Hes1 is required for the development of hematopoietic stem cells in the mouse embryo through repression of Gata2.
Abstract
Previous studies have identified Notch as a key regulator of hematopoietic stem cell (HSC) development, but the underlying downstream mechanisms remain unknown. The Notch target Hes1 is widely expressed in the aortic endothelium and hematopoietic clusters, though Hes1-deficient mice show no overt hematopoietic abnormalities. We now demonstrate that Hes is required for the development of HSC in the mouse embryo, a function previously undetected as the result of functional compensation by de novo expression of Hes5 in the aorta/gonad/mesonephros (AGM) region of Hes1 mutants. Analysis of embryos deficient for Hes1 and Hes5 reveals an intact arterial program with overproduction of nonfunctional hematopoietic precursors and total absence of HSC activity. These alterations were associated with increased expression of the hematopoietic regulators Runx1, c-myb, and the previously identified Notch target Gata2. By analyzing the Gata2 locus, we have identified functional RBPJ-binding sites, which mutation results in loss of Gata2 reporter expression in transgenic embryos, and functional Hes-binding sites, which mutation leads to specific Gata2 up-regulation in the hematopoietic precursors. Together, our findings show that Notch activation in the AGM triggers Gata2 and Hes1 transcription, and next HES-1 protein represses Gata2, creating an incoherent feed-forward loop required to restrict Gata2 expression in the emerging HSCs.
Hematopoietic stem cells (HSCs) originate during embryonic life in association with arterial vessels including the aorta in the aorta/gonad/mesonephros (AGM) region and the umbilical and vitelline arteries (Dzierzak and Speck, 2008). The first HSCs are detected around embryonic day (E) 10 of mouse development in hematopoietic clusters emerging from the ventral wall of the aorta in the AGM and are defined by their unique capacity to reconstitute hematopoiesis of immunodepleted adult recipients in transplantation assays. The Notch pathway is required for HSC development in the vertebrate embryo, upstream of Runx1 (Burns et al., 2005) and GATA-2 (Robert-Moreno et al., 2005), but is dispensable for yolk sac (YS) primitive hematopoiesis (Robert-Moreno et al., 2007; Bertrand et al., 2010; Bigas et al., 2010). Importantly, Gata2 but not Runx1 expression was totally impaired in the aorta of Jag1−/− mutant mouse embryos, which fail to generate hematopoiesis (Robert-Moreno et al., 2005, 2008). Re-expression of GATA-2 partially rescued generation of hematopoietic cells from Jag1−/− AGM cells (Robert-Moreno et al., 2008), consistent with its pivotal role in definitive embryonic and adult hematopoiesis (Tsai et al., 1994).
The Notch signaling pathway and its function in controlling cell fate diversification is conserved through evolution. Notch receptors interact with their ligands that are expressed on neighboring cells, and this interaction induces two sequential proteolytic events, which release the intracellular Notch (ICN) fragment. Once activated, Notch associates with its specific coactivator Mastermind (Mam) and the nuclear factor RBPJ to induce a transcriptional response, generally associated with the inhibition of a particular cell fate destiny from a population of equivalent cells (Kopan and Ilagan, 2009). Classical examples of Notch-dependent fate inhibition are B cell differentiation in the hematopoietic system, secretory lineage in the adult intestine, and the neural fate from neural/glial precursors. Most of these Notch responses are context dependent and associated with regulation of tissue-specific targets; however, a group of Notch target genes belonging to the Hairy/Enhancer of split (HES) family are known to mediate Notch actions in a wide range of cell types and organisms (Kageyama et al., 2007). We previously found that activation of Notch in the AGM was concomitant with the transcriptional expression of Hes1 and Hes-related family genes (Robert-Moreno et al., 2005).
HES proteins are bHLH transcriptional repressors including the Notch targets HES-1, HES-3, HES-5, HES-7, and the HES-related proteins HRT-1 and HRT-2 (Kageyama et al., 2007). In the hematopoietic system, HES-1 has a major function in normal T cell development, but it is also directly involved in the maintenance of Notch-induced T cell leukemias (Tomita et al., 1999; Espinosa et al., 2010; Wendorff et al., 2010). In addition, ectopic HES-1 inhibits differentiation of bone marrow HSCs when cultured in vitro (Kunisato et al., 2003); however, nothing is known about a putative role of HES in regulating embryonic hematopoiesis or HSC generation. In this study, we investigate the mechanisms that control HSC development downstream of Notch and demonstrate that HES repressors are essential for regulating the levels of Gata2 in the AGM precursors and generate functional hematopoietic cells.
RESULTS
Increased hematopoietic cluster emergence in Hes-deficient embryonic aortas
Hes1 is expressed in the aortic endothelium and the presumptive hematopoietic clusters of the AGM region (Fig. 1 A; Robert-Moreno et al., 2008), suggestive of a hematopoietic function. However, Hes1-null embryos contain functional HSCs and progenitors in the fetal liver that are able to reconstitute all hematopoietic lineages except T cells when transplanted into RAG2-null mice. We explored the possibility that other Hes members compensate for Hes1 deficiency during hematopoietic development, similarly to what occurs in the nervous system (Ohtsuka et al., 1999). To this purpose, we used two genetic models, the first one carrying a constitutive deletion of Hes1 and Hes5 alleles in different combinations and the second with constitutive deletion of one or two copies of Hes5 combined with Hes1-loxP alleles, which can be deleted after tamoxifen treatment when crossed with mice carrying the β-actin–CRE-ERT transgene. We first determined whether deletion of several Hes1 and Hes5 alleles in the AGM affects the expression of other members of this family of repressors. Importantly we found that Hes5, which is not detected in the endothelium nor hematopoietic clusters of wild-type AGM (Fig. 1 A), was highly induced in both tissues after genetic deletion of Hes1 (Fig. 1, B and C). In contrast, other Hes family genes such as Hrt1/Hey1 and Hrt2/Hey2 were not affected (Fig. 1 B). Notch activity but also HRT-1 and HRT-2 functions are essential to induce the arterial program (Fischer et al., 2004), which constitutes the niche for HSC generation. Thus, we tested whether combined Hes1/5 deficiency precludes arterial development in the AGM. Analysis of E10.5 embryo sections stained with the pan-endothelial/hematopoietic CD31 marker did not reveal any apparent vascular abnormality in the AGM aorta at this stage (Fig. 1 F and not depicted). Moreover, by immunostaining and quantitative RT-PCR (qRT-PCR), we found that different arterial-associated genes including EphrinB2 and SMA remained unaffected in these mutant embryos (Fig. 1, D and E). These results indicate that HES repressors are dispensable for maintaining the arterial program in the embryo. A more accurate analysis of the AGM aorta stained for CD31 (labeling endothelial and hematopoietic cells) demonstrated that Hes1−/−Hes5+/− and Hes1−/−Hes5−/− mutant embryos have larger hematopoietic cluster structures when compared with wild-type embryos (Fig. 1 F). We confirmed the hematopoietic nature of these clusters by staining with anti–C-Kit (not depicted) and CD41 (Fig. 1 F). Quantification of CD41+ cells in the AGM from different wild-type and mutant embryos demonstrated that the number of Hes1 and Hes5 deleted alleles inversely correlated with the total number of hematopoietic progenitors per AGM present in the aortic endothelium (up to 10-fold; Spearman’s correlation, P < 0.001; Fig. 1 G) and the number of enlarged clusters with more than four cells (Spearman’s correlation, P = 0.004; Fig. 1 H), indicating functional compensation between Hes1 and Hes5 in the AGM.
Figure 1.
