Abstract
A role of polyubiquitination in the activation of IκB kinase (IKK) through a proteasome-independent mechanism was first reported in 1996, but the physiological significance of this finding was not clear until 2000 when TRAF6 was found to be a ubiquitin E3 ligase that catalyzes lysine-63 (K63) polyubiquitination. Since then, several proteins known to regulate IKK have been linked to the ubiquitin pathway. These include the deubiquitination enzymes CYLD and A20 that inhibit IKK, and the ubiquitin binding proteins NEMO and TAB2 which are the regulatory subunits of IKK and TAK1 kinase complexes, respectively. Now accumulating evidence strongly supports a central role of K63 polyubiquitination in IKK activation by multiple immune and inflammatory pathways. Interestingly, recent research suggests that some alternative ubiquitin chains such as linear or K11 ubiquitin chains may also play a role in certain pathways such as the TNF pathway. Here I present a historical narrative of the discovery of the role of ubiquitin in IKK activation, review recent advances in understanding the role and mechanism of ubiquitin-mediated IKK activation, and raise some questions to be resolved in future research.
Introduction
The early 1990s was a golden era for both ubiquitin and NF-κB fields, although the two fields barely talked to each other at the time. Avram Hershko, Aaron Ciechanover, Irwin Rose and their colleagues discovered the ubiquitin pathway and elucidated the ubiquitin conjugation and deconjugation cascades that involve ubiquitin E1, E2, E3 and deubiquitination enzymes (DUBs)(1, 2). Alex Varshavsky and his colleagues proved that the ubiquitin system plays a dominant role in intracellular protein degradation in vivo(3, 4). It was also demonstrated that some prominent proteins, such as p53 and cyclin, were degraded by the ubiquitin-proteasome pathway(5, 6), thus removing the stigma that the ubiquitin system is merely a ‘garbage disposal’
By the early 1990s, David Baltimore and his colleagues had discovered NF-κB and IκB, and established the paradigm of NF-κB regulation that is still guiding our research today(7–10). The paradigm is simple and elegant: NF-κB, which consists of two subunits of the Rel family, is sequestered in the cytoplasm of most cells in association with the inhibitor IκB. Stimulation of cells with a variety of agents leads to the dissociation of IκB from NF-κB, which then enters the nucleus to turn on a battery of genes important for immune and inflammatory responses.
The question in the NF-κB field then became: how is IκB dissociated from NF-κB? Patrick Baeuerle and others found that IκBα was rapidly degraded when cells were stimulated with inflammatory cytokines(11). The degradation of IκBα correlated with NF-κB activation. Furthermore, serine protease inhibitors such as TPCK and TLCK inhibited IκBα degradation and prevented NF-κB activation. These experiments suggest that IκBα degradation allows NF-κB to enter the nucleus, and the protease for IκBα degradation might be a serine protease. However, it was later found that TPCK and TLCK also inhibited IκBα phosphorylation, so the effect of these chemicals on IκBα degradation was indirect. Nevertheless, these studies are important because they brought IκB degradation to the scene of the NF-κB field.
Tom Maniatis and his colleagues were the first to link the ubiquitin-proteasome pathway with the NF-κB pathway(12). In 1994, they showed that the processing of the NF-κB precursor p105 to p50 requires the ubiquitin-proteasome pathway. Furthermore, they found that TNFα-induced degradation of IκBα was blocked by proteasome inhibitors, which were developed by a small start-up company called MyoGenics (later renamed ProScript).
Personal and Historical Narrative
In the spring of 1994, I joined MyoGenics to help develop proteasome inhibitors as a potential treatment for muscle wasting (hence the company name). MyoGenics needed someone with expertise in the ubiquitin system at a time when only a dozen labs in the world worked on ubiquitin. My Ph.D. mentor Cecile Pickart, one of the pioneers in the ubiquitin field, recommended me to MyoGenics. My first assignment was to make polyubiquitinated conjugates for the 26S proteasome so that inhibitors could be tested. I completed this task rather quickly. Then my interest shifted to NF-κB because I had trained with Inder Verma as a postdoctoral fellow at the Salk Institute, where I was introduced to NF-κB. With some experience in both NF-κB and ubiquitin, I felt I was in a unique position to figure out how the ubiquitin-proteasome pathway regulates NF-κB. In collaboration with Tom Maniatis, who was one of the founders of the company, I showed that IκBα was polyubiquitinated in a manner that depended on phosphorylation at two serine residues (Ser32 and 36), which were independently shown by Uli Siebenlist and Dean Ballard to be important for IκBα degradation and NF-κB activation(13–15). We also showed that polyubiquitination was required for IκBα degradation by the 26S proteasome in vitro(13)
My next task was to identify the enzymes responsible for IκBα ubiquitination. Inspired by the biochemical approaches of Avram Hershko and Aaron Ciechanover, who used anion exchange chromatography to separate ubiquitin and ubiquitination enzymes into different fractions, I also prepared Fraction I (unbound) and Fraction II (bound to and eluted from the column) from HeLa cytosolic extracts (S100)(16). As my goal was to identify the E2 and E3 for IκBα, I included E1, ubiquitin, 35S-labeled IκBα, Fraction I, Fraction II and ATP in the reaction mixture. I was able to detect IκBα ubiquitination as well as its phosphorylation. Surprisingly, when I left out Fraction I or Fraction II, IκBα phosphorylation disappeared. Based on my knowledge of the ubiquitin system at the time, I knew Fraction I contained some E2s such as Ubc5. When I replaced Fraction I with Ubc5, IκBα phosphorylation was restored. A few more experiments revealed that IκBα phosphorylation depended on E1 and ubiquitin but was inhibited by methylated ubiquitin, suggesting that polyubiquitination was important. Proteasome inhibitors and a mutation of K48 of ubiquitin had no effect on IκBα phosphorylation. These results suggest an unprecedented possibility that a unique form of polyubiquitination activates an IκB kinase through a proteasome-independent mechanism. To identify the IκB kinase, I further purified Fraction II based on the IκB phosphorylation assay. Gel filtration experiments showed that the kinase activity resides in a large complex with a molecular weight corresponding to approximately 700 kDa. Based on these results, we proposed that the IκB kinase is a large, ~700 kDa complex that can be activated by polyubiquitination through a proteasome-independent mechanism that remains to be elucidated (16).
The publication of our Cell paper in 1996 immediately drew considerable skepticism in part because our claim that ubiquitination had a proteolysis-independent function in activating a protein kinase was simply too radical at the time when none of the IKK subunits was molecularly cloned yet. Admittedly, the claim was based almost entirely on in vitro biochemical experiments and we had no clue as to how ubiquitination might play a role in IKK activation in cells by any physiological stimulus. This predicament lasted for a few years until we started working on TRAF6 in my laboratory at the University of Texas Southwestern Medical Center at Dallas.
In 1999, soon after the identification of β-TrCP as the substrate-targeting subunit of the IκB ubiquitin E3 complex(17–19), we tried to understand how IKK is activated by different signaling pathways. We focused on TRAF6 because it was shown that knockout of TRAF6 in mice blocked NF-κB activation by IL-1β and LPS(20, 21). Since overexpression of TRAF6 in cells activates NF-κB(22, 23), we asked if we could use recombinant TRAF6 protein to activate IKK in HeLa cytosolic extracts. This assay, which was carried out by my postdoctoral fellow Li Deng, worked out beautifully, thus providing us a cell-free system to study how TRAF6 activates IKK(24). We then fractionated HeLa extracts and purified two factors that could support IKK activation by TRAF6. Remarkably, the first factor was a ubiquitin E2 complex consisting of Ubc13 and Uev1A. The yeast homologue of this E2 complex was shown by Cecile Pickart to catalyze the synthesis of K63 polyubiquitin chains that are important for DNA repair(25). The discovery of Ubc13/Uev1A as an IKK activator prompted us to test whether TRAF6 is a ubiquitin E3. Indeed, TRAF6 dramatically enhanced the synthesis of K63 polyubiquitin chains by Ubc13/Uev1A(24). Around that time, Alan Weissman at NIH showed that a family of proteins containing a conserved RING domain could stimulate E2-dependent ubiquitination(26). Interestingly, TRAF6 and other proteins of the TRAF family contain a highly conserved RING domain that is important for NF-κB activation(22). We found that mutations in the RING domain of TRAF6 abolished its ubiquitin ligase activity as well as its ability to activate IKK. Experiments using a panel of ubiquitin mutants containing single or multiple mutations in the lysine residues revealed that a lysine at position 63 of ubiquitin is both necessary and sufficient to support IKK activation by TRAF6(24). The discovery that TRAF6 is a ubiquitin E3 ligase and that K63 polyubiquitination is important for IKK activation immediately provides the physiological relevance of our earlier finding of a role of polyubiquitination in IKK activation.
