Abstract
Background
The increasing occurrence of livestock-associated (LA) methicillin-resistant Staphylococcus aureus (MRSA) associated with the clonal complex (CC) 398 within the past years shows the importance of standardized and comparable typing methods for the purposes of molecular surveillance and outbreak detection. Multiple-locus variable number of tandem repeats analysis (MLVA) has recently been described as an alternative and highly discriminative tool for S. aureus. However, until now the applicability of MLVA for the typing of LA-MRSA isolates from different geographic origin has not been investigated in detail. We therefore compared MLVA and S. aureus protein A (spa) typing for characterizing porcine MRSA from distinct Dutch and German farms.
Methodology/Principal Findings
Overall, 134 MRSA isolates originating from 21 different pig-farms in the Netherlands and 36 farms in Germany comprising 21 different spa types were subjected to MLVA-typing. Amplification and subsequent automated fragment sizing of the tandem repeat loci on a capillary sequencer differentiated these 134 isolates into 20 distinct MLVA types. Whereas overall MLVA and spa typing showed the same discriminatory power to type LA-MRSA (p = 0.102), MLVA was more discriminatory than spa typing for isolates associated with the prevalent spa types t011 and t034 (Simpson’s Index of Diversity 0.564 vs. 0.429, respectively; p<0.001).
Conclusion
Although the applied MLVA scheme was not more discriminatory than spa typing in general, it added valuable information to spa typing results for specific spa types (t011, t034) which are highly prevalent in the study area, i.e. Dutch-German border area. Thus, both methods may complement each other to increase the discriminatory power to resolute highly conserved clones such as CC398 (spa types t011, t034) for the detection of outbreaks and molecular surveillance of zoonotic MRSA.
Introduction
Methicillin-resistant Staphylococcus aureus (MRSA) have emerged in livestock within the past years and are increasingly detected in humans [1], [2], [3], [4]. Predominantly, livestock-associated MRSA (LA-MRSA) isolates belong to a distinct MRSA lineage characterized by the clonal complex (CC) 398 as defined by multilocus sequence typing (MLST) [3].
Livestock animals are usually only colonized with LA-MRSA CC398. However, this clonal lineage has occasionally been identified as the causative agents for wound infections in horses [5] or mastitis in cows [6]. In pigs, LA-MRSA has been associated with pathological lesions, abortion and systemic diseases [7], [8].
The impact of LA-MRSA CC398 on humans has been determined in many epidemiological studies. Direct contact to livestock is a significant risk factor for colonization with such strains leading to nasal MRSA carriage in 29% [4] to 86% [9] of pig farmers and 4.6% [10] to 45% [9] of veterinarians. Especially in areas with a high density of livestock farming, MRSA CC398 represents a significant proportion among the MRSA isolates derived from hospital inpatients [11]. Moreover, healthcare-associated outbreaks of MRSA CC398 have already been documented [12], [13]. MRSA CC398 has been discussed to be less virulent than hospital- or community-acquired MRSA [14]. Nevertheless, MRSA CC398 has shown its potential to cause human infections (e.g. pneumonia, otomastoiditis, endocarditis and bacteraemia) [15], [16].
To elucidate the molecular epidemiology of LA-MRSA CC398 and to discriminate sporadic from outbreak related cases, different typing techniques have been proposed [17], [18]. However, the lack of a standardized and feasible method with a high epidemiological resolution still hampers studies on outbreaks and transmission routes. MRSA CC398 cannot be typed by standard protocols for pulsed-field gel electrophoresis (PFGE), which separates fragments after SmaI macrorestriction [19]. Although PFGE approaches using Cfr9I as alternative restriction enzyme have been successfully applied [18], [20], these methods are laborious and expensive. Furthermore, PFGE can be problematic due to intra- and inter-laboratory reproducibility aspects. Sequence-based typing of the S. aureus protein A gene (spa) is feasible, cheaper and ensures comparability [21], [22]. However, LA-MRSA CC398 isolates are mostly associated with a very limited number of spa types including t011 and t108 as prevalent clones in the Netherlands [23] and t011, t034 and t2510 as prevalent clones in Germany [24].
A recent study by Schouls et al. has compared PFGE, MLST and spa typing with a new high throughput typing approach using multiple-locus variable number of tandem repeat analysis (MLVA) [17]. This technique yielded a high discriminatory power and a high congruence with PFGE, MLST and was twice as discriminatory as spa typing for hospital- and community-acquired MRSA. However, particularly for LA-MRSA CC398 isolates, MLVA grouped them into two dominant complexes only and did not increase the discriminatory power in comparison with spa typing. Yet, the lack of epidemiological data within the study from Schouls et al. limited its conclusion whether MLVA is suitable for differentiation of LA-MRSA of CC398. Moreover, the study did not include isolates belonging to the spa type t034 [17], which is the second most frequent spa type associated with LA-MRSA CC398 in northwestern Germany [24], [25].
