cells of infected hosts. Emerging mechanisms of viral fusion will be the basis of comparison with the less-characterized and likely more complex process of cell fusion in eukaryotes. We will focus on the proteins that drive membrane rearrangements in the transition from membrane tethering to opening and expansion of fusion pores, a process that lies at the heart of cell fusion ( Figure 1). To avoid mechanistic misinterpretations derived from emphasis on candidate fusogens that might not participate in the membrane fusion process per se, we will restrict our review to the selected group of fusogens that have been shown to directly fuse heterologous cells (Figure 1). We will discuss both similarities and differences in the structures and activities of the different fusogens, emphasizing the constraints and flexibility of the molecular mechanisms leading to cell fusion.
Figure 1.
Models for Membrane Fusion Mediated by Viral, Intracellular, and Developmental Fusogens
Fusogens are drawn embedded in the membranes after the stages of oligomerization, sorting, trafficking, and binding to the host receptors (not shown). α helices, light and dark blue cylinders; transmembrane domains, dark green; fusion peptides or loops, orange; β sheets, light green ellipses.
(A) Class I viral fusogens (such as in HIV) contain two α helices and an amphiphilic fusion peptide buried inside the protein in the prefusion state. Upon activation, the trimers undergo a conformational change leading to the formation of rigid coiled-coil structures with exposed fusion peptides at their N termini. Insertion of fusion peptides into the host membrane, concomitant with protein fold back into six-helix bundles, leads to membrane tightening, hemifusion, and pore formation (Kielian and Rey, 2006).
(B) In Class II viral fusogens (for example, in Dengue viruses), hydrophobic fusion loops emanate from a structure containing β sheets. After fusogen activation and a shift from heterodimers to homotrimers (not shown), the fusion loops are exposed and inserted into the opposing membrane. Folding back occurs presumably by interaction between different domains of the protein that drive membrane fusion (Kielian and Rey, 2006).
(C) Class III fusogens (such as in Rabies and VSV viruses) combine characteristics of Class I and II including short and discontinuous hydrophobic fusion loops that stem from a structure containing β sheets, and α helices that potentially form six-helix bundles to merge the membranes together. In contrast to class I and II fusogens, activation does not require fusion domain exposure (Heldwein et al., 2006, Weissenhorn et al., 2007).
(D) SNARE-dependent intracellular fusion. Three different t-SNAREs, each with a single α helix domain, are bundled to form the target complex. This complex binds a single α helix of v-SNARE that emanates from the vesicle membrane. Transition between loose and tight t- SNARE-v-SNARE complexes is mediated by SNARE zippering and tethering of the membranes, leading to their fusion (Jahn and Scheller, 2006).
(E) Hypothetical model of FF fusion. The extracellular domains of FF proteins (C. elegans EFF-1 and AFF-1 proteins) are comprised of juxta-membrane hypothetical globular domains (brown ellipses) and a putative cysteine-rich conserved domain that presumably forms an S-S based structure (in blue). Part of these predicted extracellular loops are reminiscent of TGF-β receptor-like domains (in red). We hypothesize that cis-dimerization of FF molecules is followed by homotypic trans-binding between dimmers. Trans-zippering of FF complexes from the two membranes induces membrane tightening, hemifusion, and fusion pore formation.
Conserved Rearrangements of Membrane Phospholipids during Fusion
Theoretical and experimental studies of membrane intermediates in diverse fusion reactions revealed a common membrane remodeling pathway apparently conserved in all membrane-fusion events, including exocytosis, protein trafficking, and viral and developmental cell fusion (Chernomordik and Kozlov, 2005, Chernomordik et al., 1993, Nakatogawa et al., 2007, Xu et al., 2005, Podbilewicz et al., 2006). One working model for the merging of biological membranes involves four simple steps: (1) Two opposed biological membranes are separated by 10–20 nm. (2) The membranes undergo deformations, causing a local approach. (3) The reaction proceeds via a stereotypic membrane lipid rearrangement, called hemifusion. In this step the outer leaflets, which face each other through an intermembrane 3–5 nm water gap, are the first to fuse, and proceed to mix their lipids. In contrast, the inner leaflets remain separated, preventing the mixing of aqueous contents (Chernomordik et al., 2006). (4) Subsequent merger of the inner leaflets marks the transition from a hemifusion intermediate to a fusion pore and contents mixing (Figure 1). Current understanding of the molecular mechanisms by which proteins fuse membranes is, to a large extent, based on the work on viral fusion.
Fusion Mediated by Diverse Viral Proteins
The entry of numerous enveloped viruses, including many human pathogens, into the host cell relies on fusion between the viral and the host cell membranes. In addition to viral-host membrane fusion, some viral fusogens mediate fusion between virus-infected cells and adjacent noninfected cells. The large numbers of organisms and cell types targeted by different viruses necessitate a diversity of both structure and function of the fusogenic arsenal. However, from the growing repertoire of enveloped viruses, only a few have been studied in depth as paradigms of virus-host cell fusion (Kielian and Rey, 2006).
In general, enveloped viruses invade cells by a pathway that involves binding of the viral particle to receptors at the host cell surface. Diverse triggers activate specific viral envelope glycoproteins that serve as fusogens. Activated fusogens fuse the viral envelope with either plasma or endosomal membranes, delivering the viral genome into the cytoplasm of the host cells.
Various Triggering Mechanisms Control Virus-Host Cell Membrane Fusion
Most viral fusogens are organized as homo- or hetero-oligomeric complexes comprising one (e.g., influenza; Earp et al., 2005, White, 1992) or up to four (e.g., herpes simplex; Subramanian and Geraghty, 2007) types of transmembrane envelope glycoproteins. Premature activation of the fusogen and incorrect fusion are suppressed either by the position of the fusogenic domains that are buried inside the protein, or by inhibitory interactions with peripheral subunits or accessory proteins. At the onset of fusion, these interactions are released to allow conformational changes of the proteins toward their fusogenic forms. Fusogen activation can be triggered by virus interaction with cell-surface receptors at neutral pH (e.g., HIV), or by virus internalization and exposure to mildly acidic pH along the endocytic pathway (e.g., influenza virus) (Earp et al., 2005). Alternatively, activation can occur via sequential receptor interactions and acidification, as in avian sarcoma and leukosis virus (Mothes et al., 2000). In the case of Ebola virus (Chandran et al., 2005) and SARS-coronavirus (Simmons et al., 2005), receptor binding induces conformational changes that allow cathepsin-mediated proteolysis within acidic endosomes, releasing fusogenic polypeptide fragments. In conclusion, the timing and location of virus-cell fusion is controlled by diverse mechanisms of fusogen activation that prevent premature nonproductive launching of the fusion machinery.
Structures and Mechanisms of Viral Fusogens
Crystallographical studies of protein fusogens from enveloped viruses have revealed many divergent structural motifs. However, these studies also uncovered a strikingly conserved hairpin structure of the postfusion conformations of viral fusogens, wherein their membrane-interacting amphiphilic peptide regions, referred to as fusion peptides, are positioned at the same end of rod-like molecules as the transmembrane domains. It is hypothesized that under fusion conditions, the viral fusogens first establish an extended conformation that delivers a bundle of exposed fusion peptides to the host cell membrane (Figures 1A–1C). Subsequent protein refolding into the final hairpin structure clamps the two membranes.
On the basis of their prefusion and postfusion structures, viral fusogens have been divided into several classes (Kielian and Rey, 2006, Weissenhorn et al., 2007).
Class I fusogens include hemagglutinins from orthomyxoviruses (e.g., influenza A, B, and C), F proteins from paramyxoviruses (e.g., parainfluenza viruses, measles virus, etc), ENV proteins from retroviruses, and fusogens of filoviruses and coronaviruses. Each class I protein is initially synthesized as an immature precursor that is proteolitically processed into two polypeptides. Ectodomains of at least some of these fusogens (e.g., influenza A hemagglutinin [HA] and HIV ENV) are organized in homotrimers that are probably oriented perpendicularly to the envelope surface. Fusion appears to require the concerted action of several HA trimers (Danieli et al., 1996, Kanaseki et al., 1997, Markovic et al., 2001, Yang et al., 2005). In contrast, fusion of HIV might require just a single ENV trimer (Yang et al., 2005). In the initial prefusion state, each trimer contains conserved fusion peptides, which are buried inside the trimeric complex. Activation induces a conformational change resulting in the formation of extended α-helical coiled-coil domains that are thought to insert the fusion peptides into the opposing membrane. In the final postfusion conformation, a six-helix bundle forms a hairpin structure that results in the colocalization of the transmembrane domains and fusion peptides at the same fused membrane (Figure 1A).