The second factor we purified from HeLa extracts turned out to be a protein kinase complex consisting of TAK1, TAB1 and TAB2(27). TAK1 was initially thought to be important for TGFβ signaling and later found to be involved in the IL-1 pathway(28, 29). We found that the TAK1 kinase complex is activated by TRAF6-catalyzed K63 polyubiquitination. After TAK1 is activated, it phosphorylates IKKβ at two serine residues within the activation loop, leading to IKK activation. Thus, TAK1 is an IKK kinase. The ubiquitin-activated TAK1 also phosphorylates MKKs (e.g, MKK6), leading to the activation of JNK and p38 kinase pathways(27).
How does K63 polyubiquitination activate TAK1? The first clue to answering this question was our observation that TAB2 and its homologue TAB3 contain a highly conserved zinc finger motif termed NZF which is a ubiquitin-binding domain(30). We found that the NZF of TAB2 and TAB3 bound preferentially to K63 polyubiquitin chains, and that mutations within this domain that disrupted ubiquitin binding also impaired the ability of TAB2 and TAB3 to activate NF-κB. These results led us to propose that the binding of TAB2 or TAB3 to K63 polyubiquitin chains results in the activation of the TAK1 kinase (Figure 1).
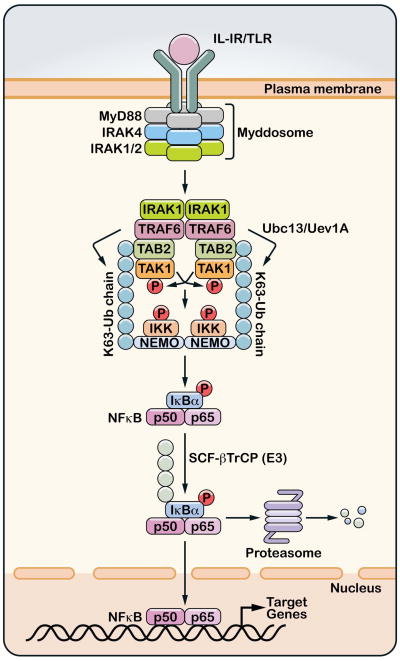
Figure 1. Ubiquitin-mediated activation of TAK1 and IKK in IL-1R/TLR pathways.
Stimulation of interleukin-1 receptor (IL-1R) or one of the Toll-like receptors (TLR) leads to the dimerization of the receptor and subsequent recruitment of the Myddosome complex, which consists of MyD88, IRAK4, IRAK1 (or IRAK2). IRAK1 is phosphorylated by IRAK4 and then associates with and activates the ubiquitin E3 ligase TRAF6. TRAF6 functions together with the ubiquitin E2 complex composed of Ubc13 and Uev1A to catalyze the synthesis of K63-linked polyubiquitin chains which are conjugated to other proteins or unanchored. Unanchored K63 polyubiquitin chains have been shown to bind the TAB2 subunit of the TAK1 kinase complex, and this binding promotes autophosphorylation of TAK1, which results in its activation. The polyubiquitin chains also bind NEMO to recruit the IKK complex, thereby facilitating the phosphorylation of IKKβ by TAK1. IKK is then activated to phosphorylate IκBα, which is polyubiquitinated by the ubiquitin ligase complex SCF-βTrCP and degraded by the 26S proteasome. NF-κB (represented by p50/p65 dimer) translocates to the nucleus to turn on the expression of many target genes.
Interestingly, NEMO, the essential regulatory subunit of the IKK complex, is also a ubiquitin-binding protein. Jonathan Ashwell’s lab and our lab independently discovered a ubiquitin-binding domain encompassing a coiled-coil region of NEMO(31, 32). Mutations within this domain that disrupt ubiquitin binding also impair IKK activation. Interestingly, several of these mutations that affect ubiquitin binding are found in human patients inflicted with ectodermal dysplasia with immunodeficiency (EDA-ID)(33). The NEMO ubiquitin binding (NUB, also known as UBAN, NOA or CoZi) domain was later shown to bind linear di-ubiquitin with even greater affinity than K63 di-ubiquitin(34, 35), raising the possibility that linear ubiquitination might also regulate IKK activity (see below). However, NEMO contains another ubiquitin-binding domain at the C-terminus, and it was shown that a NEMO fragment containing both NUB and the C-terminal ubiquitin binding domains has increased affinity for K63 polyubiquitin chains(36).
The findings that TAB2 and NEMO, which are regulatory subunits of the TAK1 and IKK complexes, respectively, are ubiquitin receptors provide strong support for the role of ubiquitination in the activation of these kinases. Further support to this model came from the discoveries by other laboratories that CYLD and A20 function as deubiquitination enzymes to inhibit IKK (reviewed by V. Dixit)(37–41). Moreover, as discussed below, recent studies by many laboratories have provided strong evidence that K63 polyubiquitination plays a critical role in signal transduction in multiple pathways, including those triggered by the T cell receptor, Toll-like receptors, RIG-I-like receptors, NOD-like receptors, and by DNA damage in the nucleus(42). More recently, we have shown that depletion of Ubc13 or replacement of endogenous ubiquitin with K63R ubiquitin in a human cell line completely abolished IKK activation by IL-1β, thus providing definitive evidence for an essential role of K63 polyubiquitination in the IL-1 pathway(43). Interestingly, we found that cells expressing K63R ubiquitin could still activate IKK in response to TNFα stimulation. Further, depletion of Ubc5 but not Ubc13 abolished IKK activation by TNFα. These results suggest that other ubiquitination events, such as alternative polyubiquitin chains, could mediate IKK activation by collaborating with or compensating for K63 polyubiquitination in the TNF pathway (see further discussion below). In this regard, it is intriguing to note that we found Ubc5 as an IKK activator 15 years ago(16). We still don’t know how Ubc5 activates IKK in cells, but the fact that Ubc5 could synthesize different types of polyubiquitin chains, and our finding that Ubc5 is involved in IKK activation by TNFα, might provide important clues to this longstanding question.
Several years ago, we ventured into the RIG-I pathway of antiviral innate immunity and discovered the mitochondrial antiviral signaling protein MAVS(44). Just as we did not expect our studies on IKK and then TRAF6 would repeatedly take us back to ubiquitination, we were surprised to find that the RIG-I - MAVS pathway is heavily regulated by ubiquitin (see below). Our inability to escape ubiquitination underscores the pervasiveness of this regulatory mechanism in cell signaling.
Role of K63 polyubiquitination in IKK activation by diverse stimuli
Interleukin-1 and Toll-like receptor pathways
IL-1 receptor and Toll-like receptors (TLRs) share an intracellular TIR domain that binds to the adaptor protein MyD88, which then recruits the IRAK family of protein kinases, including IRAK1, 2 and 4. IRAK1 binds to TRAF6, which then functions together with Ubc13/Uev1A to catalyze K63 polyubiquitination (Figure 1) (45). Several proteins, including IRAK1 and TRAF6, are modified by K63 polyubiquitin chains following IL-1 stimulation; however, there is no direct evidence that ubiquitination of any of these proteins leads to TAK1 or IKK activation. We recently showed that free polyubiquitin chains, which are not conjugated to any cellular protein, bind to the TAB2 subunit of the TAK1 kinase complex(46). This binding leads to cross autophosphorylation of TAK1 at Thr-181, resulting in TAK1 activation. In support of this model, IL-1 stimulation leads to accumulation of free polyubiquitin chains that associate with TAB2(46). After TAK1 is activated, it phosphorylates IKKβ and MKKs, leading to the activation of IKK and MAK kinases as described above. Interestingly, while activated TAK1 could directly phosphorylate MKKs, the phosphorylation of IKKβ by activated TAK1 still requires polyubiquitin chains(47). These polyubiquitin chains likely recruit both TAK1 and IKK complexes through binding to the TAB2 and NEMO subunits, thereby facilitating IKK phosphorylation by TAK1 (Figure 1).
