Abstract
The SWI/SNF-like chromatin remodeler ATRX has recently garnered renewed attention. ATRX mutations were first identified in patients bearing the syndrome after which it is named, alpha thalassemia/mental retardation, X-linked. While ATRX has long been implicated in transcriptional regulation through multiple mechanisms, recent studies have identified a role for ATRX in the regulation of histone variant deposition. In addition, current reports describe ATRX to be mutated at high percentages in multiple tumor types, suggestive of a potential ‘driver’ role in cancer. Here we discuss the numerous and seemingly diverse roles for ATRX in transcriptional regulation and histone deposition and suggest that ATRX’s effects are mediated by its regulation of histones within the chromatin template.
Keywords: ATRX, histone variants, macroH2A and H3.3, telomeres, α-globin
ATRX and its syndrome
A study of patients presenting mental retardation, developmental delay and distinctive facial features, associated with α-thalassemia, led to the mapping of mutations in a helicase encoded on the X chromosome, named ATRX.1,2 Intriguingly, ATRX syndrome mutations lie in two distinct domains: either the N-terminal ADD (ATRX-DNMT3-DNMT3L) domain,3 which contains a GATA-like domain and a histone tail binding plant homeodomain (PHD),4-6 or the C-terminal helicase region,7 characteristic of ATP-dependent chromatin remodelers (Fig. 1).8,9 Interestingly, these mutations often result in substantially reduced ATRX protein levels in patients.10 However, the mechanisms by which loss of functional ATRX results in these syndrome features, including reduced α-globin expression and mental retardation, remain unclear.11
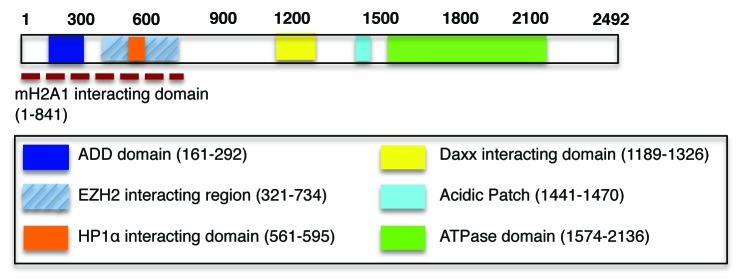
Figure 1. ATRX is a multidomain-containing chromatin remodeler. The ADD domain of ATRX (dark blue box) contains both a GATA-like domain and a PHD and “reads” the H3K9me3 modification.6,21-23 We reported the in vivo interaction between the N-terminal 841 amino acids of ATRX with macroH2A1 (dashed red line).19 ATRX interacts with HP1α via a PxVxL motif present in the indicated region (orange box).16 Of note, while the interaction between EZH2 and ATRX (hatched blue box) was detected by yeast two hybrid,17 it remains to be seen if this persists in vivo and if indeed the entire region depicted is required for this interaction.
ATRX and Condensed Chromatin
Studies over the last ~15 years have uncovered multiple roles for ATRX, some of which may appear to be contradictory. For example, the fact that ATRX mutations result in the loss of α-globin gene expression and that ATRX physically binds to the α-globin gene cluster, suggests a transcriptionally activating role for ATRX.10,12 However, its localization to telomeres, pericentric heterochromatin, and the inactive X chromosome13-15 implicate a role in the establishment and/or maintenance of transcriptionally silent chromatin. The protein interaction partners of ATRX, including HP1α, EZH2, MeCP2, and macroH2A, also implicate its role in heterochromatin structure and function (Fig. 1).16-20 Additionally, the N-terminal ADD domain of ATRX, which binds the heterochromatin-associated histone modification H3K9me3 (see below), bears homology to domains present in DNA methyltransferases,3,21-23 also suggesting transcriptional repression. Mechanistic insight into ATRX function is presently emerging and the picture is not so clear-cut—ATRX appears to have many faces in regards to its role in histone deposition and transcription, as discussed below.