Therefore, we investigated a diverse collection of LA-MRSA, mostly belonging to the CC398 lineage, from geographical distinct farms and regions in Germany and the Netherlands using the MLVA scheme by Schouls et al. The results of our study shall answer the question whether MLVA can differentiate between LA-MRSA isolates associated with the same spa types but obtained in different regions and thereby specifies the information obtained by spa typing.
Materials and Methods
Selection of Bacterial Strains
All samples were derived from pigs (nasal swabs) of 21 Dutch and 36 German farms located in different geographic regions. They were collected within the framework of the INTERREG IVa project SafeGuard (Workpackage 2.3, MRSA vet-net, III-2-03 = 025). All isolates were initially characterized by S. aureus protein A (spa) sequence-based typing as described elsewhere [22], [26]. Cluster analysis of spa types using the Based Upon Repeat Pattern (BURP) algorithm of the Ridom StaphType software (Ridom GmbH, Münster, Germany) with default parameters as recommended previously was used to assign spa types into spa-clonal complexes (spa-CC) [26], [27], [28]. Due to the high concordance of spa typing and MLST [29], spa types clustering with t011, t034 and t108, which have previously been shown to belong to the MLST clonal lineage CC398 were suspected to be part of CC398. For the remaining spa types, MLST types were assigned by comparison with data from the Spa-server (http://spaserver.ridom.de/) and by literature search.
If available, two MRSA isolates of each spa type were chosen from each farm and subjected to MLVA typing. Altogether, we analysed 134 MRSA isolated between 2009 and 2011. Among these, 66 isolates were from Dutch and 68 from German farms.
Bacterial Growth and DNA Extraction
Confirmed MRSA cultures stored at −80°C were grown overnight on Columbia blood agar plates (Heipha; Eppelheim, Germany) at 37°C in ambient atmosphere [24]. One colony was dispended in 100µl Chelex-100 solution (Bio-Rad, Hercules, CA, USA). After the heating process (10 min at 95°C), the samples were vortexed thoroughly and centrifuged at 13400 g for 3 min. The DNA-containing supernatant was either used directly or stored at −20°C until use in PCR.
MLVA
MLVA was performed as previously described by Schouls et al. [17] with some modifications. Two multiplex PCRs were prepared. Each multiplex mixture contained four different fluorescently labelled primer sets (Applied Biosystems, Foster City, CA, USA). The primer concentrations in Mastermix 1 were 0.4µM VNTR09_01f-6-FAM, VNTR09_01r, VNTR61_01f-NED, VNTR61_01r, VNTR61_02r, VNTR67_01_f-PET and VNTR67_01r and 0.2 µM VNTR61_02f-VIC and VNTR61_02f. In Mastermix 2, primer concentrations were 0.4 µM VNTR21_01f-VIC, VNTR21_01r, VNTR81_01f-NED and VNTR81_01r and 0.6 µM VNTR24_01f-PET, VNTR24_01r, VNTR63_01f-6-FAM and VNTR63_01. Both PCRs contained a Type-it Multiplex PCR Master Mix (Qiagen, Hilden, Germany) and 2 µl DNA template in a final volume of 25 µl. Amplification was performed using the following program: initial denaturation (5 min at 95°C), 28 cycles of amplification (30 sec at 95°C, 90 sec at 60°C and 30 sec at 72°C) followed by a final step of 30 min at 60°C.
After PCR, samples were diluted 1∶100 in water. 1.5 µl of the diluted samples were mixed with 15 µl of 1200 LIZ size standard (Applied Biosystems) diluted 1∶100 in HPLC-water. To denature the DNA, samples were incubated for 5 min at 95°C and then immediately frozen at −20°C for ≥3 min. Fragments were separated on an ABI Prism 3130xl Genetic Analyzer System (Applied Biosystems). The results were analysed in the GeneMapper 4.0 software (Applied Biosystems) to calculate the correct number of repeats for each VNTR locus. To calibrate sequencer-specific variation in fragment lengths, the number of repeats of reference strain S. aureus N315 strain was initially determined in silico on the basis of its genome sequence (reference sequence NC_002745; National Center for Biotechnology Information, Bethesda, MD, USA) in accordance with the MLVA protocol provided by the National Institute of Public Health and the Environment of the Netherlands (RIVM) on the MLVA home page (http://www.mlva.net/saureus/default.asp). The lengths of the determined repeats were subtracted from the fragment length of each particular locus detected in independent runs on the Genetic Analyzer. The resulting offset-size (primer plus VNTR-flanking regions) was used to create the respective bin used in the GeneMapper software. In each run, the reference strain was included to assure correct repeat determination of unknown isolates. Partial repeats were rounded to the nearest repeat number.
