Mel Greaves discusses the mechanisms underlying transformation in B cell chronic lymphocytic leukemia, including a new study suggesting a role for BCR ligation driven by recognition of an antigenic component of common yeast and fungi.
Abstract
Relatively few cancers arise in mature, differentiated cells. The propensity of mature B cells to transform has been linked to their longevity and proliferative potential, and stimulation of the B cell receptor (BCR) by cognate antigen may promote the transformation process. A study in this issue (Hoogeboom et al.) lends support to this notion, showing that cancer cells from a subset of patients with chronic lymphocytic leukemia (CLL) express a BCR specific for a sugar expressed by commensal yeast species. Another study, in contrast, suggests that B-CLL cells uniquely acquire the ability to signal in the complete absence of ligand.
Cancers of the B cell lineage, myeloma, lymphomas, and CLL, are unusual in that they originate from mature, differentiated cells. Very few other lineages of the body are at risk in this respect. Most malignant tumors are thought to derive from transformation of stem or progenitor cells (Visvader, 2011), although this view has recently been challenged (Friedmann-Morvinski et al., 2012). One plausible explanation for inherent risk or propensity to transformation in mature B cells is that they retain extensive proliferative capacity and clonal longevity, or stem cell–like function. The rationale for this is clear from the perspective of adaptive immune responses, but clonal escape for malignancy may be a trade-off. This intrinsic risk is heightened by the off-target deployment of the mutagenic lymphoid enzymes RAG1/2 and AID (Unniraman and Schatz, 2006; Pasqualucci et al., 2008).
All clones of mature B cells will have distinctive or clonotypic/idiotypic Ig-BCRs with preferential affinities for foreign infectious antigenic epitopes or, at lower affinity, cross-reactive self-antigens. The BCR provides critical signals for normal B cells (proliferation and survival) and is retained along with its inherent recognition/signaling capacity after malignant transformation (Stevenson et al., 2011). In this context, it has long been suspected that early transformation events might involve stimulation and clonal expansion by cognate antigens (Küppers, 2005). Persistent, ligand-driven clonal expansion might be expected to increase the probability of transformation via acquired mutations. The increased risk of lymphoma in patients with chronic autoimmune diseases (Ehrenfeld et al., 2001) is compatible with this notion. The more interesting question is whether, after initial transformation, the malignant or premalignant clone retains a selective advantage or proliferative response in the presence of some persistent or intermittent antigenic exposure. The best evidence for this to date comes from the extra nodal or mucosa-associated lymphoid tissue (MALT) lymphomas in which different bacterial species in different tissue sites are drivers of clonal expansion, although indirectly via T cells (Isaacson and Du, 2004). Strikingly, early-stage tumors regress with antibiotic therapy, whereas later-stage cancers with additional genetic abnormalities no longer appear to require bacterial stimulation (Wotherspoon et al., 1993; Ferreri et al., 2005; Kuo et al., 2012). Additional evidence comes from the observation that some hepatitis C virus (HCV)–associated B lymphomas bind HCV envelope proteins (Quinn et al., 2001) and that regression of lymphoma can be achieved with antiviral therapy (Hermine et al., 2002).
CLL is the most prevalent B cell malignancy, and here too, antigenic drive has been suspected (Packham and Stevenson, 2010; Chiorazzi and Ferrarini, 2011). This seems plausible with the pregerminal center unmutated (BCR) subtype of CLL, which retains low affinity for self-antigens (Hervé et al., 2005), but even more so for those germinal center IgH mutated (m) CLL that express a highly biased or stereotypic BCR repertoire, indicative of some prior and consistent or shared antigenic drive (Messmer et al., 2004; Murray et al., 2008). Two recent papers shed considerable light on this possibility.