ATRX and Histone H3 Interactions
ATRX has been linked to histone variant regulation by Choo and colleagues, who described the co-localization of ATRX and H3.3 at telomeres in mouse embryonic stem cells (ESCs).14,24 This work also reported the loss of ATRX from telomeres upon differentiation concomitant with an increase of heterochromatic histone modifications, suggesting that ATRX facilitates a euchromatic state at telomeres.14,24 Their work also showed that lysine 4 (K4) of H3.3 is critical for the in vivo interaction between ATRX and H3.3.14 Subsequently, three reports emerged describing the direct binding of the ADD domain of ATRX to H3K9me3 via biochemical and structural studies.21-23 This binding was perturbed by the transcriptionally active H3K4me3 modification, suggesting preferential association of ATRX with repressive chromatin. These studies further suggested that ATRX localization to heterochromatin is contingent upon H3K9me3, and we too have observed the in vivo association of ATRX with H3K9me3 (K.R. and E.B., unpublished data). Hence, the K4 residue reported to mediate the ATRX in vivo interaction by Choo and colleagues is likely due to K4 being a critical residue required for the ADD-H3 tail interaction.14,21Goldberg et al. demonstrated a functional interaction between H3.3 and ATRX in mouse ESCs.25 While HIRA had been characterized as a histone chaperone for H3.3,26 Goldberg et al. observed that ATRX is also involved in H3.3 deposition. Interestingly, while HIRA deposits H3.3 at transcription start sites (TSSs) and within gene bodies, ATRX deposits H3.3 at telomeres. This study uncovered that distinct factors are required for site-specific deposition of H3.3, thereby highlighting the intricacy of histone variant deposition. We anticipate much more complexity in this system not only for H3.3, but also other histone variants.ATRX has been reported to complex with the death-domain associated protein Daxx.25,27,28 Drane et al. reported an ATRX and Daxx-dependent deposition of H3.3; however, this time at pericentric heterochromatin.29 Both ATRX and Daxx were found enriched at major satellite repeats and it was suggested that H3.3 deposition at these regions was unlikely to facilitate heterochromatin formation but rather to drive transcription of pericentric repeats, thus implicating ATRX in transcriptional activation. We highlight here that recent evidence points toward Daxx as the direct histone chaperone for H3.3.29,30 In fact, two structural studies resolved Daxx in complex with an H3.3-containing nucleosome and demonstrated that glycine 90, which is unique to H3.3 (as compared with H3.1 and H3.2), is crucial for this interaction.31,32While future studies will allow a better understanding of the role of ATRX in heterochromatin regulation, a potential model begins to emerge from these studies. In the case of telomeres, ATRX assists Daxx in H3.3 deposition potentially to facilitate chromatin accessibility of highly replicating telomeres in ESCs.29,30,33 As cells differentiate, ATRX is lost from telomeres allowing heterochromatin formation to ensue.14,24 Consistent with these findings, the ATRX/Daxx dependent deposition of H3.3 at pericentric regions, mentioned above, is likely to facilitate an open environment conducive to the active transcription of pericentric repeats.29 However, Goldberg et al. reported an increase in the presence of telomere repeat containing RNA (TERRA) in the absence of ATRX, suggesting that ATRX is also involved in transcriptional repression at telomeres.25 Taken together, numerous questions arise from these studies: How does recognition of H3K9me3 via ATRX ADD domain contribute to such transcriptional activities? Does ATRX bind H3K9me3 on histone H3.3 specifically or other H3 family members as well? Does ATRX bind to heterochromatin to facilitate silencing, to potentially remodel chromatin for transcriptional activation, or to maintain distinctive but ill-defined chromatin states?
Histone Deposition at the α-globin Locus
Our interest in the regulation of macroH2A chromatin association led to the identification of ATRX association with this transcriptionally repressive H2A variant.19 In ATRX knockdown cells, which mimic the levels of ATRX protein found in ATRX syndrome patients, we observed increased levels of macroH2A1 globally and, more specifically, at telomeres and the α-globin gene cluster (proximal to human telomere 16). This corresponded to decreased α-globin expression. Furthermore, we reported an exclusive global localization pattern between ATRX and macroH2A1, suggestive of an antagonistic relationship (see Fig. 2). While ATRX is enriched at TSSs of active genes, macroH2A1 is excluded from the TSS and enriched upstream and/or in the gene bodies of inactive genes.12,19