Class II fusogens include fusion proteins of alpha- and flaviviruses that lack any clear sequence homologies but have strikingly similar structures both in prefusion and postfusion states. In their prefusion conformation, these homodimeric or heterodimeric glycoproteins lie tangentially (parallel) to the virus envelope and have “internal” rather than “terminal” fusion peptides. These fusogens have β strands and lack any coiled-coil domains. In the postfusion conformation, the proteins form homotrimers of β-structured hairpins with the fusion peptide loops and the C-terminal membrane anchors positioned at the same end of the molecule (Figure 1B). Several class II fusogens are organized in complexes and networks that appear to play a role during the fusion process (Gibbons et al., 2003, Gibbons et al., 2004).
Fusion proteins of rhabdoviruses and herpesviruses combine some of the characteristic features of class I and class II proteins and, thus, might represent a class III family of viral fusogens (Roche et al., 2006, Roche et al., 2007, Kielian and Rey, 2006, Weissenhorn et al., 2007). These proteins harbor both α helices, which presumably form a six-helix bundle to fold back the protein as with class I viral fusogens, and an elongated β sheet structure with an amphiphilic fusion peptide at its end, reminiscent of class II viral fusogens. In contrast to class I and II proteins, this fusion peptide is very short—just a few hydrophobic amino acid residues—and is discontinuous. For example, in the prefusion conformation of vesicular stomatitis virus (VSV) envelope protein G, a bipartite fusion domain is exposed rather than hidden at an oligomeric interface. In both prefusion and postfusion conformations, the membrane-interacting fusion domains and transmembrane domains are located at the same end of the homotrimeric fusogen (Figure 1C). As with class I and II fusogens, class III proteins form complexes made of several fusogenic proteins that may have cooperative functions during membrane fusion (Gaudin et al., 1996, Roche et al., 2007).
While the best-characterized protein fusogens discussed above belong to enveloped viruses, some nonstructural proteins of nonenveloped reoviruses are expressed at the plasma membrane of infected cells and apparently promote dissemination of the virus by fusing infected and noninfected cells. These fusogens, referred to as fusion-associated small transmembrane (FAST) proteins, have just 40 amino acid residues in the ectodomain, which includes a short and mildly hydrophobic domain (Cheng et al., 2005, Shmulevitz et al., 2004, Shmulevitz and Duncan, 2000). FAST proteins are much smaller than the “classical” fusogens of enveloped viruses, which are 150 to 300 amino acid residues long; they lack any extended heptad repeat structures; and they do not appear to undergo major conformational changes under the conditions of fusion. FAST proteins represent an intriguing class of viral fusogens, and future exploration of the mechanisms by which they fuse cell membranes will likely broaden our understanding of cell fusion significantly.
Even for the best-characterized viral fusogens, the specific mechanisms of action are yet to be identified. The convergence of many viral fusogens with structurally diverse prefusion conformations to a similar hairpin postfusion conformation is believed to underlie a conserved principle of action (Kielian and Rey, 2006, Weissenhorn et al., 2007). According to several proposed mechanisms, refolding of the fusogen into the final hairpin structure exerts a pulling force that bends membranes and, thus, primes them for fusion (Bentz, 2000, Kozlov and Chernomordik, 1998, Weissenhorn et al., 1997). However, some findings seem to disagree with these models. (1) Formation of a hairpin structure for class I fusogens can be separated from fusion since the former either precedes (Borrego-Diaz et al., 2003, Park et al., 2003) or follows (Markosyan et al., 2003, Markosyan et al., 2004) membrane merger. (2) FAST proteins are unlikely to establish rigid hairpin structures and also appear to be too short (∼1.5 nm) to reach the target membrane through the 10–20 nm gap between contacting membranes (Shmulevitz and Duncan, 2000). (3) The pulling force that fusogens can apply to the membranes is limited by the tightness of their membrane anchoring by fusion peptides. If binding of the peptides to the membrane is weak, as in the case of short and/or not very hydrophobic peptides, the force would detach the anchors from the membrane before the membrane bends. From this perspective, pulling membranes together by VSV G likely requires a concerted action of multiple trimers because the very short fusion peptide loops of this protein (Roche et al., 2006) restrict the force that might be delivered by each trimer.
Based on the conserved membrane rearrangements, the job of the viral protein fusogen or fusogens includes mediating a local approach between biological membranes, and then catalyzing hemifusion and its transformation into a fusion pore (Figure 1). Post-hemifusion stages of opening, and especially fusion pore expansion, require more energy and a higher number of activated fusogens than hemifusion (Chernomordik and Kozlov, 2003, Cohen and Melikyan, 2004), and represent a point of no return in the process of cell fusion. It has been hypothesized that these later stages in fusion might be driven by lateral membrane tension generated by the assembly of multiple fusogenic molecules into a relatively rigid interconnected protein coat around the fusion site (Chernomordik and Kozlov, 2003, Kozlov and Chernomordik, 2002, Leikina et al., 2004). The finding that fusion proteins located outside the direct contact zone might be involved in driving fusion pore expansion (Leikina et al., 2004, Lenz et al., 2005) substantiates the hypothesis that this fusion stage is driven by long-range membrane stresses.
It is also unclear whether fusion pore expansion at the later fusion stages relies solely on the activity of the fusogens or whether other molecules are required. Candidate molecules that may help in the membrane deformation process include force-generating cytoskeletal proteins, phospholipases, and proteins that bend and disassemble membranes and junctional complexes.
We expect future work to emphasize the functional significance of the fusion peptide-membrane interactions (Tamm, 2003) and lateral interactions between multiple fusogens (see for review Chernomordik and Kozlov, 2003). In summary, while the textbook models of viral-mediated membrane fusion are currently widely accepted (Figures 1A–1C), recent findings suggest that these are just working models that will be modified or replaced as new molecular and structural details of the pathways of viral-host cell membrane fusion are discovered.
Concluding Remarks on Viral Fusogens
Figures 1A–1C show the current consensus model for viral membrane fusion. First, viral fusogens anchor the viral and host membranes, and then fold back into hairpins to reduce the distance between the two bilayers. Second, the fusion peptides or loops and transmembrane domains of diverse fusogens interact with the membranes to initiate fusion. Diverse viral fusogens including influenza virus HA (class I) (Chernomordik and Kozlov, 2003), Sindbis virus E1 glycoprotein (class II) (Zaitseva et al., 2005), Rabies G protein (class III) (Gaudin, 2000), and FAST proteins (Top et al., 2005) are reported to catalyze fusion pore opening through the hemifusion pathway. Third, lateral interactions between adjacent fusogens drive pore expansion (Chernomordik and Kozlov, 2003). Because this pathway is controlled by the elastic properties of membrane bilayers, many models converge on the hypothesis that fusogens act by shaping the membranes and generating elastic stresses that are released by hemifusion and fusion (Kozlov and Chernomordik, 1998, Kuzmin et al., 2001, Martens et al., 2007, Zimmerberg et al., 2006).
From Pathology to Physiology: Syncytins' Activities in Placental Fusion
Various viral fusogens including HIV ENV and the FAST proteins can promote fusion between an infected cell and naive neighboring cells. This cellular fusogenic activity of viral proteins is further exemplified by the proposed role of Syncytin proteins in placental fusion. In the placenta of some, but not all, mammals, the fetal cytotrophoblasts fuse to form continuous layers of multinucleate syncytiotrophoblasts localized at the maternal-fetal interface (Huppertz et al., 2006). Several lines of evidence tie cytotrophoblast fusion in primates and some rodents with the activity of proteins from endogenous retroviruses (ERVs) that were integrated into their genomes during evolution. From the large pool of human ERV (HERV), which makes up ∼8% of the human genome (de Parseval et al., 2003, Villesen et al., 2004), 16 HERV elements have been shown to encode intact ENV proteins. Taking into account the fusogenic activity of ENV proteins in retroviruses (Figure 1A), these HERV ENV proteins are candidates for the promotion of cell fusion in humans. mRNA or protein products of three of these env genes (ENV-R, W, and FRD) are predominantly expressed in the placenta (de Parseval and Heidmann, 2005). Two of these, Syncytin-1 (encoded by the HER-W element) and Syncytin-2 (encoded by HER-FRD), have been further characterized. Antisense knockdown of syncytin-1 in primary cytotrophoblasts that undergo fusion in culture results in fusion attenuation, suggesting that Syncytin-1 accounts at least in part for trophoblast cell fusion (Frendo et al., 2003). Syncytin-1 and -2 forced expression are sufficient to mediate cell fusion in various heterologous cell types, including insect cells and liposomes (Blaise et al., 2003, Blond et al., 2000, Mi et al., 2000), indicating that Syncytin-1 and -2 act as bona fide fusogens.
