Abstract
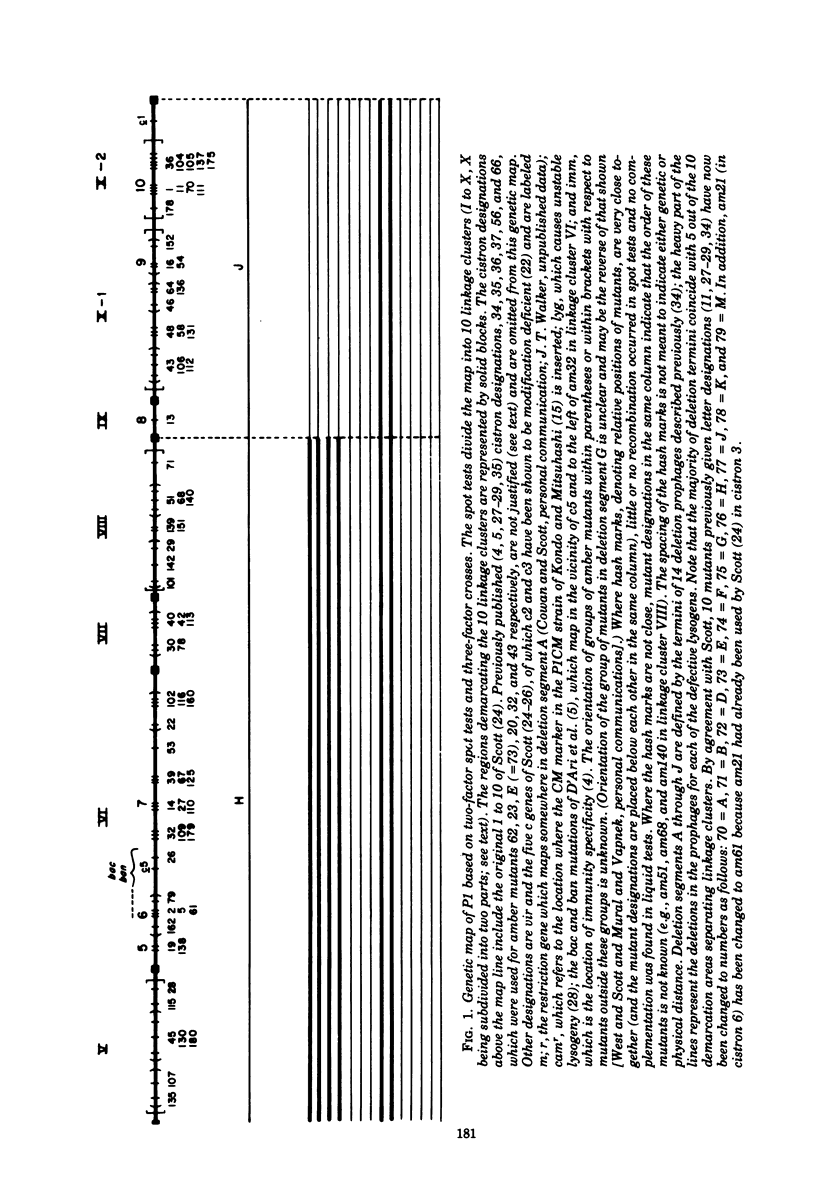
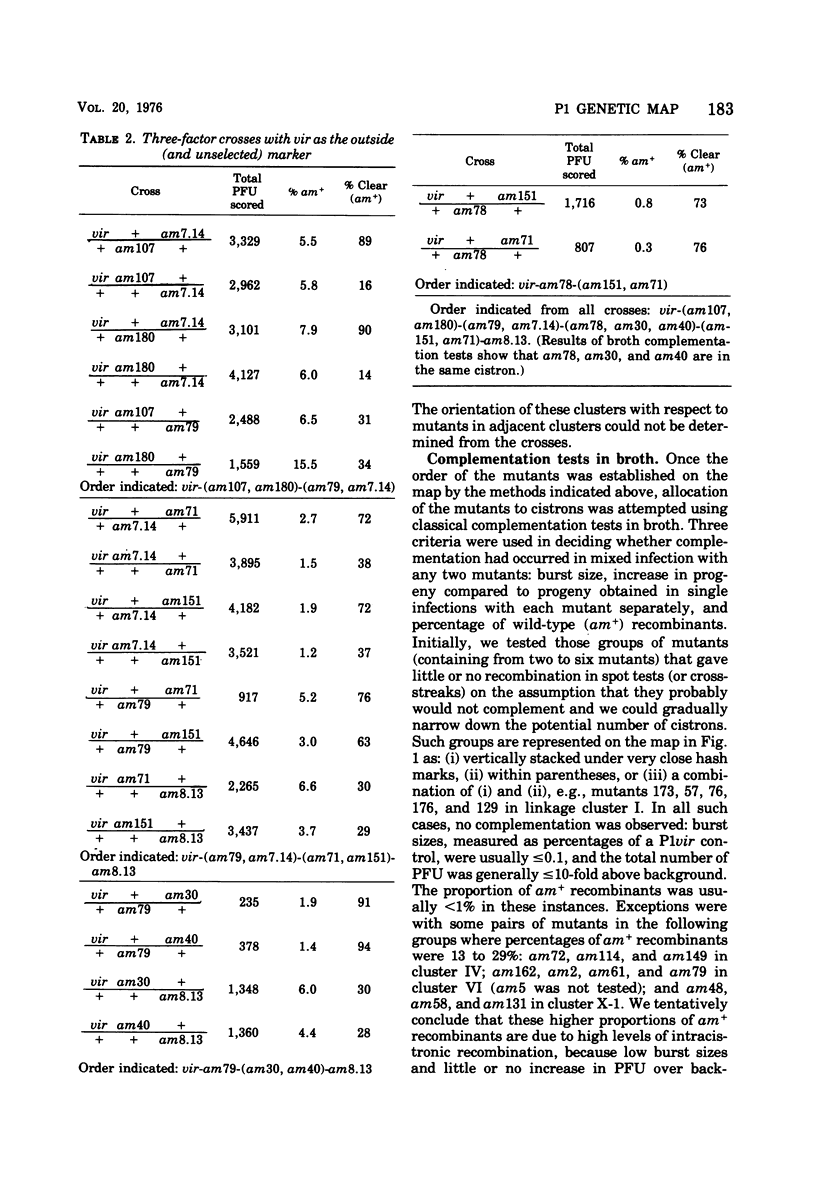
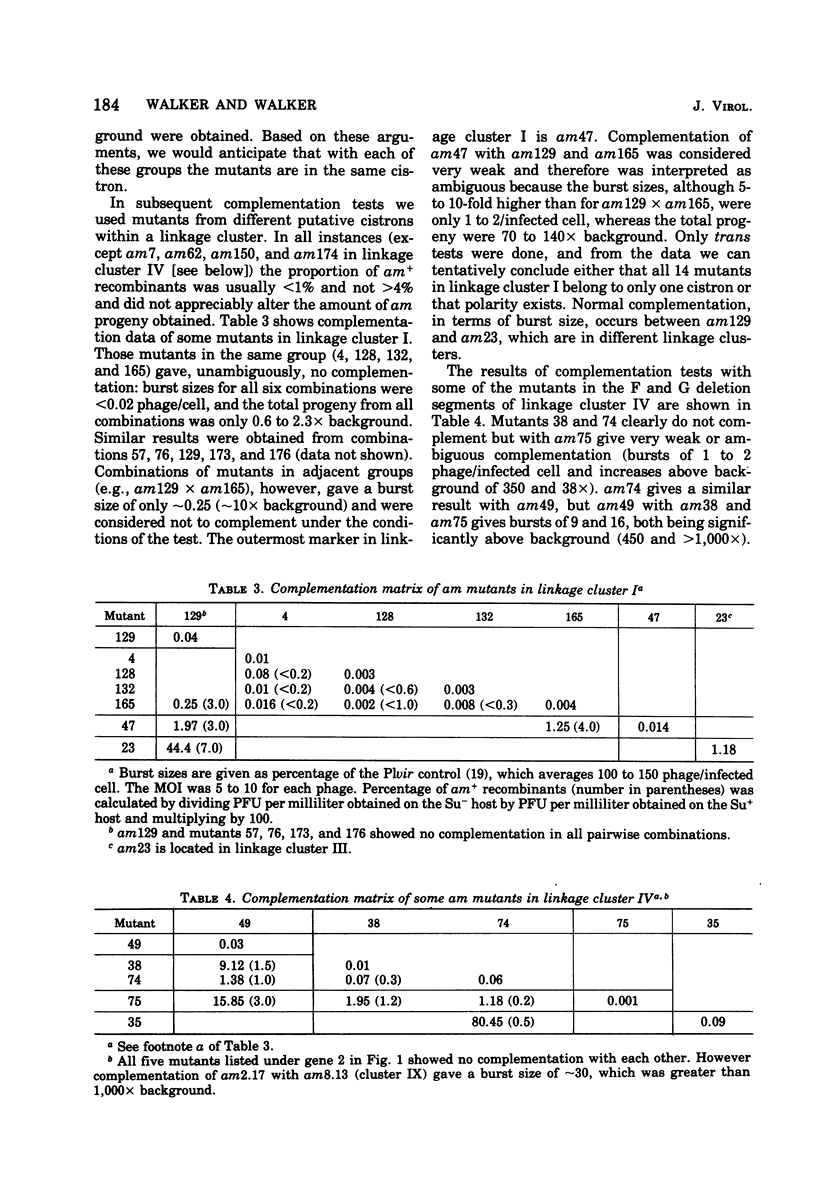
An extensive genetic map of coliphage P1 has been constructed for 113 amber mutants, using primarily a modification of the conventional complementation spot test. These spot tests failed to classify the mutants into cistrons, but when they were quantitated they permitted assignment of the mutants into 10 linkage clusters. Furthermore, a linear order could be deduced for most of the mutants within each cluster. This strongly suggested that recombination was the predominant event generating plaques and that, for the practical purpose of rapid genetic mapping, such spot tests could be considered as a series of two-factor crosses. Six of the 10 linkage clusters correlated with the P1 genetic map established by Scott (1968). The locations of the remaining four clusters were determined by three-factor crosses and by prophage deletion mapping. The nonrandom occurrence of termini for 14 deletion prophages, which we established previously (Walker and Walker, 1975), and the coincidence of these termini with five out of ten regions demarcating the linkage clusters are discussed. Complementation tests in liquid frequently gave ambiguous results. Therefore, cistron designations were not assigned.
