Parp1 can replace c-Myc to promote induced pluripotent stem cell (iPSC) generation.
Abstract
Poly(ADP-ribose) polymerase 1 (Parp1) catalyzes poly(ADP-ribosylation) (PARylation) and induces replication networks involved in multiple nuclear events. Using mass spectrometry and Western blotting, Parp1 and PARylation activity were intensively detected in induced pluripotent stem cells (iPSCs) and embryonic stem cells, but they were lower in mouse embryonic fibroblasts (MEFs) and differentiated cells. We show that knockdown of Parp1 and pharmacological inhibition of PARylation both reduced the efficiency of iPSC generation induced by Oct4/Sox2/Klf4/c-Myc. Furthermore, Parp1 is able to replace Klf4 or c-Myc to enhance the efficiency of iPSC generation. In addition, mouse iPSCs generated from Oct4/Sox2/Parp1-overexpressing MEFs formed chimeric offspring. Notably, the endogenous Parp1 and PARylation activity was enhanced by overexpression of c-Myc and repressed by c-Myc knockdown. A chromatin immunoprecipitation assay revealed a direct interaction of c-Myc with the Parp1 promoter. PAR-resin pulldown, followed by proteomic analysis, demonstrated high levels of PARylated Chd1L, DNA ligase III, SSrp1, Xrcc-6/Ku70, and Parp2 in pluripotent cells, which decreased during the differentiation process. These data show that the activation of Parp1, partly regulated by endogenous c-Myc, effectively promotes iPSC production and helps to maintain a pluripotent state by posttranslationally modulating protein PARylation.
Somatic cell reprogramming is a promising strategy for stem cell biology and regenerative medicine. Accumulated data have shown that nuclear reprogramming can be experimentally induced by three methods: nuclear transfer, cell fusion, or forced expression of transcription factors (Yamanaka and Blau, 2010). It is conceivable that mature oocytes and embryonic stem cells (ESCs) contain reprogramming factors (proteins, RNAs, lipids, and small molecules) that enable these somatic cells to undergo efficient nuclear reprogramming, a process of converting somatic cells to pluripotent states (Jullien et al., 2010; Wang et al., 2010). Recent evidence has emphasized the pivotal roles of nuclear proteins in the regulation of chromatin remodeling and epigenetic modifications during the reprogramming process (Jullien et al., 2011). However, the precise molecular mechanisms of the regulation of nuclear factors during cellular reprogramming remain uncertain.
Induced pluripotent stem cells (iPSCs) are a recently developed technology that holds promise for stem cell biology and regenerative medicine (Takahashi et al., 2007; Nakagawa et al., 2008). Nuclear reprogramming induced by transcription factors resets the epigenetic landmarks, which leads to the global reversion of the somatic epigenomes to an ESC-like state (Maherali et al., 2007; Papp and Plath, 2011). However, the mechanisms involved, particularly the posttranslational interactions and modifications, remain undetermined. Mass spectrometry (MS)–based proteomic analysis is the most powerful tool currently available for global investigation of proteome profiles in stem cell biology (Van Hoof et al., 2009; Rigbolt et al., 2011). Although the importance of nuclear proteins in epigenetic events has been addressed (Jullien et al., 2010), little was known about the involvement of functional proteins that regulate reprogramming and maintain pluripotency. Therefore, it is important to identify novel factors involved in the regulation of nuclear reprogramming using a proteomics approach to elucidate the complex molecular networks in the nucleus during the reprogramming process.
Poly(ADP-ribose) polymerase 1 (Parp1), a member of the Parp family of proteins, is a highly conserved DNA-binding protein that is abundant in the nucleus. Parp1 is a key effector of several nuclear events, such as DNA repair, replication, and transcription (Jagtap and Szabó, 2005; Kraus, 2008). It catalyzes a process called poly(ADP-ribosylation) (PARylation), in which NAD+ is used as substrate to synthesize poly(ADP-ribose) polymers with sizes varying from 2 to 200 ADP-ribose units (Krishnakumar and Kraus, 2010). This Parp1-catalyzed PARylation has been implicated in several processes, including chromatin remodeling, enhancer binding, coregulation, and insulation (Kraus, 2008). Importantly, Parp1, along with PARylation, regulates genomic methylation patterns (Caiafa et al., 2009). It was previously demonstrated that Parp1 is a regulator of Sox2 (Gao et al., 2009; Lai et al., 2012), and it is involved in the efficient generation of iPSCs (Lai et al., 2012). Recently, Doege et al. (2012) reported that Parp1 and TeT2 contribute to early-stage epigenetic modification during somatic cell reprogramming, and the induction of the Parp1 gene further promotes accessibility to the pluripotency factor Oct4. Therefore, it is conceivable that Parp1 and PARylation may be involved in the regulation of nuclear reprogramming or the maintenance of pluripotent properties in stem cells.
ESCs have the capacity of unlimited self-renewal to maintain pluripotency, express high levels of antioxidant and stress-resistant proteins, and possess prominent DNA strand break–repairing capacity (Saretzki et al., 2004). A recent study demonstrated that iPSCs (Armstrong et al., 2010), which are similar to ESCs, maintain genomic stability by elevated nonhomologous end-joining (NHEJ) activity and DNA repair efficacy (Fan et al., 2011). Notably, Parp1 and PARylation have been linked to the regulation of chromatin remodeling and genome stability (Deng, 2009). However, the posttranslational mechanisms of Parp1 and PARylation involved in regulating nuclear reprogramming are still undetermined. In this study, we compared the expression profiles of nuclear proteins between MEFs, ESCs, and iPSCs using proteomic analysis. Among these nuclear proteins, Parp1 and Parp1-mediated PARylation were consistently enhanced, which enhanced the expression of Oct4 and Nanog during the course of reprogramming, implying their pivotal roles in iPSC generation. Replacement of c-Myc with Parp-1 in the reprogramming process resulted in a similar efficiency of iPSC production. Moreover, several Parp1-associated and PARylation-interacting proteins in iPSCs, which may be involved in DNA repair and chromatin reopening, were identified. This study demonstrates an interaction between Parp1 and c-Myc, and it identifies a mechanistic role for Parp1 in nuclear reprogramming.
RESULTS
Increased Parp1 and PARylation activity in reprogramming and pluripotent cells
Recent studies using MS-based proteomic analysis have confirmed the significant similarity between the proteomic profiles of iPSCs and ESCs (Jin et al., 2011; Munoz et al., 2011; Phanstiel et al., 2011). However, these studies were conducted with whole-cell lysates and did not focus on the differential regulation of nuclear events. In our previous work, we generated mouse iPSCs by overexpressing four genes, Oct4/Sox2/Klf4/c-Myc (OSKM; Re-7 iPSC clone), or three genes (OSK without c-Myc) (Li et al., 2011). To distinguish the differences in the profiles of nuclear proteins between somatic and reprogrammed pluripotent cells, nuclear protein extracts from MEFs and Re-7 iPSCs were prepared. These extracts were then separated into five fractions by SDS-PAGE (Fig. 1 A). First, we established the differential expression profiles of these nuclear extracts using 1D liquid chromatography–tandem MS (LC-MS/MS; Table S1). Based on gene ontology (GO) database analysis, the predominant processes up-regulated in the nuclear protein profiles of iPSCs included those pertaining to RNA processing, chromatin packaging and remodeling, cell structure and motility, and protein biosynthesis, as well as those involved in mRNA transcription and DNA replication (Fig. 1 B; Table S1). Furthermore, using a statistical comparative analysis between the databases of 1D LC-MS/MS (Table S1) and 2D-differential gel electrophoresis (Fig. S1, A and B), we identified the 112 most up-regulated nuclear proteins in iPSCs compared with MEFs (Fig. S2).
Figure 1.
Parp1 and PARylation activity in the pluripotent or differentiated state. (A) Morphology and staining for ALP and SSEA-1 in Re-7 iPSCs. BF, bright field. Bars, 100 µm. Nuclear proteins from Re-7 iPSCs and MEFs were separated into five fractions by 1D-DIGE. (B) Pie chart showing the GO classification of gene functions in all nuclear proteins from iPSCs. (C) Expression of Oct4, Nanog, c-Myc, Parp1, and PARylation activity in pluripotent stem cells, including ESCs and iPSCs transfected with OSKM or OSK. (D, top) Expression of the pluripotency factors Oct4, Sox2, Klf4, c-Myc, and Parp1. (D, bottom) PARylation activity during the reprogramming process. (E) Parp1 expression and PARylation activity in EBs 6 d after differentiation. (F, left) Differentiation of Re-7 iPSCs into osteocyte-like, hepatocyte-like or neuron-like cells, confirmed by positive staining with Alizarin red, PAS, and the neuron-specific markers Nestin and MAP2, respectively. Bars, 100 µm. (F, right) Detection of Parp1, Parp2, TopII-α, Oct4, Sox2, and Klf4 proteins in Re-7 iPSCs, before and after differentiation into the indicated specific lineages. The Western blots are representative of three separate experiments with independent cell preparations.