If a VNTR locus was not detected during fragment analysis, PCR for that particular locus was repeated in a singleplex reaction with the same primer concentrations as in the multiplex-PCR. In rare cases, so-called stutter-peaks (peaks that are typically ≤1 repeat length shorter than the main peak) occurred. In that particular instance, the peak with the highest fluorescent level was used to calculate the repeat number.
Data Analysis
The geographical map and the minimum spanning tree were created with the Ridom MLVA Compare software version 0.28 (Ridom GmbH, Münster, Germany). MLVA profiles were entered into the MLVA home page (http://www.mlva.net/saureus/default.asp) of the RIVM to query the corresponding MLVA-types and complexes. To calculate the discriminatory power and concordance of the typing methods we used EpiCompare software version 1.0 (Ridom GmbH, Münster, Germany) and an internet tool (http://darwin.phyloviz.net/ComparingPartitions/index.php?link=Home).
Results
Overall, MLVA was successfully performed on 133 isolates comprising 20 different spa types (Table 1). The most frequent spa types were t011 (42.5%, n = 57), t034 (18.7%, n = 25) and t108 (14.9%, n = 20). For one isolate from the Netherlands (spa type t151), none of the eight VNTR-loci was amplified after repeated PCR. Therefore, no MLVA-profile could be determined and this isolate was excluded from further analyses.
Table 1. Distribution of spa and MLVA types among the analysed MRSA isolates.
| Spa type | Spa-CC | Presumptive MLST CC | MLVA type | MLVA-complex | No. of isolates | No. of farmsi |
| t011 | 011 | 398a | 398 | 398 | 52 | 33 |
| t011 | 011 | 398a | 3750 | NAh | 1 | 1 |
| t011 | 011 | 398a | 2215 | 398 | 1 | 1 |
| t011 | 011 | 398a | 555 | 398 | 2 | 2 |
| t011 | 011 | 398a | 713 | 398 | 1 | 1 |
| t108 | 011 | 398a | 572 | 398 | 20 | 12 |
| t034 | 011 | 398a | 569 | 398 | 14 | 10 |
| t034 | 011 | 398a | 588 | 398 | 8 | 5 |
| t034 | 011 | 398a | 591 | 398 | 2 | 1 |
| t034 | 011 | 398a | 556 | 398 | 1 | 1 |
| t6320 | 011 | 398b | 569 | 398 | 3 | 2 |
| t2346 | 011 | 398b | 565 | 398 | 4 | 2 |
| t1451 | 011 | 398a | 572 | 398 | 2 | 2 |
| t7621 | 011 | 398b | 1283 | 398 | 2 | 1 |
| t1250 | 011 | 398b | 590 | 398 | 1 | 1 |
| t1184 | 011 | 398b | 567 | 398 | 1 | 1 |
| t1606 | 011 | 398b | 572 | 398 | 1 | 1 |
| t3423 | 011 | 398b | 572 | 398 | 1 | 1 |
| t899 | singleton | 398a; 9c | 567 | 398 | 3 | 2 |
| t899 | singleton | 398a; 9c | 398 | 398 | 1 | 1 |
| t5838 | singleton | NAh | 565 | 398 | 2 | 1 |
| t2510 | excluded | 398a | 1845 | 398 | 3 | 2 |
| t1456 | excluded | 398d | 566 | 398 | 1 | 1 |
| t2383 | excluded | 398e | 568 | 398 | 1 | 1 |
| t1344 | excluded | 398d | 1845 | 398 | 1 | 1 |
| t002 | singleton | 5c | 31 | 5 | 2 | 1 |
| t015 | singleton | 45f | 45 | 45 | 1 | 1 |
| t127 | singleton | 5c; 1g | 1727 | 1 | 1 | 1 |
| t151 | singleton | NAh | NT | NT | 1 | 1 |
according to Köck et al. (2009) [24].
as determined by the Based Upon Repeat Pattern (BURP) algorithm.
according to Hasman et al. (2010) [30].
according to European Food Safety Authority (2009) [25].
according to Graveland et al. [32].
according to the Ridom SpaServer.
according to Mellmann et al. (2008) [27].
not available.
number of farms from which isolates of the respective spa type have been derived and included.
