Abstract
Accurate positioning of spindles is essential for asymmetric mitotic and meiotic cell divisions that are crucial for animal development and oocyte maturation, respectively. The predominant model for spindle positioning, termed “cortical pulling,” involves attachment of the microtubule-based motor cytoplasmic dynein to the cortex, where it exerts a pulling force on microtubules that extend from the spindle poles to the cell cortex, thereby displacing the spindle. Recent studies have addressed important details of the cortical pulling mechanism and have revealed alternative mechanisms that may be used when microtubules do not extend from the spindle to the cortex.
Mitotic and meiotic spindles are precisely positioned within eukaryotic cells for several reasons. In animal cells, spindle position determines the location of contractile ring assembly (Green et al., 2012). Thus, placing a spindle in the center of the cell will result in daughter cells of equal size, whereas positioning the spindle asymmetrically results in daughter cells of different sizes. In oocytes, the extreme asymmetrical positioning of the meiotic spindle allows expulsion of three fourths of the chromosomes into two tiny polar bodies while preserving most of the cytoplasm in the egg for the developing zygote. In polarized cells, where proteins and RNAs are asymmetrically distributed before division, the orientation of the spindle relative to the polarity axis determines whether the daughter cells will have the same or different developmental fates. An excellent review of the developmental context of spindle positioning is provided by Morin and Bellaïche (2011). In budding yeast (Slaughter et al., 2009) and plants (Rasmussen et al., 2011), the site of cytokinesis is determined before the spindle forms. In these organisms, the spindle must be oriented relative to the predetermined division plane to ensure that both daughter cells receive a complete chromosome complement.
The majority of research on the mechanisms of spindle positioning has focused on cell types that have “astral” microtubules. Astral microtubules have minus ends embedded in the spindle poles and plus ends extending outward, away from the spindle toward the cell cortex (Fig. 1, A and B). Astral microtubules have been proposed to mediate spindle positioning by generating pulling forces at the cortex or pulling forces against the cytoplasm. The minus ends of astral microtubules are embedded in the pericentriolar material that surrounds the centrioles of animal cells or in the spindle pole bodies of fungi. This attachment is essential for pulling forces on the astral microtubules to move the spindle. However, late stage oocytes of several animal phyla and all cells of flowering plants lack centrioles and lack obvious astral microtubules. Thus, these cell types have evolved alternative spindle positioning mechanisms. Here we review recent advances in both astral and nonastral spindle positioning mechanisms.
Figure 1.
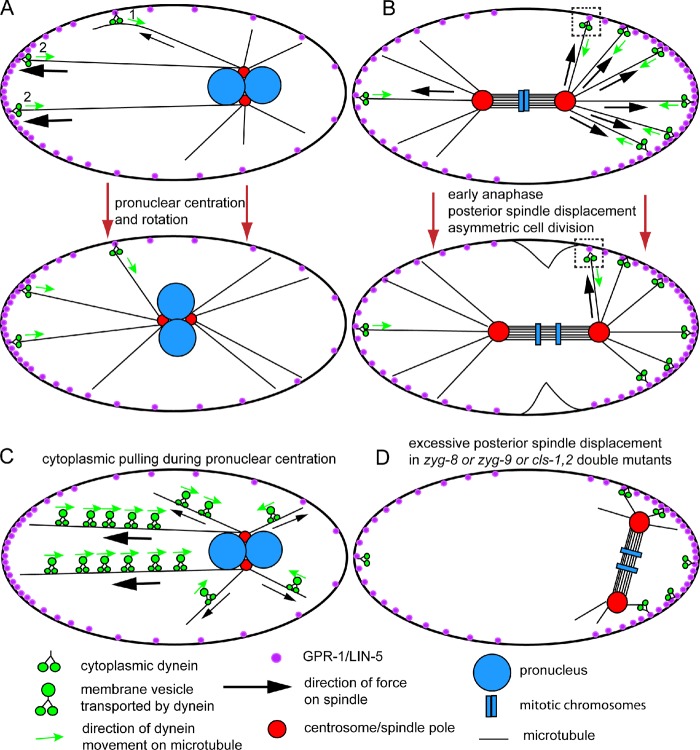
Mitotic spindle movements in the C. elegans zygote. (A) Schematic diagram of a single-celled C. elegans embryo showing cortical pulling by cytoplasmic dynein during pronuclear centration and rotation. The nuclei move toward the anterior (left) so that the spindle assembles in the center of the embryo. (B) Schematic diagram of a single-celled C. elegans embryo showing cortical pulling by dynein during anaphase. The spindle moves to the posterior (right) so that cytokinesis generates two cells of different sizes. The squares highlight a dynein molecule that pulls toward the posterior before spindle displacement, then pulls toward the anterior after spindle displacement. (C) Schematic drawing of cytoplasmic pulling that contributes to centering the pronuclei. (D) Illustration of a spindle that was centered at metaphase but in which both poles moved all the way to the posterior end of the embryo. This occurs in zyg-8 mutants (Gönczy et al., 2001), cls-1,2 (RNAi) embryos (Espiritu et al., 2012), and zyg-9(ts) mutants shifted to a nonpermissive temperature at metaphase (Bellanger et al., 2007), possibly because astral microtubules are too short to reach force generators that would pull toward the anterior.