Both ESCs and iPSCs maintain their genomic stability and pluripotency by enhancing DNA repair and NHEJ activity, and high levels of expression of DNA repair proteins, including Parp1, DNA ligIII, Rad51, and XLF, have been found in both ESCs and iPSCs (Fan et al., 2011). An elegant study provided by Doege et al. (2012) showed that Parp1 is involved in epigenetic modifications that direct subsequent transcriptional induction at pluripotency loci during somatic cell reprogramming. Using proteomic analysis (Fig. S2) and Western blotting (Fig. 1 C), we found high Parp1 expression levels in the nuclear lysates of iPSCs but not MEFs. One of the extensively characterized functions of Parp1 is the posttranslational modification of target proteins by attaching a poly(ADP-ribose) chain (PARylation; Krishnakumar and Kraus, 2010). Using poly(ADP-ribose) affinity resin to pull down the PARylated proteins, we further demonstrated that Parp1 is the most highly expressed PARylated protein in iPSCs compared with MEFs (Table 1). Therefore, we further attempted to elucidate whether Parp1 and PARylation may play a role in promoting cellular reprogramming and maintaining pluripotency. Notably, Parp1 protein, as well as Oct4, Nanog, and c-Myc, were up-regulated in both whole-cell lysates and nuclear fractions of Re-7 iPSCs (Fig. 1 C and Fig. S1 C). This up-regulation of Parp1, accompanied by increased PARylation activity, was consistently observed in iPSCs generated with OSKM (Re-7 cells) or OSK, S. Yamanaka’s (Center for iPS Cell Research and Application, Kyoto University, Kyoto, Japan and Gladstone Institute of Cardiovascular Disease, San Francisco, CA) iPSC clone (miPSCs), and ESCs (Fig. 1 C). Parp1 and PARylation, as well as these pluripotency factors, were completely undetectable in MEFs (Fig. 1 C). During the reprogramming process to convert MEFs to iPSCs, Parp1 and Oct4, Sox2, Nanog, and c-Myc were up-regulated after the transfection of OSKM, and these proteins reached maximal expression 15 d after the induction of reprogramming (Fig. 1 D, top). Increased PARylation activity was also observed during the reprogramming process (Fig. 1 D, bottom). Furthermore, we analyzed whether PARylation was influenced by the differentiation of Re-7 iPSCs. Parallel to the down-regulation of Parp1, the PARylation activity decreased significantly in iPSC-derived embryoid bodies (EBs) in a time-dependent manner (Fig. 1 E). Differentiation into different lineages was induced by specific protocols. Neuron-like, osteocyte-like (mesoderm), and hepatocyte-like (endoderm) cells were confirmed by immunofluorescence, Alizarin red, and PAS staining, respectively (Fig. 1 F, left). After differentiation of Re-7 iPSCs into different lineages with each protocol, Western blotting showed that the Parp1 protein, as well as Parp2, topoisomerase II α, Klf4, Oct4, and Sox2, was substantially down-regulated (Fig. 1 F, right). Collectively, the differential profiles of Parp1/PARylation activity during reprogramming and tridermal differentiation suggested that Parp1/PARylation may play an important role in the regulation of reprogramming efficiency and the acquisition of pluripotent properties.
Table 1.
Detection of most highly expressed PARylated proteins in iPSCs using poly(ADP-ribose)-resin pulldown
| No. | Protein name | Accession no. | Gene name | kD/pl | Pep # (unique) | PEP |
| 1 | Poly [ADP-ribose] polymerase 1 | P11103 | Parp1 | 113.10/9.05 | 70 (17) | <1.00−307 |
| 2 | FACT complex subunit SPT16 | Q92089 | Supt16h | 119.82/5.50 | 16 (16) | 5.92−148 |
| 3 | Poly [ADP-ribose] polymerase 2 | O88554 | Parp2 | 63.40/8.65 | 15 (15) | 3.28−134 |
| 4 | Chromodomain-helicase-DNA-binding protein 1-like | Q9CXF7 | Chd1l | 101.44/6.20 | 11 (11) | 1.54−54 |
| 5 | DNA ligase 3 | P97386 | Lig3 | 113.07/9.16 | 10 (10) | 3.32−104 |
| 6 | FACT complex subunit SSRP1 | Q08943 | Ssrp1 | 80.86/6.33 | 4 (4) | 9.96−9 |
| 7 | Leucine-rich repeat flightless-interacting protein 2 | Q91WK0 | Lrrfip2 | 47.15/5.54 | 3 (3) | 4.01−61 |
| 8 | x-ray repair cross-complementing protein 6 | P23475 | Xrcc6 | 69.48/6.35 | 3 (3) | 2.00−10 |
| 9 | x-ray repair cross-complementing protein 1 | Q60596 | Xrcc1 | 68.97/5.97 | 3 (3) | 8.30−7 |
| 10 | Splicing factor U2AF 35 kD subunit | Q9D883 | U2af1 | 27.82/9.09 | 2 (2) | 3.35−34 |
| 11 | Protein timeless homolog | Q9R1X4 | Timeless | 137.50/5.35 | 2 (2) | 1.11−22 |
| 12 | Nucleolar RNA helicase 2 | Q9JIK5 | Ddx21 | 93.55/9.19 | 2 (2) | 3.57−5 |
| 13 | U1 small nuclear ribonucleoprotein A | Q62189 | Snrpa | 31.84/9.81 | 2 (2) | 3.52−4 |
| 14 | Tyrosyl-DNA phosphodiesterase 1 | Q8BJ37 | Tdp1 | 68.69/7.67 | 2 (2) | 1.81−4 |
| 15 | Aprataxin and PNK-like factor | Q9D842 | Aplf | 54.97/5.05 | 2 (2) | 2.55−3 |
| 16 | Replication protein A 70 kD DNA-binding subunit | Q8VEE4 | Rpa1 | 69.04/8.13 | 2 (2) | 2.24−3 |
| 17 | Apoptotic chromatin condensation inducer in the nucleus | Q9JIX8 | Acin1 | 150.72/5.71 | 1 (1) | 3.75−10 |
| 18 | Heterogeneous nuclear ribonucleoprotein A3 | Q8BG05 | Hnrnpa3 | 39.65/9.10 | 1 (1) | 5.56−5 |
| 19 | Fragile X mental retardation protein 1 homolog | P35922 | Fmr1 | 68.99/7.27 | 1 (1) | 4.56−3 |
| 20 | Recombining binding protein suppressor of hairless | P31266 | Rbpj | 58.54/8.43 | 1 (1) | 2.56−3 |
| 21 | Splicing factor U2AF 65 kD subunit | P26369 | U2af2 | 53.52/9.19 | 1 (1) | 1.14−3 |
Posterior error probability (PEP) was obtained from statistical analysis of total peptide identification for a protein in one sample. The value essentially operates as a statistical value, and low PEP indicates high statistical significance. pl, isoelectric point.
Parp1 and PARylation regulate the efficiency of iPSC reprogramming
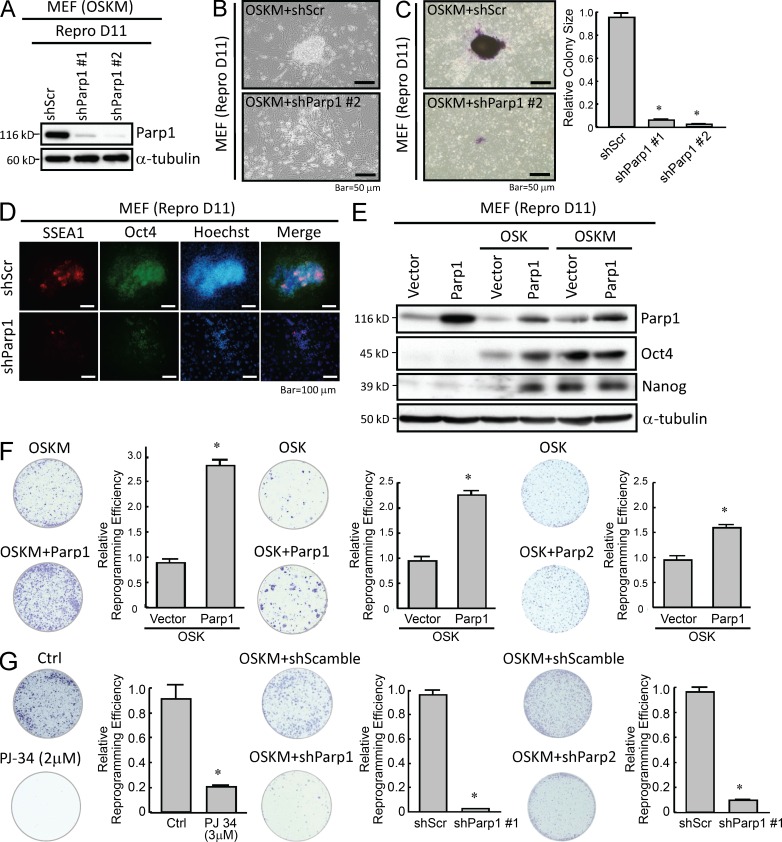
We next evaluated whether inhibition of PARylation or knockdown of Parp1 interfere with cell reprogramming. To investigate the role of Parp1 in the early phase of the reprogramming process, we first confirmed the effect of Parp1 knockdown in two MEF-derived iPSC clones 11 d after OSKM transfection by Western blotting. In the two reprogrammed clones with Parp1 knocked down, Parp1 expression was almost undetectable at day 11 after reprogramming (Fig. 2 A). The two clones of cells transfected with OSKM and shRNA against Parp1 (OSKM + shParp1) had significantly reduced self-renewal and proliferative capabilities (Fig. 2 B), formed smaller colonies, and were less positive for alkaline phosphatase (ALP) staining (Fig. 2 C) compared with cells transfected with scrambled control shRNA (OSKM + Scramble shRNA). Meanwhile, the resultant ESC markers, including Oct4 and SSEA-1, were significantly inhibited by this Parp1 knockdown (Fig. 2 D). To further validate that Parp1 facilitates cell reprogramming, we co-overexpressed Parp1 with either OSKM or OSK in MEFs using a lentiviral transfection system. Western blots confirmed the overexpression of Parp1 at day 11 after reprogramming (Fig. 2 E). We subsequently examined the effect of either Parp1 knockdown or overexpression on the efficiency of iPSC generation at day 21 after reprogramming. Parp1 overexpression significantly enhanced the reprogramming efficiency in MEFs transfected with OSKM or OSK (Fig. 2 F, left and middle, respectively). Notably, Parp2 overexpression also enhanced the reprogramming efficiency in MEFs transfected with OSK (Fig. 2 F, right), but the effect of Parp2 overexpression was significantly less than that of Parp1 overexpression. Moreover, administration of various PARylation inhibitors consistently led to reduction in the efficiency of iPSC generation induced by OSKM at day 21 after reprogramming (PJ-34: Fig. 2 G, left; ABT-888 and 3-aminobenzamide: not depicted). Parp1 knockdown by a lentivirus-delivered shRNA led to a significant inhibition of the efficiency of iPSC generation (Fig. 2 G, middle), and Parp2 knockdown also suppressed iPSC generation at a similar extent at day 21 after reprogramming (Fig. 2 G, right). Collectively, these data indicate that modulating Parp1 and PARylation activity influences the reprogramming efficiency and the pluripotent status of iPSCs, indicating that Parp1 and PARylation are crucial for nuclear reprogramming.