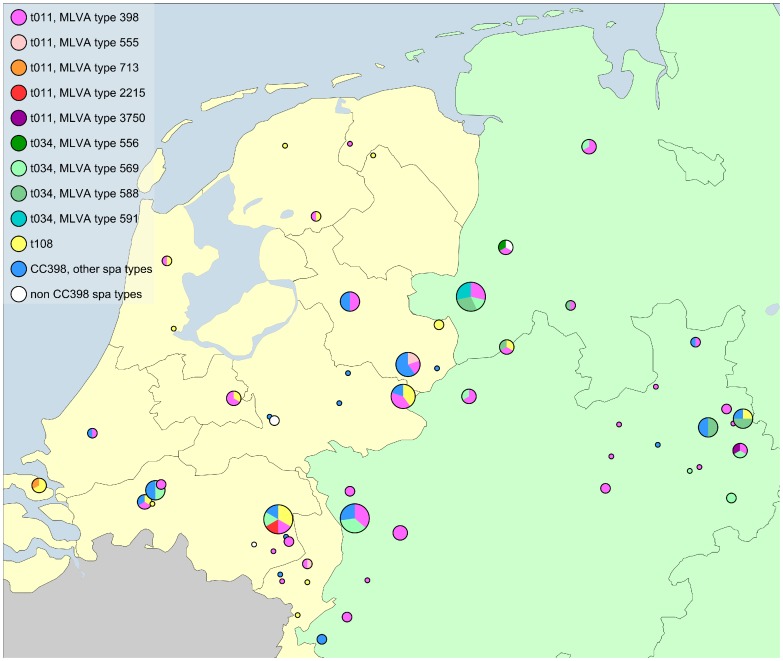
Among the 133 MLVA-typeable associated with 20 different spa types, MLVA resolved 20 different MLVA types. MLVA was able to discriminate isolates that were indistinguishable by spa typing, by distinguishing five different MLVA subtypes among spa type t011, four among spa type t034 and two among spa type t899, respectively. The spa type t108 was represented by only one MLVA type (type 572). Each of the remaining spa types t2510, t2346, t5838, t015, t1250, t1456, t2383, t6320, t002, t1451, t7621, t1184, t127, t1344, t1606 and t3423 was also characterized by solely one MLVA-type (Table 1). An overview of the geographic location of the isolates’ origin as well as the distribution of the spa types and main MLVA types is given in Fig. 1.
Figure 1. Geographic origin of the analysed isolates in Germany and the Netherlands.
One dot represents farms in one same postal code area; the size of a dot is proportional to the amount of isolates obtained from these farms. The proportion of different spa types (and MLVA types for the predominant t011 and t034 spa types) is indicated by different colours.
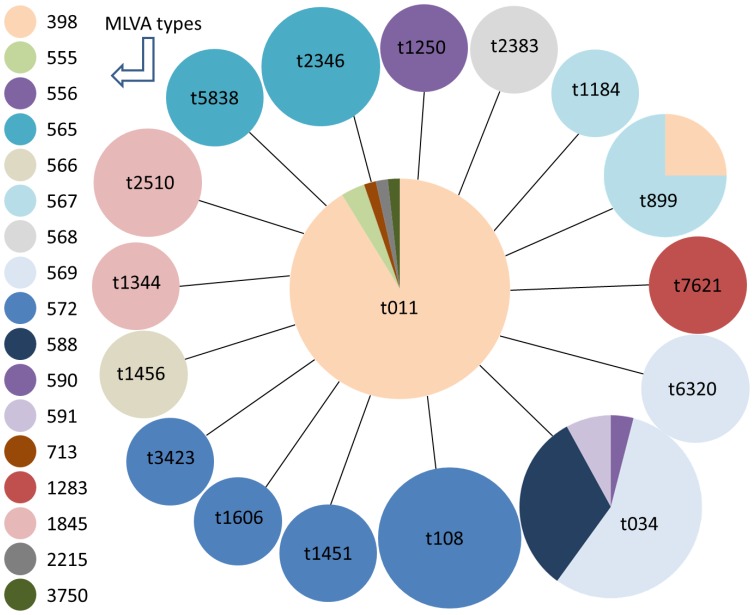
Of the 20 different spa types, 17 were assigned to CC398 according to clustering by the BURP algorithm, MLST and MLVA data (Table 1). An overview of the MLVA-types that were determined for isolates of the CC398 lineage is shown in Fig. 2. Importantly, fig. 2 shows that spa type t034 showed the greatest diversity with MLVA types 556, 569, 588 and 591 accounting for 4%, 56%, 32% and 8% of all t034 isolates, respectively. The distribution of MLVA types within spa type t011 was as follows: MLVA type 398 represented 91% of all isolates, type 555 4% and MLVA types 713, 2215 and 3750 each 2%, respectively.
Figure 2. Minimum spanning tree based on spa types for all isolates associated with LA-MRSA CC398.
Each dot represents a different spa type with the diameter of a dot being proportional to the quantity of the isolates included. Different colours within the dots indicate different MLVA types. The spa types that are associated with the same MLVA type are identically coloured.
The calculation of the Simpson’s index of diversity (ID) for all isolates characterized yielded similar IDs of 0.789 and 0.760 for by spa typing and MLVA, respectively (Table 2). Therefore, both methods had the same discriminatory power to distinguish between LA-MRSA isolates (p = 0.102). The adjusted Rand’s coefficient (AR; 95% confidence interval) was 0.790 (0.696–0.889).