Regulation on Syncytins' Activities
In contrast to viral fusogens, there are only limited data on the structure and function of Syncytins. These single transmembrane proteins harbor all the domains characteristic of class I viral fusogens, including a potential amphiphilic fusion peptide, and undergo maturation steps that are reminiscent of HIV ENV (Blond et al., 1999, Cheynet et al., 2005). Sequence analysis and partial structural data of Syncytin-1 and -2 have revealed that they share structural domains with several class I viral fusogens, strengthening the notion that these proteins originated from these fusogens and may use similar mechanisms to promote fusion (Renard et al., 2005) (Figure 1A). However, it is not known whether the fusogenic activity of Syncytins relies on dynamic conformational changes of these fusogens that are similar to the proposed fusion mechanisms of class I viral fusogens. Furthermore, the potential membrane remodeling effects of Syncytins leading to the hemifusion membrane intermediate, pore formation, and complete fusion have not yet been determined. As with fusion mediated by their viral fusogen ancestors, fusion mediated by Syncytin-1 and -2 is dependent on the binding of these proteins to cell surface receptors. Syncytin-1 binds the ASCT1 and ASCT2 receptors (Blond et al., 2000, Marin et al., 2003), whereas the receptors of Syncytin-2 remain uncharacterized (Blaise et al., 2003). An additional layer of regulation of the fusogenic activities of these proteins is provided by their placental expression. Syncytin-2 expression is restricted to the cytotrophoblasts (Malassine et al., 2007), whereas Syncytin-1 is predominantly expressed in the cytotrophoblasts and syncytial trophoblasts (Blond et al., 2000, Frendo et al., 2003, Mi et al., 2000). Hence, placental activity of Syncytins emerged, presumably, due to the organism's transcriptional networks that control Syncytins' expression. In contrast, viral tissue specificity is regulated posttranscriptionally, and is determined primarily by the distribution of appropriate receptors on target cells, and by fusogen activation.
Evolution of Placental Cell Fusion: Why Do Some Mammals Use Fusogens of Viral Origin to Mediate Cellular Fusion?
Syncytins are a unique example of convergent evolution in which two pairs of endogenous retroviral genes were independently integrated and adapted for placental function (Dupressoir et al., 2005). In rodents, Syncytins appear to be limited to the Muridae family, including the mouse, the gerbil, and the hamster (Dupressoir et al., 2005). Hence, Syncytin integration occurred after the speciation of Muridae from other rodent families some 20 million years ago (MYA). Similarly, the primate-specific distribution of Syncytin-1 and -2 suggest that they integrated specifically into the genome of the primate ancestor 20–40 MYA. Lack of Syncytins in the genomes of other mammals demonstrates that the existence of these elements is not rooted in early stages of mammalian evolution, but rather that Syncytin integration occurred independently after the speciation of rodents and primates during later mammalian evolution.
Rodents that bear Syncytins in their genomes have placenta of a three-layered structure, with one cellular and two syncytiotrophoblast layers (trichorial placenta), formed by extensive cell fusion. In contrast the placentas of other rodents are simpler, comprised of one cellular and one syncytiotrophoblast layer (monochorial placenta) that require less fusion events. Similarly, placentas of primates seem to be more complex than those of related nonprimate mammals. This raises the possibility that formation of the complex placentas of primates and these rodents necessitates fast and efficient fusion. This may occur by the preferential adoption of proficient viral-derived ENV fusogens instead of other unidentified, possibly slower, endogenous placental fusogens that act in other organisms. Although expression of Syncytins and other cellular env genes from additional HERV elements is not restricted to cells of the placenta (de Parseval and Heidmann, 2005, Mi et al., 2000), the activities of these genes cannot account for all fusion events in mammals. It appears that cellular env genes are not expressed in muscles and bones, where massive fusion events occur, suggesting that these fusions rely on the activity of as yet unidentified fusogens. Thus, nonplacental fusogens probably fuse cells via different mechanisms, suggesting that even within a single organism the mechanisms underlying cell fusion are diverse.
The FF Family: The First Eukaryotic Developmental Cell-Cell Fusogens
Although cell fusion plays key roles in the development and physiology of most eukaryotes, most developmental fusogens are still unidentified. The nematode C. elegans has emerged as a powerful system for the identification of missing fusogens thanks to the combination of genetic screening with the advantage of an invariant and completely characterized cell lineage, enabling the dynamic study of cell fusion at a single-cell resolution within a living organism (Podbilewicz and White, 1994). Identifying and characterizing developmental fusogens of C. elegans can shed light on the identity and fusion mechanism of other, still elusive, eukaryotic fusogens. In the C. elegans hermaphrodite, stereotypic fusion of about one-third of the 959 somatic cells throughout development generates 44 syncytia essential for organogenesis of various organs such as the epidermis, the pharynx, the uterus, the hymen, and the vulva (Podbilewicz, 2006). Several genetic screens for somatic fusion failure (FF) phenotypes have identified two genes that, when mutated, result in specific FF phenotypes. One gene encodes the epithelial fusion failure-1 (EFF-1) protein that is required for most cell fusion events in the epidermis, pharynx, and vulva (Mohler et al., 2002). The second gene encodes the anchor-cell fusion failure-1 (AFF-1) protein that is essential for specific fusion events leading to the formation of small syncytia, such as fusion of the anchor cell (AC) to specific uterine cells to form the nematode's hymen, and fusion of a subset of vulval cells resulting in the formation of vulval rings vulA and vulD (Sapir et al., 2007).
Further analyses of eff-1 and aff-1 mutants using cell-junction markers, plasma-membrane and cytoplasmic dyes, and transmission electron microscopy (TEM) on eff-1 mutant worms have demonstrated that the FF proteins EFF-1 and AFF-1 act specifically and independently to account for most cell fusion events in C. elegans (Gattegno et al., 2007, Mohler et al., 2002, Podbilewicz et al., 2006, Sapir et al., 2007, Shemer and Podbilewicz, 2002, Shemer et al., 2004). The fusogenic activity of FF proteins was finally demonstrated by the finding that their ectopic expression is sufficient to fuse cells that normally do not fuse in C. elegans (Shemer et al., 2004), and even more significantly, by the fusion of heterologous insect cells in culture (Podbilewicz et al., 2006, Sapir et al., 2007). Fusion of heterologous cells by FF proteins in insect cells indicates that EFF-1 and AFF-1 are bona fide fusogens and provides an experimental system to study the mechanism of their fusogenic activity, analogous to the cell culture systems used successfully for many viral fusogens.