Figure 2.
Parp1 expression and PARylation activity are essential for iPSC reprogramming. (A) Western blotting confirms the effect of Parp1 knockdown on Parp1 expression at reprogramming day 11 (Repro D11). (B) Parp1 knockdown in MEFs transfected with OSKM affects self-renewal and proliferative capabilities during the reprogramming process. (C) Reprogramming cells stained with ALP demonstrate the colony size of iPSCs with Parp1 knockdown. (D) Immunofluorescent staining for ESC markers SSEA-1 and Oct4 in iPSCs with Parp1 knockdown at Repro D11. (E) Detection of Parp1 and Oct4 expression in OSK- or OSKM-transfected MEFs overexpressing Parp1 at Repro D11. (F) Reprogramming cells stained with ALP and the relative reprogramming efficiencies of MEFs transfected with OSKM with or without Parp1 overexpression (left), OSK with or without Parp1 overexpression (middle), and OSK with or without Parp2 overexpression (right), at day 21 after reprogramming. (G, left) Reprogramming cells stained with ALP and reprogramming efficiencies of reprogramming cells (OSKM) treated with the PARylation inhibitor PJ-34, compared with cells without the inhibitor. (G, middle and right) OSKM-transfected MEFs treated with ShRNA-Parp1 (middle) or an shRNA-scrambled control (right) at day 21 after reprogramming. The Western blots are representative of three separate experiments with independent cell preparations. The ALP staining shown here are the mean ± SD of six independent experiments. *, P < 0.05 vs. parental.
Replacement of Klf-4 or c-Myc with Parp1 in OSKM reprogramming produces iPSCs and generates chimeric animals
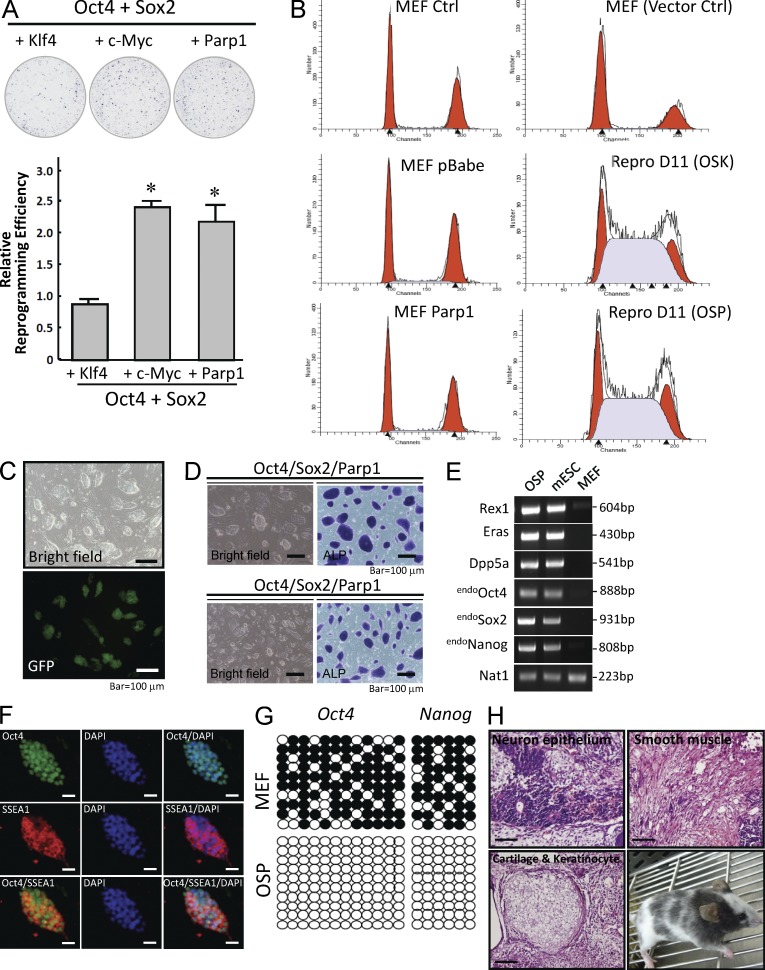
c-Myc, a proto-oncogene, is an essential factor for enhancing reprogramming efficiency, but it also increases the risk of tumorigenicity of the reprogrammed somatic cells (Nakagawa et al., 2010). Because Parp1 can increase the efficiency of iPSC generation in the OSK-transfection protocol, we investigated the potential of Parp1 to replace Klf4 and c-Myc. Remarkably, the iPSC-reprogramming efficiency of OS with Parp1 (OSP) was significantly higher than that of OSK, but it was similar to c-Myc cotransfected with OS (OSM; Fig. 3 A). We next attempted to investigate the dependence of Parp1-mediated reprogramming and iPSC generation on the cell cycle. Cell cycle analysis indicated that Parp1 overexpression showed no effect in MEFs, compared with parental MEFs or MEFs transfected with a control vector (Fig. 3 B). In addition, we observed a shift of the cell cycle to S-phase in MEFs transfected with OSK and OSP at day 11 after reprogramming (Fig. 3 B). This shift was also observed in pluripotent stem cells, including mESCs, S. Yamanaka’s iPSC clone (miPSCs), and iPSCs generated by transfection of either OSK or OSP (unpublished data), as described previously (Fujii-Yamamoto et al., 2005). These data indicate that the Parp1 effect on reprogramming efficiency and iPSC generation is cell cycle independent. Furthermore, OSP transfection activated the expression of Nanog-GFP during reprogramming in a Nanog-GFP reporter MEF clone (Fig. 3 C). The high passages of OSP-reprogrammed iPSCs were stably positive for markers of mouse ESCs, such as ALP activity (Fig. 3 D), an ESC-like gene signature (Fig. 3 E), stage-specific embryonic antigen (SSEA-1) and Oct4, and protein of stemness factors (Fig. 3 F). Bisulfite sequencing showed that the promoters of Oct4 and Nanog in OSP-iPSCs had much a lower methylation status than parental MEFs (Fig. 3 G). Importantly, 6 wk after transplantation of these iPSCs into the dorsal flanks of nude mice, we observed the formation of teratomas that contained various tissues, including neuronal epithelium (ectoderm), cartilage and keratinocytes (mesoderm), and smooth muscle (mesoderm; Fig. 3 H, top and bottom left). Furthermore, we injected these OSP-iPSCs into blastocysts that were then transplanted into the uteruses of pseudo-pregnant mice. The adult chimeras were confirmed by coat color, demonstrating that OSP-iPSCs were competent to produce adult chimeric mice (Fig. 3 H, bottom right). These observations indicate that Parp1 overexpression efficiently enhances the reprogramming of mouse somatic cells into iPSCs in the absence of c-Myc or Klf-4.
Figure 3.
Parp1 is able to replace Klf-4 or c-Myc to generate chimeric mice. (A) Comparison of reprogramming efficiencies between OSK, OSM, and OSP by ALP-positive colony counting. (B) Flow cytometry showing the cell cycle analysis in MEFs transfected with control vector or Parp1 at reprogramming day 0 (left) or day 11 (right). (C) Activation of the expression of Nanog-GFP by OSP transfection during reprogramming in a Nanog-GFP reporter MEF clone. (D) OSP-generated iPSCs can be stably cultured to at least 50 passages with high ALP activity. (E) RT-PCR showing the ESC-like gene signature in OSP-generated iPSCs at the 50th passage. (F) Immunofluorescence indicating the protein expression of pluripotency factors in OSP- iPSCs at the 50th passage. (G) Bisulfite sequencing showing the methylation profiles of the promoters of Oct4 and Nanog in MEFs and OSP-iPSCs. Open and filled circles indicate unmethylated and methylated CpG dinucleotides, respectively. (H) Ex vivo biopsies and histological analysis reveal teratoma formation in the subrenal grafts of OSP-iPSCs in nude mice. Competence of OSP-iPSCs to generate chimeric mice, as confirmed by coat color (bottom right). Bars, 100 µm. The ALP staining shown here is the mean ± SD of six independent experiments. The Western blots and other data are representative of three separate experiments with independent cell preparations.