Cortical versus cytoplasmic pulling
The most prominent model of spindle positioning involves a cortical pulling mechanism. In this model, the minus end–directed microtubule motor protein, cytoplasmic dynein, is attached to the cell cortex and exerts pulling forces on the plus ends of astral microtubules that reach the cortex. In the single-celled Caenorhabditis elegans embryo at early prophase, complexes of GPR-1,2 (G protein regulator) and LIN-5 (abnormal cell lineage), positive regulators of cytoplasmic dynein, are more concentrated at the anterior cortex of the embryo (Fig. 1 A; Park and Rose, 2008), resulting in greater net pulling force toward the anterior. This results in net movement of the pronuclei to the center of the embryo and rotation of the centrosome–pronuclear complex so that the metaphase spindle forms in the center of the embryo with its poles oriented along the anterior-posterior axis of the embryo (Fig. 1 B). During metaphase and early anaphase, GPR-1,2 and LIN-5 become more concentrated at the posterior end of the embryo, resulting in movement of the spindle toward the posterior so that cytokinesis generates daughter cells of different sizes (Fig. 1 B; Grill et al., 2001, 2003). Depending on the relative distribution of active force generators at the cortex, this mechanism can also lead to centering the spindle within the cell to allow symmetric cytokinesis as occurs in HeLa cells (Kiyomitsu and Cheeseman, 2012) and LLC-Pk1 cells (Collins et al., 2012). Cortical pulling might involve pulling on the sides of microtubules that bend as they approach the cortex (Fig. 1 A, 1) or end-on interactions that require coupling of microtubule depolymerization with pulling (Fig. 1 A, 2), as occurs at kinetochores during anaphase A (McIntosh et al., 2010).
Cortical pulling differs from a cytoplasmic pulling mechanism most clearly proposed by Kimura and Kimura (2011) and diagrammed in Fig. 1 C. In this cytoplasmic pulling mechanism, the viscous drag on membranous organelles transported toward the minus ends of astral microtubules by cytoplasmic dynein generates a force in the opposite direction, toward the cortex. Depletion of RAB-5, RAB-7, or RILP-1 (RAB-7 interacting lysosomal protein homologue) blocks dynein-dependent organelle transport and slows the velocity of pronuclear centration in C. elegans without affecting other dynein-dependent movements (Kimura and Kimura, 2011). Elimination of cortical pulling by depleting GPR-1,2 also slows pronuclear centration (Park and Rose, 2008), which suggests that cortical pulling and cytoplasmic pulling each contribute 50% of the velocity of pronuclear centration. Unlike end-on cortical pulling, cytoplasmic pulling force is proportional to the length of the astral microtubules because more organelles will be transported on a long microtubule than a short microtubule (Fig. 1 C). This generates a self-centering mechanism as the length of the astral microtubules equalize when the pronuclei reach the center of the zygote (Fig. 1 A; Hamaguchi and Hiramoto, 1986). Cytoplasmic pulling may predominate in very large zygotes where astral microtubules clearly do not reach the cortex but where both pronuclei and the mitotic spindle are centered (Mitchison et al., 2012).