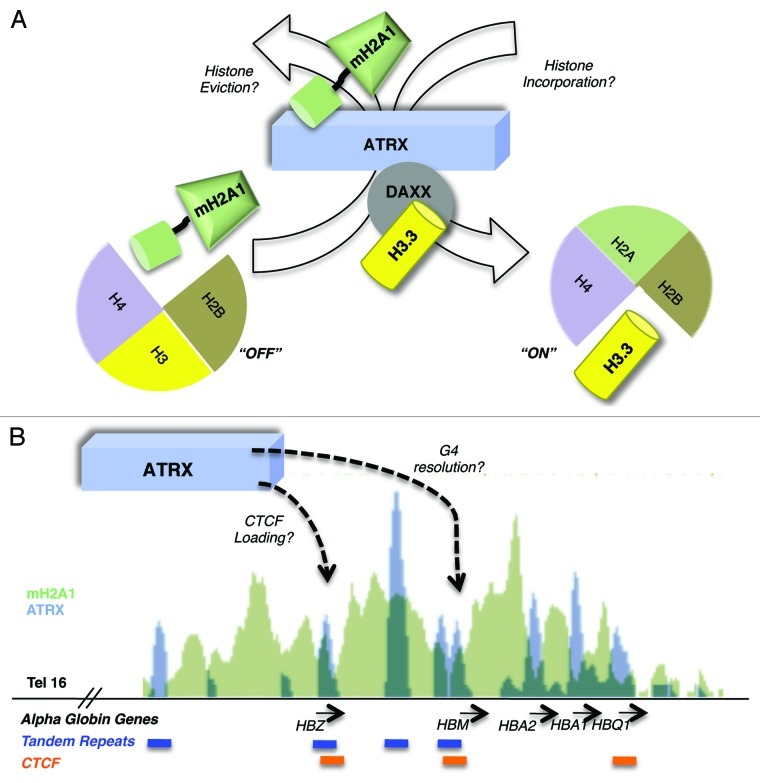
Figure 2. Hypothetical models for ATRX-mediated histone exchange and transcriptional activation of the α-globin gene cluster. (A) ATRX facilitates activation via the nucleosomal eviction of macroH2A1 and/or the Daxx mediated incorporation of H3.3 into nucleosomes potentially switching between “ON” and “OFF” chromatin states. (B) ATRX loads CTCF and resolves repressive G4 DNA structures at tandem repeats (TRs) to permit gene activation at the α-globin cluster. Shown is a 50kb UCSC genome browser snapshot of the α-globin cluster. ChIP-sequencing data plots of macroH2A1 (K562 cells, green)19 and ATRX (primary erythroblasts, blue)12 are overlaid (scales are 70 and 600 respectively). Below the panel, genes are presented in black, ATRX associated tandem repeat tracks in blue and CTCF associated sites from genome wide ChIA-PET analysis (K562 cells)43 in orange. All three CTCF sites overlap ATRX peaks and two of these occur at TRs. Of note, the ChIP-sequencing data presented were obtained from cell lines with different expression levels of α-globin (primary erythroblasts express α-globin at much higher levels than K56257) and the association of macroH2A1 and ATRX with the locus are likely reflective of the differences in α-globin transcription between these two cell types.
Our studies suggest that ATRX negatively regulates the chromatin association of macroH2A, and support the notion of ATRX as transcriptional activator. Of note, while ATRX regulates H3.3 deposition via Daxx,29,30 ATRX interacts with macroH2A in a Daxx-independent manner.19 We also reported a loss of expression of other genes outside of the α-globin cluster, suggesting that ATRX positively regulates genes in this greater genomic region in part by inhibition of deposition or through eviction/replacement of macroH2A (Fig. 2). It is now of interest to determine the mechanism by which ATRX regulates macroH2A deposition, and the H3.3 deposition changes that may occur at the α-globin cluster in the absence of ATRX. While HIRA was suggested to be the primary histone chaperone for H3.3 at genic regions in mouse ESCs,25 the α-globin cluster is occupied by ATRX when actively transcribed, such as in primary erythroblasts,12 suggesting that ATRX might mediate H3.3 deposition here.
We also observed that the N-terminal region of ATRX (1–841), containing the ADD domain, was sufficient for interaction with macroH2A (Fig. 1).19 This raises the enticing model that ATRX recognizes nucleosomes containing H3K9me3 (on H3.1 and H3.2) via its ADD domain (Fig. 1) and assists in the exchange of macroH2A for H3.3 (via Daxx) into nucleosomes at the α-globin locus, and other genomic regions such as telomeres (Fig. 2). This would promote a switch toward open chromatin, and may help to reconcile ATRX heterochromatin localization with its conflicting role in gene activation.
α-Thalassemia Syndrome Variability—G quadruplex Structures
While the regulation of histone variant deposition by ATRX supports a direct role in transcriptional activation, recent work from Higgs and colleagues suggested a novel mechanism by which ATRX supports activation.12 In a study aimed at understanding the variability of phenotypes among patients bearing the same ATRX gene mutations, ChIP-sequencing studies revealed that ATRX primarily occupies G-rich tandem repeats (TRs) which can form G quadruplex (G4) DNA structures in vivo. It was suggested that the differences in size of these TRs among patients contributes to the ranges in severity of the syndrome. For example, TRs were reported to be at or around the α-globin cluster and the absence of ATRX resulted in a distance-dependent silencing of genes (including the α-globin genes). Those genes closest to the TRs were the most severely affected. Higgs and colleagues’ model proposes that ATRX facilitates transcription, indirectly, by binding to and resolving potentially repressive G4 structures. They further hypothesized that this involves the incorporation of the variant H3.3. It would indeed be interesting to test if the incorporation of the variant H3.3 and/or the removal of macroH2A are involved in the putative helicase-mediated resolution of G4 structures (Fig. 2). While this study points toward a role for ATRX in transcriptional activation, recent reports described below may complicate this model or suggest that each genomic region has to be assessed individually for ATRX function.