Table 2. Simpson’s Indices of Diversity (ID) and concordance of MLVA and spa typing for all analysed isolates, for isolates associated with CC398 and for isolates associated with the two predominant spa types associated with CC398.
| Typing method | No. of types | Simpson’s ID (C.I 95%)1 | Adjusted Wallace’s coefficient1 | |
| All analysed isolates | spa | MLVA | ||
| MLVA | 20 | 0.789 (0.735–0.844) | 0.863(0.775–0.950) | NA |
| spa | 20 | 0.760 (0.701–0.818) | NA | 0.728(0.598–0.859) |
| All spa -types associated with CC398 | spa | MLVA | ||
| MLVA | 17 | 0.776 (0.720–0.833) | 0.860(0.770–0.949) | NA |
| spa | 17 | 0.745 (0.685–0.805) | NA | 0.724(0.591–0.856) |
| Spa -types t011 and t034 | spa | MLVA | ||
| MLVA | 9 | 0.564 (0.452–0.677) | 1.000(1.000–1.000) | NA |
| spa | 2 | 0.429 (0.350–0.508) | NA | 0.580(0.378–0.783) |
C. I. = confidence interval; NA = not applicable.
Since we were aiming to reveal the discriminatory power of MLVA to distinguish in particular among MRSA isolates associated with CC398, we repeated the calculation but included only the spa types (t011, t034, t108, t899, t1184, t1250, t1344, t1451, t1456, t1606, t2346, t2383, t2510, t3423, t5838, t6320 and t7621) that are indicative for this complex according to BURP clustering, MLST; MLVA data and literature [24], [25], [27], [30], [31], [32]. The exclusion of all non-CC398 spa types did not reveal a significant difference in the discriminatory power of both methods (MLVA: ID = 0.776; spa typing: ID = 0.745; p = 0.102). The AR was 0.786 (0.691–0.886).
To evaluate the applicability and additional value of MLVA compared to spa typing for outbreaks caused by the most prevalent spa-types associated with MRSA CC398, we therefore concentrated on the predominant spa types t011 and t034 among porcine isolates detected within the INTERREG IVa project SafeGuard and excluded all others from the calculation. This resulted in an increase of the discriminatory power of MLVA (ID = 0.564) compared to spa typing (ID = 0.429; p<0.001). As documented by the adjusted Wallace’s coefficients, in this calculation MLVA added valuable information to spa typing, since isolates being associated with one same spa type, had a probability of 0.58 (58%) to have the same MLVA type only. The AR was 0.734 (0.605–0.866).
Discussion
In this study, we evaluated a previously described MLVA scheme [17] to type LA-MRSA. Overall, we found that nearly all (133 of 134) MRSA isolates were typeable by MLVA (99.3%). The failed typing of one isolate might be due to modified DNA or chromosomal variability in S. aureus that leads to the absence or the mutation of the primer targets [33].
Schouls et al. [17] have recently shown that typing by MLVA reveals the same degree of differentiation as spa sequencing regarding LA-MRSA isolates. This finding was in contrast to other publications which had shown that MLVA schemes are usually more discriminatory compared to spa typing [33], [34]. In order to rule out the possibility that the lacking ability of MLVA to discriminate between LA-MRSA strains observed in the study by Schouls et al. [17] was due to a selection bias caused by the inclusion of epidemiologically related MRSA isolates, we have systematically chosen isolates from different locations (57 different farms in Germany and the Netherlands) and subjected them to MLVA typing. We hypothesized that typing of geographically unrelated isolates would increase the discriminatory power. As a result, we showed that even when the geographical diversity and epidemiological differentiation were ensured, MLVA was not more discriminatory than spa typing. As MRSA has only evolved a few decades ago, this might reflect the monomorphic background of this pathogen [35].
However, we found that for specific spa types, MLVA typing enabled to separate isolates that were indistinguishable by spa typing. Isolates that are associated with the highly prevalent spa types t011 and t034 were each diversified into more than one MLVA type (t011 in 5 and t034 in 4 MLVA types, respectively). While for t011, performing MLVA in addition to spa was of limited value, because 91% of all t011 isolates were associated with MLVA type 398, isolates belonging to spa type t034 demonstrated a higher diversity. This finding is of importance, because t034, which is a major spa type among European livestock [25], was not included in previous analysis [12]. This result has practical applications both for tracing LA-MRSA among livestock and for human medicine where in some regions, farmers (colonized with LA-MRSA CC398 in up to 77%) introduce these MRSA strains into regional hospitals which could facilitate nosocomial spread [11], [32], [36]. We therefore suggest that additional MLVA typing of LA-MRSA isolates assigned to spa types t034 and (sometimes) t011 can add valuable information to characterize LA-MRSA for example in healthcare-associated outbreaks. In contrast, for isolates associated with spa type t108, which is highly prevalent in the Netherlands [37], or the other spa types included in this study, additional MLVA typing did not reveal a higher degree of differentiation.