Evidence for cortical pulling.
The cortical pulling mechanism requires that microtubules extend from the spindle to the cortex and form a contiguous structure that is mechanically robust enough that the force generators do not pull the minus ends of the microtubules out of the spindle pole or cause the plasma membrane to buckle inward. Experimental evidence for this contiguous mechanical linkage comes from experiments in oocytes of the marine annelid, Chaetopterus. Insertion of a glass needle into the meiotic spindle allowed pulling the spindle away from the cortex, which caused inward buckling of the cortex. Further pulling resulted in sudden release of the spindle from the cortex, restoration of cortical shape, and concomitant disappearance of a birefringent aster extending between the spindle and cortex (Lutz et al., 1988). The second requirement for a cortical pulling mechanism is that force is generated at the cortex. Using a laser to cut the central spindle of an early anaphase C. elegans embryo, Grill et al. (2001) showed that spindle poles are pulled from outside the spindle rather than pushed from inside the spindle during posterior spindle displacement. Fragmentation of centrosomes with a laser (Grill et al., 2003) revealed that astral microtubules freed from the spindle move outward toward the cortex. Either cortical pulling or cytoplasmic pulling could explain this result; however, the asymmetric distribution of fragment velocities is controlled by proteins that are localized at the cortex, GPR-1,2 (Grill et al., 2003) and LET-99 (lethal-99; Krueger et al., 2010). In a key experiment, Redemann et al. (2010) showed that microtubule plus ends pull tubular invaginations of the plasma membrane inward when cortical stiffness is partially reduced. This experiment showed that astral microtubules are pulling on the cortex during spindle displacement, which would not occur if forces were generated by movement of cytoplasmic organelles along the sides of microtubules.
End-on versus side-on microtubule–cortex interactions.
How do microtubules interact with force generators at the cortex? Astral microtubules might polymerize to the cortex then bend along it so that cortical motors interact with the side of the microtubule to generate force (Fig. 1 A, 1). This type of interaction would be consistent with the in vitro gliding motility of microtubules generated by cytoplasmic dynein immobilized on glass coverslips (Paschal et al., 1987; Vallee et al., 1988) and is the only type of dynein-dependent cortical microtubule interaction observed in budding yeast (Adames and Cooper, 2000). Studies of C. elegans embryos, however, support end-on microtubule contacts (Fig. 1 A, 2) as the functional contact with cortical force generators for posterior displacement of the early anaphase spindle. Live imaging of YFP-tubulin in optical sections at the surface of the embryo during anaphase revealed dots rather than lines, indicating that the microtubules contacting the cortex are <200 nm in length (Labbé et al., 2003; Kozlowski et al., 2007). When short microtubule fragments are generated by ectopic katanin activity, dynein-dependent gliding of microtubule “lines” on the cortex is frequently observed (Gusnowski and Srayko, 2011), indicating that cortical dynein is capable of moving microtubules along the cortex via side-on interactions in wild-type embryos but that it does not during posterior displacement of the anaphase spindle. A likely explanation comes from the finding that astral microtubule plus ends undergo catastrophe (switch to depolymerization) on average 1.4 s after polymerizing to the cortex (Kozlowski et al., 2007). Thus astral microtubule plus ends do not have time to polymerize along the cortex to establish extensive side-on contacts. Support for this idea comes from depletion of the conserved plasma membrane protein EFA-6 (exchange factor for Arf) from the C. elegans embryo. In the absence of EFA-6, the residence time of microtubule plus ends at the cortex increases fivefold, astral microtubules form extensive lateral contacts with the cortex, and centrosomes exhibit movements consistent with excessive dynein-dependent cortical pulling (O’Rourke et al., 2010). Side-on contacts of astral microtubules with the cortex occur later during telophase in the wild-type C. elegans embryo (Kozlowski et al., 2007), but the nature of this switch has not been addressed.