Transcriptional Repressor, Transcriptional Activator or Something in Between?
The ability to visualize ATRX and Daxx proteins at an inducible multi-copy transgene array in single cells has allowed analysis of their role in regulating chromatin states.34,35 In the presence of ATRX and Daxx, the array is refractory to transcriptional activation and upon induced activation both proteins are lost, implicating them in the maintenance of a repressive chromatin state. The array displayed robust activation in the ATRX-negative U2OS osteosarcoma cell line and addition of exogenous ATRX resulted in reduced transcriptional activation. This was ascribed, at least in part, to its helicase activity, suggesting that while the more N-terminal portion of ATRX recognizes histone modifications and associates either directly or indirectly with histone variants, its C-terminal helicase domain has broader roles in regulating chromatin structure. This remodeling activity may be dependent on ATRX locus-specific protein interactions or association with histone variants.
A study performed in Drosophila melanogaster examined the ATRX homolog, XNP. Ahmad and colleagues analyzed the gain- and loss-of-function phenotypes of XNP using the fly eye, and found that both produced the same effect—de-repression of silencing.36 Taken together, it might appear that ATRX maintains a balance between the two chromatin states (heterochromatic and euchromatic) and changes in its levels, in either direction, result in an alteration of chromatin structure, possibly in a locus-specific manner. In this case, a loss of heterochromatin was observed.36 While very intriguing, a caveat here is that Drosophila XNP does not possess an ADD domain,37 which might confound these studies when applied directly to mammalian systems.
ATRX—a Regulator of Chromatin Domains?
Kernohan et al. suggested that ATRX is involved in the loading of the insulator protein, CTCF, onto the H19 gene locus in the mouse brain—in ATRX knock out cells, a decreased association of the CTCF protein was observed.18 The CTCF protein has long been described to act as a barrier between chromatin domains38-40 and hence its absence could result in aberrant spreading and/or deposition of histones with opposing transcriptional activity.41 Gamble et al. analyzed macroH2A1 genome-wide deposition pattern and determined that CTCF is enriched at macroH2A1 domain boundaries.42 Furthermore, the ENCODE genome project reveals several sites of overlap between ATRX and CTCF at the α-globin cluster (Fig. 2).12,43 We question: is the loss of H3.3 or the increased deposition of macroH2A1 a product of a defective chromatin barrier created in part by aberrant CTCF deposition at particular genomic loci in an ATRX-deficient background (Fig. 2)? Therefore, it will be of interest to study alterations to histone deposition patterns in ATRX patients. While the α-thalassemia phenotype is well characterized to occur via transcriptional alterations at the α-globin cluster, the mental retardation aspects of ATRX are poorly understood. Studying the genomic alterations of histones and factors that influence the chromatin template in ATRX patients will likely shed light on additional mechanisms involved in this syndrome.
Concluding Remarks
The numerous observations pertaining to ATRX suggest that we have only begun to uncover its roles in transcriptional regulation, and we look forward to studies that resolve the questions we raise herein. Moreover, we anticipate exciting new studies that examine ATRX in the context of tumor biology. Curiously, ATRX mutations and deletions have been reported in varied tumor types including pancreatic neuroendocrine tumors (panNETs), pediatric neuroblastoma and glioblastoma, as well as the rare α-thalassemia myelodysplasia syndrome (ATMDS).44-51 While the mechanism is yet unknown, it is likely that ATRX mutations in the context of tumor biology are distinct to those in ATRX syndrome, as patients with ATRX syndrome have not been reported to be pre-disposed to the afore mentioned tumors.44-52
Given the discussion here, we hypothesize that ATRX mutations are directly involved in tumorigenesis via alterations at the chromatin level and genome-wide analyses of histone variant deposition and histone post-translational modifications (PTMs) in the context of these tumors will elucidate whether this is indeed the case. ATRX has also been implicated in sister chromatid cohesion and mutations to ATRX potentially contribute to tumorigenesis via aberrant chromosome dynamics.53 As there are apparent roles for ATRX in telomere homeostasis,14,19,25,54 it is now of interest to determine the perturbations to histones, particularly H3.3 and macroH2A, in the absence of functional ATRX and if/how this might contribute to tumorigenesis via telomere dysfunction. Additionally, ATRX has been shown to localize to promyelocytic leukemia (PML) nuclear bodies, together with Daxx,27 and while it is unclear if there is a connection between PML bodies and histone regulation, a full understanding of the varied cellular roles of ATRX will certainly involve delineating its connection to PML bodies.