Currently, spa typing is a cheap and feasible method that is widely used for characterizing MRSA. Furuya et al. suggested in a recent study that for best resolution of the results, spa typing should be used in combination with MLVA or PFGE for short-term evolutionary studies [38]. Our results support this opinion, at least for certain spa types. Since hitherto no standardized and widely used protocol for MLVA typing exists, we suggest that spa typing of LA-MRSA should be performed to basically characterize MRSA CC398 isolates. Additional MLVA typing of isolates assigned to spa types t011 or t034 might then lead to a more detailed characterization, if this is needed for case tracing, outbreak detection, or investigations into the pathways of transmission of MRSA CC398 in healthcare facilities. In the veterinary field, MLVA typing could increase the resolution and thereby contribute to a more precise understanding of the distribution of LA-MRSA by national and international pig trading or environmental spread [39], [40], [41].
An advantage of MLVA compared to spa typing is that it is faster and cheaper to perform. Especially in those laboratories with capillary sequencing capacities both methods could complement each other, although the recent description of multiple different MLVA schemes [17], [42], [43], [44], [45] stresses the need to agree upon the use of concerted protocols when this technique is increasingly applied.
While in this study MLVA typing was not shown to be more discriminatory than single locus spa typing in general, this approach was able to subdivide LA-MRSA isolates of the highly prevalent spa types t011 and t034. Thus, in areas with a high prevalence of LA-MRSA CC398, MLVA typing offers a useful additional tool for epidemiological studies and MRSA surveillance purposes.
Funding Statement
This work was supported in part by grants from the Bundesministerium für Bildung und Forschung (BMBF, http://www.bmbf.de/), Germany “Interdisciplinary Research Network MedVet-Staph” to R.K. and K.B. (01KI1014A), by the INTERREG IVa project SafeGuard (MRSA vet-net) to R.K., P.v.d.W. and K.B. (III-2-03 = 025) and by the BMBF (01KI1020) and the Medical Faculty Münster (BD9817044) to A.M. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Voss A, Loeffen F, Bakker J, Klaassen C, Wulf M (2005) Methicillin-resistant Staphylococcus aureus in pig farming. Emerg Infect Dis 11: 1965–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wulf MW, Sorum M, van Nes A, Skov R, Melchers WJ, et al. (2008) Prevalence of methicillin-resistant Staphylococcus aureus among veterinarians: an international study. Clin Microbiol Infect 14: 29–34. [DOI] [PubMed] [Google Scholar]
- 3. Huijsdens XW, van Dijke BJ, Spalburg E, van Santen-Verheuvel MG, Heck ME, et al. (2006) Community-acquired MRSA and pig-farming. Ann Clin Microbiol Antimicrob 5: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. van den Broek IV, van Cleef BA, Haenen A, Broens EM, van der Wolf PJ, et al. (2009) Methicillin-resistant Staphylococcus aureus in people living and working in pig farms. Epidemiol Infect 137: 700–708. [DOI] [PubMed] [Google Scholar]
- 5. Cuny C, Strommenger B, Witte W, Stanek C (2008) Clusters of infections in horses with MRSA ST1, ST254, and ST398 in a veterinary hospital. Microb Drug Resist 14: 307–310. [DOI] [PubMed] [Google Scholar]
- 6. Holmes MA, Zadoks RN (2011) Methicillin Resistant S. aureus in Human and Bovine Mastitis. Journal of mammary gland biology and neoplasia. J Mammary Gland Biol Neoplasia 16: 373–382. [DOI] [PubMed] [Google Scholar]
- 7. van der Wolf PJ, Rothkamp A, Junker K, de Neeling AJ (2012) Staphylococcus aureus (MSSA) and MRSA (CC398) isolated from post-mortem samples from pigs. Vet Microbiol 158: 136–141. [DOI] [PubMed] [Google Scholar]
- 8. Meemken D, Blaha T, Tegeler R, Tenhagen BA, Guerra B, et al. (2010) Livestock associated methicillin-resistant Staphylococcus aureus (LaMRSA) isolated from lesions of pigs at necropsy in northwest Germany between 2004 and 2007. Zoonoses Public Health 57: e143–148. [DOI] [PubMed] [Google Scholar]
- 9. Cuny C, Nathaus R, Layer F, Strommenger B, Altmann D, et al. (2009) Nasal colonization of humans with methicillin-resistant Staphylococcus aureus (MRSA) CC398 with and without exposure to pigs. PLoS ONE 4: e6800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wulf M, van Nes A, Eikelenboom-Boskamp A, de Vries J, Melchers W, et al. (2006) Methicillin-resistant Staphylococcus aureus in veterinary doctors and students, the Netherlands. Emerg Infect Diseases 12: 1939–1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Köck R, Siam K, Al-Malat S, Christmann J, Schaumburg F, et al. (2011) Characteristics of hospital patients colonized with livestock-associated meticillin-resistant Staphylococcus aureus (MRSA) CC398 versus other MRSA clones. J Hosp Infect 79: 292–296. [DOI] [PubMed] [Google Scholar]