Increased hematopoietic cluster emergence in Hes-deficient embryonic aortas. (A) Transversal sections of embryos at 10.5 d showing expression of Hes5 and Hes1 by in situ hybridization (ISH). Arrowheads indicate the structures (Ao, aorta; IAHC, intra-aortic hematopoietic cluster; m, mesonephros; nt, neural tube; p2, p2 neural progenitors; SG, sympathetic ganglia). Details of IAHC and aorta in the right panels correspond to the boxed areas in the middle panels. (B) Relative expression of the indicated genes from E10.5 dissected AGMs after 3 d of 4-OH-tamoxifen treatments in explant cultures by qRT-PCR. Fold increase to the wild-type control after β2-Microglobulin and GAPDH normalization. Bars represent the mean ± SD determination from two experiments showing the fold increase versus the wild-type AGM. (C) Relative expression of Hes5 from sorted populations (endothelial, CD31−C-Kit−CD45−; hematopoietic clusters, CD31+C-Kit+) by qRT-PCR. mRNA was extracted from a pool of 10 AGMs at E10.5 in Hes1+/+ Hes5+/+ determinations and 4 AGMs in Hes1f/f Hes5+/− β-ACTIN–CRE-ERT after in vivo tamoxifen induction at E8. Bars represent the mean ± SD determination from one representative of two experiments showing the fold increase versus the wild-type AGM after β2-Microglobulin normalization. (D) Representative immunostaining of CD44 and SMA (arterial markers) in the AGM region in wild-type or Hes mutant embryos on E10.5 (A, dorsal aorta; Ao, aorta in AGM region; cv, cardinal vein; m, mesonephros; nt, neural tube). (E) Relative expression of endothelial (PECAM1), arterial markers (EphrinB2, smooth muscle actin [SMA], and CD44), and vein marker (EphB4) in E10.5 AGM after 3 d of 4-OH-tamoxifen treatment in explant culture from embryos with the indicated genotypes (n = 3 each genotype) by qRT-PCR. Bars represent the mean ± SD determination from three experiments showing the fold increase relative to the wild-type AGM. (F) Representative immunostaining of CD31 (endothelial and hematopoietic) and CD41 (hematopoietic precursors) in the AGM region in wild-type or Hes mutant embryos. Arrowheads indicate the structures. Bars: (A and D) 200 µm; (F) 50 µm. (G) Quantification of CD41+ cells per micrometer of AGM. Spearman test was used to assess the significance between wild-type allele number and CD41+ cells per micrometer. Correlation between number of wild-type alleles and CD41+ cells per micrometer was significant (**, P < 0.001; and ρ = 0.985). Bars represent the mean of counted cells in two different embryos ± SEM. (H) CD41+ clusters containing more than four cells per micrometer of AGM. Quantification of CD41+ cells per micrometer of AGM. Spearman correlation was used to assess the significance between number of wild-type alleles and CD41+ clusters containing more than four cells per micrometer (**, P = 0.005; and ρ = 0.803). Bars represent the mean of counted clusters in two different embryos ± SEM.
Occupancy of the Gata2 promoter by HES-1 and Notch/RBPJ
Next, we determined the expression levels of different hematopoietic transcription factors in the AGM clusters from wild-type and mutant embryos. We found that Runx1, c-myb, and Gata2 were significantly overexpressed in Hes-deficient embryos compared with the wild type (or deletion of one Hes allele; Fig. 2 A). Importantly, expression levels of Gata2 but not of Runx1 and c-myb in the embryonic AGM directly and significantly correlated with the number of deleted Hes1/5 alleles in the mutants (Spearman’s correlation, P < 0.001; Fig. 2 A). Increase in Gata2 levels was also detected by in situ hybridization of the whole AGM region of Hes1−/−Hes5+/− embryos. Specifically, Gata2 mRNA was up-regulated in the hematopoietic clusters, mesenchyme, mesonephros, urogenital ridge, and mesentery lining of these mutant embryos when compared with the wild type (Fig. 2 B).
Figure 2.
Occupancy of the Gata2 promoter by HES-1 and Notch/RBPJ. (A) Relative expression of the indicated genes from E10.5 AGM after 3 d of 4-OH-tamoxifen treatments in explant cultures by qRT-PCR. Fold increase relative to the wild-type control after β2-Microglobulin and GAPDH normalization. Spearman test was used to assess the significance between number of wild-type alleles and gene expression indicated on the top of the figure. Correlation between number of wild-type Hes alleles and Gata2 expression was significant (P ≤ 0.001 and ρ = 0.985) but not for c-Myb (P = 0.126 and ρ = 0.985) and Runx1 expression (P = 0.072 and ρ = 0.591). The mRNA levels of Runx1, Gata2, and c-Myb are significantly up-regulated in cells with three to four deleted compared with zero to one deleted hes alleles (Student’s t test, P < 0.05). Bars represent the mean ± SD fold increase determination from two independent experiments. (B) Representative in situ hybridization of Gata2 in the wild-type (n = 2) or Hes mutant embryos (n = 2, each genotype). Images show transversal section of E10.5 embryos in the AGM region. Arrowheads indicate the structures (Ao, aorta; IAHC, intra-aortic hematopoietic cluster; m, mesonephros; nt, neural tube). Details of IAHC in the right panels correspond to the boxed areas in the left panels. Bars: (left) 200 µm; (right) 20 µm. (C) Graphical representation of the mouse Gata2 promoter showing the putative RBPJ- or HES-binding consensus found by bioinformatic analysis. Asterisks indicate the conserved sites in the human Gata2 locus (IS, specific promoter; IG, general promoter). (D) Sequence and location of the human and mouse conserved RBPJ- and HES-binding sites relative to IS and IG promoters. (E) ChIP with the anti–HES-1, anti-ICN1, anti-RBPJ, and anti-IgG antibodies from myeloid 32D cells and analysis of the Gata2 locus by qPCR. One representative of two experiments is shown. (F and G) ChIP with the anti–HES-1, anti-ICN1, anti-RBPJ, and anti-IgG antibodies from E10.5 AGM cells and analysis of the Gata2 locus by qPCR. One representative from three experiments is shown. (E–G) Error bars represent SD from qPCR values.
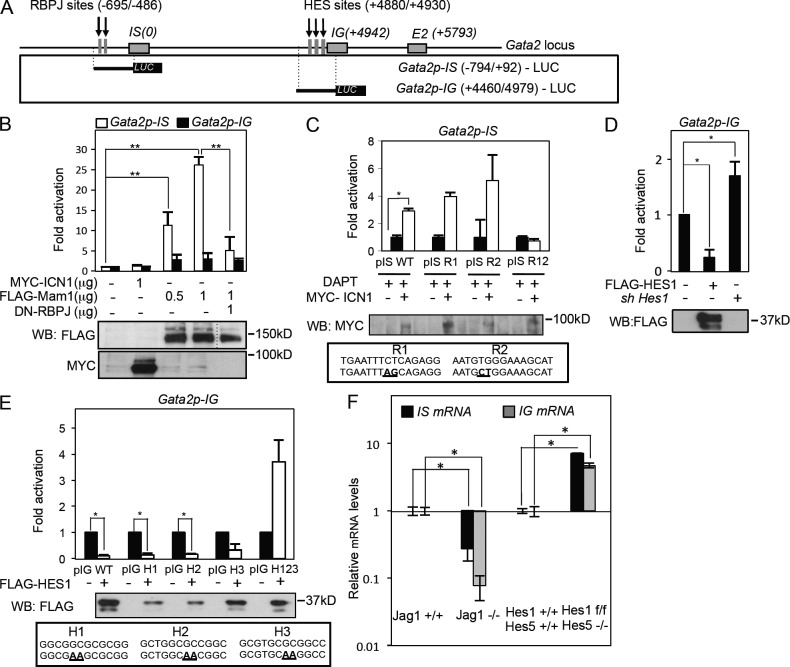
Previous data from our group indicated that Gata2 is a downstream target of Notch in the AGM (Robert-Moreno et al., 2005). By sequence analysis of the Gata2 locus using Genomatix software, we have now identified the presence of two RBPJ and three HES motifs that are conserved in both the human and mouse Gata2 gene (Fig. 2, C and D). Because HES-1 is the main Notch target and a well-characterized transcriptional repressor, we hypothesized that Notch might activate Gata2 transcription that is next antagonized by Notch-mediated induction of HES-1. To address this possibility, we first conducted chromatin immunoprecipitation (ChIP) analysis using 32D myeloid progenitor cells to determine the recruitment of RBPJ, ICN1, and HES-1 proteins to the Gata2 locus. We tested Gata2 occupancy using primers that amplify 15 regions spanning from −3033 to 5827 relative to the TSS at the IS promoter (Fig. 2 E). Of note, Gata2 can be transcribed from two different promoters (IS and IG), both yielding the same protein but using a different first untranslated exon (Minegishi et al., 1999), with IS being hematopoietic/neural specific (Nagai et al., 1994) and IG the general Gata2 promoter. Our analysis revealed RBPJ and ICN1 occupancy in the −800 to 14 region, which contains two of the three predicted RBPJ-binding sites, whereas HES-1 protein was found associated at the 3059 to 5203 region, which contains the three predicted HES-1 sites (Fig. 2 E). A similar pattern for Notch1, RBPJ, and HES-1 association was detected using dissected AGM tissue (Fig. 2, F and G) and human K562 myeloerythroid cells (not depicted). Interestingly, we failed to detect RBPJ or HES binding to the Gata2 promoter in mouse NIH-3T3 fibroblasts (not depicted). These results indicated that RBPJ/Notch proteins occupy the IS promoter, whereas HES-1 binds the IG promoter of the Gata2 gene specifically in hematopoietic cells.