Structure of FF Proteins
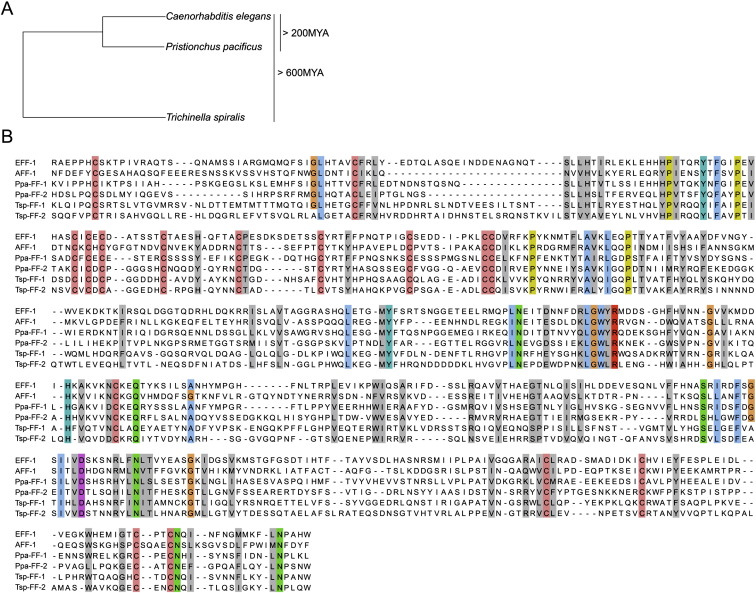
The aff-1 gene encodes one single-pass transmembrane protein, whereas the eff-1 gene encodes, by alternative splicing, two single-pass integral membrane proteins and two secreted polypeptides. Experiments in insect cell culture have demonstrated fusogenic activity for the transmembrane isoforms of FF proteins, whereas the function of the secreted forms remains uncharacterized (Podbilewicz et al., 2006, Sapir et al., 2007). Like most viral fusogens, the transmembrane forms of FF proteins possess an N-terminal signal sequence followed by a long extracellular portion, a predicted transmembrane domain, and a short intracellular tail. Sequence comparison of putative FF orthologs in several nematode species has uncovered novel motifs that do not exist in viral fusogens and might be critical for FF-mediated fusion. A striking conservation in the position and number of all 16 cysteines in the extracellular portion of FF proteins from 17 different nematode species suggests that these proteins are folded in a similar 3D structure that is essential for their fusogenic activity ( Figure 2; O.A. and B.P., unpublished data). Despite this conservation in nematodes, there is only minor sequence similarity of FFs to proteins from organisms outside the nematode phylum (Sapir et al., 2007). Most prominent is the spacing and position of nine extracellular cysteines defining a domain that exhibits similarity to the TGF-β binding domain of the activin/BMP/TGF-β type I receptor superfamily (Kirsch et al., 2000). Because this domain in FF proteins lacks critical residues that are essential for the binding of TGF-β ligands, it is unlikely that FF proteins function as TGF-β receptors (Sapir et al., 2007). This domain may emerge, however, as an interacting extracellular region conserved between nematode FF proteins and the unidentified fusogens in other phyla (Podbilewicz et al., 2006, Sapir et al., 2007).
Figure 2.
Conservation of Residues and Motifs in FF Proteins from Nematodes
(A) Phylogeny of nematode species used for the sequence alignment. Using BLAST, FF proteins were identified in 17 nematode species from different clades (O.A. and B.P., unpublished data). We aligned Caenorhabditis elegans (Ce) FF proteins with putative homologs from Pristionchus pacificus (Ppa), estimated to diverge 200 million years ago (MYA) (Gutierrez and Sommer, 2004), and with proteins from the remotest nematode wherein FF proteins have been identified so far, the parasite Trichinella spiralis (Tsp) (divergence estimated more then 600 MYA; Mitreva and Jasmer, 2006).
(B) Extracellular domains of FF proteins without signal sequences were aligned using the Jalview software (Clamp et al., 2004). Conservation of cysteines (pink), the LGWYR motifs, and partial conservation of prolines (yellow) suggests functional roles for these residues and motifs (Sapir et al., 2007). Color is shown in residues of 100% conservation and gray in cases where 50% of the physicochemical properties are conserved (Livingstone and Barton, 1993). Accession numbers: CeAFF-1: EF205023; CeEFF-1: C26D10.5 PpaFF-1: contig162.29; PpaFF-2: Contig735.1 P. pacificus california “Assembly Freeze 1.” TspFF-1: Contig1.317; TspFF-2: Contig3.96 Trichinella spiralis-1.0-contigs.
Fusion Mechanism of FF Proteins
Fusogen distribution on the two opposing membranes is one of the essential features of the FF fusion mechanism. Experiments in vivo and in heterologous culture cells indicate that expression of EFF-1 in only one of two bound cells is insufficient for fusion (Podbilewicz et al., 2006), and similar observations were made in vivo for AFF-1 (Sapir et al., 2007). The requirement for the same fusogen to be present on both fusing membranes thus defines a homotypic arrangement as a cornerstone of the FF fusion mechanism, distinguishing it from the single-membrane localization of viral and Syncytin fusogens (Figure 1; White, 2007). This homotypic machinery of FF mediated fusion is somewhat reminiscent of soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE)-dependent intracellular membrane fusion, which involves interactions among three different t-SNARE proteins localized to the surface of the target membrane and one v-SNARE protein on the opposing vesicle membrane (Bonifacino and Glick, 2004, Cai et al., 2007, Jahn et al., 2003, Jahn and Scheller, 2006). SNAREs contain domains consisting of a single α-helical amphipathic bundle that is unstructured in the nonassembled state (Figure 1D). SNARE interactions yield a hydrophobic helical core complex that is proposed to “zipper” the SNARE motifs from their N-terminal ends toward their C-terminal membrane anchors and, as a result, to clamp the membranes together (Jahn and Scheller, 2006). In addition to their vesicle fusion activity, SNARE proteins have the potential to fuse the plasma membranes of neighboring cells, as elegantly demonstrated in a system in which artificially engineered “flipped” SNAREs on the cell surface mediate hemifusion and complete cell-cell fusion (Giraudo et al., 2005, Hu et al., 2003).
Despite the similar requirement of fusogen presence in both fusing membranes, an essential distinction is that during SNARE-dependent fusion, each of the two membranes contributes a different combination of SNARE proteins (Figure 1D), while the same protein (e.g., EFF-1) is present on both membranes during FF-based fusion. Furthermore, there is no obvious long α-helical coiled-coil domain in the ectodomain region of the FF fusogens. It is possible that FF-mediated fusion occurs via a zippering mechanism like that proposed for SNAREs that leads to membrane conjunction and fusion (Figure 1E). However, this zippering possibly relies on the folding of cysteine-based structures in the extracellular domain of FF proteins and not on SNARE-like α-helical coiled-coil bundling. The novel homotypic arrangement of the fusogens identified for FF proteins may stem from the need for tight control on the execution of developmental fusion events. For example, homotypic design can prevent unregulated fusion at the edges of a multinucleate cell, allowing better control of syncytium size and shape (Podbilewicz et al., 2006). This mechanistic aspect may serve as a general theme in developmental cell fusion in multicellular organisms.
Are FF proteins the only fusogens in C. elegans? In the genome of C. elegans, there is only one more gene that encodes a protein with significant sequence similarity to the FF fusogens. Mutant analyses suggest that this gene may have, if any, only redundant functions in cell fusion. Importantly, eff-1 aff-1 double mutant analysis revealed that sperm-egg fusion seems to be normally executed in an FF-independent manner (Sapir et al., 2007). This event is facilitated, presumably, by other unidentified fusogens that may not share significant sequence similarity with the FF fusogens; thus, sperm-egg fusion may occur by a completely different mechanism.
Membrane Remodeling by Developmental Fusogens
Fusion via hemifusion emerges as a convergent point in the membrane fusion mechanism of all the diverse viral fusogens studied so far (Figure 1). The characteristic hemifusion phenotype, lipid mixing without content mixing, and fusion inhibition by hemifusion-blocking lipid lysophosphatidylcholine (LPC) (Chernomordik et al., 1993) were demonstrated for EFF-1 fusion in culture, indicating that EFF-1 proteins also fuse membranes via the hemifusion intermediate (Podbilewicz et al., 2006). The fusion of mammalian muscle cells in culture is also inhibited by LPC, suggesting a similar pathway of membrane rearrangements induced by unidentified mammalian fusogens (Reporter and Raveed, 1973; W.A. Mohler, personal communication).
In principle, different proteins might control distinct stages of the multistep fusion-through-hemifusion pathway of developmental fusion, with one protein to catalyze hemifusion, another to open a fusion pore, and still another to drive pore expansion. However, EFF-1 appears to control the entire pathway. As mentioned above, EFF-1 induces membrane rearrangement, leading to a hemifusion intermediate. EFF-1 also opens initial fusion pores, as evidenced by lack of cytoplasmic content mixing and pore formation in loss-of-function eff-1 mutants. Finally, partial activity of EFF-1 in temperature-sensitive mutant animals resulted in formation of partially expanding pores of 50–100 nm that fail to further expand (Gattegno et al., 2007, Shemer et al., 2004). Thus, EFF-1 is required first to induce membrane hemifusion at sites that are subsequently rearranged into membrane pores, and second to expand these pores to fully disassemble the contact junction to yield a syncytium (Gattegno et al., 2007, Podbilewicz et al., 2006). The requirement of many more viral glycoproteins for expansion of initial fusion pores (Chernomordik and Kozlov, 2003) raises the possibility that additional proteins are acting with EFF-1 to expand fusion pores. Recently, the role of the actin-remodeling molecules WASP and WIP in D. melanogaster muscle fusion pore expansion has been proposed (Massarwa et al., 2007) in parallel with a distinct role of these molecules in prefusion vesicle exocytosis (Kim et al., 2007). It is not known whether pore expansion in C. elegans relies solely on EFF-1 activity or requires additional proteins (for example, the C. elegans WIP and WASP orthologs).