While multiple lines of biochemical and genetic evidence strongly support the critical role of K63 polyubiquitination in the IL-1 pathway, much less is known about the role of ubiquitination in the TLR pathways. Since MyD88 and TRAF6 are essential for IKK activation by several TLRs, it is assumed that K63 polyubiquitination is important for signaling in these TLR pathways as well. However, direct evidence is lacking in most cases. Interestingly, the study of TLR4 signaling suggests an interplay between K63 and K48 polyubiquitination in controlling downstream signaling outcomes(48). Upon stimulation by LPS, TLR4 recruits MyD88 and another adaptor TRIF to induce proinflammatory cytokines and type-I interferons, respectively. Several TRAF proteins, including TRAF6 and TRAF3, are recruited to both of these adaptors. In addition, cellular inhibitors of apoptosis, cIAP1 and cIAP2, which are also RING domain ubiquitin E3 ligases, are recruited to MyD88 but not TRIF. In the MyD88 complex, TRAF6 catalyzes K63 polyubiquitination of cIAPs, which in turn promote K48 polyubiquitination and subsequent degradation of TRAF3. The degradation of TRAF3 allows cytosolic translocation of a signaling complex containing MyD88, TAK1 and MAP kinases, resulting in the activation of MAP kinases and the induction of proinflammatory genes. In the TRIF complex, which is devoid of cIAPs, TRAF3 catalyzes its own K63 polyubiquitination, which is important for inducing type-I interferons.
RIG-I-like receptor pathway
RIG-I – like receptors (RLRs) detect viral RNA in the cytoplasm and trigger signaling cascades leading to the production of type-I interferons and other antiviral molecules (Figure 2) (49, 50). RLRs consist of three members, RIG-I, MDA5 and LGP2, all of which contain an RNA helicase domain. RIG-I and MDA5 also contain N-terminal tandem CARD domains that are important for their signaling functions. LGP2 lack the CARDs and therefore serve regulatory functions. RIG-I contains a C-terminal domain (CTD) that binds to viral RNA containing 5′-triphosphate(51, 52), which is distinguished from mammalian cellular RNA that normally contains 5′ modifications such as a 5′-cap in mRNA. The binding of viral RNA to CTD of RIG-I induces a dramatic conformational change that exposes the N-terminal CARDs, which then interact with the CARD domain of the mitochondrial adaptor protein MAVS (also known as IPS-1, VISA or CARDIF)(44, 53–55). MAVS resides on the mitochondrial outer membrane and activates cytosolic protein kinases including IKK and the IKK-like kinase TBK1(44). IKK activates NF-κB, whereas TBK1 phosphorylates the transcription factor IRF3. IRF3 and NF-κB translocate to the nucleus where they function together to induce type-I interferons. Genetic experiments have shown that RIG-I is responsible for immune detection of many RNA viruses, such as influenza virus, vesicular stomatitis virus, Sendai virus and hepatitis C virus, whereas MDA5 is specifically required for interferon induction by picornaviruses (e.g., encephalomyocarditis virus)(56). MDA5 binds to long double-stranded RNA such as poly[I:C], but the nature of viral RNA ligand for MDA5 is still not well defined.
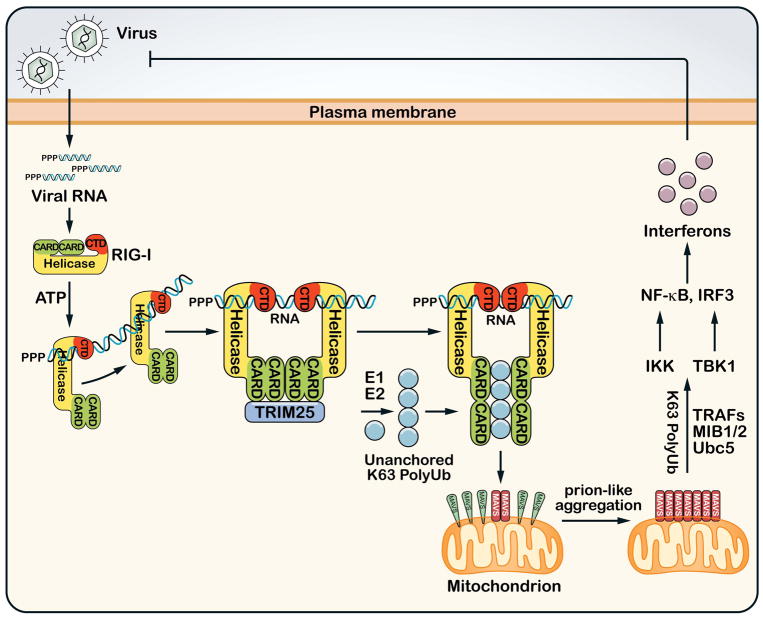
Figure 2. Ubiquitination in the RIG-I antiviral innate immunity pathway.
After infecting a host cell, RNA viruses replicate their RNA, which contains 5′-triphosphate and double-stranded segments that are recognized by the cytosolic sensor protein RIG-I. RIG-I contains an RNA helicase domain and a C-terminal domain (CTD) that bind to viral RNA. The RNA binding and ATP hydrolysis induce the translocation of RIG-I on the RNA and a conformational change that exposes the N-terminal CARD domains, which recruit the ubiquitin E3 TRIM25 to synthesize unanchored K63 polyubiquitin chains. These ubiquitin chains bind to the CARD domains of RIG-I, enabling RIG-I to induce a prion-like aggregation of MAVS on the mitochondrial membrane. The MAVS aggregates then activate IKK and TBK1 through K63 polyubiquitination that involves the E2 Ubc5 and E3s such as TRAFs or MIB1/2. IKK and TBK1 activate NF-κB and IRF3, respectively, which enter the nucleus to induce type-I interferons and other antiviral molecules.
K63 polyubiquitination plays a crucial role in signal transduction at multiple steps of the RIG-I pathway (Figure 2). Following the binding of viral RNA to the CTD of RIG-I, the N-terminal CARDs recruit the ubiquitin E3 ligase TRIM25(57). TRIM25 catalyzes K63 polyubiquitination of RIG-I at K172 within the second CARD domain. Another ubiquitin E3 ligase termed Riplet (also known as RNF135) ubiquitinates RIG-I at the C-terminus(58). We found that TRIM25 and Ubc13/Uev1A synthesize free K63 polyubiquitin chains, which bind to RIG-I CARDs(59). This binding activates RIG-I such that it promotes the aggregation of MAVS on the mitochondrial membrane(60). Remarkably, the MAVS aggregates form prion-like fibers that efficiently convert other MAVS molecules into massive aggregates that are highly potent in activating IKK and TBK1 in the cytosol. How MAVS aggregation leads to IKK and TBK1 activation is still not entirely clear, but K63 polyubiquitination again plays an important role in these processes. Using an assay in which incubation of mitochondria isolated from virus-infected cells with cytosolic extracts caused IRF3 phosphorylation and dimerization, we found that Ubc5 and K63 polyubiquitination are required for IRF3 activation by MAVS(61). TRAF proteins are candidate E3s involved in this process, as MAVS contains multiple binding sites for TRAF2, TRAF3, TRAF5 and TRAF6. However, cells lacking TRAF3, TRAF6, or both TRAF2 and TRAF5 are still capable of activating IRF3 in response to Sendai virus infection(61). A recent report showed that the mind bomb E3 ligases, MIB1 and MIB2, catalyze K63 polyubiquitination of TBK1, which activates this kinase(62).