HES-1 is a negative regulator of the Gata2 promoter in hematopoietic cells
Previously, we demonstrated that embryos with deficient Notch activity failed to express Gata2, and we presented evidence that Gata2 is a transcriptional Notch target (Robert-Moreno et al., 2005). However, ectopic expression of ICN1 in AGM cells does not increase Gata2 transcription (not depicted), which could be explained by a negative regulation of the Gata2 promoter by HES-1. To test this possibility, we analyzed different Gata2 regulatory regions (Fig. 3 A) for their capacity to respond to ectopically expressed ICN1 or HES-1. We found that Gata2p-IS, containing RBPJ sites, but not Gata2p-IG, was transcriptionally induced by the Notch coactivator Mam1 in a dose-dependent manner and repressed by a dominant-negative RBPJ (Fig. 3 B). Furthermore, Gata2p-IS promoter was activated two- to fourfold by ICN1 after inhibition of endogenous Notch activity by DAPT (Fig. 3 B and not depicted). By mutational analysis, we found that both predicted RBPJ-binding sites in the Gata2p-IS promoter were functional because only elimination of the two sites impaired the capacity of this reporter to be activated by Notch (Fig. 3 C). In similar experiments, Gata2p-IG reporter was repressed by ectopic HES-1 expression and activated in cells transduced with Hes1-specific shRNA (Fig. 3, D and E). Mutational analysis demonstrated that all three HES-binding sites were functional and sufficient to repress Gata2 reporter expression because only the combined mutation of all three sites relieved Gata2 repression by HES-1 (Fig. 3 E). As we previously showed that Jagged1 is the Notch ligand required in the AGM for the hematopoietic development (Robert-Moreno et al., 2008), we next measured the levels of IS and IG Gata2 mRNA in AGM from embryos deficient for Jagged1 by qRT-PCR and found that both transcripts are equally down-regulated in the absence of Notch activation (Fig. 3 E). Moreover, we detected a similar significant increase of both IS and IG mRNA in the AGM from Hes-deficient embryos, suggesting that Notch and HES are globally affecting the Gata2 locus. Together, these results indicate that Notch and HES coregulate expression of the Gata2 promoter in vitro in an opposite manner.
Figure 3.
HES-1 is a negative regulator of the Gata2 promoter in hematopoietic cells. (A) Scheme of the Gata2 locus indicating the conserved RBPJ- and HES-binding sites (vertical arrows). Promoter regions were used in the luciferase assay. (B) Luciferase activity from the Gata2p-IS and Gata2p-IG promoter in the presence or absence of ICN1, Mam1, or dn-RBP in HEK-293T cells (**, P ≤ 0.009). The dotted line in the Western blot (WB) indicates that intervening lanes have been spliced out. (C) Luciferase activity from the Gata2-IS promoter with indicated point mutations in the RBPJ-binding consensus (bold/underlined) measured in HEK-293T cells. DAPT incubation was used to reduce endogenous Notch activity. (D) Luciferase activity of the Gata2-IG promoter in the presence or absence of HES-1. (E) Luciferase activity from the Gata2-IG promoter with the indicated point mutations (bold/underlined) in the HES-binding consensus. (B–E) Mean ± SD of at least three independent experiments is shown in all the plots; Student’s t test was used to assess the significance (*, P < 0.05). (F) Relative expression of the indicated genes from independent determinations of fresh E10.5 AGMs in Jagged1+/+ (n = 3) and Jagged1−/− (n = 3) and E10.5 AGMs in Hes1-5 wild type (n = 2) and knockouts (n = 3) by qRT-PCR after 3 d of 4-OH-tamoxifen treatments in explant cultures. Fold increase to the wild-type control after GAPDH normalization is shown. Bars represent the mean ± SD determination; Student’s t test was used to assess the significance (*, P < 0.05).
Notch/RBPJ and HES control Gata2 expression during embryonic development
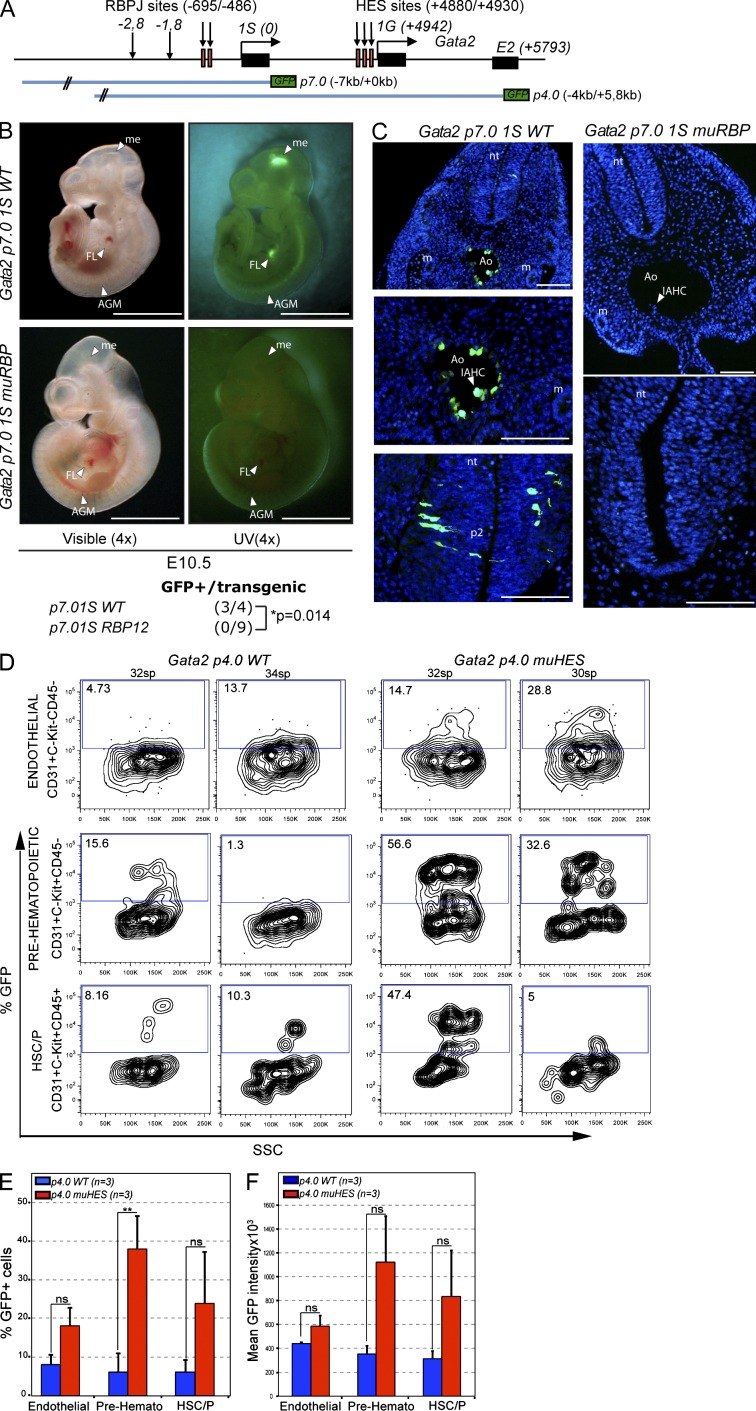
To test whether Notch and HES control Gata2 expression in the embryo, we generated point mutations for both RBPJ sites (p7.0IS RBP12) on the previously characterized Gata2-IS reporter construct (p7.0IS WT; Fig. 4 A; Minegishi et al., 1999, 2003) and microinjected mutant or wild-type DNA into fertilized oocytes. The expression patterns of GFP driven by these promoters in the transgenic F0 (founders) mice were examined at E10.5 (Fig. 4 B). In embryos carrying the wild-type reporter, we found specific expression of GFP in the dorsal aorta and neural tissue (3/4 transgene-positive embryos as determined by PCR) as previously reported (Minegishi et al., 2003). Importantly, mutations in the core of the RBPJ-binding sites abolished the expression of the reporter gene in both the aorta and the neural tissue (0/9 transgene-positive embryos by PCR). Immunostaining with anti-GFP in embryo sections confirmed the expression pattern for the Gata2 reporter in wild-type embryos and the absence of GFP in the aorta or neural tube of embryos carrying the RBPJ mutant reporter (Fig. 4 C). These results indicate that Notch/RBPJ activity is required for driving Gata2 transgene expression from the IS promoter in both hematopoietic and neural tissues. We performed comparable experiments using the p4.0 reporter with mutations of all three HES sites. This reporter contains the −4 to 5 kb sequence of the mouse Gata2 locus (including IS and IG promoter regions) and recapitulates the expression pattern of the endogenous Gata2 gene in transgenic embryos (including expression in the AGM and neural tube; Minegishi et al., 1999, 2003). Based on our in vitro data, we predicted that HES-binding site mutations might either increase the reporter levels within a particular cell population or change the cell type specificity of the reporter. Therefore, as readout for these experiments, we determined GFP levels in specific hematopoietic (CD31+/C-Kit+CD45+), prehematopoietic (CD31+C-Kit+CD45−), and endothelial (CD31+C-Kit−CD45−) cell populations from E9.5–10.5 transgenic embryos (26–35 somite pairs). Our results demonstrate that the wild-type reporter was expressed at low levels in all the AGM subpopulations. Interestingly, HES mutant reporter led to an increased percentage of GFP-positive cells that was not significant in the endothelial lineage (from 8 ± 2.6% to 18.1 ± 4.6%; P = 0.07) or in the committed HSC and progenitors (HSC/Ps; from 6 ± 3% to 24 ± 16%; P = 0.13) but reached statistical significance in the prehematopoietic population (from 6.2 ± 4.6% to 38 ± 10%; P = 0.015; Fig. 4, D and E). Moreover, hematopoietic cells carrying the mutant reporter showed increased expression levels of GFP (Fig. 4, D and F). These results demonstrate that Notch is required for Gata2 expression in the AGM, whereas HES restricts the levels of Gata2 specifically within the presumptive AGM hematopoietic cells.