c-Myc is a direct regulator of Parp1 and PARylation
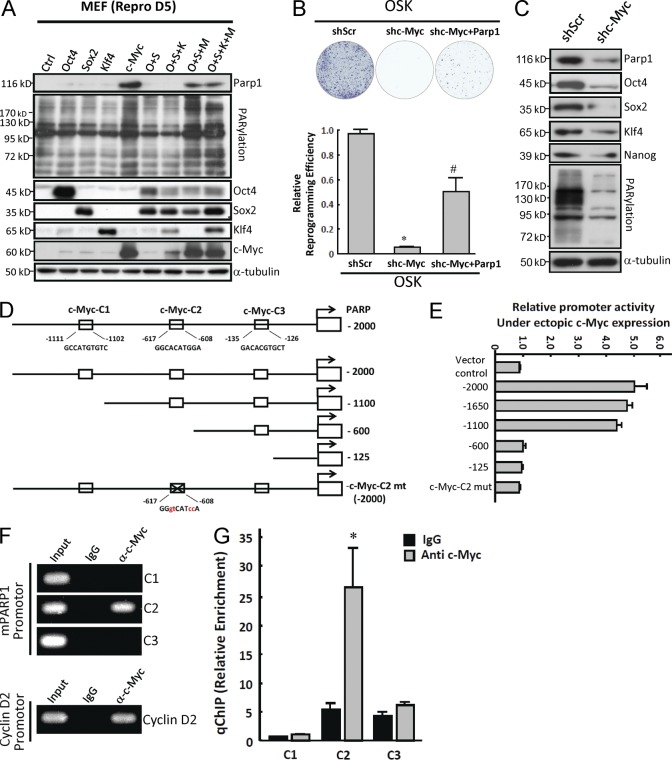
Given that Parp1 is up-regulated during reprogramming, we hypothesized that one or more of the exogenous transcription factors Oct4, Sox2, Klf4, and c-Myc may be the upstream regulators that induce Parp1 expression and PARylation activity. Therefore, we assessed the effects of forced expression of individual or combined Yamanaka’s factors on Parp1 expression and PARylation activity in MEFs (Fig. 4 A). 5 d after gene transfection, forced overexpression of c-Myc alone or transfection of OSM and OSKM resulted in substantial increases in Parp1 protein expression, as well as in PARylation activity in MEFs (Fig. 4 A). To further address whether Parp1 is the major downstream effector of c-Myc in the reprogramming process, we knocked down c-Myc and overexpressed Parp1 plus OSK in MEFs. The result suggested that overexpression of Parp1 compensates for c-Myc knockdown and allows efficient reprogramming without c-Myc (Fig. 4 B). In addition, c-Myc knockdown significantly blocked ALP activity (not depicted) and suppressed the protein-level of Parp1, Oct4, Sox2, Klf4, and Nanog, as well as PARylation activity in iPSCs (Fig. 4 C). Collectively, these results indicate that the activation of Parp1 and Parp1-related PARylation, partly regulated by c-Myc, plays a crucial role in facilitating reprogramming and maintaining the pluripotent state of stem cells.
Figure 4.
c-Myc is a key factor regulating Parp1 expression and PARylation activity. (A) Western blots showing Parp1 expression and PARylation level at day 5 after MEF cell transfection with indicated factors. (B) Reprogramming cells stained with ALP showing relative reprogramming efficiencies of cells expressing OSK and scrambled shRNA, OSK and c-Myc shRNA, or OSK, c-Myc shRNA, and Parp1. (C) Expression of Parp1, Oct4, Sox2, Klf4, and Nanog as well as PARylation activity in pluripotent stem cells treated with c-Myc shRNA. (D) Schematic representation of the reporter constructs containing serially deleted or point mutated mouse Parp1 promoter. (E) Luciferase activities of reporter constructs containing serially deleted or point mutated mouse Parp1 promoter as indicated in D. (F) ChIP analysis with IgG or c-Myc Ab for immunoprecipitation, followed by PCR using C1, C2, and C3 primer sets for Parp1 promoter and primers for cyclin D2 promoter. (G) The quantitative result in F (ChIP) with qPCR (qChIP). Input, 2% of total lysate. The Western blots are representative of three separate experiments with independent cell preparations. The ALP staining (B) shown here is the mean ± SD from six independent experiments, and other experiments (E and G) are from three independent experiments. B: *, P < 0.05 versus shScr; #, P < 0.05 versus shc-Myc. G: *, P < 0.05 versus IgG control.
We next determined whether c-Myc regulates Parp1 expression by fusing the Parp1 promoter to a luciferase reporter plasmid and coexpressing the reporter with c-Myc. Three putative c-Myc–binding sites were identified in the proximal promoter region (−2,000 to −100 base pairs) of Parp1 and deletion constructs were cloned in the luciferase reporter plasmid (Fig. 4 D). Cotransfection experiments showed that c-Myc activated the transcriptional activity of the Parp1 promoter containing three (−2,000) or two (−1,100) proximal c-Myc binding sites. In contrast, the Parp1 promoter deletion mutants lacking c-Myc-C1 and c-Myc-C2 (−600) suppressed c-Myc–activated Parp1 transcription (Fig. 4 E), indicating that C2 is an important site responding to c-MyC activity. Consistently, the Parp1 promoter construct without all three c-Myc binding sites (−125) or with point mutations in C2 (c-Myc-C2 mt) could not be stimulated by c-Myc. To investigate whether c-Myc can directly bind to the C2 c-Myc binding site in the promoter region of Parp1, chromatin immunoprecipitation (ChIP) assays were performed using C1, C2, and C3 primer sets (Fig. 4 F). The result showed that the endogenous c-Myc indeed only bound to the C2, but not C1 or C3, position of the Parp1 promoter. As a positive control, c-Myc bound to its reported target, cyclin D2 promoter. Fig. 4 G shows the result of the ChIP in Fig. 4 F with quantitative PCR (qChIP). These data strongly suggest that the Parp1 promoter region containing C2 was required for maximal activity of Parp1 in iPSCs. Together, we demonstrated that c-Myc is a direct regulator of Parp1 and PARylation.
Identification of PARylated targets and expression levels of Parp1/PARylation-associated proteins in pluripotent and differentiated states
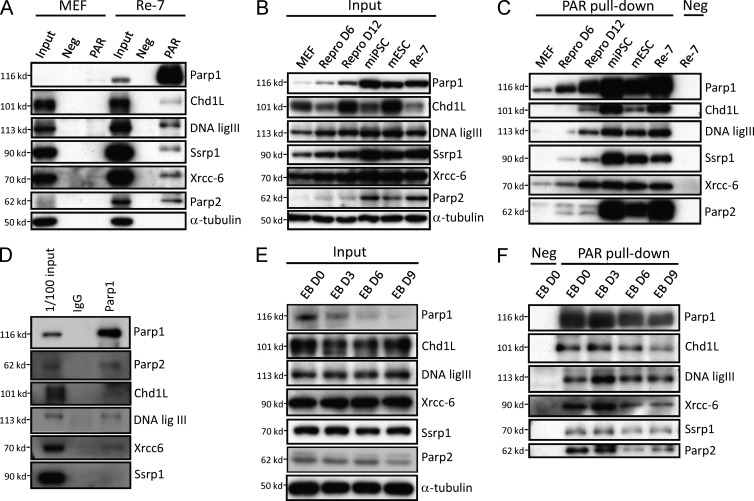
PARylation was previously considered the major catalytic function of Parp1; we therefore attempted to identify the proteins that are involved in Parp1-mediated PARylation in pluripotent stem cells. We used poly(ADP-ribose) affinity resin to pull down the PARylated proteins in iPSCs and MEFs. The PARylated proteins indentified by LC-MS/MS in iPSCs are listed in Table 1. Among these candidate proteins, Parp1, Chd1L, DNA ligase III, Ssrp1, Xrcc6, and Parp2 were identified by LC-MS/MS as having more than three peptides per protein. To confirm these results, we used Western blotting with specific antibodies to detect the expression of these candidate proteins. Compared with the input (total lysate) column, there was no detectable signal in the negative control column (resin with a mutated affinity domain), and specific signals from these six proteins were only observed in iPSCs after PAR affinity resin purification (Fig. 5 A). We attempted to further evaluate the expression profiles of these candidate proteins in the reprogramming process and pluripotent stem cells. First, we found that the total protein levels of Chd1L, DNA ligase III, Ssrp1, and Xrcc-6 were modest in MEF lysates and gradually increased in the reprogramming process (Fig. 5 B). The total protein expression of Chd1L was not correlated with pluripotency as strongly as it was with the reprogramming state (Fig. 5 B). In contrast, when PAR affinity resin was used to pull down these candidate proteins from total cell extracts, all candidate proteins were gradually PARylated during the reprogramming process, and the maximal PARylation of these proteins was found in iPSCs, S. Yamanaka’s miPSC clone (miPSC), and mESCs, but not in MEFs (Fig. 5 C). Notably, as detected by the pulldown assays, Parp2 levels in the reprogramming D6 and D12 were not significantly changed compared with Parp1 (Fig. 5, B and C). Coimmunoprecipitation further confirmed that Parp2, Chd1L, DNA ligase III, Xrcc-6, and Ssrp1 interact with Parp1 to form a complex (Fig. 5 D). To explore whether differentiation affects the expression of PARylated proteins, control (total input of cell lysate; Fig. 5 E) and PAR-resin pulldowns (Fig. 5 F) from iPSC-derived EBs were compared. In the total input, there were no significant changes of the six proteins on days 3, 6, and 9 after ED differentiation (Fig. 5 E). The expression of PARylated Parp1, Chd1L, DNA ligase III, Ssrp1, Xrcc-6, and Parp2 was decreased during the differentiation process of iPSC-derived EBs (Fig. 5 F). Notably, the expression levels of these six PARylated proteins were significantly down-regulated on day 9 after EB differentiation (Fig. 5 F). These data suggest that the cells in which PARylated levels of Chd1L, DNA ligase III, SSrp1, Xrcc-6/Ku-70, and Parp2 were increased during reprogramming were high in the pluripotent state of cells but decreased during the differentiation process.