T cell receptor pathway
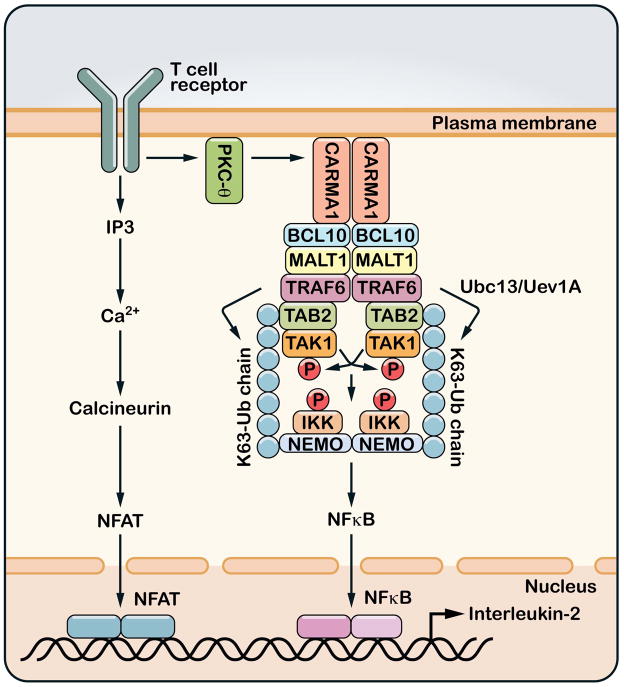
Stimulation of the T cell receptor (TCR) with a cognate MHC-bound ligand triggers a tyrosine phosphorylation cascade that results in the recruitment of protein kinase C-θ (PKCθ) to the receptor (Figure 3) (63). PKCθ phosphorylates the CARD domain protein CARMA1, which in turn recruits BCL-10 and MALT1 to TCR. MALT1 contains binding sites for TRAF2 and TRAF6. The binding of MALT1 to TRAF6 promotes TRAF6 oligomerization, which activates the E3 ligase activity of TRAF6(64). TRAF6 functions together with Ubc13/Uev1A to catalyze K63 polyubiquitination that leads to the activation of TAK1 and subsequent activation of IKK and NF-κB. In support of this model, deletion of Ubc13 or TAK1 in mouse thymocytes abolishes IKK activation by TCR stimulation(65–68). However, TRAF6-deficient T cells are hyper-activated as a result of their resistance to suppression by regulatory T cells(69). It is possible that other E3 ligases such as TRAF2 or MIB2 could compensate for the loss of TRAF6 in maintaining T cell activation(64, 70).
Figure 3. Ubiquitination and T cell activation.
Upon engagement of MHC-bound peptides, T cell receptors (TCR) trigger a cascade of tyrosine phosphorylation events that lead to the activation of protein kinase C-θ (PKCθ). PKCθ then phosphorylates the membrane-associated protein CARMA1, which in turn recruits BCL10 and MALT1. MALT1 binds to TRAF6 and perhaps other ubiquitin E3 ligases. The binding of MALT1 to TRAF6 induces TRAF6 oligomerization and activates its E3 ligase activity, which then catalyzes K63 polyubiquitination to activate TAK1 and IKK. T cell receptor signaling also activates the calcineurin – NFAT pathway through increasing the intracellular concentration of calcium. NFAT, NF-κB and other transcription factors cooperate in the nucleus to induce the production of interleukin-2 (IL-2).
Roles of K63 and alternative ubiquitin linkages in IKK activation by TNFα
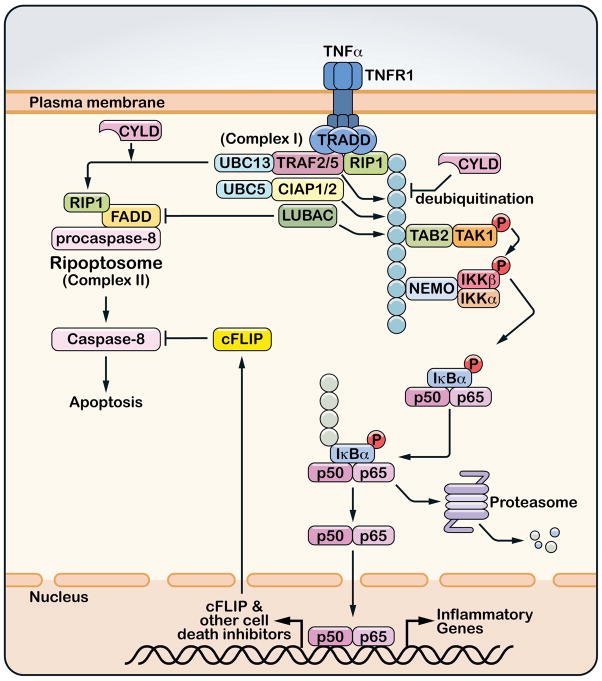
The TNFα pathway is one of the most intensely studied signaling pathways in part because of the pivotal role of TNFα in a wide spectrum of autoimmune and inflammatory diseases(71). Two TNF receptors, TNFR1 and TNFR2, mediate the functions of TNFα. TNFR1 is expressed in most mammalian cells and is more extensively studied than TNFR2, which exhibits a more restricted expression pattern, predominantly in lymphocytes. Upon binding to TNFα, TNFR1 recruits a large number of proteins, including the adaptor TRADD, the protein kinase RIP1, several ubiquitin E3 ligases such as TRAF2, TRAF5, cIAP1, cIAP2 and LUBAC, the deubiquitination enzymes A20 and CYLD, and the cell death proteins FADD and procaspase-8 (Figure 4) (45). Concomitant with the assembly of the receptor complex, RIP1 is rapidly polyubiquitinated, which is important for recruiting the TAK1 and IKK complexes, leading to the activation of NF-κB and MAP kinases(31). Subsequent to the rapid activation of IKK and NF-κB, the TNF receptor complex dissociates, and some of the proteins are released from the receptor to form a cytosolic complex consisting of FADD, RIP1 and procaspase-8 (72). Within this complex, procaspase-8 cleaves itself to form mature caspase-8, which then cleaves caspase-3 to initiate apoptosis. Caspase-8 also cleaves RIP1, thereby preventing RIP1 from activating RIP3, which would otherwise trigger necrosis(73–75). When caspase 8 is absent or inhibited, RIP1 phosphorylates RIP3, leading to necrosis, which is highly inflammatory. In most cells, TNFα does not induce apoptosis or necrosis because NF-κB is activated before caspase 8 can execute the cell death program. NF-κB induces the expression of cell survival factors, including apoptosis inhibitors such as cIAPs and c-FLIP. c-FLIP binds to caspase-8 and blocks the initiation of apoptosis (Figure 4).
Figure 4. Roles of ubiquitination in TNFα-induced NF-κB activation and cell death.
The binding of TNFα to its receptor (TNFR1) induces the trimerization of the receptor and recruitment of a protein complex (complex I) that includes the adaptor protein TRADD, the protein kinase RIP1 and ubiquitin E3 ligases TRAF2, TRAF5, cIAP1, cIAP2 and LUBAC. Some or all of these E3s catalyze polyubiquitination of RIP1, which recruits and activates the TAK1 and IKK complexes. Deubiquitination of RIP1 by CYLD not only inhibits TAK1 and IKK activation, but also facilitates the formation of a cytoplasmic complex (complex II or Ripoptosome) consisting of RIP1, FADD and procaspase-8. Within this complex, pro-caspase 8 auto-cleaves to generate mature caspase-8, which then initiates apoptosis. However, caspase-8 is normally inhibited by caspase inhibitors such as c-FLIP, which is induced by NF-κB. Ubiquitin E3 ligases inhibit cell death indirectly by activating NF-κB and/or directly by blocking the formation of Ripoptosome.
The TNF-dependent recruitment of multiple ubiquitin ligases and deubiquitination enzymes implies that ubiquitination might be important for regulating inflammation and cell death in this pathway. Indeed, knockout of both TRAF2 and TRAF5 severely impairs IKK activation by TNFα(76). However, despite the overall structural similarity between TRAF2 and TRAF6, TRAF2 lacks key residues in its RING domain that are required for binding Ubc13(77). Interestingly, it was recently shown that sphingosine-1-phosphate (S1P) binds to TRAF2 and renders TRAF2 competent in catalyzing K63 polyubiquitination of RIP1(78). cIAP1 and cIAP2 also catalyze polyubiquitination of RIP1; however, the ubiquitin chains synthesized by cIAPs and Ubc5 are not restricted to the K63 linkage(43, 79, 80). Depletion of cIAP1 and cIAP2 also impairs IKK activation by TNFα.
