Inactivation of Llgl1 enhances HSC self-renewal and fitness and is associated with unfavorable outcome in human AML.
Abstract
A unique characteristic of hematopoietic stem cells (HSCs) is the ability to self-renew. Several genes and signaling pathways control the fine balance between self-renewal and differentiation in HSCs and potentially also in leukemia stem cells. Recently, studies have shed light on developmental molecules and evolutionarily conserved signals as regulators of stem cells in hematopoiesis and leukemia. In this study, we provide evidence that the cell fate determinant Llgl1 (lethal giant larvae homolog 1) plays an important role in regulation of HSCs. Loss of Llgl1 leads to an increase in HSC numbers that show increased repopulation capacity and competitive advantage after transplantation. This advantage increases upon serial transplantation or when stress is applied to HSCs. Llgl1−/− HSCs show increased cycling but neither exhaust nor induce leukemia in recipient mice. Llgl1 inactivation is associated with transcriptional repression of transcription factors such as KLF4 (Krüppel-like factor 4) and EGR1 (early-growth-response 1) that are known inhibitors of HSC self-renewal. Decreased Llgl1 expression in human acute myeloid leukemia (AML) cells is associated with inferior patient survival. Thus, inactivation of Llgl1 enhances HSC self-renewal and fitness and is associated with unfavorable outcome in human AML.
Evolutionarily conserved signaling pathways and cell fate determinants, such as RNA-binding proteins or polarity regulators, have been recently described as effectors in hematopoietic stem cell (HSC) biology (Wu et al., 2007; Hope et al., 2010; Ito et al., 2010; Kharas et al., 2010). However, only a few members of the polarity network have been investigated to date, and the exact role of these genes in HSCs and leukemia is not well understood. Several genes and signaling pathways control the fine balance between self-renewal and differentiation in HSCs and potentially also in leukemia stem cells (LSCs; Heidel et al., 2011). For example, knockdown of select polarity-associated genes leads to enhanced (Prox1) or decreased (Pard6a, Prkcz, and Msi2) repopulation potential of HSCs in vivo. Llgl1 (lethal giant larvae homolog 1) is a highly conserved gene, known to be involved in regulation of cell polarity, proliferation, and cell division. It connects the polarity nodes “Par complex” (Pard6, Pard3, and Prkcz/Prkci) and “Scribble complex” (Dlg, Scrib, and Llgl1), components of which have been shown to influence the repopulation capacity of HSCs (Humbert et al., 2006). This network has been reported to interact with several stem cell relevant pathways, such as canonical Wnt signaling, Notch signaling, and Hippo pathway signaling (Humbert et al., 2006; Menéndez et al., 2010). Recently, atypical protein kinases (λ and ζ) have been reported to be dispensable in HSCs (Sengupta et al., 2011). However, few other polarity proteins have been investigated for a role in hematopoietic cells. In Drosophila melanogaster, deletion of Llgl1 results in neoplastic proliferation and loss of polarity of imaginal epithelia and neuroblasts (Gateff, 1978). In Drosophila, Llgl1 was the first in vivo example of a gene in which loss of function resulted in tumor formation and is therefore distinct from most other transforming oncogenes, which are frequently overexpressed (Gateff, 1978). Pronounced similarities with Drosophila Llgl1 mutants were also found in Llgl1-KO mice. Llgl1−/− mice presented at birth with severe brain dysplasia caused by the loss of cell polarity, increased self-renewal, and decreased differentiation of neural progenitor cells (Klezovitch et al., 2004). In clinical samples, loss of Llgl1 expression is associated with a variety of epithelial and mesenchymal cancers (Ohali et al., 2004; Schimanski et al., 2005; Kuphal et al., 2006; Lu et al., 2009). Recently, mutations of Llgl2, a close human homologue of Llgl1, have been found in progression from severe congenital neutropenia to acute myeloid leukemia (AML; Beekman et al., 2012).
In this project, we investigated a potential role for Llgl1 in HSCs via genetic inactivation of Llgl1. Loss of Llgl1 leads to expansion of HSCs in steady-state hematopoiesis and to enhanced repopulation capacity after serial transplantation or stress. Llgl1 loss influences several known transcriptional activators involved in HSC self-renewal, and decreased LLGL1 expression is associated with poor prognosis in human AML.