Figure 5.
Parp1 maintains the PARylation of chromatin-remodeling proteins in pluripotent stem cells. (A) PARylated proteins in iPSCs were prepared by PARylated protein pulldown assays and analyzed by LC-MS/MS. Western blotting confirms the expression of these identified PARylated proteins (Neg: negative resin with mutant affinity domain; PAR: PAR affinity resin). (B) In the total lysates (input), Western blotting shows the expression levels of Parp1, Parp2, Chd1L, DNA ligase III, Ssrp1, and Xrcc6 in pluripotent stem cells and during the reprogramming process. (C) Using PAR affinity resin with pulldown assays, evaluation of the expression profiles of the identified PARylated proteins in MEFs, reprogramming D6 cells, reprogramming D12 cells, miPSCs, mESCs, and Re-7 iPSCs (Con: negative resin with a mutant affinity domain). (D) Coimmunoprecipitation showed that that Parp1 interacts with Parp2, Chd1L, DNA ligase III, Xrcc-6, and Ssrp1. (E) The total protein from iPSC-derived EBs were evaluated. (F) The protein expression of Parp1, Chd1L, DNA ligase III, Ssrp1, Xrcc-6, and Parp2 during the differentiation process of iPSC-derived EBs. The Western blots are representative of three separate experiments with independent cell preparations.
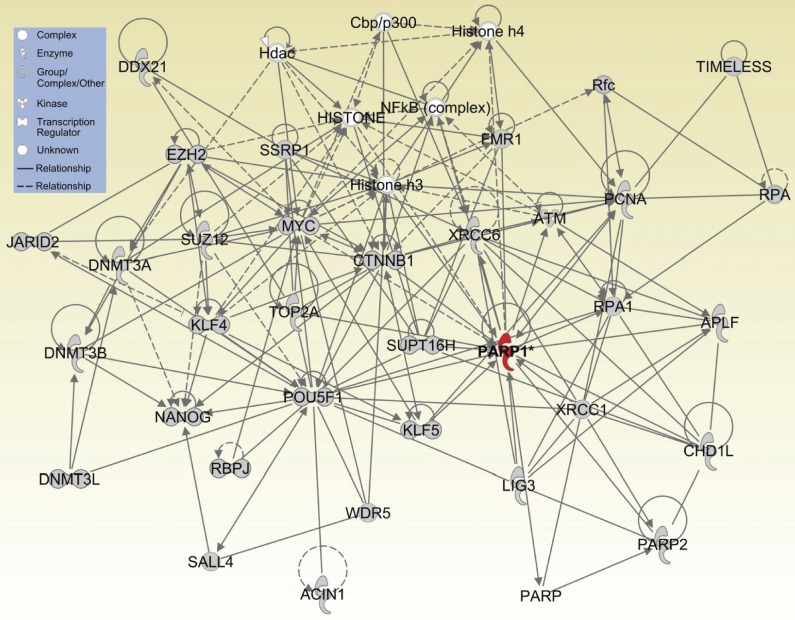
In addition, we further explored the roles of Parp1 in posttranslational modification to gain additional insights into the functional consequences of differential patterns of PARylated proteins in pluripotent stem cells. Using gene network analysis with the IPA software package to construct network modules, we found that Parp1 may be a key factor regulating the pathways related to DNA repair, chromatin modification, the polycomb complex, and histone modification. Remarkably, the bioinformatic analysis revealed that Parp1-PARylated proteins interacted significantly with Oct4, Nanog, c-Myc, Klf4, CTNNB1, WDR5, SUZ12, EZH2, DNMT3A/B, and JARID2 in the core network of nuclear reprogramming and pluripotent status (Fig. 6).
Figure 6.
A protein-interaction network in nuclear reprogramming and pluripotency maintenance by Parp1-interacting and Parp1-PARylated proteins using IPA analysis.
DISCUSSION
Nuclear reprogramming is the process of converting somatic cells to a pluripotent state and involves nuclear proteins (Jullien et al., 2010). However, the difference between the nuclear protein profiles of somatic and pluripotent stem cells throughout the reprogramming process has not been clearly defined. Using a proteomic approach, we compared the nuclear protein expression profiles among MEFs, ESCs, and iPSCs, and we identified Parp1 as a pivotal regulator of nuclear reprogramming and pluripotency. Recently, the deficiency of Parp1 was shown to result in reduced iPSC reprogramming efficiency and abnormal ESC gene expression (Lai et al., 2012). Our data demonstrated that the expression of Parp1 and PARylation was increased during reprogramming and decreased upon differentiation. Parp1 replaced Klf4 or c-Myc in promoting iPSC production and generating chimeric mice with Oct4/Sox2-transfected cells (Fig. 3). We further showed that c-Myc directly binds to the Parp1 promoter to enhance its expression, resulting in increased PARylation activity. The reduced reprogramming efficiency of MEFs transfected with OSK plus RNAi against c-Myc was rescued by ectopic Parp1 (Fig. 4). Finally, we demonstrated that Parp1 interacted with several DNA repair- and chromatin remodeling-associated proteins, which were highly expressed and PARylated in reprogrammed and pluripotent cells (Fig. 5). These data indicate that the activation of Parp1 and PARylation, partly through endogenous c-Myc, effectively promotes nuclear reprogramming and the maintenance of pluripotency.
The oncogene c-Myc has been implicated in the regulatory networks of ESCs and cancer cells (Kim et al., 2010). c-Myc can indirectly increase Parp1 activity through decreasing BIN1, a nucleocytoplasmic adaptor protein that binds Parp1 and suppresses its catalytic activity (Pyndiah et al., 2011). Carbone et al. (2008) also demonstrated that Parp1 and PARylation modulate the induction of c-Myc in serum-stimulated quiescent fibroblasts. However, whether Parp1 is also a regulator of c-Myc in pluripotent stem cells still remained undetermined. Our results indicated that forced expression of c-Myc alone, OSM, or OSKM significantly up-regulated Parp1 expression and PARylation activity. Notably, endogenous c-Myc can directly bind to the Parp1 promoter (Fig. 4), which is a predicted c-Myc binding element, and subsequently activate Parp1 protein expression. Knockdown of endogenous c-Myc blocked reprogramming and pluripotency, suppressed Parp1, inactivated PARylation, and promoted differentiation in iPSCs and ESCs (Fig. 4). Our data demonstrate a direct interaction between c-Myc and Parp1 and provide a new insight into the role of c-Myc in modulating Parp1 expression and downstream signaling. This finding supports the idea that endogenous c-Myc promotes reprogramming by its upstream regulation of Parp1 and subsequent PARylation.
PARylation of proteins by Parp1 is one of the earliest responses to chromatin remodeling, transcriptional regulation, and cell death, and it is required for genome stability (Haber, 2000; Amé et al., 2004; Caldecott, 2008; Nakagawa et al., 2008). Our immunoprecipitation results show that Parp1 interacted with DNA repair and chromatin-remodeling proteins, including Chd1L, DNA ligase III, SSrp1, Xrcc-6/Ku70, and Parp2 in pluripotent iPSCs and ESCs. Ssrp1 is a subunit of the FACT complex, which is regulated by Parp1 through PARylation, and this modification promotes chromatin remodeling (Huang et al., 2006). Notably, DNA ligase III, Xrcc1, Xrcc6, and Ssrp1 are involved in early embryonic development, and knockdown of these genes causes embryonic lethality (Tebbs et al., 1999; Puebla-Osorio et al., 2006; Fattah et al., 2008; Kumari et al., 2009). Chromodomain helicase DNA-binding protein 1-like (Chd1L) is a SNF2 family member whose macrodomain can be PARylated by Parp1, and this PARylation has been linked to DNA repair pathways (Ahel et al., 2009; Deng, 2009; Gaspar-Maia et al., 2009). Nevertheless, dysregulation of Chd1L leads to overextended chromatin, making the DNA accessible to mutagens and increasing the incidence of cancer (Deng, 2009). In addition, the fact that epithelial characteristics are required for efficient nuclear reprogramming (Li et al., 2010) and that Parp1 attenuates Smad-specific gene responses, including the TGF-β–induced epithelial-to-mesenchymal transition (Lönn et al., 2010), further suggests the involvement of Parp1 in promoting reprogramming.
Gao et al. (2009) first demonstrated that Parp1 is a novel cofactor of Oct4 and Sox2, as well as a regulator of FGF4 expression, and directly interacts with and PARylates Sox2 during ESC differentiation (Lai et al., 2012). Notably, Lai et al. (2012) demonstrated that the Sox2–Parp1 interaction regulated by Parp1-PARylation is required for ESC differentiation and that auto-PARylation of Parp1 can be activated by the FGF/ERK pathway. Our immunoprecipitation data showed that Parp1 interacts with Sox2 in iPSC induction (D11; unpublished data), and treatment with the PARylation inhibitor PJ34 suppressed the Sox2–Parp1 interaction during the reprogramming process. Consistent with the findings by Lai et al. (2012), we also found that knockdown of Parp1, Parp2, or pharmacological inhibition of PARylation significantly inhibited the reprogramming efficiency (Fig. 2). In addition, Doege, et al. (2012) recently reported that Parp1 and TeT2 serve essential roles in the regulation of epigenetic markers and the chromatin state at Nanog and Essrb during somatic cell reprogramming. Importantly, Parp1 induction further promotes Oct4 reprogramming factor binding to the pluripotency loci of Nanog and Essrb. Our results demonstrated that increased Parp1 and PARylation modulate c-Myc–regulated reprogramming. Moreover, Parp1 increased the Oct4 and Nanog expression levels in OSK-transfected reprogramming cells (Fig. 2 D). Bioinformatic analysis further indicated that Parp1 and Parp1-PARylated proteins interacted significantly with Oct4 and Nanog (Fig. 6). Given that increased Oct4 and Nanog expression are key factors regulating the efficiency of reprogramming (Miyamoto et al., 2011), these data suggested another mechanism in which Parp1 enhances the reprogramming process. Therefore, elucidating the fundamental mechanisms of Parp1-related epigenetic regulation involved in embryonic development, stem-like properties, and pluripotent programming is necessary for the validation of our results in the future.