A third ubiquitin E3 ligase complex, LUBAC, which is composed of HOIP, HOIL-1 and Sharpin, is also recruited to TNFR1(81). LUBAC and Ubc5 catalyze linear polyubiquitination of NEMO in vitro(82). Depletion of LUBAC subunits by RNAi or gene knockout in mice partially impairs NF-κB activation. An important role of LUBAC in the TNF pathway is exemplified by the strong inflammatory phenotypes of a naturally occurring mouse mutant called cpdm (chronic proliferative dermatitis), which is deficient in Sharpin expression(83). Several groups showed that Sharpin-deficient cells failed to activate IKK, and suggested that the NF-κB defect in cpdm mice causes cell death, which then triggers the inflammatory diseases (84–86). However, others reported that IKK or NF-κB activation was largely normal or even hyperactivated in these cells(87). Moreover, the inflammatory phenotypes in cpdm mice can be rescued by crossing cpdm mice with IL-1 receptor accessory protein (IL-1RAcP)-deficient mice, suggesting that enhanced IL-1 signaling is an important cause of the inflammatory diseases(88). The cpdm phenotypes can also be rescued by treating the mice with the proteasome inhibitor bortezomib, which is known to inhibit NF-κB(88). Thus, the inflammation in cpdm mice is likely due to excessive rather than attenuated NF-κB activation. It is possible that Sharpin inhibits TNFα-induced cell death through both NF-κB – dependent and –independent mechanisms. Consistent with potentially complex functions of Sharpin, a recent study showed that Sharpin is an inhibitor of integrin activation(89). Thus, further studies are required to understand how Sharpin deficiency leads to inflammatory diseases.
It is interesting to note that all ubiquitin E3 ligases recruited to TNFR1 inhibit cell death, whereas CYLD has the opposite effect (Figure 4). A key target of these E3s and CYLD is RIP1(90). Indeed, RIP1 polyubiquitination leads to NF-κB activation and promotes cell survival, whereas deubiquitination of RIP1 by CYLD promotes the assembly of the cytosolic complex termed Ripoptosome, which leads to cell death (91, 92). In light of the involvement of multiple ubiquitin E3s in the TNFR1 complex, perhaps it is not surprising that the ubiquitin chains on RIP1 are complex. In addition to K63 polyubiquitin chains, other ubiquitin linkages, including K11 and linear chains, have been detected on RIP1(31, 84, 93, 94). These findings might explain why depletion of Ubc13 or replacement of endogenous ubiquitin with K63R mutant did not block IKK activation by TNFα(43). It is likely that multiple E3 ligases, including TRAFs, cIAPs and LUBAC, cooperate with or compensate for each other to regulate NF-κB and control cell death in the TNF pathway.
Perspectives – conclusions and outstanding questions
When we stumbled upon the role of ubiquitin in activating IκB kinase 15 years ago, I thought it was interesting but probably just a mechanism only relevant to some obscure or niche pathways. However, research in the past decade has brought ubiquitination to mainstream NF-κB pathways, including immune and inflammatory responses. The discovery of increasing numbers of ubiquitin E2s (e.g, Ubc13, Ubc5), E3s (e.g, TRAFs, cIAPs, LUBAC), DUBs (e.g, CYLD, A20), and ubiquitin receptors (e.g, NEMO, TAB2/3, RIG-I) in diverse NF-κB signaling pathways underscores the central role of ubiquitination in IKK activation. Of course, the role of ubiquitination is not limited to IKK activation – ubiquitin-mediated degradation of IκB and processing of NF-κB precursors (p100 and p105) are crucial to NF-κB activation(42). Through activation of TAK1, ubiquitination also regulates MAP kinases in many immune and inflammatory pathways. Emerging evidence also suggests that ubiquitination plays an important role in the activation of other kinases such as TBK1 and AKT(61, 62, 95). Beyond NF-κB and protein kinases, K63 polyubiquitination has also been shown to regulate diverse physiological processes, including DNA damage repair, vesicle trafficking, and chromatin dynamics(96).
The high degree of conservation of K63 polyubiquitination enzymes such as Ubc13/Uev1A from yeast to human implies essential functions of this type of polyubiquitin chains. Other types of polyubiquitin chains also exist in eukaryotic cells. Proteomics studies in yeast suggest that with the exception of K63 ubiquitin chains, all other ubiquitin chains appear to function in targeting proteins to the proteasome(97). However, this does not rule out the possibility that alternative ubiquitin chains could have regulatory functions independent of proteolysis. Recent studies suggest that K11 and linear ubiquitin chains could function in some NF-κB signaling pathways(82, 94). Both K11 and linear ubiquitin chains could also target protein degradation by the proteasome in cells(98–100). It is not clear how a protein is fated to degradation or to perform a regulatory function after it is modified by certain ubiquitin chains. Even K63 polyubiquitin chains could target a protein for degradation by the proteasome in vitro(101), but in cells K63 polyubiquitinated proteins are largely spared from the proteasome, probably because they bind to certain ubiquitin receptors, which protect the ubiquitin chains from the proteasome and DUBs.
An outstanding issue concerning ubiquitination and IKK activation is the identification of physiological ubiquitination targets that are important for IKK activation. In most cases, a putative ubiquitination target was identified by overexpression of a protein containing mutations at certain lysine residues. This can be misleading, as lysine mutation can have unintended consequences, such as inactivation of an enzyme activity (e.g, a kinase) or alteration of protein folding. Many putative ubiquitin modifications have not been verified by mass spectrometry. Ultimately, a bona fide ubiquitination target should be verified by genetic complementation experiments (e.g, mouse knock-in) and by direct biochemical assays (e.g, ubiquitin modification of a protein alters its activity). Recently, knock-in mouse models carrying point mutations that selectively disrupt ubiquitin binding of some proteins (e.g., ABIN1 and optineurin) have been generated, providing strong evidence for the role of polyubiquitin binding in the functions of these proteins(102, 103). We expect that similar approaches will be taken to validate putative ubiquitination targets in the coming years (assuming that the knock-in mutation only impairs ubiquitin conjugation).
The difficulty in detecting ubiquitination of an endogenous protein is due in large part to the very low stoichiometry of signal-dependent ubiquitin modification. Recent advances in quantitative mass spectrometry should greatly facilitate the discovery of physiological ubiquitination targets. However, when a very small fraction of a protein is ubiquitinated, it presents a challenge to obtain direct evidence that the ubiquitin modification alters the protein activity. This is further complicated by the recent findings in our lab and others that free K63 polyubiquitin chains can directly activate TAK1, IKK and RIG-I (46, 59, 104). The idea that free polyubiquitin chains could function as a second messenger-like molecule may explain how signaling could proceed robustly when a minuscule fraction of proteins is modified by ubiquitin (or not modified at all), but also poses a problem of how the signaling specificity is achieved. One possible solution to the specificity question is the fact that free ubiquitin chains are very labile, due to the presence of abundant DUBs in the cell. When the ubiquitin chains bind to specific ubiquitin receptors they are protected from DUBs. Thus, the proximity of a ubiquitin chain producer (E3 such as TRAF6 or TRIM25) to a ubiquitin receptor (e.g, TAK1/TAB2 or RIG-I) allows the ubiquitin chains to be delivered to the receptor to initiate signaling (Figure 1 and 2); excess ubiquitin chains are degraded by DUBs, avoiding the activation of irrelevant targets. It should be noted that most ubiquitination reactions generate far more unanchored ubiquitin chains than ubiquitin chains anchored on a protein target, thus it is not necessary to invoke a mechanism whereby a ubiquitin chain is conjugated to a target and then taken off by a DUB to generate unanchored ubiquitin chains. Clearly, endogenous free K63 ubiquitin chains can be isolated from cells and these chains have potent activity in vitro. However, more work is needed to determine the role of these ubiquitin chains in cell signaling in vivo.