Full text
PDF







Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bertani L. E., Bertani G. Genetics of P2 and related phages. Adv Genet. 1971;16:199–237. doi: 10.1016/s0065-2660(08)60359-4. [DOI] [PubMed] [Google Scholar]
- Botstein D., Chan R. K., Waddell C. H. Genetics of bacteriophage P22. II. Gene order and gene function. Virology. 1972 Jul;49(1):268–282. doi: 10.1016/s0042-6822(72)80028-x. [DOI] [PubMed] [Google Scholar]
- CAMPBELL A. Sensitive mutants of bacteriophage lambda. Virology. 1961 May;14:22–32. doi: 10.1016/0042-6822(61)90128-3. [DOI] [PubMed] [Google Scholar]
- Chesney R. H., Scott J. R. Superinfection immunity and prophage repression in phage P1. II. Mapping of the immunity-difference and ampicillin-resistance loci of P1 and phi amp. Virology. 1975 Oct;67(2):375–384. doi: 10.1016/0042-6822(75)90439-0. [DOI] [PubMed] [Google Scholar]
- D'Ari R., Jaffé-Brachet A., Touati-Schwartz D., Yarmolinsky M. B. A dnaB analog specified by bacteriophage P1. J Mol Biol. 1975 May 25;94(3):341–366. doi: 10.1016/0022-2836(75)90207-7. [DOI] [PubMed] [Google Scholar]
- EDGAR R. S., DENHARDT G. H., EPSTEIN R. H. A COMPARATIVE GENETIC STUDY OF CONDITIONAL LETHAL MUTATIONS OF BACTERIOPHAGE T4D. Genetics. 1964 Apr;49:635–648. doi: 10.1093/genetics/49.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figurski D. H., Christensen J. R. Functional characterization of the genes of bacteriophage T1. Virology. 1974 Jun;59(2):397–407. doi: 10.1016/0042-6822(74)90453-x. [DOI] [PubMed] [Google Scholar]
- Henderson D., Weil J. Recombination-deficient deletions in bacteriophage lambda and their interaction with chi mutations. Genetics. 1975 Feb;79(2):143–174. doi: 10.1093/genetics/79.2.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson H. E., McCorquodale D. J. Genetic and physiological studies of bacteriophage t5 I. An expanded genetic map of t5. J Virol. 1971 May;7(5):612–618. doi: 10.1128/jvi.7.5.612-618.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertman I., Scott J. R. Recombination of phage P1 in recombination deficient hosts. Virology. 1973 Jun;53(2):468–470. doi: 10.1016/0042-6822(73)90227-4. [DOI] [PubMed] [Google Scholar]
- Horii Z., Clark A. J. Genetic analysis of the recF pathway to genetic recombination in Escherichia coli K12: isolation and characterization of mutants. J Mol Biol. 1973 Oct 25;80(2):327–344. doi: 10.1016/0022-2836(73)90176-9. [DOI] [PubMed] [Google Scholar]
- Howe M. M., Bade E. G. Molecular biology of bacteriophage mu. Science. 1975 Nov 14;190(4215):624–632. doi: 10.1126/science.1103291. [DOI] [PubMed] [Google Scholar]
- Ikeda H., Tomizawa J. Prophage P1, and extrachromosomal replication unit. Cold Spring Harb Symp Quant Biol. 1968;33:791–798. doi: 10.1101/sqb.1968.033.01.091. [DOI] [PubMed] [Google Scholar]
- KONDO E., MITSUHASHI S. DRUG RESISTANCE OF ENTERIC BACTERIA. IV. ACTIVE TRANSDUCING BACTERIOPHAGE P1 CM PRODUCED BY THE COMBINATION OF R FACTOR WITH BACTERIOPHAGE P1. J Bacteriol. 1964 Nov;88:1266–1276. doi: 10.1128/jb.88.5.1266-1276.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam S. T., Stahl M. M., McMilin K. D., Stahl F. W. Rec-mediated recombinational hot spot activity in bacteriophage lambda. II. A mutation which causes hot spot activity. Genetics. 1974 Jul;77(3):425–433. doi: 10.1093/genetics/77.3.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine M. Replication and lysogeny with phage P22 in Salmonella typhimurium. Curr Top Microbiol Immunol. 1972;58:135–156. doi: 10.1007/978-3-642-65357-5_4. [DOI] [PubMed] [Google Scholar]