In conclusion, Parp1 and PARylation, partly activated by endogenous c-Myc, may act as the major regulator in reprogramming and the maintenance of stem cell pluripotency. Further studies aimed at identifying the PARylation complex, Parp1-related posttranslational modifications, and Parp1’s cellular functions are critical for a better understanding of the core networks involved in nuclear reprogramming and iPSC research.
MATERIALS AND METHODS
Generation of iPSC lines and cell culture.
C57BL/6 mice were used in this study and all procedures involving animals were approved by the Animals Committee of the Taipei Veterans General Hospital. Mouse iPSCs were generated from MEFs derived from 13.5-d-old embryos of C57/B6 mice. The iPSCs were reprogrammed by the transduction of retroviral vectors encoding four transcription factors (Oct-4/Sox2/Klf4/c-Myc; OSKM), or three transcription factors (Oct-4/Sox2/Klf4; OSK), as described previously (Li et al., 2011). A total of 12 clones (Re-1 to Re-12; OSKM) were selected and established. The clone of Re-7 iPSCs had been stably passed to >100 passages with high pluripotency. Therefore, Re-7 iPSCs were selected and widely used in this study. In brief, undifferentiated iPSCs were routinely cultured and expanded on mitotically inactivated MEFs (50,000 cells/cm2) in 6-well culture plates (BD) in the presence of 0.3% leukemia inhibitory factor in an iPSC medium consisting of DMEM (Sigma-Aldrich) supplemented with 15% FBS (Invitrogen), 100 mM MEM nonessential amino acids (Sigma-Aldrich), 0.55 mM 2-mercaptoethanol (Gibco), and antibiotics (Invitrogen). Every 3–4 d, colonies were detached with 0.2% collagenase IV (Invitrogen), dissociated into single cells with 0.025% trypsin (Sigma-Aldrich) and 0.1% chicken serum (Invitrogen) in PBS, and replated onto MEFs. For EB formation, iPSCs were dissociated into a single cell suspension by 0.25% trypsin-EDTA and plated onto nonadherent culture dishes in DMEM with 15% FBS, 100 mM MEM nonessential amino acids, 0.55 mM 2-mercaptoethanol, and antibiotics at a density of 2 × 106 cells/100 mm plate. After 4 d in floating culture, EBs were transferred onto gelatin-coated plates and maintained in the same medium for 24 h. EBs were then assigned for in vitro differentiation into tridermal lineages as previously described (Li et al., 2011).
ALP activity, Alizarin red, and PAS staining.
For detecting the AP activity of cells on original plates, cells were fixed with 80% alcohol, and then fixed cells stained using the Blue Alkaline Phosphatase Substrate kit III (Vector Laboratories) according to the manufacturer’s instructions. Alizarin red staining and PAS staining was performed as previously described (Li et al., 2011).
1D gel electrophoresis (SDS-PAGE) and in-gel digestion.
Of every cell type, 107 cells of extracted nuclear proteins from each condition were denatured by boiling at 95°C for 10 min. 1D gel electrophoresis was as described previously (Liu et al., 2012) and performed with 10% SDS-PAGE. After the gel was stained using VisPRO 5 min protein stain kit (VP01-500; Visual Protein), every lane was cut off into 10 equal sections, followed by reduction with β-mercaptoethanol (1% vol/vol) in 25 mM ammonium bicarbonate at room temperature in the dark for 20 min, and alkylation with 5% vol/vol 4-vinylpyridine in 25 mM ammonium bicarbonate for 20 min. Digestion was then done with 0.1% vol/vol proteomics grade modified trypsin (Sigma-Aldrich) in 25 mM ammonium bicarbonate at 37°C overnight. Extracts of trypsin-digested peptides were dried in a SpeedVac concentrator (Jouan, RC1022; Thermo Fisher Scientific).
LC-MS/MS analysis.
LC-MS/MS analysis was performed through the application of LTQ Orbitrap (Thermo Fisher Scientific) as described. In brief, each sample of digested peptides was reconstituted to 20 µl of 0.1% formic acid. Peptides were first injected in and separated by the nanoflow HPLC (Agilent 1100; Agilent Technologies) with a C18 column (75 µm ID × 360 µm OD × 15 cm; Agilent Technologies), and became ionized particles once passed through the succeeding nanospray tip (New Objective). In operating HPLC, the flow rate was at 0.4 µl/min after a splitter. LC gradient for the LC-MS/MS system ramped from 2–40% ACN in 120 min, and the system was performed under the setting of automated data-dependent acquisition, with a mode of 200–2000 m/z full scan for the maximum three most intense peaks from each Orbitrap MS scan. Peptides with +2 or +3 charge state were further subjected to CID. Spectra were obtained in raw data files with Xcalibur (version 2.0 SR2). Protein identification was accomplished via TurboSEQUEST (Thermo Fisher Scientific) using the UniProt database. A protein was confirmed once three peptides with Xcorr >2.5 were matched in sequencing.
PARylated protein purification.
PARylated proteins contain poly-ADP-ribose which has a high affinity with macrodomains. The PAR affinity resin set (Tulip)–conjugated Af1521 macrodomains were used to pull down the PARylated proteins. Protein were extracted from cells by lysis buffer (e.g., 50 mM Tris, pH 8, 200 mM NaCl, 1 mM EDTA, 1% Triton X-100, 10% glycerol, 1 mM DTT, 0.5% deoxycholate, and protease inhibitors) and incubated with resin overnight in 4°C. After incubation, resin was washed with lysis buffer three times and 1× SDS-sample buffer was added at 95°C for 10 min to dissociated proteins.
GO, pathway, and network analysis.
The filtered genes and proteins were subjected to GO enrichment analysis using the AltAnalyze bundled module GO-Elite (GenMAPP). GO-Elite implements an overrepresentation statistical inference that can identify significantly enriched GO categories with nuclear proteins. GO terms with a z-score >2, a permutation of P < 0.01, and three or more regulated proteins for each GO term were reported as significant. Canonical pathway and gene interaction network analyses were conducted using the Ingenuity Pathway Analysis (IPA) web tool. IPA constructs hypothetical protein interaction clusters on the foundation of a regularly updated Ingenuity Pathways Knowledge Base, which is a database that consists of millions of individual relationships between proteins collected from the public literature. Every gene interaction is supported by evidence extracted from the underlying publications, structured using an ontology, and stored in the database. To further explore the potential relationship between the target-PARylated proteins (listed in Table 1) and stemness genes or other factors in regulating cellular reprogramming, the bioinformatics analysis of the IPA web tool was used. First, we uploaded a dataset (containing the PARylated proteins identified in the proteomic experiment, Table 1) into the IPA web. Then we chose the “new core analysis” option to generate protein–protein interaction networks. We further used “experimentally observed” to limit the confidence, set the reference as “ingenuity knowledge base (gene only),” and output 10 networks (70 molecules per network). Hypothetical networks in Fig. 6 were generated from these target-PARylated proteins, ESC-related proteins, and other candidate proteins from the database by IPA analysis. The solid line is representative of the direct relationship of protein-to-protein interaction supported by the literature database, whereas the dashed line is representative of the indirect interaction supported by the literature database.
Quantitative PCR and RT-PCR for marker genes.
Reverse transcription reactions were performed using SuperScript III reverse transcription (Invitrogen). cDNA was used in the following quantitative PCR (qPCR) and RT-PCR. qPCR was performed with Power SYBR Green PCR Master Mix (Applied Biosystems) according to manufacturer’s instructions. Signals were detected with 7900HT Fast Real-Time PCR system (Applied Biosystems). Primer sequences are listed in Table S2.
shLuc and shParp1 and shParp2 expression construct and lentiviral transduction.
The stable ablation of Parp1 and Parp2 in MEFs was obtained using shRNA probes for the mouse gene Parp1 and Parp2 in Table S3. Control cells stably expressed shLuc (pLKO.1-shLuc). Cells were infected with shRNA lentiviral vector generated using a three-plasmid-based lentiviral system (all plasmids are available from the RNAi Consortium [TRC]). Lentivirus production was performed by transfection of 293T cells at 5 × 106 cells/10 cm plate using Lipofectamine 2000 (LF2000; Invitrogen). Supernatants were collected 48 h after transfection and then were filtered. Subconfluent cells were infected with lentiviral vector in the presence of 8 mg/ml polybrene (Sigma-Aldrich). Infected cells were selected with 2 mg/ml puromycin until control uninfected cells were completely dead. Immunoblotting was used to confirm the knockdown efficiency of shParp1 and shParp2 (Chen et al., 2011).
Luciferase activity assay.