The two distinct activities of cytoplasmic dynein, end-on pulling and walking along the side of a microtubule, have been genetically separated in C. elegans. Cortical pulling forces during early anaphase require the redundant cortical dynein activators GPR-1 and -2 (Grill et al., 2003), which bind to LIN-5 (Gotta et al., 2003). GPR-1,2/LIN-5 is anchored in the plasma membrane via the myristoyl and palmitoyl lipid modifications of the redundant Gα proteins GOA-1 and GPA-16 (Gotta et al., 2003; Park and Rose, 2008; Kotak et al., 2012). The complex of GPR-1 and LIN-5 interacts with the dynein light chain DYRB-1 (Couwenbergs et al., 2007) and the dynein regulator LIS-1 (human lissencephaly gene related; Nguyen-Ngoc et al., 2007). GPR-1 and -2, however, are not required for dynein-dependent gliding of severed microtubule fragments along the cortex (Gusnowski and Srayko, 2011), dynein-dependent transport of membranous organelles along the sides of microtubules (Kimura and Kimura, 2011), dynein-dependent positioning of the acentriolar C. elegans meiotic spindle (van der Voet et al., 2009), or dynein-dependent centration of the male pronucleus (Kimura and Kimura, 2011). These GPR-independent activities of cytoplasmic dynein likely do not require end-on pulling.
Recently, end-on pulling by cytoplasmic dynein has been reconstituted in vitro with a purified preparation of artificially dimerized budding yeast cytoplasmic dynein. Laan et al. (2012) immobilized purified cytoplasmic dynein on microfabricated barriers and observed the interaction of centrosome-nucleated microtubules as they approached these dynein-coated barriers. Microtubule plus ends hitting a dynein-coated barrier switched to catastrophe with high frequency but the microtubule depolymerization rate after the catastrophe was reduced. The result was an extended period of interaction between the depolymerizing plus end and the dynein-coated barrier. Plus-end depolymerization pulled the centrosome toward the barrier, and in similar reactions the pulling force was measured as high as 5 pN. Side-on interactions with the barrier were not observed. Whereas ATP was required for this end-on pulling, it is not clear if the energy source is ATP hydrolysis–driven stepping by dynein or if the energy source is GTP hydrolysis–driven depolymerization of the microtubule. In the latter case, ATP might only be required to prevent rigor binding of dynein to the microtubule. Indeed, an artificial rigor binding of a streptavidin-coated bead to a depolymerizing biotinylated microtubule plus end resulted in a pulling force that is restricted to an extremely short distance (Grishchuk et al., 2005). In vitro reconstitution of pulling force coupled to depolymerizing microtubule plus ends was first demonstrated with beads coated with kinesin-1 or a nonmotile kinesin chimera (NK350; Lombillo et al., 1995). Like barrier-bound cytoplasmic dynein, kinesin-coated beads slowed the depolymerization rate of microtubule plus ends, whereas NK350-coated beads enhanced the depolymerization rate of bound plus ends. ATP enhanced depolymerization-coupled pulling for both kinesin-1 and NK350, just as it did for barrier-bound cytoplasmic dynein. The in vitro pulling reaction reconstituted by Laan et al. (2012) seems unlikely to be the same reaction that pulls on plus ends in the anaphase budding yeast cell, as these are through side-on interactions (Adames and Cooper, 2000). In vitro reconstitution of a GPR/LIN-5–dependent, end-on pulling reaction with purified metazoan cytoplasmic dynein may reveal mechanisms acting on spindles in vivo.