Regulation of FF Fusion
In vivo studies have revealed that there is a strict correlation between the expression of FF mRNAs, induced in fusion-fated cells shortly before fusion, and the occurrence of cell fusion. Transcription factors from different families (e.g., homeobox, GATA, Zn fingers, FOS-1) positively and negatively control FF protein expression and hence FF-mediated fusion (Cassata et al., 2005, Margalit et al., 2007, Sapir et al., 2007, Shemer and Podbilewicz, 2002, Alper and Kenyon, 2001, Alper and Kenyon, 2002, Koh et al., 2002, Koh and Rothman, 2001, Podbilewicz, 2006). In addition to transcriptional control, posttranslational regulation of FF proteins is suggested by their dynamic localization in the fusing cells. In an elegant study, del Campo et al. (2005) showed that EFF-1 is specifically and dynamically enriched at the contact zone between two fusing cells in vivo. In addition, possible posttranslational modulators of FF fusion have been identified: fus-1, which encodes a subunit of the Vacuolar-ATPase complex that was proposed to function as a proton pump, represses EFF-1-mediated fusion (Kontani et al., 2005). nsf-1, which encodes the NEM-sensitive factor (NSF) that presumably functions in vesicle trafficking, may promote AFF-1-mediated fusion of the AC (Choi et al., 2006). This possible connection between FUS-1 and NSF-1 proteins and FFs was not further elucidated. Currently, there is no indication that FF-mediated fusion is regulated by conformational changes of these proteins like the trigger-dependent restructuring of viral fusogens. Although this mode of regulation might exist, it seems that transcriptional control combined with dynamic subcellular localization of the proteins might be a central mechanism to regulate and specify FF-mediated cell fusion within the organism.
Conclusions and Future Directions
Based on the current review, we ask four general questions that summarize what we know and what we still have to learn about cell fusion.
How Do Viral Fusogens Fuse Membranes?
We still do not know the molecular mechanisms by which even the best-characterized viral fusogens unite membranes. However, there are five principles that appear to be shared by most of the studied viruses (Figure 1): First, the prefusion structures of oligomeric viral glycoproteins are localized to one of the fusing membranes (the virus). Second, fusion triggering involves processing, receptor binding, exposure to low pH, or some combination thereof. Third, triggering elicits diverse conformational changes involving coiled-coils and/or β sheets exposing and inserting amphiphilic fusion domains into the opposing membrane. Fourth, diverse viral fusogens establish remarkably similar hairpin rod-like structures. Fifth, hairpin formation, interactions between the fusion domains and membranes, and, likely, lateral protein-protein interactions drive membrane rearrangements into a hemifusion intermediate followed by fusion pore formation and expansion.
Can We Use the Motifs of Viral Fusogens to Find Developmental Fusogens?
No; because of the diversity between the genuine fusogens at the level of primary sequences, the 3D structures, the triggering events, and the conformational changes during fusion, it appears unlikely that structural motifs can be used as reliable earmarks of a fusogen. Thus, the strategies used to find bona fide fusogens need to be adapted to each developmental system.
How Similar Are the Mechanisms of Viral and Developmental Fusion?
The structures of the fusogens, activation mechanisms, and requirements in one or both membranes (homotypic/heterotypic) are very diverse. However, the membrane pathway of fusion through hemifusion appears to be conserved between all classes of viral, intracellular SNARE, and developmental FF fusogens.
Will the Paradigms for FF and Syncytin Cell Fusion Help to Identify and Characterize Fusogens for Gametes, Myoblasts, Macrophages, and Trophoblasts?
Although the missing developmental fusogens may not be identified by sequence similarity with FFs or Syncytins, the discovery and characterization of the homotypic FF and the heterotypic Syncytin fusion mechanisms can serve as general guidelines for the identification and characterization of missing developmental fusogens. Syncytins may represent an intermediate transition from viral to developmental fusogens, combining a typical class I viral fusion mechanism with cellular transcriptional regulation during development. Deciphering the evolution of the fusion mechanism employed by Syncytins, and their role in vivo, are two intriguing topics for future research. Sperm-egg fusion may represent a heterotypic Syncytin-like fusion mechanism. As with virus-host fusion, fertilization involves fusion between heterotypic cells with different genetic programs.
FF proteins exhibit a different fusion mechanism, which relies on homotypic interactions between FF proteins anchored on the two opposing fusing membranes. We hypothesize that developmental fusion in muscles and bones will be regulated mainly by transcription, will require cooperativity of the fusogens, and will rely on homotypic interactions to restrict syncytium boundaries. One of the main challenges for the future is to find the missing developmental fusogens and determine the conserved and divergent mechanisms of cell membrane fusion.
Acknowledgments
We thank Michael Kozlov, Evgenia Leikina, Kamran Melikov, Meital Oren-Suissa, Eyal D. Schejter, Ben-Zion Shilo, three anonymous reviewers for critically reading the manuscript, and William A. Mohler for sharing unpublished results. Research in our laboratories was supported by grants from the Israel Science Foundation (B.P.) and by the Intramural Research Program of the National Institute of Child Health and Human Development, National Institutes of Health (L.V.C.).
Contributor Information
Benjamin Podbilewicz, Email: podbilew@tx.technion.ac.il.
Leonid V. Chernomordik, Email: chernoml@mail.nih.gov.
References
- Alper S., Kenyon C. REF-1, a protein with two bHLH domains, alters the pattern of cell fusion in C. elegans by regulating Hox protein activity. Development. 2001;128:1793–1804. doi: 10.1242/dev.128.10.1793. [DOI] [PubMed] [Google Scholar]
- Alper S., Kenyon C. The zinc finger protein REF-2 functions with the Hox genes to inhibit cell fusion in the ventral epidermis of C. elegans. Development. 2002;129:3335–3348. doi: 10.1242/dev.129.14.3335. [DOI] [PubMed] [Google Scholar]
- Bentz J. Membrane fusion mediated by coiled coils: a hypothesis. Biophys. J. 2000;78:886–900. doi: 10.1016/S0006-3495(00)76646-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaise S., de Parseval N., Benit L., Heidmann T. Genomewide screening for fusogenic human endogenous retrovirus envelopes identifies syncytin 2, a gene conserved on primate evolution. Proc. Natl. Acad. Sci. USA. 2003;100:13013–13018. doi: 10.1073/pnas.2132646100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blond J.L., Beseme F., Duret L., Bouton O., Bedin F., Perron H., Mandrand B., Mallet F. Molecular characterization and placental expression of HERV-W, a new human endogenous retrovirus family. J. Virol. 1999;73:1175–1185. doi: 10.1128/jvi.73.2.1175-1185.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blond J.L., Lavillette D., Cheynet V., Bouton O., Oriol G., Chapel-Fernandes S., Mandrand B., Mallet F., Cosset F.L. An envelope glycoprotein of the human endogenous retrovirus HERV-W is expressed in the human placenta and fuses cells expressing the type D mammalian retrovirus receptor. J. Virol. 2000;74:3321–3329. doi: 10.1128/jvi.74.7.3321-3329.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino J.S., Glick B.S. The mechanisms of vesicle budding and fusion. Cell. 2004;116:153–166. doi: 10.1016/s0092-8674(03)01079-1. [DOI] [PubMed] [Google Scholar]