- Lindahl G. On the control of transcription in bacteriophage P2. Virology. 1971 Dec;46(3):620–633. doi: 10.1016/0042-6822(71)90065-1. [DOI] [PubMed] [Google Scholar]
- Morse D. E., Primakoff P. Relief of polarity in E. coli by "suA". Nature. 1970 Apr 4;226(5240):28–31. doi: 10.1038/226028a0. [DOI] [PubMed] [Google Scholar]
- Rosner J. L. Modification-deficient mutants of bacteriophage P1. I. Restriction by P1 cryptic lysogens. Virology. 1973 Mar;52(1):213–222. doi: 10.1016/0042-6822(73)90410-8. [DOI] [PubMed] [Google Scholar]
- Rosner J. L. Specialized transduction of pro genes by coliphage P1: structure of a partly diploid P1-pro prophage. Virology. 1975 Sep;67(1):42–55. doi: 10.1016/0042-6822(75)90401-8. [DOI] [PubMed] [Google Scholar]
- Scott J. R. A new gene controlling lysogeny in phage P1. Virology. 1972 Apr;48(1):282–283. doi: 10.1016/0042-6822(72)90139-0. [DOI] [PubMed] [Google Scholar]
- Scott J. R. A turbid plaque-forming mutant of phage P1 that cannot lysogenize Escherichia coli. Virology. 1974 Dec;62(2):344–349. doi: 10.1016/0042-6822(74)90397-3. [DOI] [PubMed] [Google Scholar]
- Scott J. R. Clear plaque mutants of phage P1. Virology. 1970 May;41(1):66–71. doi: 10.1016/0042-6822(70)90054-1. [DOI] [PubMed] [Google Scholar]
- Scott J. R. Genetic studies on bacteriophage P1. Virology. 1968 Dec;36(4):564–574. doi: 10.1016/0042-6822(68)90188-8. [DOI] [PubMed] [Google Scholar]
- Scott J. R. Phage Pl cryptic. II. Location and regulation of prophage genes. Virology. 1973 Jun;53(2):327–336. doi: 10.1016/0042-6822(73)90210-9. [DOI] [PubMed] [Google Scholar]
- Scott J. R. Superinfection immunity and prophage repression in phage P1. Virology. 1975 May;65(1):173–178. doi: 10.1016/0042-6822(75)90017-3. [DOI] [PubMed] [Google Scholar]
- Stahl F. W., Murray N. E., Nakata A., Crasemann J. M. Intergenic cis-trans position effects in bacteriophage T4. Genetics. 1966 Jul;54(1):223–232. doi: 10.1093/genetics/54.1.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streisinger G., Emrich J., Stahl M. M. Chromosome structure in phage t4, iii. Terminal redundancy and length determination. Proc Natl Acad Sci U S A. 1967 Feb;57(2):292–295. doi: 10.1073/pnas.57.2.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tye B. K., Huberman J. A., Botstein D. Non-random circular permutation of phage P22 DNA. J Mol Biol. 1974 Jan 5;85(4):501–528. doi: 10.1016/0022-2836(74)90312-x. [DOI] [PubMed] [Google Scholar]
- Walker D. H., Jr, Anderson T. F. Morphological variants of coliphage P1. J Virol. 1970 Jun;5(6):765–782. doi: 10.1128/jvi.5.6.765-782.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker D. H., Jr, Walker J. T. Genetic studies of coliphage P1. I. Mapping by use of prophage deletions. J Virol. 1975 Sep;16(3):525–534. doi: 10.1128/jvi.16.3.525-534.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker D. H., Jr, Walker J. T. Genetic studies of coliphage P1. II. Relatedness to P7. J Virol. 1976 Jul;19(1):271–274. doi: 10.1128/jvi.19.1.271-274.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]