Figure 4.
Notch/RBPJ and HES-1 regulate Gata2 promoter in vivo during embryonic development. (A) Schematic representation of the Gata2 promoter constructs analyzed in vivo. (B) Representative brightfield (left) and fluorescent (right) micrographs of embryos containing the wild-type or RBPJ mutant Gata2p7.0IS construct at E10.5. Arrowheads indicate the structures (FL, fetal liver; me, mesencephalon). Summary of the data from at least three independent experiments is indicated at the bottom. Fisher test was used to assess the significance. (C) Transversal section of the AGM region and neural tube (nt) of E10.5 embryos containing the indicated constructs showing expression of GFP as detected by immunostaining. Nuclear staining by DAPI is represented in blue. Arrowheads indicate the structures (Ao, aorta; IAHC, intra-aortic hematopoietic cluster; m, mesonephros; p2, neural tube p2 domain). Bars: (B) 2 mm; (C) 200 µm. (D) Flow cytometry analysis of GFP in the indicated CD31+ subpopulation of E10.5 AGM cells from two representative embryos containing the p4.0 wild-type or mutant for HES sites. (E and F) Mean percentage (E) and mean intensity (F) of GFP+ cells in the endothelial (CD31+C-Kit−CD45−), prehematopoietic (CD31+C-Kit+CD45−), and HSC/P (CD31+C-Kit+CD45+) populations from three transgene-positive embryos from at least two independent experiments (**, P = 0.01). Error bars represent SEM.
AGM hematopoietic clusters in Hes-deficient embryos lack functional progenitors and HSCs
We have shown that Hes deficiency results in enlarged hematopoietic clusters in the developing AGM. To test the functionality of these mutant hematopoietic cells, we used the mouse line containing the tamoxifen-inducible β-actin–CRE-ERT transgene in different combinations of the loxP-Hes1 and the Hes5 deleted alleles. E10.5 AGMs of the different genotypes were dissected and cultured as explants for 3 d in the presence of 4-OH-tamoxifen to induce the deletion. Dissociated AGM cells were then plated in methylcellulose to determine the number of hematopoietic progenitors by colony-forming cell (CFC) assays. We found that AGM cells containing three deleted Hes alleles showed significantly different number of functional progenitors (CFC) when compared with those carrying one or two deleted alleles (Fig. 5 A). These results suggest that a minimum threshold of HES levels (two wild-type alleles) is required to maintain the functionality of hematopoietic progenitors. Interestingly, CFC assay of E10.5 YS cells revealed that progenitors contained in the extraembryonic tissue are not significantly affected in the β-actin–CRE-ERT loxP-Hes1 R26-YFPf/+ Hes5+/− embryos (not depicted), in agreement with previous findings showing the Notch-independent formation of YS progenitors (Robert-Moreno et al., 2007; Bertrand et al., 2010). Analysis of the colonies by PCR confirmed the deletion of the inducible alleles after tamoxifen treatment (Fig. 5 A). Furthermore, hematopoietic progenitors (defined as CD34+C-Kit+) isolated from constitutive Hes1−/−Hes5−/− embryos were unable to produce sca1+C-Kit+ progenitors and CD45+ hematopoietic cells when cultured on OP9 stroma, compared with Hes5−/− cells (Fig. 5 B).
Figure 5.
AGM hematopoietic clusters in Hes-deficient embryos lack functional progenitors and HSCs. (A) Determination of CFU-C hematopoietic progenitors obtained from E10.5 AGM after 3 d of explant culture in the presence of 4-OH-tamoxifen to induce Hes1 deletion. Kruskal–Wallis and Mann–Whitney U tests were used to assess the significance. Mean ± SEM from three or more AGM explants from at least three independent experiments is shown. PCRs from isolated colonies to test Hes1 deletion are shown. (B) Flow cytometry analysis from CD34+C-KIT+ sorted cells from E9.5 and E10.5 embryos with the indicated genotypes cultured on OP9 with factors (SCF+IL3+Epo+G-CSF) for 10 d. Dot plots are representative of three independent experiments. (C) Spleen colonies from 4-OH-tamoxifen–treated E10.5 AGM explants from the indicated genotypes injected in irradiated SCID-Beige mice. Mean of (CFU-S11) from three or more injected animals is shown. Student’s t test was used to assess significance. Error bars represent the mean of colonies in the spleens analyzed. (D) Representative spleen images (top) and PCR from dissected spleen colonies to test for Hes1 deletion (bottom) are shown. (E) Flow cytometry analysis at 2 mo of SCID-Beige transplanted animals. Each mouse was transplanted with 2 ee AGMs after 3 d of 4-OH-tamoxifen treatments in explant culture. Two representative controls (n = 4) and two Hes1f/fHes5+/− R26-YFPf/+ β-actin–CRE-ERT (n = 3) are shown. Controls with >1% of YFP cells were considered as reconstituted. (F) Quantification of Ki67 and TUNEL within the CD31-positive cells from immunostainings (n = 2 each genotype) of Ki67/CD31 and TUNEL/CD31 in the AGM region in wild-type or Hes1f/fHes5−/− β-actin–CRE-ERT mutant embryos after in vivo tamoxifen induction at day E8. Error bars represent the mean of positive cells from the different embryos analyzed. (G) Gating strategy to analyze the different subpopulations by flow cytometry analysis of fresh E10.5 AGMs with functional HSC phenotype (zero to one Hes deleted alleles; n = 4) or with nonfunctional HSC phenotype (three to four Hes deleted alleles; n = 3) after in vivo 4-OH-tamoxifen induction at E8. (H) Bars represent the mean percentage ± SD of positive cells of the indicated populations.
To measure the number and functionality of Hes-deficient hematopoietic progenitors in an in vivo system, we determined the capacity of dissociated AGM cells from tamoxifen-treated explants of different genotypes to generate spleen colonies (CFU-S11) in lethally irradiated mice. As Hes1f/fHes5−/− mutant animals were obtained on a mixed background, we used immunodeficient SCID-Beige mice as recipients for the transplants to avoid graft rejection. In these experiments, we obtained a mean number of 16.6 ± 1.4 CFU-S11 from one wild-type E10.5 AGM explant (1 embryo equivalent [1 ee]), similarly to that obtained from Hes5−/− AGM explants (Fig. 5, C and D; and not depicted). In contrast, the number of colonies obtained from AGM explants with all four Hes alleles deleted was comparable with the noninjected/irradiated controls (4 ± 0.3; Fig. 5, C and D), and the few colonies detected did not contain Hes1 deletion (Fig. 5 D, bottom). We next determined the existence of functional HSCs after induction of Hes1 deletion in E10.5 AGM explants from the β-actin–CRE-ERT loxP-Hes1 R26-YFPf/+ Hes5+/− background compared with the β-actin–CRE-ERT R26-YFPf/+. To increase the repopulating capacity of E10.5 AGM, we injected 2 ee per each SCID-Beige recipient together with spleen cells as a support. In these experiments, we did not detect any HSC activity from Hes-deficient AGM cells (three alleles deleted), whereas all mice transplanted with control cells showed hematopoietic multilineage reconstitution after 8 wk (n = 4, considering >1% of donor cells in peripheral blood; Fig. 5 E and not depicted).