RESULTS AND DISCUSSION
Inactivation of Llgl1 increases HSC number and fitness in vivo
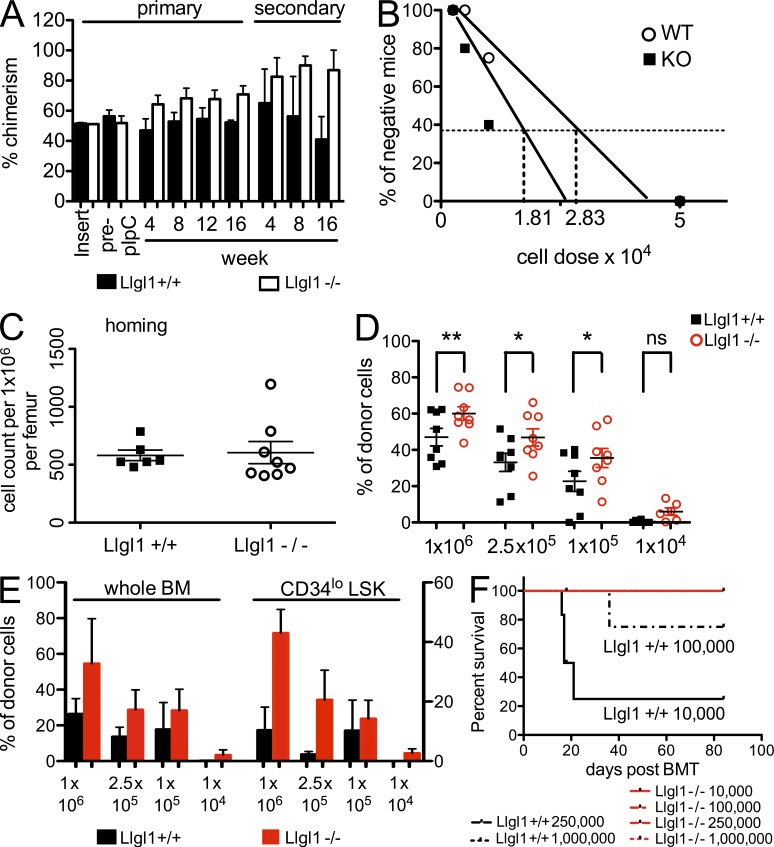
As disruption of evolutionarily conserved polarity regulators may impact HSC biology, we aimed to investigate the role of Llgl1 in steady-state hematopoiesis. We injected poly-I–poly-C peptide (pIpC) into Llgl1loxP/loxP Mx-Cre+ mice or the corresponding littermate controls (Llgl1+/loxP Mx-Cre+, Llgl1+/+ Mx-Cre+, or Llgl1loxP/loxP Mx-Cre− mice) to induce genetic deletion of Llgl1. Over the next 16 wk, we monitored peripheral blood counts and did not find any significant abnormalities after conditional deletion of Llgl1 except a mild lymphocytosis 4 wk after pIpC (Fig. 1 A). Efficient excision of Llgl1 was confirmed in the BM by PCR. However, immunophenotypic analysis of the BM 16 wk after pIpC treatment revealed a significant increase in long-term HSCs (LT-HSCs; lineage−, Sca-1+, Kit+ [LSK], CD34lo, Flk2−). This increase was also found in the lineage−, Sca-1+, Kit+, CD34lo, Flk2−, CD48− (SLAM+) population (Fig. 1, B and C), which is the most highly enriched LT-HSC population described so far (LT-HSC frequency 1:2; Kiel et al., 2005). When whole BM cells were transplanted into primary recipient mice in a competitive fashion at a ratio of 1:1 (and also 9:1 [not depicted]; control vs. test BM) on week 16, we found that Llgl1−/− cells competed significantly better than Llgl1+/+ controls as indicated by increased chimerism (Fig. 1 D). This data suggests a competitive advantage of ∼10-fold when compared with Llgl1+/+ cells. The competitive advantage in peripheral blood chimerism increased in secondary (Fig. 1 E) and tertiary recipient mice (Fig. 1 F). This increase in chimerism was also detectable in total BM (Fig. 1, G and H, left) or HSC (CD34lo LSK cells; Fig. 1, G and H, right) populations. In tertiary recipients, the HSC pool was almost exclusively repopulated with cells from the Llgl−/− mice, whereas in BM cells of the myeloid lineage, cells from Llgl1+/+ control mice almost disappeared (Fig. 1 I). This gain in self-renewal capacity could potentially lead to the development of leukemia; however, none of the mice developed leukemia during serial transplantation up to 12 mo after conditional deletion of Llgl1 (not depicted).
Figure 1.
Loss of Llgl1 increases the amount and competitive advantage of HSCs. (A) Peripheral blood counts after conditional deletion of Llgl1 during steady-state hematopoiesis. (B and C) Numbers of HSCs (CD34lo, Flk2− LSK and CD34lo, Flk2− CD48− SLAM+ LSK) 16 wk after deletion. (D and G) Llgl1−/− cells competed better against WT competitor cells (Llgl1+/+ littermate controls) in peripheral blood (D)– and BM-HSC chimerism (G; two independent cohorts; n > 4 per cohort). (E, F, H, and I) Increase of competitive advantage over time throughout serial transplantations in total BM and HSCs of secondary (E and H) and tertiary (F and I) recipient mice (secondary and tertiary recipients received total unfractionated BM from primary recipient mice). (E and H) Horizontal lines represent the mean of each group. Error bars indicate the standard deviation. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
This phenotype might be explained by improved homing of Llgl1 deleted cells or by transplantation of increased HSC numbers. Therefore, we transplanted “undeleted” (CD45.1) BM cells derived from Llgl1loxP/loxP Mx-Cre+ mice or Llgl1loxP/loxP Mx-Cre− mice competitively (1:1) into C57BL/6 recipients and assured equal engraftment levels after 4 wk (Fig. 2 A). This ensured that even without differences in homing or cell numbers, Llgl1−/− cells would still compete better in this experiment. Mice were then treated with pIpC to inactivate Llgl1. In this setting, a competitive advantage for Llgl1−/− BM cells could still be detected and increased after transplantation into secondary recipient mice (Fig. 2 A).
Figure 2.
Enhanced HSC fitness after genetic inactivation of Llgl1 is independent of engraftment and detectable after ex vivo culture. (A) Conditional deletion of Llgl1 4 wk after transplantation of previously undeleted cells. Competitive advantage of Llgl1−/− cells (compared with WT controls) in vivo in primary (left) and secondary (right) recipient mice (two independent cohorts, n > 3 per cohort) is shown. (B) Limiting dilution assay. Increased frequency of HSCs in Llgl1−/− cohorts compared with WT controls 14 wk after transplantation is shown. (C) Homing of LSK cells in lethally irradiated recipient mice. (D and E) Repopulation capacity of Llgl1−/− cells after ex vivo culture of HSCs for 5 d. (D) Peripheral blood chimerism at 4 wk. *, P < 0.05; **, P < 0.01. (E) BM and HSC chimerism 16 wk after transplantation (two independent cohorts; >7 mice/dilution). (F) Radioprotection of lethally irradiated recipient mice (survival after ex vivo culture of transplanted HSCs). Error bars indicate the standard deviation.