- 12.Wulf MW, Markestein A, van der Linden FT, Voss A, Klaassen C, et al. (2008) First outbreak of methicillin-resistant Staphylococcus aureus ST398 in a Dutch hospital, June 2007. Euro Surveill 13. [PubMed]
- 13.Fanoy E, Helmhout LC, van der Vaart WL, Weijdema K, van Santen-Verheuvel MG, et al.. (2009) An outbreak of non-typeable MRSA within a residential care facility. Euro Surveill 14. [PubMed]
- 14. van Rijen MM, Bosch T, Heck ME, Kluytmans JA (2009) Meticillin-resistant Staphylococcus aureus epidemiology and transmission in a Dutch hospital. Journal Hosp Infect 72: 299–306. [DOI] [PubMed] [Google Scholar]
- 15. Wulf MW, Verduin CM, van Nes A, Huijsdens X, Voss A (2011) Infection and colonization with methicillin resistant Staphylococcus aureus ST398 versus other MRSA in an area with a high density of pig farms. Eur J Clin Microbiol Infect Dis 31: 61–65. [DOI] [PubMed] [Google Scholar]
- 16. Köck R, Becker K, Cookson B, van Gemert-Pijnen JE, Harbarth S, et al. (2010) Methicillin-resistant Staphylococcus aureus (MRSA): burden of disease and control challenges in Europe. Euro Surveill 15: 19688. [DOI] [PubMed] [Google Scholar]
- 17. Schouls LM, Spalburg EC, van Luit M, Huijsdens XW, Pluister GN, et al. (2009) Multiple-locus variable number tandem repeat analysis of Staphylococcus aureus: comparison with pulsed-field gel electrophoresis and spa-typing. PLoS ONE 4: e5082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bosch T, de Neeling AJ, Schouls LM, van der Zwaluw KW, Kluytmans JA, et al. (2010) PFGE diversity within the methicillin-resistant Staphylococcus aureus clonal lineage ST398. BMC Microbiol 10: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bens CC, Voss A, Klaassen CH (2006) Presence of a novel DNA methylation enzyme in methicillin-resistant Staphylococcus aureus isolates associated with pig farming leads to uninterpretable results in standard pulsed-field gel electrophoresis analysis. J Clin Microbiol 44: 1875–1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Argudin MA, Rodicio MR, Guerra B (2010) The emerging methicillin-resistant Staphylococcus aureus ST398 clone can easily be typed using the Cfr9I SmaI-neoschizomer. Lett Appl Microbiol 50: 127–130. [DOI] [PubMed] [Google Scholar]
- 21. Frenay HM, Theelen JP, Schouls LM, Vandenbroucke-Grauls CM, Verhoef J, et al. (1994) Discrimination of epidemic and nonepidemic methicillin-resistant Staphylococcus aureus strains on the basis of protein A gene polymorphism. J Clin Microbiol 32: 846–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Harmsen D, Claus H, Witte W, Rothganger J, Turnwald D, et al. (2003) Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J Clin Microbiol 41: 5442–5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huijsdens XW, Bosch T, van Santen-Verheuvel MG, Spalburg E, Pluister GN, et al. (2009) Molecular characterisation of PFGE non-typable methicillin-resistant Staphylococcus aureus in The Netherlands, 2007. Euro Surveill 14. [DOI] [PubMed]
- 24. Köck R, Harlizius J, Bressan N, Laerberg R, Wieler LH, et al. (2009) Prevalence and molecular characteristics of methicillin-resistant Staphylococcus aureus (MRSA) among pigs on German farms and import of livestock-related MRSA into hospitals. Eur J Clin Microbiol Infect Dis 28: 1375–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. European Food Safety Authority (2009) Analysis of the baseline survey on the prevalence of methicillin-resistant Staphylococcus aureus (MRSA) in holdings with breeding pigs, in the EU, Part A: MRSA prevalence estimates. EFSA Journal 2009 7(11): 1376. [Google Scholar]
- 26. Mellmann A, Friedrich AW, Rosenkotter N, Rothganger J, Karch H, et al. (2006) Automated DNA sequence-based early warning system for the detection of methicillin-resistant Staphylococcus aureus outbreaks. PLoS Med 3: e33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mellmann A, Weniger T, Berssenbrugge C, Keckevoet U, Friedrich AW, et al. (2008) Characterization of clonal relatedness among the natural population of Staphylococcus aureus strains by using spa sequence typing and the BURP (based upon repeat patterns) algorithm. J Clin Microbiol 46: 2805–2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mellmann A, Weniger T, Berssenbrugge C, Rothganger J, Sammeth M, et al. (2007) Based Upon Repeat Pattern (BURP): an algorithm to characterize the long-term evolution of Staphylococcus aureus populations based on spa polymorphisms. BMC Microbiol 7: 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Strommenger B, Kettlitz C, Weniger T, Harmsen D, Friedrich AW, et al. (2006) Assignment of Staphylococcus isolates to groups by spa typing, SmaI macrorestriction analysis, and multilocus sequence typing. J Clin Microbiol 44: 2533–2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hasman H, Moodley A, Guardabassi L, Stegger M, Skov RL, et al. (2010) Spa type distribution in Staphylococcus aureus originating from pigs, cattle and poultry. Vet Microbiol 141: 326–331. [DOI] [PubMed] [Google Scholar]