- Borrego-Diaz E., Peeples M.E., Markosyan R.M., Melikyan G.B., Cohen F.S. Completion of trimeric hairpin formation of influenza virus hemagglutinin promotes fusion pore opening and enlargement. Virology. 2003;316:234–244. doi: 10.1016/j.virol.2003.07.006. [DOI] [PubMed] [Google Scholar]
- Byrne R.D., Garnier-Lhomme M., Han K., Dowicki M., Michael N., Totty N., Zhendre V., Cho A., Pettitt T.R., Wakelam M.J. PLCgamma is enriched on poly-phosphoinositide-rich vesicles to control nuclear envelope assembly. Cell. Signal. 2007;19:913–922. doi: 10.1016/j.cellsig.2006.10.011. [DOI] [PubMed] [Google Scholar]
- Cai H., Reinisch K., Ferro-Novick S. Coats, tethers, Rabs, and SNAREs work together to mediate the intracellular destination of a transport vesicle. Dev. Cell. 2007;12:671–682. doi: 10.1016/j.devcel.2007.04.005. [DOI] [PubMed] [Google Scholar]
- Cassata G., Shemer G., Morandi P., Donhauser R., Podbilewicz B., Baumeister R. ceh-16/engrailed patterns the embryonic epidermis of Caenorhabditis elegans. Development. 2005;132:739–749. doi: 10.1242/dev.01638. [DOI] [PubMed] [Google Scholar]
- Chandran K., Sullivan N.J., Felbor U., Whelan S.P., Cunningham J.M. Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science. 2005;308:1643–1645. doi: 10.1126/science.1110656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E.H., Olson E.N. Unveiling the mechanisms of cell-cell fusion. Science. 2005;308:369–373. doi: 10.1126/science.1104799. [DOI] [PubMed] [Google Scholar]
- Chen E.H., Grote E., Mohler W., Vignery A. Cell-cell fusion. FEBS Lett. 2007;581:2181–2193. doi: 10.1016/j.febslet.2007.03.033. [DOI] [PubMed] [Google Scholar]
- Cheng L.T., Plemper R.K., Compans R.W. Atypical fusion peptide of Nelson Bay virus fusion-associated small transmembrane protein. J. Virol. 2005;79:1853–1860. doi: 10.1128/JVI.79.3.1853-1860.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernomordik L.V., Kozlov M.M. Protein-lipid interplay in fusion and fission of biological membranes. Annu. Rev. Biochem. 2003;72:175–207. doi: 10.1146/annurev.biochem.72.121801.161504. [DOI] [PubMed] [Google Scholar]
- Chernomordik L.V., Kozlov M.M. Membrane hemifusion: crossing a chasm in two leaps. Cell. 2005;123:375–382. doi: 10.1016/j.cell.2005.10.015. [DOI] [PubMed] [Google Scholar]
- Chernomordik L.V., Vogel S.S., Sokoloff A., Onaran H.O., Leikina E.A., Zimmerberg J. Lysolipids reversibly inhibit Ca(2+)-, GTP- and pH-dependent fusion of biological membranes. FEBS Lett. 1993;318:71–76. doi: 10.1016/0014-5793(93)81330-3. [DOI] [PubMed] [Google Scholar]
- Chernomordik L.V., Zimmerberg J., Kozlov M.M. Membranes of the world unite! J. Cell Biol. 2006;175:201–207. doi: 10.1083/jcb.200607083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheynet V., Ruggieri A., Oriol G., Blond J.L., Boson B., Vachot L., Verrier B., Cosset F.L., Mallet F. Synthesis, assembly, and processing of the Env ERVWE1/syncytin human endogenous retroviral envelope. J. Virol. 2005;79:5585–5593. doi: 10.1128/JVI.79.9.5585-5593.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J., Richards K.L., Cinar H.N., Newman A.P. N-ethylmaleimide sensitive factor is required for fusion of the C. elegans uterine anchor cell. Dev. Biol. 2006;297:87–102. doi: 10.1016/j.ydbio.2006.04.471. [DOI] [PubMed] [Google Scholar]
- Clamp M., Cuff J., Searle S.M., Barton G.J. The Jalview Java alignment editor. Bioinformatics. 2004;20:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
- Cohen F.S., Melikyan G.B. The energetics of membrane fusion from binding, through hemifusion, pore formation, and pore enlargement. J. Membr. Biol. 2004;199:1–14. doi: 10.1007/s00232-004-0669-8. [DOI] [PubMed] [Google Scholar]
- Danieli T., Pelletier S.L., Henis Y.I., White J.M. Membrane fusion mediated by the influenza virus hemagglutinin requires the concerted action of at least three hemagglutinin trimers. J. Cell Biol. 1996;133:559–569. doi: 10.1083/jcb.133.3.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Parseval N., Heidmann T. Human endogenous retroviruses: from infectious elements to human genes. Cytogenet. Genome Res. 2005;110:318–332. doi: 10.1159/000084964. [DOI] [PubMed] [Google Scholar]
- de Parseval N., Lazar V., Casella J.F., Benit L., Heidmann T. Survey of human genes of retroviral origin: identification and transcriptome of the genes with coding capacity for complete envelope proteins. J. Virol. 2003;77:10414–10422. doi: 10.1128/JVI.77.19.10414-10422.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Campo J.J., Opoku-Serebuoh E., Isaacson A.B., Scranton V.L., Tucker M., Han M., Mohler W.A. Fusogenic activity of EFF-1 is regulated via dynamic localization in fusing somatic cells of C. elegans. Curr. Biol. 2005;15:413–423. doi: 10.1016/j.cub.2005.01.054. [DOI] [PubMed] [Google Scholar]
- Dupressoir A., Marceau G., Vernochet C., Benit L., Kanellopoulos C., Sapin V., Heidmann T. Syncytin-A and syncytin-B, two fusogenic placenta-specific murine envelope genes of retroviral origin conserved in Muridae. Proc. Natl. Acad. Sci. USA. 2005;102:725–730. doi: 10.1073/pnas.0406509102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earp L.J., Delos S.E., Park H.E., White J.M. The many mechanisms of viral membrane fusion proteins. Curr. Top. Microbiol. Immunol. 2005;285:25–66. doi: 10.1007/3-540-26764-6_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frendo J.L., Olivier D., Cheynet V., Blond J.L., Bouton O., Vidaud M., Rabreau M., Evain-Brion D., Mallet F. Direct involvement of HERV-W Env glycoprotein in human trophoblast cell fusion and differentiation. Mol. Cell. Biol. 2003;23:3566–3574. doi: 10.1128/MCB.23.10.3566-3574.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gattegno T., Mittal A., Valansi C., Nguyen K.C., Hall D.H., Chernomordik L.V., Podbilewicz B. Genetic control of fusion pore expansion in the epidermis of Caenorhabditis elegans. Mol. Biol. Cell. 2007;18:1153–1166. doi: 10.1091/mbc.E06-09-0855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudin Y. Rabies virus-induced membrane fusion pathway. J. Cell Biol. 2000;150:601–612. doi: 10.1083/jcb.150.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudin Y., Raux H., Flamand A., Ruigrok R.W. Identification of amino acids controlling the low-pH-induced conformational change of rabies virus glycoprotein. J. Virol. 1996;70:7371–7378. doi: 10.1128/jvi.70.11.7371-7378.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons D.L., Erk I., Reilly B., Navaza J., Kielian M., Rey F.A., Lepault J. Visualization of the target-membrane-inserted fusion protein of Semliki Forest virus by combined electron microscopy and crystallography. Cell. 2003;114:573–583. doi: 10.1016/s0092-8674(03)00683-4. [DOI] [PubMed] [Google Scholar]
- Gibbons D.L., Vaney M.C., Roussel A., Vigouroux A., Reilly B., Lepault J., Kielian M., Rey F.A. Conformational change and protein-protein interactions of the fusion protein of Semliki Forest virus. Nature. 2004;427:320–325. doi: 10.1038/nature02239. [DOI] [PubMed] [Google Scholar]