ATRX has been directly linked to repression in some cases, while in others it appears to be intimately associated with gene activation. Work from Berube and colleagues reported phosphorylation-dependent changes to ATRX localization during the cell cycle.55 In particular, ATRX was present at the nuclear matrix in interphase, but observed along chromosomes during mitosis when it interacts with HP1α. The authors suggested dual roles for ATRX in gene expression (interphase) and chromosome segregation (mitosis), as distinguished by its PTM profiles. Perhaps attempts at delineating ATRX function in transcription will lead to the identification of differentially modified forms of ATRX that have specific binding partners, thereby conferring distinct functions.
While an intriguing extrapolation that requires testing, the possibility that ATRX regulates a more global genome association of CTCF (than just the HG19 locus18) is particularly enticing. ATRX might then be involved in a greater organization of the genome than has been previously thought potentially via regulation of chromatin domains and higher order chromatin structure.56
Finally, we imagine that genome-wide localization studies of ATRX, as well as biochemical analyses in different cell types (e.g., erythroid, neuronal, tumor cells) will add to our growing knowledge about this peculiar helicase and how its function goes awry in disease. Does ATRX localize to the same genomic regions in erythroid cells as it does in neuronal cell types? Does ATRX interact with Daxx (and thereby H3.3) and macroH2A in all cell types or does it exhibit tissue specificity? In ATRX mutated tumors that retain nuclear ATRX expression or express truncated forms of the protein, what does the epigenomic landscape look like? How are histone variants and histone PTMs altered, if at all, in the absence of functional ATRX? These questions are indeed of critical and timely importance.
Acknowledgments
The authors thank Luis Felipe Duarte, David Valle-Garcia and Dan Hasson for critical reading of this article. We apologize to those whose work could not be referenced directly due to space constraints. This work was supported by The Ellison Medical Foundation New Scholar Award to E.B.
Glossary
Abbreviations:
- ChIP
chromatin immunoprecipitation
- TSS
transcription start site
- ESC
embryonic stem cell
- TR
tandem repeat
- G4
G quadruplex
- PTM
post-translational modification
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/epigenetics/article/23271
References
- 1.Gibbons RJ, Wilkie AO, Weatherall DJ, Higgs DR. A newly defined X linked mental retardation syndrome associated with alpha thalassaemia. J Med Genet. 1991;28:729–33. doi: 10.1136/jmg.28.11.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stayton CL, Dabovic B, Gulisano M, Gecz J, Broccoli V, Giovanazzi S, et al. Cloning and characterization of a new human Xq13 gene, encoding a putative helicase. Hum Mol Genet. 1994;3:1957–64. doi: 10.1093/hmg/3.11.1957. [DOI] [PubMed] [Google Scholar]
- 3.Aapola U, Kawasaki K, Scott HS, Ollila J, Vihinen M, Heino M, et al. Isolation and initial characterization of a novel zinc finger gene, DNMT3L, on 21q22.3, related to the cytosine-5-methyltransferase 3 gene family. Genomics. 2000;65:293–8. doi: 10.1006/geno.2000.6168. [DOI] [PubMed] [Google Scholar]
- 4.Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol. 2007;14:1025–40. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baker LA, Allis CD, Wang GG. PHD fingers in human diseases: disorders arising from misinterpreting epigenetic marks. Mutat Res. 2008;647:3–12. doi: 10.1016/j.mrfmmm.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Argentaro A, Yang JC, Chapman L, Kowalczyk MS, Gibbons RJ, Higgs DR, et al. Structural consequences of disease-causing mutations in the ATRX-DNMT3-DNMT3L (ADD) domain of the chromatin-associated protein ATRX. Proc Natl Acad Sci U S A. 2007;104:11939–44. doi: 10.1073/pnas.0704057104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gibbons RJ, McDowell TL, Raman S, O’Rourke DM, Garrick D, Ayyub H, et al. Mutations in ATRX, encoding a SWI/SNF-like protein, cause diverse changes in the pattern of DNA methylation. Nat Genet. 2000;24:368–71. doi: 10.1038/74191. [DOI] [PubMed] [Google Scholar]