A milestone in the convergence of the ubiquitin and NF-κB fields is marked by the first joint Keystone meeting on NF-κB and ubiquitin to be held in 2012. Research on ubiquitination in the NF-κB pathways has produced several new conceptual advances that move both fields forward. Continued cross-fertilization between these fields should be fruitful for many years to come.
Acknowledgments
I thank Eric Olson for critically reading the manuscript and Jose Cabrera for graphics. Research in my laboratory is supported by grants from NIH, the Welch Foundation, and Cancer Prevention and Research Institute of Texas.
References
- 1.Hershko A. Ubiquitin: roles in protein modification and breakdown. Cell. 1983;34:11–2. doi: 10.1016/0092-8674(83)90131-9. [DOI] [PubMed] [Google Scholar]
- 2.Pickart CM. Ubiquitin enters the new millennium. Mol Cell. 2001;8:499–504. doi: 10.1016/s1097-2765(01)00347-1. [DOI] [PubMed] [Google Scholar]
- 3.Ciechanover A, Finley D, Varshavsky A. Ubiquitin dependence of selective protein degradation demonstrated in the mammalian cell cycle mutant ts85. Cell. 1984;37:57–66. doi: 10.1016/0092-8674(84)90300-3. [DOI] [PubMed] [Google Scholar]
- 4.Finley D, Ciechanover A, Varshavsky A. Thermolability of ubiquitin-activating enzyme from the mammalian cell cycle mutant ts85. Cell. 1984;37:43–55. doi: 10.1016/0092-8674(84)90299-x. [DOI] [PubMed] [Google Scholar]
- 5.Scheffner M, Werness BA, Huibregtse JM, Levine AJ, Howley PM. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63:1129–36. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 6.Glotzer M, Murray AW, Kirschner MW. Cyclin is degraded by the ubiquitin pathway. Nature. 1991;349:132–8. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- 7.Sen R, Baltimore D. Multiple nuclear factors interact with the immunoglobulin enhancer sequences. Cell. 1986;46:705–16. doi: 10.1016/0092-8674(86)90346-6. [DOI] [PubMed] [Google Scholar]
- 8.Sen R, Baltimore D. Inducibility of kappa immunoglobulin enhancer-binding protein Nf-kappa B by a posttranslational mechanism. Cell. 1986;47:921–8. doi: 10.1016/0092-8674(86)90807-x. [DOI] [PubMed] [Google Scholar]
- 9.Baeuerle PA, Baltimore D. Activation of DNA-binding activity in an apparently cytoplasmic precursor of the NF-kappa B transcription factor. Cell. 1988;53:211–7. doi: 10.1016/0092-8674(88)90382-0. [DOI] [PubMed] [Google Scholar]
- 10.Baeuerle PA, Baltimore D. NF-kappa B: ten years after. Cell. 1996;87:13–20. doi: 10.1016/s0092-8674(00)81318-5. [DOI] [PubMed] [Google Scholar]
- 11.Henkel T, Machleidt T, Alkalay I, Kronke M, Ben-Neriah Y, Baeuerle PA. Rapid proteolysis of I kappa B-alpha is necessary for activation of transcription factor NF-kappa B. Nature. 1993;365:182–5. doi: 10.1038/365182a0. [DOI] [PubMed] [Google Scholar]
- 12.Palombella VJ, Rando OJ, Goldberg AL, Maniatis T. The ubiquitin-proteasome pathway is required for processing the NF-kappa B1 precursor protein and the activation of NF-kappa B. Cell. 1994;78:773–85. doi: 10.1016/s0092-8674(94)90482-0. [DOI] [PubMed] [Google Scholar]
- 13.Chen Z, et al. Signal-induced site-specific phosphorylation targets I kappa B alpha to the ubiquitin-proteasome pathway. Genes Dev. 1995;9:1586–97. doi: 10.1101/gad.9.13.1586. [DOI] [PubMed] [Google Scholar]
- 14.Brown K, Gerstberger S, Carlson L, Franzoso G, Siebenlist U. Control of I kappa B-alpha proteolysis by site-specific, signal-induced phosphorylation. Science. 1995;267:1485–8. doi: 10.1126/science.7878466. [DOI] [PubMed] [Google Scholar]
- 15.Brockman JA, et al. Coupling of a signal response domain in I kappa B alpha to multiple pathways for NF-kappa B activation. Mol Cell Biol. 1995;15:2809–18. doi: 10.1128/mcb.15.5.2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen ZJ, Parent L, Maniatis T. Site-specific phosphorylation of IkappaBalpha by a novel ubiquitination-dependent protein kinase activity. Cell. 1996;84:853–62. doi: 10.1016/s0092-8674(00)81064-8. [DOI] [PubMed] [Google Scholar]
- 17.Spencer E, Jiang J, Chen ZJ. Signal-induced ubiquitination of IkappaBalpha by the F-box protein Slimb/beta-TrCP. Genes Dev. 1999;13:284–94. doi: 10.1101/gad.13.3.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yaron A, et al. Identification of the receptor component of the IkappaBalpha-ubiquitin ligase. Nature. 1998;396:590–4. doi: 10.1038/25159. [DOI] [PubMed] [Google Scholar]
- 19.Winston JT, Strack P, Beer-Romero P, Chu CY, Elledge SJ, Harper JW. The SCFbeta-TRCP-ubiquitin ligase complex associates specifically with phosphorylated destruction motifs in IkappaBalpha and beta-catenin and stimulates IkappaBalpha ubiquitination in vitro. Genes Dev. 1999;13:270–83. doi: 10.1101/gad.13.3.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lomaga MA, et al. TRAF6 deficiency results in osteopetrosis and defective interleukin-1, CD40, and LPS signaling. Genes Dev. 1999;13:1015–24. doi: 10.1101/gad.13.8.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Naito A, et al. Severe osteopetrosis, defective interleukin-1 signalling and lymph node organogenesis in TRAF6-deficient mice. Genes Cells. 1999;4:353–62. doi: 10.1046/j.1365-2443.1999.00265.x. [DOI] [PubMed] [Google Scholar]
- 22.Ishida T, et al. Identification of TRAF6, a novel tumor necrosis factor receptor-associated factor protein that mediates signaling from an amino-terminal domain of the CD40 cytoplasmic region. J Biol Chem. 1996;271:28745–8. doi: 10.1074/jbc.271.46.28745. [DOI] [PubMed] [Google Scholar]
- 23.Cao Z, Xiong J, Takeuchi M, Kurama T, Goeddel DV. TRAF6 is a signal transducer for interleukin-1. Nature. 1996;383:443–6. doi: 10.1038/383443a0. [DOI] [PubMed] [Google Scholar]
- 24.Deng L, et al. Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell. 2000;103:351–61. doi: 10.1016/s0092-8674(00)00126-4. [DOI] [PubMed] [Google Scholar]
- 25.Hofmann RM, Pickart CM. Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell. 1999;96:645–53. doi: 10.1016/s0092-8674(00)80575-9. [DOI] [PubMed] [Google Scholar]
- 26.Lorick KL, Jensen JP, Fang S, Ong AM, Hatakeyama S, Weissman AM. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc Natl Acad Sci U S A. 1999;96:11364–9. doi: 10.1073/pnas.96.20.11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang C, Deng L, Hong M, Akkaraju GR, Inoue J, Chen ZJ. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature. 2001;412:346–51. doi: 10.1038/35085597. [DOI] [PubMed] [Google Scholar]
- 28.Yamaguchi K, et al. Identification of a member of the MAPKKK family as a potential mediator of TGF-beta signal transduction. Science. 1995;270:2008–11. doi: 10.1126/science.270.5244.2008. [DOI] [PubMed] [Google Scholar]
- 29.Ninomiya-Tsuji J, Kishimoto K, Hiyama A, Inoue J, Cao Z, Matsumoto K. The kinase TAK1 can activate the NIK-I kappaB as well as the MAP kinase cascade in the IL-1 signalling pathway. Nature. 1999;398:252–6. doi: 10.1038/18465. [DOI] [PubMed] [Google Scholar]
- 30.Kanayama A, et al. TAB2 and TAB3 activate the NF-kappaB pathway through binding to polyubiquitin chains. Mol Cell. 2004;15:535–48. doi: 10.1016/j.molcel.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 31.Ea CK, Deng L, Xia ZP, Pineda G, Chen ZJ. Activation of IKK by TNFalpha requires site-specific ubiquitination of RIP1 and polyubiquitin binding by NEMO. Mol Cell. 2006;22:245–57. doi: 10.1016/j.molcel.2006.03.026. [DOI] [PubMed] [Google Scholar]