Another interesting contrast between end-on versus side-on cortical pulling reactions is suggested by differences in the dependence on cortical F-actin. F-actin is required for cortical rigidity to prevent end-on microtubule contacts from pulling membrane tubules inward instead of moving the spindle pole outward (Redemann et al., 2010). Side-on pulling by cortical dynein in budding yeast, however, does not require F-actin (Theesfeld et al., 1999; Heil-Chapdelaine et al., 2000a). The curvature of microtubules gliding on the bud cortex indicates that the microtubule is engaged with multiple dynein molecules distributed over several microns of cortex, and this distribution of force might allow effective pulling against a less rigid cortex. Alternatively, rigidity of the yeast plasma membrane might be mediated by oligomers of BAR domain proteins like Num1 (nuclear migration; Tang et al., 2012) or eisosomes (Walther et al., 2006; Olivera-Couto et al., 2011), by osmotic pressure, or by attachment of the plasma membrane to the cell wall.
Why the spindle is not pulled all the way to the cortex with more active force generators.
What prevents the C. elegans centrosome–pronuclear complex from moving all the way to the anterior cortex where the concentration of cortical force generators is highest during prophase (Fig. 1 A), and what prevents the spindle from moving all the way to the posterior cortex, which has the highest concentration of active force generators during anaphase (Fig. 1, B and D)? Increasing the concentration of GPR-1,2/LIN-5 at the anterior cortex causes pronuclei to move further toward the anterior, but they still do not crash into the anterior cortex (Panbianco et al., 2008). During metaphase/anaphase (Fig. 1 B), weak cortical pulling on the anterior aster might oppose strong pulling on the posterior aster. Supporting this idea, spindle severing results in the posterior aster moving further posterior than when the spindle is intact (Grill et al., 2001), but the posterior pole still does not move all the way to the posterior cortex. Monopolar spindles move toward the posterior in a GPR1,2-dependent manner but then reverse direction and oscillate along the anterior-posterior axis (Krueger et al., 2010). Laan et al. (2012) found that centrosome-nucleated microtubule asters could accurately self-center within microchambers whose walls are coated with dynein. Their mathematical modeling suggested that self-centering was achieved because of a balance between cortical pulling by dynein and pushing by polymerizing microtubules (Dogterom and Yurke, 1997) that have not yet engaged a dynein molecule. Thus, cortical pushing by astral microtubules might buffer cortical pulling in vivo. Grill and Hyman (2005) suggested another simple solution. When a spindle pole moves to the posterior, it passes a subset of cortical force generators that were initially pulling toward the posterior (Fig. 1 B, box). After passage of the spindle pole, these force generators pull toward the anterior. Failure in any of these buffering mechanisms might explain why both spindle poles move to the posterior cortex (Fig. 1 D) in mutants that have either short astral microtubules or fewer astral microtubules reaching the cortex (Gönczy et al., 2001; Bellanger et al., 2007; Espiritu et al., 2012).
Recent experiments in HeLa and LLC-Pk1 cells have revealed a more sophisticated feedback mechanism that effectively centers the mitotic spindle. Kiyomitsu and Cheeseman (2012) found that HeLa cell mitotic spindles oscillate back and forth along their pole-to-pole axis. They found that dynein/dynactin formed a lateral crescent on the cortex when the spindle was far from that lateral cortex (Fig. 2 A). As the spindle moved toward the dynein crescent, the dynein crescent disappeared as the spindle approached and a new crescent appeared on the opposite lateral cortex (Fig. 2 C). They found that polo kinase 1 (Plk1), which is concentrated on spindle poles, causes dynein/dynactin to dissociate from the LGN–NuMA–Gαi complex (Leu-Gly-Asn repeat enriched protein–nuclear mitotic apparatus protein–Gα; homologues of GPR-1,2–LIN-5–Gα), which explains why the dynein crescent disappears once the spindle pole gets close to the cortex. They also found that the GTP-Ran gradient produced by chromosome-bound RCC1 (regulator of chromosome condensation) was responsible for inhibiting LGN–NuMA association with the cortex, and hence dynein, above the central spindle (Fig. 2). The RCC1 pathway explains why the spindles move only along their pole–pole axis. The two pathways combine to center the spindle in two different axes. Similar spindle oscillations with dynein/dynactin crescents forming only when the spindle pole is far from the cortex were reported in LLC-Pk1 epithelial cells (Collins et al., 2012). Because spindles are small relative to the two-dimensional flattened area of these cells, the role of this pathway in centering spindles to allow symmetric cytokinesis is much more obvious than in HeLa cells.