- 8.Picketts DJ, Higgs DR, Bachoo S, Blake DJ, Quarrell OW, Gibbons RJ. ATRX encodes a novel member of the SNF2 family of proteins: mutations point to a common mechanism underlying the ATR-X syndrome. Hum Mol Genet. 1996;5:1899–907. doi: 10.1093/hmg/5.12.1899. [DOI] [PubMed] [Google Scholar]
- 9.Smith CL, Peterson CL. ATP-dependent chromatin remodeling. Curr Top Dev Biol. 2005;65:115–48. doi: 10.1016/S0070-2153(04)65004-6. [DOI] [PubMed] [Google Scholar]
- 10.Higgs DR, Garrick D, Anguita E, De Gobbi M, Hughes J, Muers M, et al. Understanding alpha-globin gene regulation: Aiming to improve the management of thalassemia. Ann N Y Acad Sci. 2005;1054:92–102. doi: 10.1196/annals.1345.012. [DOI] [PubMed] [Google Scholar]
- 11.Gibbons R. Alpha thalassaemia-mental retardation, X linked. Orphanet J Rare Dis. 2006;1:15. doi: 10.1186/1750-1172-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Law MJ, Lower KM, Voon HP, Hughes JR, Garrick D, Viprakasit V, et al. ATR-X syndrome protein targets tandem repeats and influences allele-specific expression in a size-dependent manner. Cell. 2010;143:367–78. doi: 10.1016/j.cell.2010.09.023. [DOI] [PubMed] [Google Scholar]
- 13.McDowell TL, Gibbons RJ, Sutherland H, O’Rourke DM, Bickmore WA, Pombo A, et al. Localization of a putative transcriptional regulator (ATRX) at pericentromeric heterochromatin and the short arms of acrocentric chromosomes. Proc Natl Acad Sci U S A. 1999;96:13983–8. doi: 10.1073/pnas.96.24.13983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wong LH, McGhie JD, Sim M, Anderson MA, Ahn S, Hannan RD, et al. ATRX interacts with H3.3 in maintaining telomere structural integrity in pluripotent embryonic stem cells. Genome Res. 2010;20:351–60. doi: 10.1101/gr.101477.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baumann C, De La Fuente R. ATRX marks the inactive X chromosome (Xi) in somatic cells and during imprinted X chromosome inactivation in trophoblast stem cells. Chromosoma. 2009;118:209–22. doi: 10.1007/s00412-008-0189-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lechner MS, Schultz DC, Negorev D, Maul GG, Rauscher FJ., 3rd The mammalian heterochromatin protein 1 binds diverse nuclear proteins through a common motif that targets the chromoshadow domain. Biochem Biophys Res Commun. 2005;331:929–37. doi: 10.1016/j.bbrc.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 17.Cardoso C, Timsit S, Villard L, Khrestchatisky M, Fontès M, Colleaux L. Specific interaction between the XNP/ATR-X gene product and the SET domain of the human EZH2 protein. Hum Mol Genet. 1998;7:679–84. doi: 10.1093/hmg/7.4.679. [DOI] [PubMed] [Google Scholar]
- 18.Kernohan KD, Jiang Y, Tremblay DC, Bonvissuto AC, Eubanks JH, Mann MR, et al. ATRX partners with cohesin and MeCP2 and contributes to developmental silencing of imprinted genes in the brain. Dev Cell. 2010;18:191–202. doi: 10.1016/j.devcel.2009.12.017. [DOI] [PubMed] [Google Scholar]
- 19.Ratnakumar K, Duarte LF, LeRoy G, Hasson D, Smeets D, Vardabasso C, et al. ATRX-mediated chromatin association of histone variant macroH2A1 regulates α-globin expression. Genes Dev. 2012;26:433–8. doi: 10.1101/gad.179416.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nan X, Hou J, Maclean A, Nasir J, Lafuente MJ, Shu X, et al. Interaction between chromatin proteins MECP2 and ATRX is disrupted by mutations that cause inherited mental retardation. Proc Natl Acad Sci U S A. 2007;104:2709–14. doi: 10.1073/pnas.0608056104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iwase S, Xiang B, Ghosh S, Ren T, Lewis PW, Cochrane JC, et al. ATRX ADD domain links an atypical histone methylation recognition mechanism to human mental-retardation syndrome. Nat Struct Mol Biol. 2011;18:769–76. doi: 10.1038/nsmb.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dhayalan A, Tamas R, Bock I, Tattermusch A, Dimitrova E, Kudithipudi S, et al. The ATRX-ADD domain binds to H3 tail peptides and reads the combined methylation state of K4 and K9. Hum Mol Genet. 2011;20:2195–203. doi: 10.1093/hmg/ddr107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eustermann S, Yang JC, Law MJ, Amos R, Chapman LM, Jelinska C, et al. Combinatorial readout of histone H3 modifications specifies localization of ATRX to heterochromatin. Nat Struct Mol Biol. 2011;18:777–82. doi: 10.1038/nsmb.2070. [DOI] [PubMed] [Google Scholar]