Although these results indicate that E10.5 Hes-deficient embryos (deletion induced in vivo at E8.5) do not contain functional HSC/Ps, their AGMs contained higher numbers of CD41+ and C-Kit+ cluster cells (Fig. 1, F–H; and not depicted). To further understand how Hes deficiency contributes to this particular phenotype, we determined by immunofluorescence the number of TUNEL- and Ki67-positive cells in the CD31+ cells of sections from E10.5 wild-type and β-actin–CRE-ERT loxP-Hes1Hes5−/− embryos treated with tamoxifen at E8.5. We found that CD31+ cells of the AGM contain a significantly higher number of Ki67+ cells and very low but comparable numbers of TUNEL+ cells (Fig. 5 F). Most importantly, a detailed analysis of the different subpopulations contained within the CD31+ population of the AGM (hematopoietic clusters plus endothelium) demonstrated that whereas the C-Kit+CD41+CD45− (pre-HSC) population was increased upon Hes deletion, the C-Kit+CD41+CD45+ population that defines the committed HSCs was significantly decreased (Fig. 5, G and H). These results suggest that Hes deficiency induces higher proliferation in the AGM clusters while preventing prehematopoietic cells to reach the HSC stage.
Reduction of Notch signaling partially rescues the Hes-dependent hematopoietic defect
To investigate the putative connection between overexpression of Gata2 and overproduction and nonfunctional hematopoietic cells in the AGM, we aimed to reduce Gata2 levels in the Hes-deficient AGM cells. Because Gata2 expression is dependent on Notch activity, we anticipated that incubation of Hes-deficient cells with limited concentrations of Notch/γ-secretase inhibitors should lead to a decrease of Gata2 levels that would be compatible with the hematopoietic development.
After testing different DAPT concentrations, we found that incubation of Hes-deficient AGM cells in liquid cultures containing 0.05 µM DAPT resulted in a substantial decrease of Gata2 levels, resembling the levels detected in DMSO-treated wild-type AGM cells (Fig. 6 A). In these conditions, Hes-deficient AGM cultures produced increased numbers of CD45+ cells compared with the DMSO-treated Hes-deficient cultures (Fig. 6 B). Conversely, incubation of wild-type AGM cells with the same dose of DAPT (or higher) resulted in a decrease in CD45+ cell production as previously published (Robert-Moreno et al., 2008). These results indicate that the positive hematopoietic effect of DAPT observed in Hes-deficient cells is not directly mediated by changes in Notch activity but by a downstream target of Notch and Hes. Based on this data, we propose the existence of a permissive threshold of Gata2 levels that is acquired by the combined action of Notch and Hes repressors that allow hematopoietic development in the AGM (see model in Fig. 6 C). Together, these results indicate that HES proteins are responsible for the control of Gata2 levels downstream of Notch signaling, which is essentail for the generation of HSCs during embryonic development.
Figure 6.
Reduction of Notch signaling partially rescues Hes-dependent hematopoietic defect. (A) Wild-type and Hes1f/fHes5−/− β-actin–CRE-ERT E10.5 dissected AGMs after in vivo tamoxifen induction at E8.5, disrupted with collagenase, and plated in liquid culture in the presence of cytokines (IL6, IL3, and SCF), DMSO, or 0.05 µM DAPT; mRNA was obtained to quantify the levels of Gata2 after 24 h. Bars represent the mean ± SD determination (n = 3 each condition). (B) In parallel, cultures were analyzed after 6 d by flow cytometry to quantify the amount of cells CD45+ (wild type, n = 4; Hes1f/fHes5−/− β-actin–CRE-ERT, n = 10). (C) In the presence of HES-1, Gata2 levels are limited to the just-right dose to generate functional HSC/P. (D) In the absence of Hes1, Gata2 levels cannot be repressed and result in the generation of a great number of nonfunctional progenitors.
DISCUSSION
Deciphering the molecular events that control hematopoietic development is a crucial issue for understanding how functional HSCs are produced and maintained. In this work, we focused on the role of HES proteins, which are the principal mediators of the Notch pathway in multiple systems.
Fetal liver of Hes1-deficient embryos had been previously analyzed, and no major defects on HSC activity were found (Tomita et al., 1999), although a direct measure of HSC activity was not performed. Importantly, here we demonstrate that Hes5 (which is functionally equivalent to Hes1) is expressed de novo in the aorta of Hes1 mutants, which suggests the possibility of functional compensation between both proteins. In agreement with this, we found that cumulative deletion of Hes1 and Hes5 alleles leads to the generation of nonfunctional hematopoietic progenitors in the developing AGM and the total absence of HSC activity. In addition, we demonstrate that the Gata2 gene is directly activated by Notch/RBPJ but repressed by HES-1 specifically in the C-Kit+ cell population of the emerging AGM clusters. Thus, in Hes-deficient mutants, Gata2 is up-regulated, and the number of hematopoietic cluster cells increased, but they only contained nonfunctional hematopoietic cells, in agreement with previous work demonstrating that Gata2 levels need to be tightly regulated to generate and maintain functional HSCs (Ling et al., 2004; Rodrigues et al., 2005). Our experiments show that limiting Notch activity in the absence of HES can decrease the levels of Gata2 and partially restore the hematopoietic production from AGM cells. Mechanistically, one possibility is that overexpression of Gata2 in AGM cells imposes an impairment to cell cycle entry in the hematopoietic progenitor populations, as it was demonstrated in the cord blood HSCs (Tipping et al., 2009); however, our results indicate that the Hes-deficient endothelium contains more proliferative cells, suggesting that AGM subpopulations may show specific response to different GATA-2 levels.
Analysis of the Gata2 regulatory regions led to the identification of two functional DNA-binding sites for Notch/RBPJ and three for HES, and we demonstrated their relevance in the activation and repression of this gene in the embryonic aorta, the niche of nascent HSCs. As Hes is robustly induced by Notch, activation of this pathway in the hematopoietic progenitors might induce both positive and negative signals in the Gata2 gene, resulting in the so-called type I incoherent feed-forward loop (I1-FFL; Mangan and Alon, 2003), which from our predictions would limit Gata2 activation in this tissue (Fig. 6 A). These types of circuits have previously been found to regulate Notch-dependent cell fate decisions in Drosophila (Krejcí et al., 2009). Here, we propose that after Notch activation, likely through Jagged1, Gata2 and Hes genes start to be transcribed until HES protein reaches the threshold for repressing the Gata2 promoter. Then Gata2 transcription decreases, and its mRNA and protein concentration drops, resulting in a pulse-like dynamic that it is required to achieve limited/transient (producing functional progenitors) rather than high/sustained activation (producing nonfunctional progenitors; see model in Fig. 6 B). I1-FFLs are also known to regulate biphasic responses where the output levels (in this case Gata2) depend on the input dose (Notch intensity; Kim et al., 2008). By computer modeling, it has been demonstrated that I1-FFLs regulate time- or dose-dependent biphasic behaviors in a mutually exclusive manner (Kim et al., 2008), and further work is needed to address whether Gata2 regulation by Notch and HES follows one of these two models. In this work, we have dissected the effects of a single Notch pathway mediator (HES) during the generation of HSCs. However, this is an extremely simplified picture of the Notch-dependent regulatory networks occurring during this process, and many other elements and pathways will be sequentially incorporated into the final model. One example is the restriction imposed by the availability of Notch ligands to the Notch-responding cell that will depend on other upstream signals such as WNT (Estrach et al., 2006; Rodilla et al., 2009), the specificity of Notch for its ligands that is modulated by Fringe-mediated glycosylation (Koch et al., 2001), or other interactions with the niche, which will be crucial to achieve the proper intensity of Notch signal in individual AGM cells. Another variable is HES-1 protein stability, which is regulated by the JAK–STAT pathway and directly impacts HES oscillation (Yoshiura et al., 2007). In addition, Hes1 transcription itself is controlled by a negative autoregulatory feedback loop (Hirata et al., 2002). Finally, a complex network of interactions that involves FLI1 and SCL and operates downstream of Notch and GATA-2 in the HSC and hematopoietic precursors has already been modeled (Narula et al., 2010). In summary, we have here deciphered the first mechanism downstream of Notch signaling that regulates HSC development.