To assess whether HSC frequency contributed to the competitive advantage, we performed limiting dilution experiments using whole BM cells shortly after deletion of Llgl1. BM of Llgl1−/− mice and Llgl1+/+ littermates (50,000, 10,000, 5,000, and 2,500 cells) were injected into lethally irradiated (11 Gy) C57BL/6 recipients along with 500,000 (CD45.1/2 double positive) competitor cells from age-matched controls. BM chimerism was analyzed at week 14. Calculating the stem cell frequencies, the Llgl1 WT cohort showed a frequency of 1/28,346 (95% CI 0.0000146 to 0.0000855) and the Llgl1 KO cohort 1/18,156 (95% CI 0.0000253 to 0.0001198; Fig. 2 B). This indicates that cell numbers (through expansion of HSCs) do contribute to the increase in chimerism, at least to some extent. Besides that, homing of LSK cells, as detected 16 h after injection into lethally irradiated mice, was not different between Llgl1−/− cells compared with Llgl1+/+ controls (Fig. 2 C). Therefore, a potential difference in homing capacity cannot fully explain the enhanced repopulation capacity of Llgl1−/− HSCs.
Next, we aimed to investigate distinct stressors of HSCs to determine the extent to which Llgl1 loss appears to be protective for HSCs. Ex vivo culture of HSCs is known to decrease stem cell numbers, viability, and repopulation capacity (Perry et al., 2011). Augmentation of ex vivo culture is therefore an attractive therapeutic approach in transplantation biology. Stress was applied by ex vivo culture of Llgl1−/− or Llgl1+/+ BM cells for 5 d. Transplantation of these cells into primary recipient mice at limiting dilutions revealed a significantly increased percentage of repopulating Llgl1−/− donor cells in the peripheral blood compared with WT cells (Fig. 2 D) 4 wk after transplantation. Analysis of BM and stem cell fractions displayed a similar degree of enhanced repopulation capacity 16 wk after transplantation (Fig. 2 E). These experiments confirm enhanced function of Llgl1−/− HSCs that appears to increase after stress and eventually leads to radioprotection of lethally irradiated recipient mice. Survival was increased for Llgl1−/− versus Llgl1+/+ at levels of 105 cells (100 vs. 71.4%) and 104 cells (100 vs. 33.3%; (Fig. 2 F).
Llgl1 protects quiescence of LT-HSCs
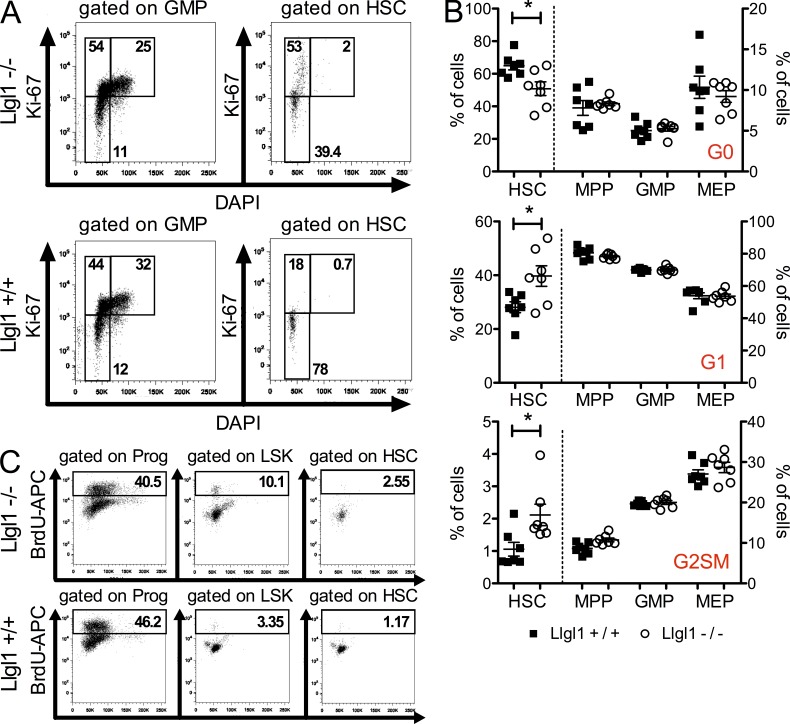
As loss of Llgl1 was associated with an increase of LT-HSC numbers in vivo, we assessed whether changes in cell cycle status might occur after Llgl1 deletion. In most cases where more HSCs are found to be actively cycling, this ultimately leads to exhaustion of the HSC pool. However, two recent gene KO models have been published showing increased (Rathinam et al., 2011) or unchanged (Park, C.S., et al. 2011. American Society of Hematology Annual Meeting and Exposition. Abstr. 861.) cell cycle activity in HSCs while conferring enhanced fitness and competitive advantage to stem cells. Genetic inactivation of the E3 ubiquitin ligase Itch led to increased cell cycle activity of HSCs through activation of Notch signaling without affecting the long-term repopulation activity of HSCs (Rathinam et al., 2011). Deletion of the transcription factor Klf4 (Krüppel-like factor 4) did not cause any changes in cell cycle activity but also led to improved HSC capacity through protection from apoptosis (Park, C.S., et al. 2011. American Society of Hematology Annual Meeting and Exposition. Abstr. 861.). Thus, we investigated whether conditional inactivation of Llgl1 would influence cell cycle status in HSCs, multipotent progenitors (MPPs), and more differentiated progenitors such as common myeloid progenitors (CMPs), granulocyte macrophage progenitors (GMPs), or megakaryocyte erythroid progenitors (MEPs). Ki-67 staining in steady-state hematopoiesis revealed a significant increase in the number of cycling Llgl1−/− HSCs as compared with Llgl1+/+ HSCs (Fig. 3, A and B). This effect was not evident in MPPs, GMPs, or MEPs (Fig. 3 B). This cell cycle analysis was performed 16 wk after pIpC injection. To investigate these effects at an earlier time point, we injected mice with pIpC and then assessed BrdU incorporation 3 wk later. To avoid stem cell toxicity by BrdU incorporation, we analyzed the BM of injected mice early (18 h) after BrdU injection. Consistent with our findings using Ki-67 staining, more Llgl1−/− HSCs were cycling than Llgl1+/+ controls (Fig. 3 C). This indicates increased cell cycle activity of HSCs upon inactivation of Llgl1 without exhaustion of the stem cell pool. Thus, the stem cell compartment rather increased and gained repopulation capacity. This could be an indicator of an increase in symmetric (vs. asymmetric) stem cell divisions, leading to an increase of the stem cell number.