Figure 2.
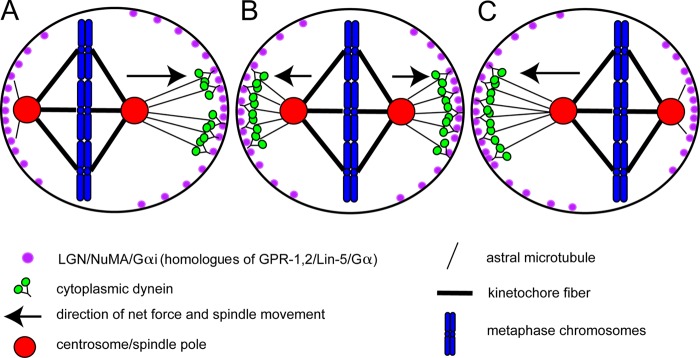
How to center a spindle. Schematic diagram of a metaphase HeLa cell where the spindle oscillates along its pole–pole axis to maintain a centered position to allow symmetric cytokinesis. (A) When the left spindle pole is close to the cortex, Plk1 on the pole (red) causes dynein (green) to dissociate from LGN–NuMA complexes (purple; human homologues of GPR-1,2/LIN-5). (B and C) The spindle moves to the right because of the higher concentration of LGN–NuMA–dynein complexes on the right cortex. When chromosomes are close to the cortex as in A, the GTP-Ran gradient from the chromosomes causes dissociation of LGN/NuMA from the cortex. This second system centers the spindle in the axis perpendicular to the pole–pole axis.
There are normal situations where movement of one spindle pole all the way to the cortex occurs. This exaggerated movement that results in one set of astral microtubules being splayed onto the cortex is observed in fourth cleavage sea urchin embryos (Holy and Schatten, 1991) and for the female meiotic spindles of Chaetopterus (Lutz et al., 1988) and Spisula solidissima (Pielak et al., 2004).
The budding yeast model.
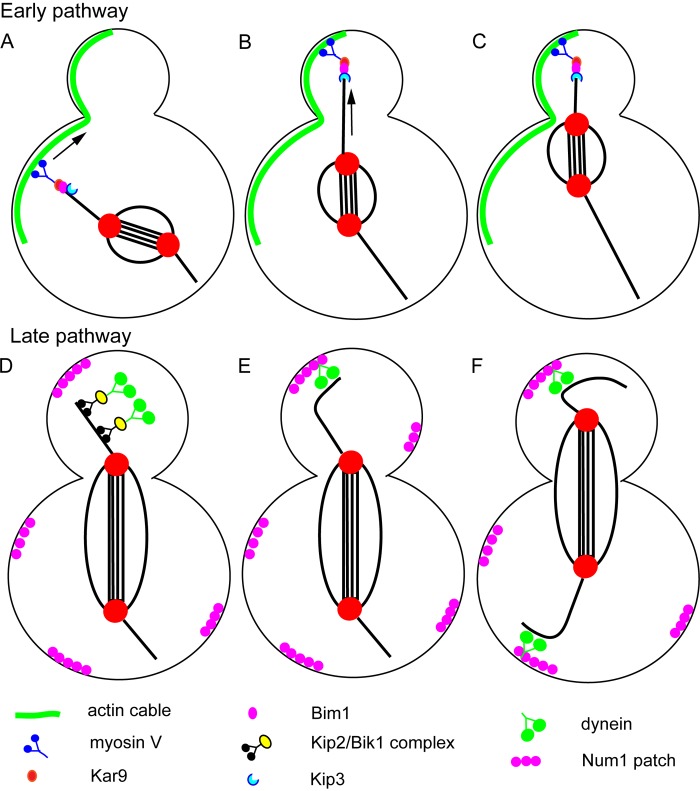
The S. cerevisiae mitotic spindle is positioned relative to a preformed bud neck by two sequential and partially redundant cortical pulling pathways to ensure that chromosomes are deposited into both mother and daughter cells. Spindle positioning is also monitored by a budding yeast–specific checkpoint (Caydasi et al., 2010). Deletion of genes in any one positioning pathway results in a viable yeast strain, whereas double mutants bearing deletions of genes in both positioning pathways or of genes in one positioning pathway plus the checkpoint results in lethality (Miller et al., 1998). In the early pathway, the microtubule plus-end tip tracking protein, Bim1 (binding to microtubules; EB1 homologue), binds to the yeast-specific adaptor protein Kar9 (karyogamy; Korinek et al., 2000; Lee et al., 2000; Miller et al., 2000), which binds to the yeast myosin V (Myo2; Yin et al., 2000). Myosin V transports the growing microtubule plus end toward the bud tip on polarized actin cables (Hwang et al., 2003). This results in a unique “sweeping” or “pivoting” of the growing astral microtubule toward the bud neck (Fig. 3 A; Adames and Cooper, 2000). Because Bim1 only binds to growing microtubule plus ends (Zimniak et al., 2009), this mechanism alone cannot bring the spindle to the bud neck because the polymerizing astral microtubule would push the spindle