MATERIALS AND METHODS
Animals.
CD1, C57B6/J wild-type, and Jagged1−/− (Xue et al., 1999), Hes1−/−;Hes5−/− (Ohtsuka et al., 1999), Hes1f/f (Imayoshi et al., 2008), and β-actin–CRE-ERT (Hayashi and McMahon, 2002) strains were used. R26-RYFPf/f (strain name: B6.129X1-GT(ROSA)26Sortm1(EYFP)Cos/J) was obtained from the Jackson Laboratory. C57BL6/J oocytes were used in F0 assays. SCID-Beige animals were used as recipients in CFU-S11 and long-term reconstitution experiments. Animals were kept under pathogen-free conditions, and all procedures were approved by the Animal Care Committee of the Parc de Recerca Biomèdica de Barcelona (regulation of Generalitat de Catalunya). Embryos were obtained from timed pregnant females and staged by somite counting: E10.5 (31–40 sp). The detection of the vaginal plug was designated as day 0.5. Mice and embryos were all genotyped by PCR.
qRT–PCR.
Total RNA was extracted with a QIAGEN kit, and RT-First Strand cDNA Synthesis kit (GE Healthcare) was used to produce cDNA. qRT-PCR was performed in a LightCycler480 system using SYBR Green I Master kit (Roche). Primers used are in Table S1.
Promoter analysis, site-directed mutagenesis, and luciferase assays.
Human and mouse Gata2 promoters were analyzed using Genomatix software. For luciferase assays, Gata2p-IG was previously described (Minegishi et al., 1998), which covers from −482 of IG transcript to exon 2. Gata2p-IS was generated cloning the region from −794 to 93 of exon IS into the pGL3 vector and verified by sequencing. The primers used were Fwd (5′-AAAACTCGAGGGTGCTACAAGGATGTGGTTGC-3′) and Rev (5′-AAAAAAGCTTGCTATGGTTGAGGTGATCTTAGGC-3′). Mutations were introduced into wild-type plasmids using the QuikChange Site-Directed Mutagenesis kit from Agilent Technologies according to the manufacturer’s instructions. The mutations introduced and primers used are specified in Table S2. All mutations were verified by DNA sequencing.
Luciferase reporter assays were performed in HEK-293T cells. Cells were seeded in 12-well plates at a density of 5 × 104 cells/well. Equal amounts (150 ng) of the different Gata2 reporter constructs ICN1, Mastermind, dn-Rbpj, shHes1 (MISSION, TRCN0000018989), or irrelevant DNA and 150 ng CMV–β-Galactosidase plasmids were transfected in triplicate wells. Cells were transfected using polyethylenimine (PEI; Polysciences, Inc.). To reduce endogenous ICN1 levels, HEK-293T cells were treated with γ-secretase inhibitor, DAPT (EMD), at 50 µM final concentration for 72 h before transfection and during the assay. Luciferase was measured after 48 h of transfection according to the manufacturer’s instructions (Luciferase Assay System; Promega). Expression levels of transfected proteins were verified by Western blot.
ChIP assay.
Chromatin was obtained from a pool of 40 dissected AGMs at E10.5 or 32D myeloid progenitor cells line. ChIP was performed as previously described (Aguilera et al., 2004) with minor modifications. In brief, cross-linked chromatin from E10.5 dissected AGMs or 32D cell line was sonicated for 10 min, medium-power, 0.5-interval, with a Bioruptor (Diagenode) and precipitated with anti-RBPJ (Chu and Bresnick, 2004), anti-ICN1 (Abcam), and anti-HES1 (H-20 from Santa Cruz Biotechnology, Inc. and N terminus from Aviva Systems Biology). After cross-linkage reversal, DNA was used as a template for PCR. RT-PCR was performed with SYBR Green I Master (Roche) in a LightCycler480 system. Primers used are in Table S3.
Cell culture, viral particle production, and viral infection.
32D cells were cultured in Iscove’s and 10% FBS supplemented with 10% WeHi-IL3–conditioned media. HEK-293T cells were seeded at a density of 5 × 105 cells/well and transfected with ICN1-IRES-GFP with PEI. Hematopoietic cells from disrupted AGMs were incubated in Iscove’s, 10% FBS, 10% IL-3, and 10% stem cell factor (SCF)–conditioned medium plus 0.2 µg/ml IL6 and 0.1 µg/ml Flt3. To assess hematopoietic potential of sorted populations from Hes1−/−Hes5−/− mutant mice, 500 CD34+C-Kit+ cells were FACS purified and added to confluent cultures of OP9 stromal cells and cultured for up to 10 d in the presence of 100 ng/ml IL-3, 100 U/ml SCF, 100 ng/ml G-CSF, and 200 ng/ml erythropoietin (Epo). Supplemented differentiation medium was replaced every 4 d. Cultures were maintained for up to 12 d. Cells were harvested by pipetting and examined for hematopoietic development by flow cytometry.
Flow cytometry analysis and sorting.
The AGM region was dissected from E10.5 embryos and dissociated in 0.12% collagenase (Sigma-Aldrich) in PBS supplemented with 10% FBS for 30 min at 37°C. The cells were analyzed by flow cytometry in a FACSCalibur (BD) or FACSAria (BD) and FlowJo software (Tree Star). Dead cells were excluded by 7-aminoactinomycin-D staining (Molecular Probes) in the FACSCalibur analysis and with Hoechst or DAPI in the FACSAria analysis. The YSs and lower trunk regions were harvested from individually dissected embryos washed of maternal blood (Fraser et al., 2002). The anterior regions were used for genotyping (Ohtsuka et al., 1999). YSs and lower trunk regions were dissociated with 0.1% wt/vol collagenase types I and IV (Sigma-Aldrich) for 1 h at 30°C. Cells were then blocked with normal mouse serum and incubated with fluorescence-conjugated antibodies. For flow cytometric analysis, cells were resuspended in Hank’s buffered Saline Solution (Gibco) containing propidium iodide (Sigma-Aldrich) for dead cell exclusion. Flow cytometry was performed using a FACSVantage (BD) with CellQuest (BD), and data were analyzed using the FlowJo software package. The antibodies CD31-PE, Sca1-PE, CD45-FITC, Ter119-PE, CD45-PerCPCy5.5, and CD117-APC were purchased from BD and CD41-PeCy7 from eBioscience. VE-cadherin–APC was generated in-house.
Immunostaining.
E10.5 embryos were fixed overnight in 4% paraformaldehyde (Sigma-Aldrich) at 4°C and frozen in Tissue-Tek (Sakura). 5-µm sections were fixed with −20°C methanol for 15 min and block permeabilized in 10% FBS, 0.3% Surfact-AmpsX100 (Thermo Fisher Scientific), and 5% nonfat milk in PBS for 90 min at 4°C. Samples were stained with CD31 (PECAM1 at 1:50; BD) or CD41 (1:50; BD) or C-Kit (CD117 at 1:50; BD) in 10% FBS, 5% nonfat milk in PBS overnight, and HRP-conjugated secondary antibody (Dako) at 1:100 for 90 min and developed with Cy3 or coupled tyramide (PerkinElmer). Anti-GFP (MBL) was used at 1:100, and Alexa Fluor 488–conjugated donkey anti–rabbit (1:500; Invitrogen) was used as a secondary antibody. DeadEnd Fluorimetric System (Promega) and Ki67 (1:500; Novocastra) were also used. Sections were mounted in Vectashield medium with DAPI.
In situ hybridization.
For in situ hybridization, precisely timed embryos were fixed overnight at 4°C in 4% paraformaldehyde and embedded in OCT (Tissue-Tek). Embryos were sectioned in an RM2135 cryostat (Leica) at 16 µm. Digoxigenin-labeled probes were generated from a Bluescript plasmid (Agilent Technologies) containing a partial mouse Gata2 cDNA (nt 170–871) and from plasmid containing Hes1 cDNA and Hes5 cDNA. Images were acquired with a BX-60 (Olympus). Representative images were edited on Photoshop CS4 software (Adobe).
Transgenic mice: F0 assays.