In this issue, Hoogeboom et al. (2013) report that a small fraction (4/82) of mCLL selected with IGHV3-7–encoded BCRs had unusually short CDR3 sequences (designated V3-7Sh). Additionally, these four cases had the same specific replacement mutations (Y37H and S4OH) and near identical IGKV2-24–encoded light chains. The possibility that all four cases were antigen selected was strongly suggested by two further observations. First, recombinant soluble IgM from these cells bound to 4 commensal yeast species (out of a total of 33 microbial species screened) and showed high-affinity binding to β-(1,6)-glucan. Site-directed mutation experiments demonstrated that reactivity was dependent on a glutamic acid at position 106 in the CDR3 region and on additional somatic mutations in IGHV3-7. Second, CLL cells from three patients proliferated in response to the β-(1,6)-glucan pustulan. Other mB-CLL cells and other recombinant BCRs did not show this specificity.
The authors conclude that β-(1,6)-glucan is the sole (or primary) cognate antigen for the CLL clones in these four patients and that the reactivity of CLL cells is compatible with the notion that common fungal pathogens might drive clonal expansion as a critical component of the step-wise transformation process. This is perhaps the best evidence to date for microbial drive in CLL.
These data, as with MALT lymphomas and bacterial infection, raise the prospect of treatment with antimicrobials. The difficulty here lies in the prospect that there could be multiple different fungal, bacteria, or viral species involved in driving CLL in different patients and, as in MALT lymphomas, that later mutations could obviate the need for cognate ligand drive. Nevertheless, this important lead could encourage the search for recognition of other microbial ligands in CLL.
Coincidentally, Dühren-von Minden et al. (2012) report a rather different story: namely that recombinant BCRs from CLL (17/17 cases tested) are capable of ligand (or antigen)-independent signaling (Ca2+ flux in a cell reporter assay using cells from mice lacking endogenous BCRs and pre-BCRs). In contrast, BCRs from myeloma, mantle cell lymphoma, follicular lymphoma, and marginal cell lymphomas (15 cases in total) required cross-linking for signaling. The observation was replicated in TCL1 transgenic mice, a murine model for CLL. The authors generated mutants to demonstrate that particular amino acids within the framework region 2 (FR2) VH domains are required for autonomous signaling (in concert with HCDR3), perhaps via some inter-BCR aggregation or binding. The BCR is, in effect, functioning as an oncogenic driver.
These findings are reminiscent of data from other types of cancer in which ligand-dependent signaling of proliferation is subverted by the acquisition, via mutation, of ligand-independent self-aggregation or dimerization and downstream intracellular signaling, mimicking physiological stimulation of receptors, e.g., EGFR/ErbB in breast cancer and glioblastoma (Weinberg, 2007) and androgen receptors in prostate cancer (Trapman and Cleutjens, 1997). As cancer clones expand and compete, “natural” ligand availability may be limited, and there may be selective pressure favoring mutants that can co-opt autonomous signaling either from receptor molecules (BCRs) themselves or their downstream effectors.
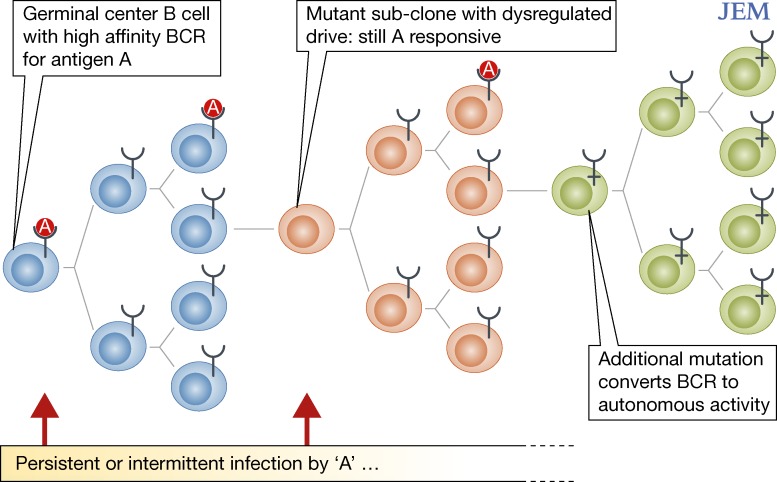
These two contrasting observations of CLL may not be mutually contradictory. First, some degree of autonomous signaling could be augmented by ligand (antigen)-driven response. B-CLL clones could originate as antigen dependent but evolve to become more autonomous if the critical BCR regions are mutated (Fig. 1). Alternatively, autonomous versus ligand-dependent BCR signaling could reflect different routes for clonal expansion after initial transformation. As in MALT lymphomas, these new data suggest that any therapeutic intervention in CLL aimed at microbial ligands might need to be instigated early. Alternatively, and perhaps more practically, signals downstream of the BCR, including SYK, BTK, and PI3K, provide viable therapeutic targets, irrespective of how the BCR is activated, and are currently the focus of clinical trials (Friedberg et al., 2010; Burger, 2012).