- 32.Wu CJ, Conze DB, Li T, Srinivasula SM, Ashwell JD. Sensing of Lys 63-linked polyubiquitination by NEMO is a key event in NF-kappaB activation [corrected] Nat Cell Biol. 2006;8:398–406. doi: 10.1038/ncb1384. [DOI] [PubMed] [Google Scholar]
- 33.Israel A. NF-kappaB activation: Nondegradative ubiquitination implicates NEMO. Trends Immunol. 2006;27:395–7. doi: 10.1016/j.it.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 34.Lo YC, et al. Structural Basis for Recognition of Diubiquitins by NEMO. Mol Cell. 2009 doi: 10.1016/j.molcel.2009.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rahighi S, et al. Specific recognition of linear ubiquitin chains by NEMO is important for NF-kappaB activation. Cell. 2009;136:1098–109. doi: 10.1016/j.cell.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 36.Laplantine E, et al. NEMO specifically recognizes K63-linked poly-ubiquitin chains through a new bipartite ubiquitin-binding domain. Embo J. 2009 doi: 10.1038/emboj.2009.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kovalenko A, Chable-Bessia C, Cantarella G, Israel A, Wallach D, Courtois G. The tumour suppressor CYLD negatively regulates NF-kappaB signalling by deubiquitination. Nature. 2003;424:801–5. doi: 10.1038/nature01802. [DOI] [PubMed] [Google Scholar]
- 38.Trompouki E, Hatzivassiliou E, Tsichritzis T, Farmer H, Ashworth A, Mosialos G. CYLD is a deubiquitinating enzyme that negatively regulates NF-kappaB activation by TNFR family members. Nature. 2003;424:793–6. doi: 10.1038/nature01803. [DOI] [PubMed] [Google Scholar]
- 39.Brummelkamp TR, Nijman SM, Dirac AM, Bernards R. Loss of the cylindromatosis tumour suppressor inhibits apoptosis by activating NF-kappaB. Nature. 2003;424:797–801. doi: 10.1038/nature01811. [DOI] [PubMed] [Google Scholar]
- 40.Wertz IE, et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature. 2004;430:694–9. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- 41.Boone DL, et al. The ubiquitin-modifying enzyme A20 is required for termination of Toll-like receptor responses. Nat Immunol. 2004;5:1052–60. doi: 10.1038/ni1110. [DOI] [PubMed] [Google Scholar]
- 42.Skaug B, Jiang X, Chen ZJ. The role of ubiquitin in NF-kappaB regulatory pathways. Annu Rev Biochem. 2009;78:769–96. doi: 10.1146/annurev.biochem.78.070907.102750. [DOI] [PubMed] [Google Scholar]
- 43.Xu M, Skaug B, Zeng W, Chen ZJ. A ubiquitin replacement strategy in human cells reveals distinct mechanisms of IKK activation by TNFalpha and IL-1beta. Mol Cell. 2009;36:302–14. doi: 10.1016/j.molcel.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–82. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 45.Liu S, Chen ZJ. Expanding role of ubiquitination in NF-kappaB signaling. Cell Res. 2011;21:6–21. doi: 10.1038/cr.2010.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xia ZP, et al. Direct activation of protein kinases by unanchored polyubiquitin chains. Nature. 2009;461:114–9. doi: 10.1038/nature08247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Skaug B, Chen J, Du F, He J, Ma A, Chen ZJ. Direct, Noncatalytic Mechanism of IKK Inhibition by A20. Mol Cell. 2011;44:559–71. doi: 10.1016/j.molcel.2011.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tseng PH, Matsuzawa A, Zhang W, Mino T, Vignali DA, Karin M. Different modes of ubiquitination of the adaptor TRAF3 selectively activate the expression of type I interferons and proinflammatory cytokines. Nat Immunol. 2010;11:70–5. doi: 10.1038/ni.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yoneyama M, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–7. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 50.Yoneyama M, Fujita T. RNA recognition and signal transduction by RIG-I-like receptors. Immunol Rev. 2009;227:54–65. doi: 10.1111/j.1600-065X.2008.00727.x. [DOI] [PubMed] [Google Scholar]
- 51.Hornung V, et al. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–7. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 52.Pichlmair A, et al. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 53.Kawai T, et al. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6:981–8. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 54.Meylan E, et al. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–72. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 55.Xu LG, Wang YY, Han KJ, Li LY, Zhai Z, Shu HB. VISA Is an Adapter Protein Required for Virus-Triggered IFN-beta Signaling. Mol Cell. 2005;19:727–40. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 56.Kato H, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–5. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 57.Gack MU, et al. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446:916–20. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- 58.Oshiumi H, Matsumoto M, Hatakeyama S, Seya T. Riplet/RNF135, a RING finger protein, ubiquitinates RIG-I to promote interferon-beta induction during the early phase of viral infection. J Biol Chem. 2009;284:807–17. doi: 10.1074/jbc.M804259200. [DOI] [PubMed] [Google Scholar]
- 59.Zeng W, et al. Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell. 2010;141:315–30. doi: 10.1016/j.cell.2010.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hou F, Sun L, Zheng H, Skaug B, Jiang QX, Chen ZJ. MAVS Forms Functional Prion-like Aggregates to Activate and Propagate Antiviral Innate Immune Response. Cell. 2011;146:448–61. doi: 10.1016/j.cell.2011.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zeng W, Xu M, Liu S, Sun L, Chen ZJ. Key role of Ubc5 and lysine-63 polyubiquitination in viral activation of IRF3. Mol Cell. 2009;36:315–25. doi: 10.1016/j.molcel.2009.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li S, Wang L, Berman M, Kong YY, Dorf ME. Mapping a dynamic innate immunity protein interaction network regulating type I interferon production. Immunity. 2011;35:426–40. doi: 10.1016/j.immuni.2011.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Blonska M, Lin X. CARMA1-mediated NF-kappaB and JNK activation in lymphocytes. Immunol Rev. 2009;228:199–211. doi: 10.1111/j.1600-065X.2008.00749.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sun L, Deng L, Ea CK, Xia ZP, Chen ZJ. The TRAF6 ubiquitin ligase and TAK1 kinase mediate IKK activation by BCL10 and MALT1 in T lymphocytes. Mol Cell. 2004;14:289–301. doi: 10.1016/s1097-2765(04)00236-9. [DOI] [PubMed] [Google Scholar]
- 65.Yamamoto M, et al. Cutting Edge: Pivotal function of Ubc13 in thymocyte TCR signaling. J Immunol. 2006;177:7520–4. doi: 10.4049/jimmunol.177.11.7520. [DOI] [PubMed] [Google Scholar]
- 66.Liu HH, Xie M, Schneider MD, Chen ZJ. Essential role of TAK1 in thymocyte development and activation. Proc Natl Acad Sci U S A. 2006;103:11677–82. doi: 10.1073/pnas.0603089103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sato S, et al. TAK1 is indispensable for development of T cells and prevention of colitis by the generation of regulatory T cells. Int Immunol. 2006;18:1405–11. doi: 10.1093/intimm/dxl082. [DOI] [PubMed] [Google Scholar]
- 68.Wan YY, Chi H, Xie M, Schneider MD, Flavell RA. The kinase TAK1 integrates antigen and cytokine receptor signaling for T cell development, survival and function. Nat Immunol. 2006;7:851–8. doi: 10.1038/ni1355. [DOI] [PubMed] [Google Scholar]
- 69.King CG, et al. TRAF6 is a T cell-intrinsic negative regulator required for the maintenance of immune homeostasis. Nat Med. 2006;12:1088–92. doi: 10.1038/nm1449. [DOI] [PubMed] [Google Scholar]