- 24.Wong LH, Ren H, Williams E, McGhie J, Ahn S, Sim M, et al. Histone H3.3 incorporation provides a unique and functionally essential telomeric chromatin in embryonic stem cells. Genome Res. 2009;19:404–14. doi: 10.1101/gr.084947.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goldberg AD, Banaszynski LA, Noh KM, Lewis PW, Elsaesser SJ, Stadler S, et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell. 2010;140:678–91. doi: 10.1016/j.cell.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116:51–61. doi: 10.1016/S0092-8674(03)01064-X. [DOI] [PubMed] [Google Scholar]
- 27.Xue Y, Gibbons R, Yan Z, Yang D, McDowell TL, Sechi S, et al. The ATRX syndrome protein forms a chromatin-remodeling complex with Daxx and localizes in promyelocytic leukemia nuclear bodies. Proc Natl Acad Sci U S A. 2003;100:10635–40. doi: 10.1073/pnas.1937626100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tang J, Wu S, Liu H, Stratt R, Barak OG, Shiekhattar R, et al. A novel transcription regulatory complex containing death domain-associated protein and the ATR-X syndrome protein. J Biol Chem. 2004;279:20369–77. doi: 10.1074/jbc.M401321200. [DOI] [PubMed] [Google Scholar]
- 29.Drané P, Ouararhni K, Depaux A, Shuaib M, Hamiche A. The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes Dev. 2010;24:1253–65. doi: 10.1101/gad.566910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lewis PW, Elsaesser SJ, Noh KM, Stadler SC, Allis CD. Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc Natl Acad Sci U S A. 2010;107:14075–80. doi: 10.1073/pnas.1008850107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Elsässer SJ, Huang H, Lewis PW, Chin JW, Allis CD, Patel DJ. DAXX envelops a histone H3.3-H4 dimer for H3.3-specific recognition. Nature. 2012;491:560–5. doi: 10.1038/nature11608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu CP, Xiong C, Wang M, Yu Z, Yang N, Chen P, et al. Structure of the variant histone H3.3-H4 heterodimer in complex with its chaperone DAXX. Nat Struct Mol Biol. 2012;19:1287–92. doi: 10.1038/nsmb.2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Elsässer SJ, Huang H, Lewis PW, Chin JW, Allis CD, Patel DJ. DAXX envelops a histone H3.3-H4 dimer for H3.3-specific recognition. Nature. 2012;491:560–5. doi: 10.1038/nature11608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rafalska-Metcalf IU, Powers SL, Joo LM, LeRoy G, Janicki SM. Single cell analysis of transcriptional activation dynamics. PLoS One. 2010;5:e10272. doi: 10.1371/journal.pone.0010272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Newhart A, Rafalska-Metcalf IU, Yang T, Negorev DG, Janicki SM. Single cell analysis of Daxx and ATRX-dependent transcriptional repression. J Cell Sci. 2012 doi: 10.1242/jcs.110148. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schneiderman JI, Sakai A, Goldstein S, Ahmad K. The XNP remodeler targets dynamic chromatin in Drosophila. Proc Natl Acad Sci U S A. 2009;106:14472–7. doi: 10.1073/pnas.0905816106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bassett AR, Cooper SE, Ragab A, Travers AA. The chromatin remodelling factor dATRX is involved in heterochromatin formation. PLoS One. 2008;3:e2099. doi: 10.1371/journal.pone.0002099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bell AC, West AG, Felsenfeld G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999;98:387–96. doi: 10.1016/S0092-8674(00)81967-4. [DOI] [PubMed] [Google Scholar]
- 39.Gerasimova TI, Corces VG. Chromatin insulators and boundaries: effects on transcription and nuclear organization. Annu Rev Genet. 2001;35:193–208. doi: 10.1146/annurev.genet.35.102401.090349. [DOI] [PubMed] [Google Scholar]