Gata2 promoter inserts from plasmids (p4.0, p4.0 muHes, p7.0, and p7.0 muRBP) were purified by NACS PREPAC (Gibco), adjusted to 5–10 ng/ml, and injected into mouse oocytes using standard strategies. Transgene integration was confirmed by PCR from genomic DNA from embryonic tissues. The wild-type constructs p4.0 (Kobayashi-Osaki et al., 2005) and p7.0 (Minegishi et al., 1999) were previously described, and point mutations were introduced into these plasmids.
AGM explant culture.
The AGM region was dissected and cultured in explants for 3 d as previously described (Medvinsky and Dzierzak, 1996). In brief, AGMs were deposited on nylon filters (EMD Millipore) placed on metallic supports and cultured in myeloid long-term culture medium (STEMCELL Technologies) supplemented with 10 µM hydrocortisone (Sigma-Aldrich) in an air-liquid interphase. The induction of β-actin–CRE-ERT transgene was performed by adding 4-OH-tamoxifen (Sigma-Aldrich) every 36 h to the explant medium at a final concentration of 1 µM.
Hematopoietic colony assay (CFU-C) and CFU-S11 and long-term reconstitution assay.
The explanted AGMs were digested in 0.12% collagenase (Sigma-Aldrich) in PBS supplemented with 10% FBS for 20 min at 37°C and used for hematopoietic colony assay (CFU-C), CFU-S11, and long-term reconstitution assay experiments. In CFU-C assay, 20,000 cells/AGM were plated in duplicates in M-3434 semisolid medium (STEMCELL Technologies). After 7 d, the presence of hematopoietic colonies was scored under the microscope. In CFU-S11 experiments, whole disrupted AGMs were injected intravenously in SCID-Beige recipients. SCID-Beige recipients were previously irradiated at 2.5 Gy. After 11 d, the presence of colonies in the spleen was scored under the stereoscope. Some colonies were picked and lysed in 0.01 M Tris, pH, 7.4, 0.15 M NaCl, 0.01 M EDTA, and 0.5% SDS. From this lysate, Hes1 deletion was detected in both types of colonies, CFU-C and CFU-S11, by PCR (primers in Table S4). In long-term reconstitution assay experiments, whole disrupted AGMs were injected intravenously in SCID-Beige recipients, with 500,000 white spleen cells from a SCID-Beige animal as support cells. SCID-Beige recipients were previously irradiated at 2.5 Gy.
Statistics.
Statistical analyses were performed using Excel 2011 (Microsoft) and SPSS. In Fig. 3 (B–D), Fig. 4 E, and Fig. 5 D, statistical significance was assessed using two-tailed Student’s t tests. In Fig. 1 (F and G) and Fig. 2 A, statistical significance was assessed using the Spearman test. In Fig. 4 A, statistical significance was assessed using the Fisher test. In Fig. 5 A, Kruskal–Wallis and Mann–Whitney U tests were used to assess the significance. In all tests, p-values < 0.05 were considered significant. In all experiments: *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
Online supplemental material.
Table S1 lists mouse RT-PCR primers. Table S2 lists mutagenesis primers. Table S3 lists mouse ChIP primers. Table S4 lists Hes1 lox-P sites to detect deleted bands. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20120993/DC1.
Supplementary Material
Acknowledgments
We thank J. González (Bigas laboratory), C. Vink (Dzierzak laboratory), A. Maas (Erasmus University, Rotterdam, Netherlands), and Y. Kawatani and H. Suda (Shimizu and Yamamoto laboratory) for technical assistance. We would like to thank J. Sharpe (Centre for Genomic Regulation, Barcelona, Spain) and D. Sprinzak (Tel Aviv University, Tel Aviv, Israel) for helpful genetic circuit consultation and members of the Bigas laboratory for critically reading the manuscript and helpful discussions.
J. Guiu is a recipient of Formación de Personal Investigador BES-2008-005708; L. Espinosa is an investigator of the Instituto de Salud Carlos III program (02/30279). This research was funded by the Ministerio de Economía y Competitividad (SAF2007-60080, PLE2009-0111, and SAF2010-15450), Red Temática de Investigación Cooperativa en Cáncer (RD06/0020/0098), Agència de Gestió d’Ajuds Universitaris i de Recerca (2009SGR-23 and CONES2010-0006 to A. Bigas), and Grants-in-Aids for Scientific Research from the Japanese Ministry of Education, Culture, Sports, Science and Technology (to R. Shimizu and M. Yamamoto).
There are no competing financial interests to declare.
Footnotes
Abbreviations used:
- AGM
- aorta/gonad/mesonephros
- CFC
- colony-forming cell
- ChIP
- chromatin immunoprecipitation
- ee
- embryo equivalent
- HSC
- hematopoietic stem cell
- HSC/P
- HSC and progenitor
- I1-FFL
- type I incoherent feed-forward loop
- SCF
- stem cell factor
- YS
- yolk sac
References
- Aguilera C., Hoya-Arias R., Haegeman G., Espinosa L., Bigas A. 2004. Recruitment of IkappaBalpha to the hes1 promoter is associated with transcriptional repression. Proc. Natl. Acad. Sci. USA. 101:16537–16542 10.1073/pnas.0404429101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand J.Y., Cisson J.L., Stachura D.L., Traver D. 2010. Notch signaling distinguishes 2 waves of definitive hematopoiesis in the zebrafish embryo. Blood. 115:2777–2783 10.1182/blood-2009-09-244590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigas A., Robert-Moreno A., Espinosa L. 2010. The Notch pathway in the developing hematopoietic system. Int. J. Dev. Biol. 54:1175–1188 10.1387/ijdb.093049ab [DOI] [PubMed] [Google Scholar]
- Burns C.E., Traver D., Mayhall E., Shepard J.L., Zon L.I. 2005. Hematopoietic stem cell fate is established by the Notch-Runx pathway. Genes Dev. 19:2331–2342 10.1101/gad.1337005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu J., Bresnick E.H. 2004. Evidence that C promoter-binding factor 1 binding is required for Notch-1-mediated repression of activator protein-1. J. Biol. Chem. 279:12337–12345 10.1074/jbc.M311510200 [DOI] [PubMed] [Google Scholar]
- Dzierzak E., Speck N.A. 2008. Of lineage and legacy: the development of mammalian hematopoietic stem cells. Nat. Immunol. 9:129–136 10.1038/ni1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa L., Cathelin S., D’Altri T., Trimarchi T., Statnikov A., Guiu J., Rodilla V., Inglés-Esteve J., Nomdedeu J., Bellosillo B., et al. 2010. The Notch/Hes1 pathway sustains NF-κB activation through CYLD repression in T cell leukemia. Cancer Cell. 18:268–281 10.1016/j.ccr.2010.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrach S., Ambler C.A., Lo Celso C., Hozumi K., Watt F.M. 2006. Jagged 1 is a beta-catenin target gene required for ectopic hair follicle formation in adult epidermis. Development. 133:4427–4438 10.1242/dev.02644 [DOI] [PubMed] [Google Scholar]
- Fischer A., Schumacher N., Maier M., Sendtner M., Gessler M. 2004. The Notch target genes Hey1 and Hey2 are required for embryonic vascular development. Genes Dev. 18:901–911 10.1101/gad.291004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser S.T., Ogawa M., Yu R.T., Nishikawa S., Yoder M.C., Nishikawa S. 2002. Definitive hematopoietic commitment within the embryonic vascular endothelial-cadherin(+) population. Exp. Hematol. 30:1070–1078 10.1016/S0301-472X(02)00887-1 [DOI] [PubMed] [Google Scholar]
- Hayashi S., McMahon A.P. 2002. Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: a tool for temporally regulated gene activation/inactivation in the mouse. Dev. Biol. 244:305–318 10.1006/dbio.2002.0597 [DOI] [PubMed] [Google Scholar]
- Hirata H., Yoshiura S., Ohtsuka T., Bessho Y., Harada T., Yoshikawa K., Kageyama R. 2002. Oscillatory expression of the bHLH factor Hes1 regulated by a negative feedback loop. Science. 298:840–843 10.1126/science.1074560 [DOI] [PubMed] [Google Scholar]