- 70.Stempin CC, Chi L, Giraldo-Vela JP, High AA, Hacker H, Redecke V. The E3 ubiquitin ligase mind bomb-2 (MIB2) protein controls B-cell CLL/lymphoma 10 (BCL10)-dependent NF-kappaB activation. J Biol Chem. 2011;286:37147–57. doi: 10.1074/jbc.M111.263384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen G, Goeddel DV. TNF-R1 signaling: a beautiful pathway. Science. 2002;296:1634–5. doi: 10.1126/science.1071924. [DOI] [PubMed] [Google Scholar]
- 72.Micheau O, Tschopp J. Induction of TNF receptor I-mediated apoptosis via two sequential signaling complexes. Cell. 2003;114:181–90. doi: 10.1016/s0092-8674(03)00521-x. [DOI] [PubMed] [Google Scholar]
- 73.He S, et al. Receptor interacting protein kinase-3 determines cellular necrotic response to TNF-alpha. Cell. 2009;137:1100–11. doi: 10.1016/j.cell.2009.05.021. [DOI] [PubMed] [Google Scholar]
- 74.Zhang DW, et al. RIP3, an energy metabolism regulator that switches TNF-induced cell death from apoptosis to necrosis. Science. 2009;325:332–6. doi: 10.1126/science.1172308. [DOI] [PubMed] [Google Scholar]
- 75.Cho YS, et al. Phosphorylation-driven assembly of the RIP1-RIP3 complex regulates programmed necrosis and virus-induced inflammation. Cell. 2009;137:1112–23. doi: 10.1016/j.cell.2009.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tada K, et al. Critical roles of TRAF2 and TRAF5 in tumor necrosis factor-induced NF-kappa B activation and protection from cell death. J Biol Chem. 2001;276:36530–4. doi: 10.1074/jbc.M104837200. [DOI] [PubMed] [Google Scholar]
- 77.Yin Q, Lamothe B, Darnay BG, Wu H. Structural basis for the lack of E2 interaction in the RING domain of TRAF2. Biochemistry. 2009;48:10558–67. doi: 10.1021/bi901462e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Alvarez SE, et al. Sphingosine-1-phosphate is a missing cofactor for the E3 ubiquitin ligase TRAF2. Nature. 2010;465:1084–8. doi: 10.1038/nature09128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bertrand MJ, et al. cIAP1 and cIAP2 facilitate cancer cell survival by functioning as E3 ligases that promote RIP1 ubiquitination. Mol Cell. 2008;30:689–700. doi: 10.1016/j.molcel.2008.05.014. [DOI] [PubMed] [Google Scholar]
- 80.Mahoney DJ, et al. Both cIAP1 and cIAP2 regulate TNFalpha-mediated NF-kappaB activation. Proc Natl Acad Sci U S A. 2008;105:11778–83. doi: 10.1073/pnas.0711122105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Haas TL, et al. Recruitment of the linear ubiquitin chain assembly complex stabilizes the TNF-R1 signaling complex and is required for TNF-mediated gene induction. Mol Cell. 2009;36:831–44. doi: 10.1016/j.molcel.2009.10.013. [DOI] [PubMed] [Google Scholar]
- 82.Tokunaga F, et al. Involvement of linear polyubiquitylation of NEMO in NF-kappaB activation. Nat Cell Biol. 2009;11:123–32. doi: 10.1038/ncb1821. [DOI] [PubMed] [Google Scholar]
- 83.Seymour RE, et al. Spontaneous mutations in the mouse Sharpin gene result in multiorgan inflammation, immune system dysregulation and dermatitis. Genes Immun. 2007;8:416–21. doi: 10.1038/sj.gene.6364403. [DOI] [PubMed] [Google Scholar]
- 84.Gerlach B, et al. Linear ubiquitination prevents inflammation and regulates immune signalling. Nature. 2011;471:591–6. doi: 10.1038/nature09816. [DOI] [PubMed] [Google Scholar]
- 85.Ikeda F, et al. SHARPIN forms a linear ubiquitin ligase complex regulating NF-kappaB activity and apoptosis. Nature. 2011;471:637–41. doi: 10.1038/nature09814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tokunaga F, et al. SHARPIN is a component of the NF-kappaB-activating linear ubiquitin chain assembly complex. Nature. 2011;471:633–6. doi: 10.1038/nature09815. [DOI] [PubMed] [Google Scholar]
- 87.Zak DE, et al. Systems analysis identifies an essential role for SHANK-associated RH domain-interacting protein (SHARPIN) in macrophage Toll-like receptor 2 (TLR2) responses. Proc Natl Acad Sci U S A. 2011;108:11536–41. doi: 10.1073/pnas.1107577108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liang Y, Seymour RE, Sundberg JP. Inhibition of NF-kappaB signaling retards eosinophilic dermatitis in SHARPIN-deficient mice. J Invest Dermatol. 2011;131:141–9. doi: 10.1038/jid.2010.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rantala JK, et al. SHARPIN is an endogenous inhibitor of beta1-integrin activation. Nat Cell Biol. 2011;13:1315–24. doi: 10.1038/ncb2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang L, Du F, Wang X. TNF-alpha induces two distinct caspase-8 activation pathways. Cell. 2008;133:693–703. doi: 10.1016/j.cell.2008.03.036. [DOI] [PubMed] [Google Scholar]
- 91.O’Donnell MA, Legarda-Addison D, Skountzos P, Yeh WC, Ting AT. Ubiquitination of RIP1 regulates an NF-kappaB-independent cell-death switch in TNF signaling. Curr Biol. 2007;17:418–24. doi: 10.1016/j.cub.2007.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bertrand MJ, Vandenabeele P. The Ripoptosome: death decision in the cytosol. Mol Cell. 2011;43:323–5. doi: 10.1016/j.molcel.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 93.Newton K, et al. Ubiquitin chain editing revealed by polyubiquitin linkage-specific antibodies. Cell. 2008;134:668–78. doi: 10.1016/j.cell.2008.07.039. [DOI] [PubMed] [Google Scholar]
- 94.Dynek JN, et al. c-IAP1 and UbcH5 promote K11-linked polyubiquitination of RIP1 in TNF signalling. Embo J. 2010;29:4198–209. doi: 10.1038/emboj.2010.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yang WL, et al. The E3 ligase TRAF6 regulates Akt ubiquitination and activation. Science. 2009;325:1134–8. doi: 10.1126/science.1175065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chen ZJ, Sun LJ. Nonproteolytic functions of ubiquitin in cell signaling. Mol Cell. 2009;33:275–86. doi: 10.1016/j.molcel.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 97.Xu P, et al. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 2009;137:133–45. doi: 10.1016/j.cell.2009.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jin L, Williamson A, Banerjee S, Philipp I, Rape M. Mechanism of ubiquitin-chain formation by the human anaphase-promoting complex. Cell. 2008;133:653–65. doi: 10.1016/j.cell.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Inn KS, et al. Linear ubiquitin assembly complex negatively regulates RIG-I- and TRIM25-mediated type I interferon induction. Mol Cell. 2011;41:354–65. doi: 10.1016/j.molcel.2010.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhao S, Ulrich HD. Distinct consequences of posttranslational modification by linear versus K63-linked polyubiquitin chains. Proc Natl Acad Sci U S A. 2010;107:7704–9. doi: 10.1073/pnas.0908764107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hofmann RM, Pickart CM. In vitro assembly and recognition of Lys-63 polyubiquitin chains. J Biol Chem. 2001;276:27936–43. doi: 10.1074/jbc.M103378200. [DOI] [PubMed] [Google Scholar]
- 102.Gleason CE, Ordureau A, Gourlay R, Arthur JS, Cohen P. Polyubiquitin binding to optineurin is required for optimal activation of TANK-binding kinase 1 and production of interferon beta. J Biol Chem. 2011;286:35663–74. doi: 10.1074/jbc.M111.267567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Nanda SK, et al. Polyubiquitin binding to ABIN1 is required to prevent autoimmunity. J Exp Med. 2011;208:1215–28. doi: 10.1084/jem.20102177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pertel T, et al. TRIM5 is an innate immune sensor for the retrovirus capsid lattice. Nature. 2011;472:361–5. doi: 10.1038/nature09976. [DOI] [PMC free article] [PubMed] [Google Scholar]