- 40.Cuddapah S, Jothi R, Schones DE, Roh TY, Cui K, Zhao K. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res. 2009;19:24–32. doi: 10.1101/gr.082800.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Splinter E, Heath H, Kooren J, Palstra RJ, Klous P, Grosveld F, et al. CTCF mediates long-range chromatin looping and local histone modification in the beta-globin locus. Genes Dev. 2006;20:2349–54. doi: 10.1101/gad.399506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gamble MJ, Frizzell KM, Yang C, Krishnakumar R, Kraus WL. The histone variant macroH2A1 marks repressed autosomal chromatin, but protects a subset of its target genes from silencing. Genes Dev. 2010;24:21–32. doi: 10.1101/gad.1876110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li G, Fullwood MJ, Xu H, Mulawadi FH, Velkov S, Vega V, et al. ChIA-PET tool for comprehensive chromatin interaction analysis with paired-end tag sequencing. Genome Biol. 2010;11:R22. doi: 10.1186/gb-2010-11-2-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu XY, Gerges N, Korshunov A, Sabha N, Khuong-Quang DA, Fontebasso AM, et al. Frequent ATRX mutations and loss of expression in adult diffuse astrocytic tumors carrying IDH1/IDH2 and TP53 mutations. Acta Neuropathol. 2012;124:615–25. doi: 10.1007/s00401-012-1031-3. [DOI] [PubMed] [Google Scholar]
- 45.Jiao Y, Shi C, Edil BH, de Wilde RF, Klimstra DS, Maitra A, et al. DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science. 2011;331:1199–203. doi: 10.1126/science.1200609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Steensma DP, Higgs DR, Fisher CA, Gibbons RJ. Acquired somatic ATRX mutations in myelodysplastic syndrome associated with alpha thalassemia (ATMDS) convey a more severe hematologic phenotype than germline ATRX mutations. Blood. 2004;103:2019–26. doi: 10.1182/blood-2003-09-3360. [DOI] [PubMed] [Google Scholar]
- 47.Jiao Y, Killela PJ, Reitman ZJ, Rasheed AB, Heaphy CM, de Wilde RF, et al. Frequent ATRX, CIC, and FUBP1 mutations refine the classification of malignant gliomas. Oncotarget. 2012;3:709–22. doi: 10.18632/oncotarget.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gibbons RJ, Pellagatti A, Garrick D, Wood WG, Malik N, Ayyub H, et al. Identification of acquired somatic mutations in the gene encoding chromatin-remodeling factor ATRX in the alpha-thalassemia myelodysplasia syndrome (ATMDS) Nat Genet. 2003;34:446–9. doi: 10.1038/ng1213. [DOI] [PubMed] [Google Scholar]
- 49.Cheung NK, Zhang J, Lu C, Parker M, Bahrami A, Tickoo SK, et al. St Jude Children’s Research Hospital–Washington University Pediatric Cancer Genome Project Association of age at diagnosis and genetic mutations in patients with neuroblastoma. JAMA. 2012;307:1062–71. doi: 10.1001/jama.2012.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schwartzentruber J, Korshunov A, Liu XY, Jones DT, Pfaff E, Jacob K, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482:226–31. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- 51.Molenaar JJ, Koster J, Zwijnenburg DA, van Sluis P, Valentijn LJ, van der Ploeg I, et al. Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature. 2012;483:589–93. doi: 10.1038/nature10910. [DOI] [PubMed] [Google Scholar]
- 52.Gibbons RJ. α-Thalassemia, mental retardation, and myelodysplastic syndrome. Cold Spring Harb Perspect Med. 2012;2 doi: 10.1101/cshperspect.a011759. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ritchie K, Seah C, Moulin J, Isaac C, Dick F, Bérubé NG. Loss of ATRX leads to chromosome cohesion and congression defects. J Cell Biol. 2008;180:315–24. doi: 10.1083/jcb.200706083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Heaphy CM, de Wilde RF, Jiao Y, Klein AP, Edil BH, Shi C, et al. Altered telomeres in tumors with ATRX and DAXX mutations. Science. 2011;333:425. doi: 10.1126/science.1207313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bérubé NG, Smeenk CA, Picketts DJ. Cell cycle-dependent phosphorylation of the ATRX protein correlates with changes in nuclear matrix and chromatin association. Hum Mol Genet. 2000;9:539–47. doi: 10.1093/hmg/9.4.539. [DOI] [PubMed] [Google Scholar]
- 56.Degner SC, Verma-Gaur J, Wong TP, Bossen C, Iverson GM, Torkamani A, et al. CCCTC-binding factor (CTCF) and cohesin influence the genomic architecture of the Igh locus and antisense transcription in pro-B cells. Proc Natl Acad Sci U S A. 2011;108:9566–71. doi: 10.1073/pnas.1019391108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Benz EJ, Jr., Murnane MJ, Tonkonow BL, Berman BW, Mazur EM, Cavallesco C, et al. Embryonic-fetal erythroid characteristics of a human leukemic cell line. Proc Natl Acad Sci U S A. 1980;77:3509–13. doi: 10.1073/pnas.77.6.3509. [DOI] [PMC free article] [PubMed] [Google Scholar]