- 31. Fessler AT, Kadlec K, Hassel M, Hauschild T, Eidam C, et al. (2011) Characterization of Methicillin-resistant Staphylococcus aureus Isolates from Food and Food Products of Poultry Origin in Germany. Appl Environ Microbiol 77: 7151–7157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Graveland H, Wagenaar JA, Heesterbeek H, Mevius D, Van Duijkeren E, et al. (2010) Methicillin Resistant Staphylococcus aureus ST398 in Veal Calf Farming: Human MRSA Carriage Related with Animal Antimicrobial Usage and Farm Hygiene. PLoS One. 2010 5(6): e10990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pourcel C, Hormigos K, Onteniente L, Sakwinska O, Deurenberg RH, et al. (2009) Improved multiple-locus variable-number tandem-repeat assay for Staphylococcus aureus genotyping, providing a highly informative technique together with strong phylogenetic value. J Clin Microbiol 47: 3121–3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Malachowa N, Sabat A, Gniadkowski M, Krzyszton-Russjan J, Empel J, et al. (2005) Comparison of multiple-locus variable-number tandem-repeat analysis with pulsed-field gel electrophoresis, spa typing, and multilocus sequence typing for clonal characterization of Staphylococcus aureus isolates. J Clin Microbiol 43: 3095–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Achtman M (2012) Insights from genomic comparisons of genetically monomorphic bacterial pathogens. Philos Trans R Soc Lond B Biol Sci 367: 860–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Köck R, Loth B, Koksal M, Schulte-Wulwer J, Harlizius J, et al. (2012) Persistence of Nasal Colonization with Livestock-Associated Methicillin-Resistant Staphylococcus aureus in Pig Farmers after Holidays from Pig Exposure. Appl Environ Microbiol 78: 4046–4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. de Neeling AJ, van den Broek MJ, Spalburg EC, van Santen-Verheuvel MG, Dam-Deisz WD, et al. (2007) High prevalence of methicillin resistant Staphylococcus aureus in pigs. Vet Microbiol 122: 366–372. [DOI] [PubMed] [Google Scholar]
- 38. Furuya D, Tsuji N, Kuribayashi K, Tanaka M, Hosono Y, et al. (2010) Evaluation of spa typing for the classification of clinical methicillin-resistant Staphylococcus aureus isolates. Jpn J Infect Dis 63: 364–367. [PubMed] [Google Scholar]
- 39. Vanderhaeghen W, Hermans K, Haesebrouck F, Butaye P (2010) Methicillin-resistant Staphylococcus aureus (MRSA) in food production animals. Epidemiol Infect 138: 606–625. [DOI] [PubMed] [Google Scholar]
- 40. Broens EM, Graat EA, van der Wolf PJ, van de Giessen AW, van Duijkeren E, et al. (2011) MRSA CC398 in the pig production chain. Prev Vet Med 98: 182–189. [DOI] [PubMed] [Google Scholar]
- 41. Espinosa-Gongora C, Broens EM, Moodley A, Nielsen JP, Guardabassi L (2012) Transmission of MRSA CC398 strains between pig farms related by trade of animals. Vet Rec 170: 564. [DOI] [PubMed] [Google Scholar]
- 42. Holmes A, Edwards GF, Girvan EK, Hannant W, Danial J, et al. (2010) Comparison of two multilocus variable-number tandem-repeat methods and pulsed-field gel electrophoresis for differentiating highly clonal methicillin-resistant Staphylococcus aureus isolates. J Clin Microbiol 48: 3600–3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sabat A, Krzyszton-Russjan J, Strzalka W, Filipek R, Kosowska K, et al. (2003) New method for typing Staphylococcus aureus strains: multiple-locus variable-number tandem repeat analysis of polymorphism and genetic relationships of clinical isolates. J Clin Microbiol 41: 1801–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tenover FC, Vaughn RR, McDougal LK, Fosheim GE, McGowan JE Jr (2007) Multiple-locus variable-number tandem-repeat assay analysis of methicillin-resistant Staphylococcus aureus strains. J Clin Microbiol 45: 2215–2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sobral D, Schwarz S, Bergonier D, Brisabois A, Fessler AT, et al. (2012) High Throughput Multiple Locus Variable Number of Tandem Repeat Analysis (MLVA) of Staphylococcus aureus from Human, Animal and Food Sources. PLoS ONE 7: e33967. [DOI] [PMC free article] [PubMed] [Google Scholar]