Figure 3.
Llgl1 protects quiescence of LT-HSCs. (A and B) HSC cell cycle in steady-state hematopoiesis after genetic inactivation of Llgl1. (B) Distribution of cell cycle phases in HSCs (34lo LSK), MPPs, GMPs, or MEPs (2 cohorts, n = 6) after genetic inactivation of Llgl1. Error bars indicate the standard deviation. *, P < 0.05. (C) Short-term BrdU incorporation confirms stem cell cycling in vivo.
Llgl1 inactivation leads to transcriptional repression of KLF4 and EGR1 (early-growth-response 1)
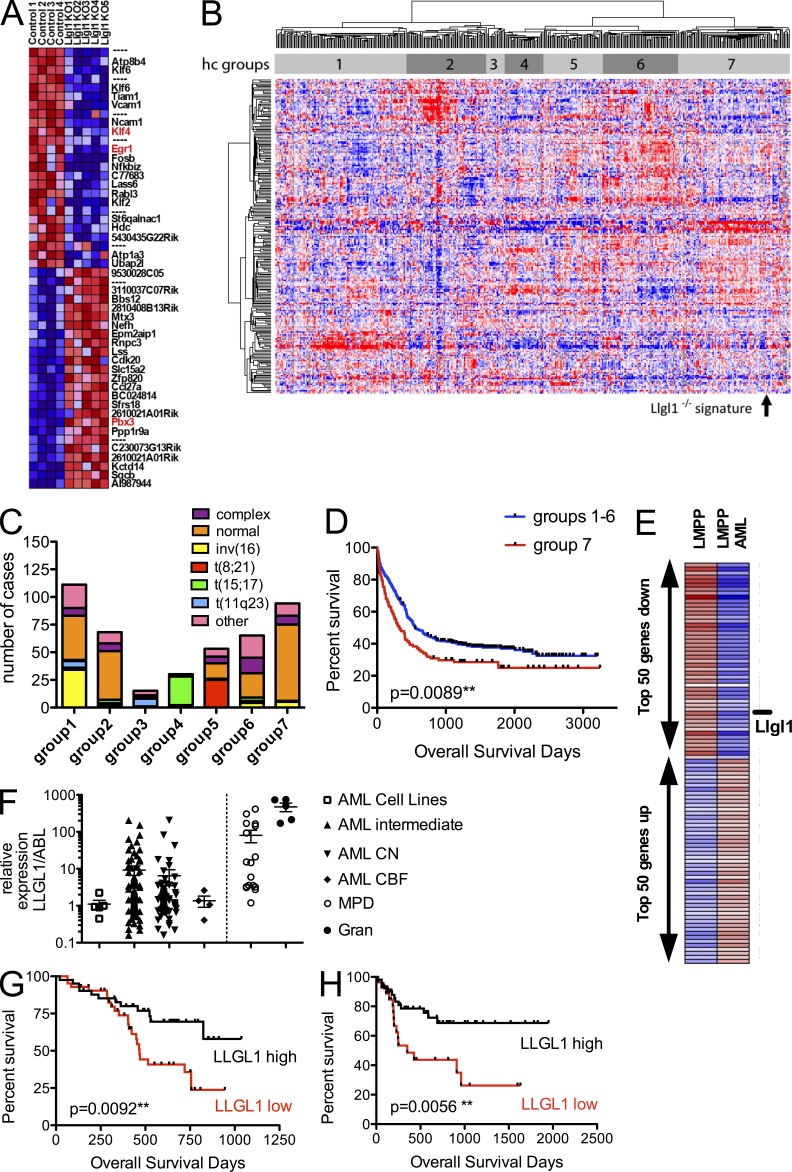
To identify potential downstream effectors of Llgl1 loss, we performed gene expression profiling (GEP) on HSCs (Flk2− LSK) 10 wk after conditional deletion of Llgl1 in steady-state hematopoiesis. The top 50 deregulated genes are displayed in Fig. 4 A. Several of the genes that change expression in the absence of Llgl1 have already been implicated in stem cell biology. The most prominent effectors are Klf4 and Egr1 (Fig. 4 A). Loss of Klf4 leads to increased stem cell numbers, self-renewal capacity, and survival of stressed HSCs (Park, C.S., et al. 2011. American Society of Hematology Annual Meeting and Exposition. Abstr. 861.). This advantage increases over serial transplantations. Moreover, KLF4 has recently been shown to be a repressed target in CDX2-induced AML development (Faber et al., 2012). Egr-1 revealed significant increase of HSCs in the BM of the respective KO mice (Min et al., 2008). However, enhanced cycling and proliferation of Egr1−/− HSCs eventually led to exhaustion of the HSC pool. In our model, Llgl1−/− HSCs do not exhaust although more HSCs are in the cell cycle. This may be caused by compensatory effects, for example, via up-regulation of other target genes such as Hox genes (e.g., Pbx3; Fig. 4 A), tyrosine kinases (e.g., Yes, associated with Hippo-signaling), or other enzymes (e.g., the oxidosqualene cyclase Lss) that are known mediators of stem cell self-renewal and protection (Faber et al., 2009; Mejia-Pous et al., 2011; Tamm et al., 2011; Li et al., 2012).
Figure 4.