STO cells were grown in 24-well tissue culture dishes to 70% confluence and then cotransfected with 0.2 µg pMXs and pMXs-c-Myc in the presence of 0.2 µg pGL3-PARP promoter firefly luciferase or PARP promoter mutants and 10 ng SV40 Renilla luciferase plasmids (Promega). 24 h after transfection, cells were harvested in 100 µl of reporter lysis buffer and then subjected to a dual luciferase assay according to the manufacturer’s protocol (Dual-Luciferase Reporter Assay System; Promega). Firefly luciferase activity was normalized to Renilla luciferase activity, and data are represented as the mean and standard deviation of three independent experiments, each performed in triplicate.
ChIP and site-directed mutagenesis of mouse PARP promoter mutants.
The study protocol of ChIP was according to the manufacturer’s instructions (EZ ChIP kit; EMD Millipore) using anti–c-Myc (Santa Cruz Biotechnology, Inc.) antibodies. The mouse PARP promoter with deletion or point mutations clones were created by site-directed mutagenesis according to the manufacturer’s instructions (Phusion Site-Directed Mutagenesis kit; Finnzymes), and all mutants were amplified using PARP (−2,000) as the template. −2,000 fragment constructs were amplified with the primer pairs as indicated in Table S3. Amplified fragments were further amplified with suitable forward primers with MluI cutting sequences and HIndIII cutting sequences in the reverse primers. Then, the restriction cutting site–added PCR products were subcloned in pGL3 luciferase reporter plasmids. DNA eluted from precipitated complexes for ChIP assays was amplified the fragment of PARP promoter using the primers shown in Table S3. As a positive control for ChIP study, c-Myc bound to its reported target, cyclin D2 promoter (Bouchard et al., 1999).
Western blot analysis and immunofluorescence staining.
Western blotting was performed as previously described (Li et al., 2011). The primary antibodies are listed in Table S4. For immunostaining, cells were cultured on coverslips, fixed with 4% paraformaldehyde, and permeabilized with 0.5% Triton X-100. Cells were stained with monoclonal anti-SSEA1 antibody (Abcam) and anti-Oct4 monoclonal antibody (Cell Signaling Technology), and then incubated with fluorophore-labeled secondary antibodies (Jackson ImmunoResearch Laboratories), Hoechst, and DAPI (Sigma-Aldrich) before visualized under a microscope (Olympus).
Chimera mouse production by blastocyst injection.
The introduction of mouse iPS cells (derived from C57BL/6J strain, black coat color) into mouse blastocysts-derived from C57BL/6J-Tyrc2J strain (albino) was followed as previously described with some modifications (Sung et al., 2006). The adult chimeras were confirmed by coat color, demonstrating that iPSCs were competent to produce adult chimeric mice. This study was assisted by the Transgenic Mouse Model Core Facility, Academic Sinica, Taiwan.
Statistical analyses.
Results are reported as mean ± SD. Statistical analysis was performed using a Student’s t test or a one-way or two-way ANOVA, followed by Turkey’s test, as appropriate. The survival rate analysis was performed using a log-rank test. Results were considered statistically significant at P < 0.05.
Online supplemental material.
Fig. S1 shows the establishment of the differential profiling of nuclear proteins from pluripotent stem cells and MEFs by MS-based proteomics. Fig. S2 is the list of the 112 most up-regulated nuclear proteins in iPSCs compared with MEFs using a statistical analysis between the databases of 1D LC-MS/MS and 2D-DIGE. Table S1 shows the differential expression profiles of the nuclear extracts using 1D LC-MS/MS. Compared with MEFs, the predominant candidates of nuclear protein profiles up-regulated in iPSCs (Re-7) are listed. Table S2 shows the sequences of the primers for quantitative RT-PCR and RT-PCR. Table S3 shows the sequences of ShRNA and the primers for Parp1 promoter constructs. Table S4 is the list of antibody information. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20121044/DC1.
Supplementary Material
Acknowledgments
This study was assisted in part by the Division of Experimental Surgery of the Department of Surgery and the Animal Center of Taipei Veterans General Hospital. We thank Dr. Cheng-Chung Liao (Proteomic Research Center, National Yang-Ming University) for his outstanding proteomic analysis, Dr. Yu-Tai Wang and Dr. Ueng-Cheng Yang (Center for Systems and Synthetic Biology, National Yang-Ming University) for bioinformatics analysis, and Dr. Pei-Chan Hsieh and Dr. Hsin-Yang Li (School of Medicine, National Yang-Ming University) and Li-Ing Sung (Institute of Biotechnology, National Taiwan University) for their valuable assistance in experimental technology.
This study was funded by the National Science Council (NSC100/101-2120-M-002-011, 101-2325-B-010-009, 100/101-2321-B-010-020, 101-2628-B-010-002, 99-2632-B-039-001-MY3, 100-2321-B-039-004), Taipei Veterans General Hospital (Stem Cell Project E99-101), Yen-Tjing-Ling Medical Foundation (CI-99/100), The Department of Health Cancer Center Research of Excellence (DOH101-TD-C-111-007), National Health Research Institutes (NHRI-EX102-10258SI), and The Genomic Center Project and Cancer Center Project of National Yang-Ming University (Ministry of Education, Aim for the Top University Plan), Taiwan.
The authors declare that they have no competing financial interests.
Footnotes
Abbreviations used:
- ALP
- alkaline phosphatase
- ChIP
- chromatin immunoprecipitation
- EB
- embryoid body
- ESC
- embryonic stem cell
- GO
- gene ontology
- iPSC
- induced pluripotent stem cell
- LC-MS/MS
- liquid chromatography–tandem MS
- MS
- mass spectrometry
- Parp1
- Poly(ADP-ribose) polymerase 1
- PARylation
- poly(ADP-ribosylation)
References
- Ahel D., Horejsí Z., Wiechens N., Polo S.E., Garcia-Wilson E., Ahel I., Flynn H., Skehel M., West S.C., Jackson S.P., et al. 2009. Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science. 325:1240–1243 10.1126/science.1177321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amé J.C., Spenlehauer C., de Murcia G. 2004. The PARP superfamily. Bioessays. 26:882–893 10.1002/bies.20085 [DOI] [PubMed] [Google Scholar]
- Armstrong L., Tilgner K., Saretzki G., Atkinson S.P., Stojkovic M., Moreno R., Przyborski S., Lako M. 2010. Human induced pluripotent stem cell lines show stress defense mechanisms and mitochondrial regulation similar to those of human embryonic stem cells. Stem Cells. 28:661–673 10.1002/stem.307 [DOI] [PubMed] [Google Scholar]
- Bouchard C., Thieke K., Maier A., Saffrich R., Hanley-Hyde J., Ansorge W., Reed S., Sicinski P., Bartek J., Eilers M. 1999. Direct induction of cyclin D2 by Myc contributes to cell cycle progression and sequestration of p27. EMBO J. 18:5321–5333 10.1093/emboj/18.19.5321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caiafa P., Guastafierro T., Zampieri M. 2009. Epigenetics: poly(ADP-ribosyl)ation of PARP-1 regulates genomic methylation patterns. FASEB J. 23:672–678 10.1096/fj.08-123265 [DOI] [PubMed] [Google Scholar]
- Caldecott K.W. 2008. Single-strand break repair and genetic disease. Nat. Rev. Genet. 9:619–631 [DOI] [PubMed] [Google Scholar]
- Carbone M., Rossi M.N., Cavaldesi M., Notari A., Amati P., Maione R. 2008. Poly(ADP-ribosyl)ation is implicated in the G0-G1 transition of resting cells. Oncogene. 27:6083–6092 10.1038/onc.2008.221 [DOI] [PubMed] [Google Scholar]
- Chen L.H., Loong C.C., Su T.L., Lee Y.J., Chu P.M., Tsai M.L., Tsai P.H., Tu P.H., Chi C.W., Lee H.C., Chiou S.H. 2011. Autophagy inhibition enhances apoptosis triggered by BO-1051, an N-mustard derivative, and involves the ATM signaling pathway. Biochem. Pharmacol. 81:594–605 10.1016/j.bcp.2010.12.011 [DOI] [PubMed] [Google Scholar]
- Deng W. 2009. PARylation: strengthening the connection between cancer and pluripotency. Cell Stem Cell. 5:349–350 10.1016/j.stem.2009.09.002 [DOI] [PubMed] [Google Scholar]
- Doege C.A., Inoue K., Yamashita T., Rhee D.B., Travis S., Fujita R., Guarnieri P., Bhagat G., Vanti W.B., Shih A., et al. 2012. Early-stage epigenetic modification during somatic cell reprogramming by Parp1 and Tet2. Nature. 488:652–655 10.1038/nature11333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J., Robert C., Jang Y.Y., Liu H., Sharkis S., Baylin S.B., Rassool F.V. 2011. Human induced pluripotent cells resemble embryonic stem cells demonstrating enhanced levels of DNA repair and efficacy of nonhomologous end-joining. Mutat. Res. 713:8–17 10.1016/j.mrfmmm.2011.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fattah F.J., Lichter N.F., Fattah K.R., Oh S., Hendrickson E.A. 2008. Ku70, an essential gene, modulates the frequency of rAAV-mediated gene targeting in human somatic cells. Proc. Natl. Acad. Sci. USA. 105:8703–8708 10.1073/pnas.0712060105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii-Yamamoto H., Kim J.M., Arai K., Masai H. 2005. Cell cycle and developmental regulations of replication factors in mouse embryonic stem cells. J. Biol. Chem. 280:12976–12987 10.1074/jbc.M412224200 [DOI] [PubMed] [Google Scholar]