Figure 1.
Putative pattern of clonal evolution and antigen dependence in CLL. Three potential mechanisms may underlie microbial drive in the transformation of B cells in CLL. Germinal center B cells with high-affinity BCRs (blue) may be chronically stimulated by microbial antigens, increasing the probability that the cell acquires transforming mutations. After transformation, subclones of antigen-specific B cells may acquire mutations that confer a selective proliferative advantage in the presence of antigen (orange). Finally, antigen-specific B cells may acquire mutations that render BCR signaling completely independent of antigenic ligation (green). The “Y”-shaped symbols indicate BCRs, and “A” indicates nominal antigens derived from microbial infection.
Intriguingly, the recently described recurrent mutations in CLL (Puente et al., 2011; Wang et al., 2011) include MYD88, a gene which encodes a downstream component of Toll-like receptor (TCR) signaling. These receptors are expressed and biologically active in CLL cells (Muzio et al., 2009), suggesting an additional route of microbial stimulation or co-stimulation and its subversion by acquired mutations. Finally, in the context of the etiology and pathogenesis of CLL, this particular blood cell cancer occurs in familial pedigrees and has a long suspected component of inherited susceptibility (Gunz et al., 1975). Recent genome-wide association studies (GWAS) have identified many of the gene variants involved, some of which encode proteins likely to regulate B cell function (Crowther-Swanepoel et al., 2010; Slager et al., 2012). The challenge over the next period will be to build up a composite picture of clonal evolution in CLL in which constitutive and acquired gene variants modulate the way mature B cells respond to microenvironmental signals, including those from microbial ligands for the BCR.
References
- Burger J.A. 2012. Inhibiting B-cell receptor signaling pathways in chronic lymphocytic leukemia. Curr. Hematol. Malig. Rep. 7:26–33 10.1007/s11899-011-0104-z [DOI] [PubMed] [Google Scholar]
- Chiorazzi N., Ferrarini M. 2011. Cellular origin(s) of chronic lymphocytic leukemia: cautionary notes and additional considerations and possibilities. Blood. 117:1781–1791 10.1182/blood-2010-07-155663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowther-Swanepoel D., Broderick P., Di Bernardo M.C., Dobbins S.E., Torres M., Mansouri M., Ruiz-Ponte C., Enjuanes A., Rosenquist R., Carracedo A., et al. 2010. Common variants at 2q37.3, 8q24.21, 15q21.3 and 16q24.1 influence chronic lymphocytic leukemia risk. Nat. Genet. 42:132–136 10.1038/ng.510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dühren-von Minden M., Übelhart R., Schneider D., Wossning T., Bach M.P., Buchner M., Hofmann D., Surova E., Follo M., Köhler F., et al. 2012. Chronic lymphocytic leukaemia is driven by antigen-independent cell-autonomous signalling. Nature. 489:309–312 10.1038/nature11309 [DOI] [PubMed] [Google Scholar]
- Ehrenfeld M., Abu-Shakra M., Buskila D., Shoenfeld Y. 2001. The dual association between lymphoma and autoimmunity. Blood Cells Mol. Dis. 27:750–756 10.1006/bcmd.2001.0442 [DOI] [PubMed] [Google Scholar]