Llgl1 acts upstream of transcriptional activators of self-renewal capacity, and its repression is associated with leukemia development and an adverse prognosis in CN-AML. (A) GEP on HSCs (Flk2− LSK) after conditional deletion of Llgl1 in steady-state hematopoiesis (14-wk-old mice, 10 wk after pIpC administration). The top 25 up- or down-regulated genes (by fold change) are displayed. (B) Hierarchical clustering of 436 AML cases along with the Llgl1−/− signature. hc groups, hierarchical cluster–defined groups. (C) Clustering based on the overlap for the top 200 genes reveals a significant grouping into karyotype-associated groups (P < 0.001). The Llgl1−/− signature clusters with a CN-AML predominated subgroup. (D) The Llgl1−/− signature–associated subgroup is associated with inferior outcome (P = 0.0089). (E) GEP during development of LMPP and GMP into their malignant counterparts reveals Llgl1 to be the top down-regulated cell fate determinant. Expression of Llgl1 is reduced in LMPP-AML compared with LMPP and in GMP-AML compared with GMP. (F) Primary AML samples show a high variability in Llgl1 expression. AML patient samples reveal a decreased expression in AML when compared with the MPD cases or normal granulocytes. Error bars indicate the standard deviation. (G) Low expression of Llgl1 as measured by Affymetrix gene arrays was associated with decreased survival in 83 karyotypic normal AML (CN-AML) patients below the age of 60 yr (AMLSG). (H) This finding could be confirmed in an independent CN-AML cohort (n = 80; OSHO study group) by qRT-PCR (all patient samples measured in triplicate).
Loss of Llgl1 is involved in disease development and influences prognosis of human AML
Decreased Llgl1 expression has been previously shown to be associated with increased metastatic potential, malignant phenotype, or inferior survival in a variety of solid tumors (Ohali et al., 2004; Schimanski et al., 2005; Kuphal et al., 2006). Given that Llgl1 restrains self-renewal in HSCs and that leukemia is characterized by aberrant activation of self-renewal, we assessed for a role in human AML. First, we asked whether the gene expression signature revealed by genetic inactivation of Llgl1 (Llgl1−/− signature) in HSCs was associated with any genetic subtypes of primary AML. Indeed, clustering based on the Llgl1−/− signature (Fig. 4 B) grouped AML cases into subgroups significantly associated with cytogenetic abnormalities (Fig. 4, B and C). Especially core-binding factor leukemias (t(8;21) and inv(16)) or t(15;17) clustered into specific subgroups. Moreover, the Llgl1−/− signature was most strongly correlated with group 7 based on hierarchical clustering. This group contained mostly cytogenetically normal leukemias (Fig. 4, B and C). Interestingly, this group (enriched for the Llgl1−/− signature) also correlated with decreased survival as compared with the other leukemia subgroups (Fig. 4 D). To confirm this potential relevance for disease biology and prognosis in cytogenetically normal AML (CN-AML), we first screened for differences in Llgl1 expression levels using a recently published GEP dataset. This dataset described the coexistence of lymphoid-primed MPP (LMPP)–like and GMP-like LSCs in human AML (Goardon et al., 2011). Of interest, the top differentially expressed cell fate determinate was Llgl1, and its loss was associated with both LMPP-like and GMP-like LSC signatures when compared with their normal counterparts (Fig. 4 E and not depicted). As we found the Llgl1−/− signature to be prognostically relevant in a group of predominantly CN-AML, we asked whether repression of Llgl1 itself could be associated with outcome. Therefore, we investigated two independent patient cohorts treated for CN-AML in two different AML study groups by GEP and quantitative PCR, respectively. We analyzed 83 patients below the age of 60 with CN-AML (AMLSG study group) using gene expression analysis. Here, we dichotomized the Llgl1 expression data and compared survival of the high expressing (upper 50%) with the low expressing (lower 50%) cohort. This analysis revealed significantly decreased overall survival in the low expressing group (**, P = 0.0092; Fig. 4 G). This is an important indicator for Llgl1 playing a role in disease biology of CN-AML. Multivariable analysis using a COX regression model proved Llgl1 expression to be of prognostic relevance independent of known prognostic factors such as FLT3-ITD, NPM1, or CEBPa mutation status. To confirm the GEP data, we analyzed Llgl1 expression by quantitative RT-PCR (qRT-PCR) and found a wide range of expression throughout CN-AML samples (Fig. 4 F). Using an independent cohort of 80 patients treated for CN-AML in the OSHO study group with standard chemotherapy we screened LLGL1 expression by qRT-PCR. We were able to confirm a significantly inferior survival in the LLGL1lo cohort (**, P = 0.0056; Fig. 4 H).
In summary, we provide evidence that decreased Llgl1 expression is associated with a poor prognosis in human AML. Conditional deletion of Llgl1 in vivo leads to expansion of LT-HSCs, increased fitness, and competitive advantage of HSCs. These data support a role for stem cell–associated properties as critical for human leukemia development and treatment response.
MATERIALS AND METHODS
Primary patient samples and human leukemia cell lines.
For gene expression analysis (GEP), 83 primary AML patient samples with normal karyotype (CN-AML) were obtained within the AML trials (AMLSG07-04) of the German–Austrian AMLSG study group. Samples for qRT-PCR analysis of Llgl1 expression were obtained within the AML trials (OSHO-2003) of the East German Study Group (OSHO) including the Hematology Tumor Bank Magdeburg (HTM) and approved by the respective local ethics committee (Ethics Committee of the Otto-von-Guericke University Magdeburg and Ethics Committee of the University of Ulm). Human leukemia cell lines were purchased from the American Type Culture Collection or DSMZ.
LLGL1 expression analysis.