- Imayoshi I., Shimogori T., Ohtsuka T., Kageyama R. 2008. Hes genes and neurogenin regulate non-neural versus neural fate specification in the dorsal telencephalic midline. Development. 135:2531–2541 10.1242/dev.021535 [DOI] [PubMed] [Google Scholar]
- Kageyama R., Ohtsuka T., Kobayashi T. 2007. The Hes gene family: repressors and oscillators that orchestrate embryogenesis. Development. 134:1243–1251 10.1242/dev.000786 [DOI] [PubMed] [Google Scholar]
- Kim D., Kwon Y.K., Cho K.H. 2008. The biphasic behavior of incoherent feed-forward loops in biomolecular regulatory networks. Bioessays. 30:1204–1211 10.1002/bies.20839 [DOI] [PubMed] [Google Scholar]
- Kobayashi-Osaki M., Ohneda O., Suzuki N., Minegishi N., Yokomizo T., Takahashi S., Lim K.C., Engel J.D., Yamamoto M. 2005. GATA motifs regulate early hematopoietic lineage-specific expression of the Gata2 gene. Mol. Cell. Biol. 25:7005–7020 10.1128/MCB.25.16.7005-7020.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch U., Lacombe T.A., Holland D., Bowman J.L., Cohen B.L., Egan S.E., Guidos C.J. 2001. Subversion of the T/B lineage decision in the thymus by lunatic fringe-mediated inhibition of Notch-1. Immunity. 15:225–236 10.1016/S1074-7613(01)00189-3 [DOI] [PubMed] [Google Scholar]
- Kopan R., Ilagan M.X. 2009. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 137:216–233 10.1016/j.cell.2009.03.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krejcí A., Bernard F., Housden B.E., Collins S., Bray S.J. 2009. Direct response to Notch activation: signaling crosstalk and incoherent logic. Sci. Signal. 2:ra1 10.1126/scisignal.2000140 [DOI] [PubMed] [Google Scholar]
- Kunisato A., Chiba S., Nakagami-Yamaguchi E., Kumano K., Saito T., Masuda S., Yamaguchi T., Osawa M., Kageyama R., Nakauchi H., et al. 2003. HES-1 preserves purified hematopoietic stem cells ex vivo and accumulates side population cells in vivo. Blood. 101:1777–1783 10.1182/blood-2002-07-2051 [DOI] [PubMed] [Google Scholar]
- Ling K.W., Ottersbach K., van Hamburg J.P., Oziemlak A., Tsai F.Y., Orkin S.H., Ploemacher R., Hendriks R.W., Dzierzak E. 2004. GATA-2 plays two functionally distinct roles during the ontogeny of hematopoietic stem cells. J. Exp. Med. 200:871–882 10.1084/jem.20031556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangan S., Alon U. 2003. Structure and function of the feed-forward loop network motif. Proc. Natl. Acad. Sci. USA. 100:11980–11985 10.1073/pnas.2133841100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medvinsky A., Dzierzak E. 1996. Definitive hematopoiesis is autonomously initiated by the AGM region. Cell. 86:897–906 10.1016/S0092-8674(00)80165-8 [DOI] [PubMed] [Google Scholar]
- Minegishi N., Ohta J., Suwabe N., Nakauchi H., Ishihara H., Hayashi N., Yamamoto M. 1998. Alternative promoters regulate transcription of the mouse GATA-2 gene. J. Biol. Chem. 273:3625–3634 10.1074/jbc.273.6.3625 [DOI] [PubMed] [Google Scholar]
- Minegishi N., Ohta J., Yamagiwa H., Suzuki N., Kawauchi S., Zhou Y., Takahashi S., Hayashi N., Engel J.D., Yamamoto M. 1999. The mouse GATA-2 gene is expressed in the para-aortic splanchnopleura and aorta-gonads and mesonephros region. Blood. 93:4196–4207 [PubMed] [Google Scholar]
- Minegishi N., Suzuki N., Yokomizo T., Pan X., Fujimoto T., Takahashi S., Hara T., Miyajima A., Nishikawa S., Yamamoto M. 2003. Expression and domain-specific function of GATA-2 during differentiation of the hematopoietic precursor cells in midgestation mouse embryos. Blood. 102:896–905 10.1182/blood-2002-12-3809 [DOI] [PubMed] [Google Scholar]
- Nagai T., Harigae H., Ishihara H., Motohashi H., Minegishi N., Tsuchiya S., Hayashi N., Gu L., Andres B., Engel J.D., et al. 1994. Transcription factor GATA-2 is expressed in erythroid, early myeloid, and CD34+ human leukemia-derived cell lines. Blood. 84:1074–1084 [PubMed] [Google Scholar]
- Narula J., Smith A.M., Gottgens B., Igoshin O.A. 2010. Modeling reveals bistability and low-pass filtering in the network module determining blood stem cell fate. PLOS Comput. Biol. 6:e1000771 10.1371/journal.pcbi.1000771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuka T., Ishibashi M., Gradwohl G., Nakanishi S., Guillemot F., Kageyama R. 1999. Hes1 and Hes5 as notch effectors in mammalian neuronal differentiation. EMBO J. 18:2196–2207 10.1093/emboj/18.8.2196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert-Moreno A., Espinosa L., de la Pompa J.L., Bigas A. 2005. RBPjkappa-dependent Notch function regulates Gata2 and is essential for the formation of intra-embryonic hematopoietic cells. Development. 132:1117–1126 10.1242/dev.01660 [DOI] [PubMed] [Google Scholar]
- Robert-Moreno A., Espinosa L., Sanchez M.J., de la Pompa J.L., Bigas A. 2007. The notch pathway positively regulates programmed cell death during erythroid differentiation. Leukemia. 21:1496–1503 10.1038/sj.leu.2404705 [DOI] [PubMed] [Google Scholar]
- Robert-Moreno A., Guiu J., Ruiz-Herguido C., López M.E., Inglés-Esteve J., Riera L., Tipping A., Enver T., Dzierzak E., Gridley T., et al. 2008. Impaired embryonic haematopoiesis yet normal arterial development in the absence of the Notch ligand Jagged1. EMBO J. 27:1886–1895 10.1038/emboj.2008.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodilla V., Villanueva A., Obrador-Hevia A., Robert-Moreno A., Fernández-Majada V., Grilli A., López-Bigas N., Bellora N., Albà M.M., Torres F., et al. 2009. Jagged1 is the pathological link between Wnt and Notch pathways in colorectal cancer. Proc. Natl. Acad. Sci. USA. 106:6315–6320 10.1073/pnas.0813221106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues N.P., Janzen V., Forkert R., Dombkowski D.M., Boyd A.S., Orkin S.H., Enver T., Vyas P., Scadden D.T. 2005. Haploinsufficiency of GATA-2 perturbs adult hematopoietic stem-cell homeostasis. Blood. 106:477–484 10.1182/blood-2004-08-2989 [DOI] [PubMed] [Google Scholar]
- Tipping A.J., Pina C., Castor A., Hong D., Rodrigues N.P., Lazzari L., May G.E., Jacobsen S.E., Enver T. 2009. High GATA-2 expression inhibits human hematopoietic stem and progenitor cell function by effects on cell cycle. Blood. 113:2661–2672 10.1182/blood-2008-06-161117 [DOI] [PubMed] [Google Scholar]
- Tomita K., Hattori M., Nakamura E., Nakanishi S., Minato N., Kageyama R. 1999. The bHLH gene Hes1 is essential for expansion of early T cell precursors. Genes Dev. 13:1203–1210 10.1101/gad.13.9.1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai F.Y., Keller G., Kuo F.C., Weiss M., Chen J., Rosenblatt M., Alt F.W., Orkin S.H. 1994. An early haematopoietic defect in mice lacking the transcription factor GATA-2. Nature. 371:221–226 10.1038/371221a0 [DOI] [PubMed] [Google Scholar]
- Wendorff A.A., Koch U., Wunderlich F.T., Wirth S., Dubey C., Brüning J.C., MacDonald H.R., Radtke F. 2010. Hes1 is a critical but context-dependent mediator of canonical Notch signaling in lymphocyte development and transformation. Immunity. 33:671–684 10.1016/j.immuni.2010.11.014 [DOI] [PubMed] [Google Scholar]
- Xue Y., Gao X., Lindsell C.E., Norton C.R., Chang B., Hicks C., Gendron-Maguire M., Rand E.B., Weinmaster G., Gridley T. 1999. Embryonic lethality and vascular defects in mice lacking the Notch ligand Jagged1. Hum. Mol. Genet. 8:723–730 10.1093/hmg/8.5.723 [DOI] [PubMed] [Google Scholar]
- Yoshiura S., Ohtsuka T., Takenaka Y., Nagahara H., Yoshikawa K., Kageyama R. 2007. Ultradian oscillations of Stat, Smad, and Hes1 expression in response to serum. Proc. Natl. Acad. Sci. USA. 104:11292–11297 10.1073/pnas.0701837104 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.