- Ferreri A.J.M., Ponzoni M., Guidoboni M., De Conciliis C., Resti A.G., Mazzi B., Lettini A.A., Demeter J., Dell’Oro S., Doglioni C., et al. 2005. Regression of ocular adnexal lymphoma after Chlamydia psittaci-eradicating antibiotic therapy. J. Clin. Oncol. 23:5067–5073 10.1200/JCO.2005.07.083 [DOI] [PubMed] [Google Scholar]
- Friedberg J.W., Sharman J., Sweetenham J., Johnston P.B., Vose J.M., Lacasce A., Schaefer-Cutillo J., De Vos S., Sinha R., Leonard J.P., et al. 2010. Inhibition of Syk with fostamatinib disodium has significant clinical activity in non-Hodgkin lymphoma and chronic lymphocytic leukemia. Blood. 115:2578–2585 10.1182/blood-2009-08-236471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedmann-Morvinski D., Bushong E.A., Ke E., Soda Y., Marumoto T., Singer O., Ellisman M.H., Verma I.M. 2012. Dedifferentiation of neurons and astrocytes by oncogenes can induce gliomas in mice. Science. 338:1080–1084 10.1126/science.1226929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunz F.W., Gunz J.P., Veale A.M., Chapman C.J., Houston I.B. 1975. Familial leukaemia: a study of 909 families. Scand. J. Haematol. 15:117–131 10.1111/j.1600-0609.1975.tb01063.x [DOI] [PubMed] [Google Scholar]
- Hermine O., Lefrère F., Bronowicki J.P., Mariette X., Jondeau K., Eclache-Saudreau V., Delmas B., Valensi F., Cacoub P., Brechot C., et al. 2002. Regression of splenic lymphoma with villous lymphocytes after treatment of hepatitis C virus infection. N. Engl. J. Med. 347:89–94 10.1056/NEJMoa013376 [DOI] [PubMed] [Google Scholar]
- Hervé M., Xu K., Ng Y.S., Wardemann H., Albesiano E., Messmer B.T., Chiorazzi N., Meffre E. 2005. Unmutated and mutated chronic lymphocytic leukemias derive from self-reactive B cell precursors despite expressing different antibody reactivity. J. Clin. Invest. 115:1636–1643 10.1172/JCI24387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogeboom R., van Kessel K.P.M., Hochstenbach F., Wormhoudt T.A., Reinten R.J.A., Wagner K., Kater A.P., Guikema J.E.J., Bende R.J., van Noesel C.J.M. 2013. A mutated B cell chronic lymphocytic leukemia subset that recognizes and responds to fungi. J. Exp. Med. 210:59–70 10.1084/jem.20121801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacson P.G., Du M.-Q. 2004. MALT lymphoma: from morphology to molecules. Nat. Rev. Cancer. 4:644–653 10.1038/nrc1409 [DOI] [PubMed] [Google Scholar]
- Kuo S.-H., Yeh K.H., Wu M.S., Lin C.W., Hsu P.N., Wang H.P., Chen L.T., Cheng A.L. 2012. Helicobacter pylori eradication therapy is effective in the treatment of early-stage H pylori-positive gastric diffuse large B-cell lymphomas. Blood. 119:4838–4844 10.1182/blood-2012-01-404194 [DOI] [PubMed] [Google Scholar]
- Küppers R. 2005. Mechanisms of B-cell lymphoma pathogenesis. Nat. Rev. Cancer. 5:251–262 10.1038/nrc1589 [DOI] [PubMed] [Google Scholar]
- Messmer B.T., Albesiano E., Efremov D.G., Ghiotto F., Allen S.L., Kolitz J., Foa R., Damle R.N., Fais F., Messmer D., et al. 2004. Multiple distinct sets of stereotyped antigen receptors indicate a role for antigen in promoting chronic lymphocytic leukemia. J. Exp. Med. 200:519–525 10.1084/jem.20040544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray F., Darzentas N., Hadzidimitriou A., Tobin G., Boudjogra M., Scielzo C., Laoutaris N., Karlsson K., Baran-Marzsak F., Tsaftaris A., et al. 2008. Stereotyped patterns of somatic hypermutation in subsets of patients with chronic lymphocytic leukemia: implications for the role of antigen selection in leukemogenesis. Blood. 111:1524–1533 10.1182/blood-2007-07-099564 [DOI] [PubMed] [Google Scholar]