LLGL1 and ABL as endogenous control for each sample were analyzed in triplicate on the same 96-well plate using a 7300HT real-time PCR machine (Applied Biosystems). Expression levels were determined by SYBR-green using the following primers: Llgl_Q-fw, 5′-CTGCACTCCTCTGCAATCAC-3′; Llgl_Q-re, 5′-GCTCAAGGCACTGGAGACA-3′; ABL_Q-fw, 5′-TGCCCAGAGAAGGTCTATGAA-3′; and ABL_Q-re, 5′-CCACTTCGTCTGAGATACTGGA-3′. 12.5 µl of 2× SensiMixPlus SYBR (Quantace), 5.5 µl H2O, and 1 µl of each primer (10-nM stock solution) were added to 5 µl cDNA for a total reaction volume of 25 µl. We used the mean expression level of AML cell lines investigated as an arbitrary threshold and compared the lower expressing patient samples with higher expressing samples using quantitative PCR analysis. MOLM-13, a human AML cell line (FAB M5a) carrying an FLT3-length mutation, was used as an internal standard. Relative expression of Llgl1 was calculated in comparison with MOLM-13 levels (μΔΔCt = μΔCtpatient − μΔCtMOLM-13) to obtain relative quantification RQ = 2−μΔΔCt. Expression levels ≥1 were considered to be Llgl1hi, whereas expression levels <1 were considered to be Llgl1lo. Survival analysis was conducted accordingly using Prism version 5.00 for Windows (GraphPad Software).
Mouse experiments.
All mice were housed under pathogen-free conditions in the accredited Animal Research facility at Boston Children’s Hospital or the Animal Research Facility of the Otto-von-Guericke University Medical Faculty, Magdeburg. All experiments were conducted after approval by the Institutional Animal Care and Use Committee and the Landesverwaltungsamt Sachsen-Anhalt (42502–2-1054 UniMD). Mice harboring a floxed (flanked with loxP sites) allele of Llgl1 have been generated as previously described (Klezovitch et al., 2004). Exon 2 (the exon downstream from the exon with the first ATG codon) was flanked by LoxP sequences, and the β-geo selectable marker was removed by transient expression of Cre-recombinase in the embryonic stem cells. These mice have been backcrossed more than eight generations into a C57BL/6 background.
BM transplantation assays.
For transplantation, 106 BM cells of 6–8-wk-old Llgl1−/− or Llgl1+/+ (CD45.1) littermates and 106 (CD45.1/2) competitor cells (derived from intercrossing CD45.1 animals with CD45.2 animals purchased from Charles River) were transplanted via lateral tail vein injection into lethally irradiated (1,100 cGy, split-dose) 6–8-wk-old (CD45.2) C57BL/6 mice (The Jackson Laboratory).
For serial transplantation experiments, whole BM of primary recipient mice was harvested, and 2 × 106 whole BM cells were injected into lethally irradiated secondary recipients. Tertiary recipient mice were injected as well with 2 × 106 whole BM cells, harvested from secondary recipient mice.
Ex vivo expansion of HSCs.
We cultured whole BM cells of (CD45.1) Llgl1+/+ and Llgl1−/− cells for 5 d in vitro using STEMSpan medium (Cell Signaling Technology) supplemented with 50 ng/µl stem cell factor. As erythrolysis is suspected to severely inhibit functional HSC expansion, cells were not exposed to any erythrolysis procedure. Instead, PBMCs were isolated from mouse BM using Histopaque 1077 (Sigma-Aldrich). Cells were washed and resuspended in HSC expansion media. On day 6, different dilutions (106, 2.5 × 105, 105, and 104) were transplanted into lethally irradiated CD45.2 recipient mice. Recipients were monitored for survival and analyzed for peripheral blood chimerism at week 4, 8, 12, and 16 after transplantation. HSC and whole BM chimerism was determined at week 16.
Flow cytometry.
For immunophenotype analysis, peripheral blood cells, BM, or spleen cells were resuspended in PBS/1% FBS after erythrocyte lysis (PharmLyse; BD). Unless otherwise stated, the following antibodies were used. Sorting and analysis of LSK cells or Sca-1+ cells were performed as previously described (Krivtsov et al., 2009). Biotinylated antibodies against Gr-1 (RB6-8C5), B220 (RA3-6B2), CD19 (6D5), CD3 (145-2C11), CD4 (GK1.5), CD8 (53-6.7), TER119, and IL7Ra (A7R34; all from BioLegend) were used for lineage staining. An APC-Cy7– or Qdot605-labeled streptavidin antibody (BD) was used for secondary staining together with an APC–anti-KIT (clone 2B8) and a PE-Cy7– or Pacific Blue–anti–Sca-1 antibody (clone E13-161.7).
Cells were analyzed using an LSRII or FACSCanto II (BD) cytometer. Analysis was performed using FlowJo software (Tree Star). A Fix & Perm kit (Invitrogen) was used for cell cycle analysis with Ki-67 according to the manufacturer’s protocol.
In vivo BrdU incorporation assay.
Llgl1−/− or Llgl1+/+ mice were injected i.p. with 180 µl BrdU/PBS solution (10 mg/ml; BrdU kit; BD). 18 h after injection, BM cells were harvested and stained with lineage and secondary antibodies as indicated above.
RNA amplification and gene expression array.
RNA was isolated from 104 sorted HSCs (Lineage−, Kit+, Sca-1+, Flk2−) using TRIzol (Invitrogen). RNA was amplified using the Ovation Pico WTA system (Nugen) and labeled using the Encore Biotin Module (Nugen). 5 µg of amplified and labeled DNA was hybridized to Affymetrix 430 2.A mouse microarrays.
Statistics and analysis of gene expression data.
For survival analysis, Kaplan–Meier curves were plotted using Prism version 5.00. Differences between survival distributions were analyzed using the logrank test. Statistical analyses were performed using the Student’s t test (normal distribution) or Mann–Whitney U test (when normal distribution was not given). P < 0.05 was considered statistically significant (P < 0.05 indicated as *, P < 0.01 indicated as **, and P < 0.001 indicated as ***). Published gene expression datasets (GSE16432 [Wilson et al., 2009] and GSE15434 [Kohlmann et al., 2010]) were downloaded from GEO. Primary gene expression data were provided by Goardon et al. (2011). Gene expression data were normalized and analyzed as previously reported (Lück et al., 2011).