- Giraudo C.G., Hu C., You D., Slovic A.M., Mosharov E.V., Sulzer D., Melia T.J., Rothman J.E. SNAREs can promote complete fusion and hemifusion as alternative outcomes. J. Cell Biol. 2005;170:249–260. doi: 10.1083/jcb.200501093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez A., Sommer R.J. Evolution of dnmt-2 and mbd-2-like genes in the free-living nematodes Pristionchus pacificus, Caenorhabditis elegans and Caenorhabditis briggsae. Nucleic Acids Res. 2004;32:6388–6396. doi: 10.1093/nar/gkh982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heldwein E.E., Lou H., Bender F.C., Cohen G.H., Eisenberg R.J., Harrison S.C. Crystal structure of glycoprotein B from herpes simplex virus 1. Science. 2006;313:217–220. doi: 10.1126/science.1126548. [DOI] [PubMed] [Google Scholar]
- Hu C., Ahmed M., Melia T.J., Sollner T.H., Mayer T., Rothman J.E. Fusion of cells by flipped SNAREs. Science. 2003;300:1745–1749. doi: 10.1126/science.1084909. [DOI] [PubMed] [Google Scholar]
- Huppertz B., Bartz C., Kokozidou M. Trophoblast fusion: fusogenic proteins, syncytins and ADAMs, and other prerequisites for syncytial fusion. Micron. 2006;37:509–517. doi: 10.1016/j.micron.2005.12.011. [DOI] [PubMed] [Google Scholar]
- Jahn R., Scheller R.H. SNAREs–engines for membrane fusion. Nat. Rev. Mol. Cell Biol. 2006;7:631–643. doi: 10.1038/nrm2002. [DOI] [PubMed] [Google Scholar]
- Jahn R., Lang T., Sudhof T.C. Membrane fusion. Cell. 2003;112:519–533. doi: 10.1016/s0092-8674(03)00112-0. [DOI] [PubMed] [Google Scholar]
- Kanaseki T., Kawasaki K., Murata M., Ikeuchi Y., Ohnishi S. Structural features of membrane fusion between influenza virus and liposome as revealed by quick-freezing electron microscopy. J. Cell Biol. 1997;137:1041–1056. doi: 10.1083/jcb.137.5.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerbel R.S., Lagarde A.E., Dennis J.W., Donaghue T.P. Spontaneous fusion in vivo between normal host and tumor cells: possible contribution to tumor progression and metastasis studied with a lectin-resistant mutant tumor. Mol. Cell. Biol. 1983;3:523–538. doi: 10.1128/mcb.3.4.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kielian M., Rey F.A. Virus membrane-fusion proteins: more than one way to make a hairpin. Nat. Rev. Microbiol. 2006;4:67–76. doi: 10.1038/nrmicro1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Shilagardi K., Zhang S., Hong S.N., Sens K.L., Bo J., Gonzalez G.A., Chen E.H. A critical function for the actin cytoskeleton in targeted exocytosis of prefusion vesicles during myoblast fusion. Dev. Cell. 2007;12:571–586. doi: 10.1016/j.devcel.2007.02.019. [DOI] [PubMed] [Google Scholar]
- Kirsch T., Sebald W., Dreyer M.K. Crystal structure of the BMP-2-BRIA ectodomain complex. Nat. Struct. Biol. 2000;7:492–496. doi: 10.1038/75903. [DOI] [PubMed] [Google Scholar]
- Koh K., Rothman J.H. ELT-5 and ELT-6 are required continuously to regulate epidermal seam cell differentiation and cell fusion in C. elegans. Development. 2001;128:2867–2880. doi: 10.1242/dev.128.15.2867. [DOI] [PubMed] [Google Scholar]
- Koh K., Peyrot S.M., Wood C.G., Wagmaister J.A., Maduro M.F., Eisenmann D.M., Rothman J.H. Cell fates and fusion in the C. elegans vulval primordium are regulated by the EGL-18 and ELT-6 GATA factors–apparent direct targets of the LIN-39 Hox protein. Development. 2002;129:5171–5180. doi: 10.1242/dev.129.22.5171. [DOI] [PubMed] [Google Scholar]
- Kontani K., Moskowitz I.P., Rothman J.H. Repression of cell-cell fusion by components of the C. elegans vacuolar ATPase complex. Dev. Cell. 2005;8:787–794. doi: 10.1016/j.devcel.2005.02.018. [DOI] [PubMed] [Google Scholar]
- Kozlov M.M., Chernomordik L.V. A mechanism of protein-mediated fusion: coupling between refolding of the influenza hemagglutinin and lipid rearrangements. Biophys. J. 1998;75:1384–1396. doi: 10.1016/S0006-3495(98)74056-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov M.M., Chernomordik L.V. The protein coat in membrane fusion: lessons from fission. Traffic. 2002;3:256–267. doi: 10.1034/j.1600-0854.2002.030403.x. [DOI] [PubMed] [Google Scholar]
- Kuzmin P.I., Zimmerberg J., Chizmadzhev Y.A., Cohen F.S. A quantitative model for membrane fusion based on low-energy intermediates. Proc. Natl. Acad. Sci. USA. 2001;98:7235–7240. doi: 10.1073/pnas.121191898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leikina E., Mittal A., Cho M.S., Melikov K., Kozlov M.M., Chernomordik L.V. Influenza hemagglutinins outside of the contact zone are necessary for fusion pore expansion. J. Biol. Chem. 2004;279:26526–26532. doi: 10.1074/jbc.M401883200. [DOI] [PubMed] [Google Scholar]
- Lemaire I., Falzoni S., Leduc N., Zhang B., Pellegatti P., Adinolfi E., Chiozzi P., Di Virgilio F. Involvement of the purinergic P2X7 receptor in the formation of multinucleated giant cells. J. Immunol. 2006;177:7257–7265. doi: 10.4049/jimmunol.177.10.7257. [DOI] [PubMed] [Google Scholar]
- Lenz O., Dittmar M.T., Wagner A., Ferko B., Vorauer-Uhl K., Stiegler G., Weissenhorn W. Trimeric membrane-anchored gp41 inhibits HIV membrane fusion. J. Biol. Chem. 2005;280:4095–4101. doi: 10.1074/jbc.M411088200. [DOI] [PubMed] [Google Scholar]
- Livingstone C.D., Barton G.J. Protein sequence alignments: a strategy for the hierarchical analysis of residue conservation. Comput. Appl. Biosci. 1993;9:745–756. doi: 10.1093/bioinformatics/9.6.745. [DOI] [PubMed] [Google Scholar]
- Malassine A., Blaise S., Handschuh K., Lalucque H., Dupressoir A., Evain-Brion D., Heidmann T. Expression of the fusogenic HERV-FRD Env glycoprotein (syncytin 2) in human placenta is restricted to villous cytotrophoblastic cells. Placenta. 2007;28:185–191. doi: 10.1016/j.placenta.2006.03.001. [DOI] [PubMed] [Google Scholar]
- Margalit A., Neufeld E., Feinstein N., Wilson K.L., Podbilewicz B., Gruenbaum Y. Barrier to autointegration factor blocks premature cell fusion and maintains adult muscle integrity in C. elegans. J. Cell Biol. 2007;178:661–673. doi: 10.1083/jcb.200704049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin M., Lavillette D., Kelly S.M., Kabat D. N-linked glycosylation and sequence changes in a critical negative control region of the ASCT1 and ASCT2 neutral amino acid transporters determine their retroviral receptor functions. J. Virol. 2003;77:2936–2945. doi: 10.1128/JVI.77.5.2936-2945.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markosyan R.M., Cohen F.S., Melikyan G.B. HIV-1 envelope proteins complete their folding into six-helix bundles immediately after fusion pore formation. Mol. Biol. Cell. 2003;14:926–938. doi: 10.1091/mbc.E02-09-0573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markosyan R.M., Bates P., Cohen F.S., Melikyan G.B. A study of low pH-induced refolding of Env of avian sarcoma and leukosis virus into a six-helix bundle. Biophys. J. 2004;87:3291–3298. doi: 10.1529/biophysj.104.047696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markovic I., Leikina E., Zhukovsky M., Zimmerberg J., Chernomordik L.V. Synchronized activation and refolding of influenza hemagglutinin in multimeric fusion machines. J. Cell Biol. 2001;155:833–844. doi: 10.1083/jcb.200103005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens S., Kozlov M.M., McMahon H.T. How synaptotagmin promotes membrane fusion. Science. 2007;316:1205–1208. doi: 10.1126/science.1142614. [DOI] [PubMed] [Google Scholar]
- Massarwa R., Carmon S., Shilo B.Z., Schejter E.D. WIP/WASp-based actin-polymerization machinery is essential for myoblast fusion in Drosophila. Dev. Cell. 2007;12:557–569. doi: 10.1016/j.devcel.2007.01.016. [DOI] [PubMed] [Google Scholar]
- Mi S., Lee X., Li X.-p., Veldman G.M., Finnerty H., Racie L., LaVallie E., Tang X.-Y., Edouard P., Howes S. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature. 2000;403:785–789. doi: 10.1038/35001608. [DOI] [PubMed] [Google Scholar]