- Gao F., Kwon S.W., Zhao Y., Jin Y. 2009. PARP1 poly(ADP-ribosyl)ates Sox2 to control Sox2 protein levels and FGF4 expression during embryonic stem cell differentiation. J. Biol. Chem. 284:22263–22273 10.1074/jbc.M109.033118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar-Maia A., Alajem A., Polesso F., Sridharan R., Mason M.J., Heidersbach A., Ramalho-Santos J., McManus M.T., Plath K., Meshorer E., Ramalho-Santos M. 2009. Chd1 regulates open chromatin and pluripotency of embryonic stem cells. Nature. 460:863–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber J.E. 2000. Partners and pathwaysrepairing a double-strand break. Trends Genet. 16:259–264 10.1016/S0168-9525(00)02022-9 [DOI] [PubMed] [Google Scholar]
- Huang J.Y., Chen W.H., Chang Y.L., Wang H.T., Chuang W.T., Lee S.C. 2006. Modulation of nucleosome-binding activity of FACT by poly(ADP-ribosyl)ation. Nucleic Acids Res. 34:2398–2407 10.1093/nar/gkl241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagtap P., Szabó C. 2005. Poly(ADP-ribose) polymerase and the therapeutic effects of its inhibitors. Nat. Rev. Drug Discov. 4:421–440 10.1038/nrd1718 [DOI] [PubMed] [Google Scholar]
- Jin J., Kwon Y.W., Paek J.S., Cho H.J., Yu J., Lee J.Y., Chu I.S., Park I.H., Park Y.B., Kim H.S., Kim Y. 2011. Analysis of differential proteomes of induced pluripotent stem cells by protein-based reprogramming of fibroblasts. J. Proteome Res. 10:977–989 10.1021/pr100624f [DOI] [PubMed] [Google Scholar]
- Jullien J., Astrand C., Halley-Stott R.P., Garrett N., Gurdon J.B. 2010. Characterization of somatic cell nuclear reprogramming by oocytes in which a linker histone is required for pluripotency gene reactivation. Proc. Natl. Acad. Sci. USA. 107:5483–5488 10.1073/pnas.1000599107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jullien J., Pasque V., Halley-Stott R.P., Miyamoto K., Gurdon J.B. 2011. Mechanisms of nuclear reprogramming by eggs and oocytes: a deterministic process? Nat. Rev. Mol. Cell Biol. 12:453–459 10.1038/nrm3140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Woo A.J., Chu J., Snow J.W., Fujiwara Y., Kim C.G., Cantor A.B., Orkin S.H. 2010. A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell. 143:313–324 10.1016/j.cell.2010.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus W.L. 2008. Transcriptional control by PARP-1: chromatin modulation, enhancer-binding, coregulation, and insulation. Curr. Opin. Cell Biol. 20:294–302 10.1016/j.ceb.2008.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnakumar R., Kraus W.L. 2010. The PARP side of the nucleus: molecular actions, physiological outcomes, and clinical targets. Mol. Cell. 39:8–24 10.1016/j.molcel.2010.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari A., Mazina O.M., Shinde U., Mazin A.V., Lu H. 2009. A role for SSRP1 in recombination-mediated DNA damage response. J. Cell. Biochem. 108:508–518 10.1002/jcb.22280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Y.S., Chang C.W., Pawlik K.M., Zhou D., Renfrow M.B., Townes T.M. 2012. SRY (sex determining region Y)-box2 (Sox2)/poly ADP-ribose polymerase 1 (Parp1) complexes regulate pluripotency. Proc. Natl. Acad. Sci. USA. 109:3772–3777 10.1073/pnas.1108595109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H.Y., Chien Y., Chen Y.J., Chen S.F., Chang Y.L., Chiang C.H., Jeng S.Y., Chang C.M., Wang M.L., Chen L.K., et al. 2011. Reprogramming induced pluripotent stem cells in the absence of c-Myc for differentiation into hepatocyte-like cells. Biomaterials. 32:5994–6005 [DOI] [PubMed] [Google Scholar]
- Li R., Liang J., Ni S., Zhou T., Qing X., Li H., He W., Chen J., Li F., Zhuang Q., et al. 2010. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell. 7:51–63 10.1016/j.stem.2010.04.014 [DOI] [PubMed] [Google Scholar]
- Liu M., Lee D.F., Chen C.T., Yen C.J., Li L.Y., Lee H.J., Chang C.J., Chang W.C., Hsu J.M., Kuo H.P., et al. 2012. IKKα activation of NOTCH links tumorigenesis via FOXA2 suppression. Mol. Cell. 45:171–184 10.1016/j.molcel.2011.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lönn P., van der Heide L.P., Dahl M., Hellman U., Heldin C.H., Moustakas A. 2010. PARP-1 attenuates Smad-mediated transcription. Mol. Cell. 40:521–532 10.1016/j.molcel.2010.10.029 [DOI] [PubMed] [Google Scholar]
- Maherali N., Sridharan R., Xie W., Utikal J., Eminli S., Arnold K., Stadtfeld M., Yachechko R., Tchieu J., Jaenisch R., et al. 2007. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 1:55–70 10.1016/j.stem.2007.05.014 [DOI] [PubMed] [Google Scholar]
- Miyamoto K., Pasque V., Jullien J., Gurdon J.B. 2011. Nuclear actin polymerization is required for transcriptional reprogramming of Oct4 by oocytes. Genes Dev. 25:946–958 10.1101/gad.615211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz J., Low T.Y., Kok Y.J., Chin A., Frese C.K., Ding V., Choo A., Heck A.J. 2011. The quantitative proteomes of human-induced pluripotent stem cells and embryonic stem cells. Mol. Syst. Biol. 7:550 10.1038/msb.2011.84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa M., Koyanagi M., Tanabe K., Takahashi K., Ichisaka T., Aoi T., Okita K., Mochiduki Y., Takizawa N., Yamanaka S. 2008. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat. Biotechnol. 26:101–106 10.1038/nbt1374 [DOI] [PubMed] [Google Scholar]
- Nakagawa M., Takizawa N., Narita M., Ichisaka T., Yamanaka S. 2010. Promotion of direct reprogramming by transformation-deficient Myc. Proc. Natl. Acad. Sci. USA. 107:14152–14157 10.1073/pnas.1009374107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp B., Plath K. 2011. Reprogramming to pluripotency: stepwise resetting of the epigenetic landscape. Cell Res. 21:486–501 10.1038/cr.2011.28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phanstiel D.H., Brumbaugh J., Wenger C.D., Tian S., Probasco M.D., Bailey D.J., Swaney D.L., Tervo M.A., Bolin J.M., Ruotti V., et al. 2011. Proteomic and phosphoproteomic comparison of human ES and iPS cells. Nat. Methods. 8:821–827 10.1038/nmeth.1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puebla-Osorio N., Lacey D.B., Alt F.W., Zhu C. 2006. Early embryonic lethality due to targeted inactivation of DNA ligase III. Mol. Cell. Biol. 26:3935–3941 10.1128/MCB.26.10.3935-3941.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyndiah S., Tanida S., Ahmed K.M., Cassimere E.K., Choe C., Sakamuro D. 2011. c-MYC suppresses BIN1 to release poly(ADP-ribose) polymerase 1: a mechanism by which cancer cells acquire cisplatin resistance. Sci. Signal. 4:ra19 10.1126/scisignal.2001556 [DOI] [PubMed] [Google Scholar]
- Rigbolt K.T., Prokhorova T.A., Akimov V., Henningsen J., Johansen P.T., Kratchmarova I., Kassem M., Mann M., Olsen J.V., Blagoev B. 2011. System-wide temporal characterization of the proteome and phosphoproteome of human embryonic stem cell differentiation. Sci. Signal. 4:rs3 10.1126/scisignal.2001570 [DOI] [PubMed] [Google Scholar]
- Saretzki G., Armstrong L., Leake A., Lako M., von Zglinicki T. 2004. Stress defense in murine embryonic stem cells is superior to that of various differentiated murine cells. Stem Cells. 22:962–971 10.1634/stemcells.22-6-962 [DOI] [PubMed] [Google Scholar]
- Sung L.Y., Gao S., Shen H., Yu H., Song Y., Smith S.L., Chang C.C., Inoue K., Kuo L., Lian J., et al. 2006. Differentiated cells are more efficient than adult stem cells for cloning by somatic cell nuclear transfer. Nat. Genet. 38:1323–1328 10.1038/ng1895 [DOI] [PubMed] [Google Scholar]
- Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. 2007. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 131:861–872 10.1016/j.cell.2007.11.019 [DOI] [PubMed] [Google Scholar]
- Tebbs R.S., Flannery M.L., Meneses J.J., Hartmann A., Tucker J.D., Thompson L.H., Cleaver J.E., Pedersen R.A. 1999. Requirement for the Xrcc1 DNA base excision repair gene during early mouse development. Dev. Biol. 208:513–529 10.1006/dbio.1999.9232 [DOI] [PubMed] [Google Scholar]
- Van Hoof D., Muñoz J., Braam S.R., Pinkse M.W., Linding R., Heck A.J., Mummery C.L., Krijgsveld J. 2009. Phosphorylation dynamics during early differentiation of human embryonic stem cells. Cell Stem Cell. 5:214–226 10.1016/j.stem.2009.05.021 [DOI] [PubMed] [Google Scholar]
- Wang S., Kou Z., Jing Z., Zhang Y., Guo X., Dong M., Wilmut I., Gao S. 2010. Proteome of mouse oocytes at different developmental stages. Proc. Natl. Acad. Sci. USA. 107:17639–17644 10.1073/pnas.1013185107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka S., Blau H.M. 2010. Nuclear reprogramming to a pluripotent state by three approaches. Nature. 465:704–712 10.1038/nature09229 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.