Acknowledgments
We thank Ms. S. Frey and Dr. T. Schnoeder (Otto-von-Guericke University, Magdeburg, Germany) for technical assistance and Dr. D. Strand (University Hospital Mainz, Mainz, Germany) for scientific advice during the first steps of this project. We thank A. Fenske (Otto-von-Guericke University) for his support with the BM transplantation experiments and Dr. R. Mathieu (Boston Children’s Hospital, Boston, MA) and Dr. R. Hartig (Otto-von-Guericke University) for their support with cell sorting.
This work was supported by a grant of the “Else Kröner-Fresenius-Stiftung” (2012_A152 to F.H. Heidel). F.H. Heidel was a Mildred-Scheel fellow (2009-2011) of the German Cancer Aid (DKH D/08/00661, Deutsche Krebshilfe e.V.). Use of primary patient samples was supported by a grant from the Jose-Carreras-Foundation to F.H. Heidel (SP 12/04; in support of the Hematology Tumor Bank Magdeburg). Moreover, funding was provided by the German Cancer Aid (Deutsche Krebshilfe TP6, “Oncogene Networks in AML“ to T. Fischer), a grant by the German Research Council (DFG; SFB854 TP20 T. Fischer), and a National Cancer Institute grant (#CA66996 to S.A. Armstrong). S.A. Armstrong is a Leukemia and Lymphoma Society Scholar, and L. Bullinger was supported in part by the German Research Foundation (Heisenberg-Stipendium BU 1339/3-1).
The authors have no financial conflicts of interest.
Footnotes
Abbreviations used:
- AML
- acute myeloid leukemia
- CN-AML
- cytogenetically normal AML
- GEP
- gene expression profiling
- GMP
- granulocyte macrophage progenitor
- HSC
- hematopoietic stem cell
- LMPP
- lymphoid-primed MPP
- LSC
- leukemia stem cell
- LT-HSC
- long-term HSC
- MEP
- megakaryocyte erythroid progenitor
- MPD
- myeloproliferative disease
- MPP
- multipotent progenitor
- pIpC
- poly-I–poly-C peptide
- qRT-PCR
- quantitative RT-PCR
References
- Beekman R., Valkhof M.G., Sanders M.A., van Strien P.M., Haanstra J.R., Broeders L., Geertsma-Kleinekoort W.M., Veerman A.J., Valk P.J., Verhaak R.G., et al. 2012. Sequential gain of mutations in severe congenital neutropenia progressing to acute myeloid leukemia. Blood. 119:5071–5077 10.1182/blood-2012-01-406116 [DOI] [PubMed] [Google Scholar]
- Faber J., Krivtsov A.V., Stubbs M.C., Wright R., Davis T.N., van den Heuvel-Eibrink M., Zwaan C.M., Kung A.L., Armstrong S.A. 2009. HOXA9 is required for survival in human MLL-rearranged acute leukemias. Blood. 113:2375–2385 10.1182/blood-2007-09-113597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber K., Bullinger L., Ragu C., Garding A., Mertens D., Miller C., Martin D., Walcher D., Döhner K., Döhner H., et al. 2012. CDX2-driven leukemogenesis involves KLF4 repression and deregulated PPARγ signaling. J. Clin. Invest. 10.1172/JCI64745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gateff E. 1978. Malignant neoplasms of genetic origin in Drosophila melanogaster. Science. 200:1448–1459 10.1126/science.96525 [DOI] [PubMed] [Google Scholar]
- Goardon N., Marchi E., Atzberger A., Quek L., Schuh A., Soneji S., Woll P., Mead A., Alford K.A., Rout R., et al. 2011. Coexistence of LMPP-like and GMP-like leukemia stem cells in acute myeloid leukemia. Cancer Cell. 19:138–152 10.1016/j.ccr.2010.12.012 [DOI] [PubMed] [Google Scholar]
- Heidel F.H., Mar B.G., Armstrong S.A. 2011. Self-renewal related signaling in myeloid leukemia stem cells. Int. J. Hematol. 94:109–117 10.1007/s12185-011-0901-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope K.J., Cellot S., Ting S.B., MacRae T., Mayotte N., Iscove N.N., Sauvageau G. 2010. An RNAi screen identifies Msi2 and Prox1 as having opposite roles in the regulation of hematopoietic stem cell activity. Cell Stem Cell. 7:101–113 10.1016/j.stem.2010.06.007 [DOI] [PubMed] [Google Scholar]
- Humbert P.O., Dow L.E., Russell S.M. 2006. The Scribble and Par complexes in polarity and migration: friends or foes? Trends Cell Biol. 16:622–630 10.1016/j.tcb.2006.10.005 [DOI] [PubMed] [Google Scholar]
- Ito T., Kwon H.Y., Zimdahl B., Congdon K.L., Blum J., Lento W.E., Zhao C., Lagoo A., Gerrard G., Foroni L., et al. 2010. Regulation of myeloid leukaemia by the cell-fate determinant Musashi. Nature. 466:765–768 10.1038/nature09171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharas M.G., Lengner C.J., Al-Shahrour F., Bullinger L., Ball B., Zaidi S., Morgan K., Tam W., Paktinat M., Okabe R., et al. 2010. Musashi-2 regulates normal hematopoiesis and promotes aggressive myeloid leukemia. Nat. Med. 16:903–908 10.1038/nm.2187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiel M.J., Yilmaz O.H., Iwashita T., Yilmaz O.H., Terhorst C., Morrison S.J. 2005. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 121:1109–1121 10.1016/j.cell.2005.05.026 [DOI] [PubMed] [Google Scholar]