- Mitreva, M., and Jasmer, D.P. (2006). Biology and genome of Trichinella spiralis. In WormBook, The C. elegans Research Community, ed. (WormBook). 10.1895/wormbook.1.124.1 http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- Mohler W.A., Shemer G., del Campo J., Valansi C., Opoku-Serebuoh E., Scranton V., Assaf N., White J.G., Podbilewicz B. The type I membrane protein EFF-1 is essential for developmental cell fusion in C. elegans. Dev. Cell. 2002;2:355–362. doi: 10.1016/s1534-5807(02)00129-6. [DOI] [PubMed] [Google Scholar]
- Mothes W., Boerger A.L., Narayan S., Cunningham J.M., Young J.A. Retroviral entry mediated by receptor priming and low pH triggering of an envelope glycoprotein. Cell. 2000;103:679–689. doi: 10.1016/s0092-8674(00)00170-7. [DOI] [PubMed] [Google Scholar]
- Nakatogawa H., Ichimura Y., Ohsumi Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell. 2007;130:165–178. doi: 10.1016/j.cell.2007.05.021. [DOI] [PubMed] [Google Scholar]
- Oren-Suissa M., Podbilewicz B. Cell fusion during development. Trends Cell Biol. 2007;17:537–546. doi: 10.1016/j.tcb.2007.09.004. [DOI] [PubMed] [Google Scholar]
- Palovuori R., Eskelinen S. Role of vinculin in the maintenance of cell-cell contacts in kidney epithelial MDBK cells. Eur. J. Cell Biol. 2000;79:961–974. doi: 10.1078/0171-9335-00120. [DOI] [PubMed] [Google Scholar]
- Park H.E., Gruenke J.A., White J.M. Leash in the groove mechanism of membrane fusion. Nat. Struct. Biol. 2003;10:1048–1053. doi: 10.1038/nsb1012. [DOI] [PubMed] [Google Scholar]
- Podbilewicz, B. (2006). Cell fusion. In WormBook, The C. elegans Research Community, ed. (WormBook). doi:10.1895/wormbook.1.52.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- Podbilewicz B., White J.G. Cell fusions in the developing epithelia of C. elegans. Dev. Biol. 1994;161:408–424. doi: 10.1006/dbio.1994.1041. [DOI] [PubMed] [Google Scholar]
- Podbilewicz B., Leikina E., Sapir A., Valansi C., Suissa M., Shemer G., Chernomordik L.V. The C. elegans developmental fusogen EFF-1 mediates homotypic fusion in heterologous cells and in vivo. Dev. Cell. 2006;11:471–481. doi: 10.1016/j.devcel.2006.09.004. [DOI] [PubMed] [Google Scholar]
- Renard M., Varela P.F., Letzelter C., Duquerroy S., Rey F.A., Heidmann T. Crystal structure of a pivotal domain of human syncytin-2, a 40 million years old endogenous retrovirus fusogenic envelope gene captured by primates. J. Mol. Biol. 2005;352:1029–1034. doi: 10.1016/j.jmb.2005.07.058. [DOI] [PubMed] [Google Scholar]
- Reporter M., Raveed D. Plasma membranes: isolation from naturally fused and lysolecithin-treated muscle cells. Science. 1973;181:863–865. doi: 10.1126/science.181.4102.863. [DOI] [PubMed] [Google Scholar]
- Roche S., Bressanelli S., Rey F.A., Gaudin Y. Crystal structure of the low-pH form of the vesicular stomatitis virus glycoprotein G. Science. 2006;313:187–191. doi: 10.1126/science.1127683. [DOI] [PubMed] [Google Scholar]
- Roche S., Rey F.A., Gaudin Y., Bressanelli S. Structure of the prefusion form of the vesicular stomatitis virus glycoprotein G. Science. 2007;315:843–848. doi: 10.1126/science.1135710. [DOI] [PubMed] [Google Scholar]
- Sapir A., Choi J., Leikina E., Avinoam O., Valansi C., Chernomordik L.V., Newman A.P., Podbilewicz B. AFF-1, a FOS-1-regulated fusogen, mediates fusion of the anchor cell in C. elegans. Dev. Cell. 2007;12:683–698. doi: 10.1016/j.devcel.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shemer G., Podbilewicz B. LIN-39/Hox triggers cell division and represses EFF-1/Fusogen-dependent vulval cell fusion. Genes Dev. 2002;16:3136–3141. doi: 10.1101/gad.251202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shemer G., Podbilewicz B. The story of cell fusion: big lessons from little worms. Bioessays. 2003;25:672–682. doi: 10.1002/bies.10301. [DOI] [PubMed] [Google Scholar]
- Shemer G., Suissa M., Kolotuev I., Nguyen K.C.Q., Hall D.H., Podbilewicz B. EFF-1 is sufficient to initiate and execute tissue-specific cell fusion in C. elegans. Curr. Biol. 2004;14:1587–1591. doi: 10.1016/j.cub.2004.07.059. [DOI] [PubMed] [Google Scholar]
- Shmulevitz M., Duncan R. A new class of fusion-associated small transmembrane (FAST) proteins encoded by the non-enveloped fusogenic reoviruses. EMBO J. 2000;19:902–912. doi: 10.1093/emboj/19.5.902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmulevitz M., Corcoran J., Salsman J., Duncan R. Cell-cell fusion induced by the avian reovirus membrane fusion protein is regulated by protein degradation. J. Virol. 2004;78:5996–6004. doi: 10.1128/JVI.78.11.5996-6004.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons G., Gosalia D.N., Rennekamp A.J., Reeves J.D., Diamond S.L., Bates P. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc. Natl. Acad. Sci. USA. 2005;102:11876–11881. doi: 10.1073/pnas.0505577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian R.P., Geraghty R.J. Herpes simplex virus type 1 mediates fusion through a hemifusion intermediate by sequential activity of glycoproteins D, H, L, and B. Proc. Natl. Acad. Sci. USA. 2007;104:2903–2908. doi: 10.1073/pnas.0608374104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamm L.K. Hypothesis: spring-loaded boomerang mechanism of influenza hemagglutinin-mediated membrane fusion. Biochim. Biophys. Acta. 2003;1614:14–23. doi: 10.1016/s0005-2736(03)00159-7. [DOI] [PubMed] [Google Scholar]
- Top D., de Antueno R., Salsman J., Corcoran J., Mader J., Hoskin D., Touhami A., Jericho M.H., Duncan R. Liposome reconstitution of a minimal protein-mediated membrane fusion machine. EMBO J. 2005;24:2980–2988. doi: 10.1038/sj.emboj.7600767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villesen P., Aagaard L., Wiuf C., Pedersen F.S. Identification of endogenous retroviral reading frames in the human genome. Retrovirology. 2004;1:32. doi: 10.1186/1742-4690-1-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissenhorn W., Dessen A., Harrison S.C., Skehel J.J., Wiley D.C. Atomic structure of the ectodomain from HIV-1 gp41. Nature. 1997;387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- Weissenhorn W., Hinz A., Gaudin Y. Virus membrane fusion. FEBS Lett. 2007;581:2150–2155. doi: 10.1016/j.febslet.2007.01.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J.M. Membrane fusion. Science. 1992;258:917–924. doi: 10.1126/science.1439803. [DOI] [PubMed] [Google Scholar]
- White J.M. The first family of cell-cell fusion. Dev. Cell. 2007;12:667–668. doi: 10.1016/j.devcel.2007.04.009. [DOI] [PubMed] [Google Scholar]
- Xu Y., Zhang F., Su Z., McNew J.A., Shin Y.K. Hemifusion in SNARE-mediated membrane fusion. Nat. Struct. Mol. Biol. 2005;12:417–422. doi: 10.1038/nsmb921. [DOI] [PubMed] [Google Scholar]
- Yang X., Kurteva S., Ren X., Lee S., Sodroski J. Stoichiometry of envelope glycoprotein trimers in the entry of human immunodeficiency virus type 1. J. Virol. 2005;79:12132–12147. doi: 10.1128/JVI.79.19.12132-12147.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaitseva E., Mittal A., Griffin D.E., Chernomordik L.V. Class II fusion protein of alphaviruses drives membrane fusion through the same pathway as class I proteins. J. Cell Biol. 2005;169:167–177. doi: 10.1083/jcb.200412059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerberg J., Akimov S.A., Frolov V. Synaptotagmin: fusogenic role for calcium sensor? Nat. Struct. Mol. Biol. 2006;13:301–303. doi: 10.1038/nsmb0406-301. [DOI] [PubMed] [Google Scholar]