- Muzio M., Scielzo C., Bertilaccio M.T., Frenquelli M., Ghia P., Caligaris-Cappio F. 2009. Expression and function of toll like receptors in chronic lymphocytic leukaemia cells. Br. J. Haematol. 144:507–516 10.1111/j.1365-2141.2008.07475.x [DOI] [PubMed] [Google Scholar]
- Packham G., Stevenson F. 2010. The role of the B-cell receptor in the pathogenesis of chronic lymphocytic leukaemia. Semin. Cancer Biol. 20:391–399 10.1016/j.semcancer.2010.08.004 [DOI] [PubMed] [Google Scholar]
- Pasqualucci L., Bhagat G., Jankovic M., Compagno M., Smith P., Muramatsu M., Honjo T., Morse H.C., III, Nussenzweig M.C., Dalla-Favera R. 2008. AID is required for germinal center-derived lymphomagenesis. Nat. Genet. 40:108–112 10.1038/ng.2007.35 [DOI] [PubMed] [Google Scholar]
- Puente X.S., Pinyol M., Quesada V., Conde L., Ordóñez G.R., Villamor N., Escaramis G., Jares P., Beà S., González-Díaz M., et al. 2011. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature. 475:101–105 10.1038/nature10113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn E.R., Chan C.H., Hadlock K.G., Foung S.K., Flint M., Levy S. 2001. The B-cell receptor of a hepatitis C virus (HCV)-associated non-Hodgkin lymphoma binds the viral E2 envelope protein, implicating HCV in lymphomagenesis. Blood. 98:3745–3749 10.1182/blood.V98.13.3745 [DOI] [PubMed] [Google Scholar]
- Slager S.L., Skibola C.F., Di Bernardo M.C., Conde L., Broderick P., McDonnell S.K., Goldin L.R., Croft N., Holroyd A., Harris S., et al. 2012. Common variation at 6p21.31 (BAK1) influences the risk of chronic lymphocytic leukemia. Blood. 120:843–846 10.1182/blood-2012-03-413591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson F.K., Krysov S., Davies A.J., Steele A.J., Packham G. 2011. B-cell receptor signaling in chronic lymphocytic leukemia. Blood. 118:4313–4320 10.1182/blood-2011-06-338855 [DOI] [PubMed] [Google Scholar]
- Trapman J., Cleutjens K.B.J.M. 1997. Androgen-regulated gene expression in prostate cancer. Semin. Cancer Biol. 8:29–36 10.1006/scbi.1997.0050 [DOI] [PubMed] [Google Scholar]
- Unniraman S., Schatz D.G. 2006. AID and Igh switch region-Myc chromosomal translocations. DNA Repair (Amst.). 5:1259–1264 10.1016/j.dnarep.2006.05.019 [DOI] [PubMed] [Google Scholar]
- Visvader J.E. 2011. Cells of origin in cancer. Nature. 469:314–322 10.1038/nature09781 [DOI] [PubMed] [Google Scholar]
- Wang L., Lawrence M.S., Wan Y., Stojanov P., Sougnez C., Stevenson K., Werner L., Sivachenko A., DeLuca D.S., Zhang L., et al. 2011. SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. N. Engl. J. Med. 365:2497–2506 10.1056/NEJMoa1109016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg R.A. 2007. The Biology of Cancer. Garland Science, New York: 796 pp [Google Scholar]
- Wotherspoon A.C., Doglioni C., Diss T.C., Pan L., Moschini A., de Boni M., Isaacson P.G. 1993. Regression of primary low-grade B-cell gastric lymphoma of mucosa-associated lymphoid tissue type after eradication of Helicobacter pylori. Lancet. 342:575–577 10.1016/0140-6736(93)91409-F [DOI] [PubMed] [Google Scholar]