- Klezovitch O., Fernandez T.E., Tapscott S.J., Vasioukhin V. 2004. Loss of cell polarity causes severe brain dysplasia in Lgl1 knockout mice. Genes Dev. 18:559–571 10.1101/gad.1178004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlmann A., Bullinger L., Thiede C., Schaich M., Schnittger S., Döhner K., Dugas M., Klein H.U., Döhner H., Ehninger G., Haferlach T. 2010. Gene expression profiling in AML with normal karyotype can predict mutations for molecular markers and allows novel insights into perturbed biological pathways. Leukemia. 24:1216–1220 10.1038/leu.2010.73 [DOI] [PubMed] [Google Scholar]
- Krivtsov A.V., Wang Y., Feng Z., Armstrong S.A. 2009. Gene expression profiling of leukemia stem cells. Methods Mol. Biol. 538:231–246 10.1007/978-1-59745-418-6_11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuphal S., Wallner S., Schimanski C.C., Bataille F., Hofer P., Strand S., Strand D., Bosserhoff A.K. 2006. Expression of Hugl-1 is strongly reduced in malignant melanoma. Oncogene. 25:103–110 10.1038/sj.onc.1209008 [DOI] [PubMed] [Google Scholar]
- Li Z., Huang H., Li Y., Jiang X., Chen P., Arnovitz S., Radmacher M.D., Maharry K., Elkahloun A., Yang X., et al. 2012. Up-regulation of a HOXA-PBX3 homeobox-gene signature following down-regulation of miR-181 is associated with adverse prognosis in patients with cytogenetically abnormal AML. Blood. 119:2314–2324 10.1182/blood-2011-10-386235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X., Feng X., Man X., Yang G., Tang L., Du D., Zhang F., Yuan H., Huang Q., Zhang Z., et al. 2009. Aberrant splicing of Hugl-1 is associated with hepatocellular carcinoma progression. Clin. Cancer Res. 15:3287–3296 10.1158/1078-0432.CCR-08-2078 [DOI] [PubMed] [Google Scholar]
- Lück S.C., Russ A.C., Botzenhardt U., Paschka P., Schlenk R.F., Döhner H., Fulda S., Döhner K., Bullinger L. 2011. Deregulated apoptosis signaling in core-binding factor leukemia differentiates clinically relevant, molecular marker-independent subgroups. Leukemia. 25:1728–1738 10.1038/leu.2011.154 [DOI] [PubMed] [Google Scholar]
- Mejia-Pous C., Damiola F., Gandrillon O. 2011. Cholesterol synthesis-related enzyme oxidosqualene cyclase is required to maintain self-renewal in primary erythroid progenitors. Cell Prolif. 44:441–452 10.1111/j.1365-2184.2011.00771.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menéndez J., Pérez-Garijo A., Calleja M., Morata G. 2010. A tumor-suppressing mechanism in Drosophila involving cell competition and the Hippo pathway. Proc. Natl. Acad. Sci. USA. 107:14651–14656 10.1073/pnas.1009376107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min I.M., Pietramaggiori G., Kim F.S., Passegué E., Stevenson K.E., Wagers A.J. 2008. The transcription factor EGR1 controls both the proliferation and localization of hematopoietic stem cells. Cell Stem Cell. 2:380–391 10.1016/j.stem.2008.01.015 [DOI] [PubMed] [Google Scholar]
- Ohali A., Avigad S., Zaizov R., Ophir R., Horn-Saban S., Cohen I.J., Meller I., Kollender Y., Issakov J., Yaniv I. 2004. Prediction of high risk Ewing’s sarcoma by gene expression profiling. Oncogene. 23:8997–9006 10.1038/sj.onc.1208060 [DOI] [PubMed] [Google Scholar]
- Perry J.M., He X.C., Sugimura R., Grindley J.C., Haug J.S., Ding S., Li L. 2011. Cooperation between both Wnt/beta-catenin and PTEN/PI3K/Akt signaling promotes primitive hematopoietic stem cell self-renewal and expansion. Genes Dev. 25:1928–1942 10.1101/gad.17421911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathinam C., Matesic L.E., Flavell R.A. 2011. The E3 ligase Itch is a negative regulator of the homeostasis and function of hematopoietic stem cells. Nat. Immunol. 12:399–407 10.1038/ni.2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimanski C.C., Schmitz G., Kashyap A., Bosserhoff A.K., Bataille F., Schäfer S.C., Lehr H.A., Berger M.R., Galle P.R., Strand S., Strand D. 2005. Reduced expression of Hugl-1, the human homologue of Drosophila tumour suppressor gene lgl, contributes to progression of colorectal cancer. Oncogene. 24:3100–3109 10.1038/sj.onc.1208520 [DOI] [PubMed] [Google Scholar]
- Sengupta A., Duran A., Ishikawa E., Florian M.C., Dunn S.K., Ficker A.M., Leitges M., Geiger H., Diaz-Meco M., Moscat J., Cancelas J.A. 2011. Atypical protein kinase C (aPKCzeta and aPKClambda) is dispensable for mammalian hematopoietic stem cell activity and blood formation. Proc. Natl. Acad. Sci. USA. 108:9957–9962 10.1073/pnas.1103132108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamm C., Böwer N., Annerén C. 2011. Regulation of mouse embryonic stem cell self-renewal by a Yes-YAP-TEAD2 signaling pathway downstream of LIF. J. Cell Sci. 124:1136–1144 10.1242/jcs.075796 [DOI] [PubMed] [Google Scholar]
- Wilson A., Laurenti E., Trumpp A. 2009. Balancing dormant and self-renewing hematopoietic stem cells. Curr. Opin. Genet. Dev. 19:461–468 10.1016/j.gde.2009.08.005 [DOI] [PubMed] [Google Scholar]
- Wu M., Kwon H.Y., Rattis F., Blum J., Zhao C., Ashkenazi R., Jackson T.L., Gaiano N., Oliver T., Reya T. 2007. Imaging hematopoietic precursor division in real time. Cell Stem Cell. 1:541–554 10.1016/j.stem.2007.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]