back into the mother cell. To prevent the astral microtubules from becoming too long, the kinesin-8 family member, Kip3, passively tracks the plus end until the plus end reaches the bud cortex (Fig. 3 A). Kip3 then switches to a plus-end depolymerase and shortens the astral microtubule to pull the spindle to the bud neck (Fig. 3, B and C; Gupta et al., 2006). Purified Kip3 has unique biochemical properties that contribute to its in vivo function. In vitro, Kip3 accumulates at plus-end tips via its plus end–directed motor activity. Longer microtubules accumulated more Kip3 at their plus ends than short microtubules simply because there are more lateral binding sites on a long microtubule and Kip3 switches to a plus-end depolymerase only when the microtubule has reached a threshold length (Varga et al., 2009). Plus-end pulling by the fission yeast homologue of Kip3 has also been reconstituted in vitro (Grissom et al., 2009). More recent work suggests that Kip3-dependent cortical pulling requires the concerted action of the cortical protein, Bud6 (BUD site selection), and the plus end tracker, Bim1, as well as cytoplasmic dynein (ten Hoopen et al., 2012).
Figure 3.
Spindle positioning in budding yeast. Schematic diagram of the two sequential spindle positioning pathways of budding yeast. In the early pathway (A–C), myosin V transports the plus end of an astral microtubule toward the bud tip on a polarized actin cable. Once the plus end has reached the bud cortex, the plus-end depolymerase, KIP3, is activated to allow pulling of the spindle pole toward the bud neck. In the late pathway (D–F), the plus end–directed microtubule motor Kip2 transports dynein to the plus ends of microtubules via the adaptor protein Bik1 (D). Dynein can be targeted to plus ends by two additional Bik1-dependent mechanisms (see text). When dynein reaches the bud cortex on a polymerizing microtubule plus end (E), contact with the cortical protein Num1 allows dynein to pull the spindle toward the bud (F). During early anaphase (D and E), dynein is not loaded onto microtubules in the mother cell. During late anaphase (F), dynein is loaded on microtubules in the mother cell to prevent movement of the spindle all the way into the bud.
The late pathway is initiated when the yeast-specific dynein inhibitor She1 (sensitivity to high expression) is removed from astral microtubules at the metaphase–anaphase transition (Woodruff et al., 2009). She1 appears to act specifically by preventing recruitment of dynactin to microtubules (Bergman et al., 2012) and by inhibiting dynein motility (Markus et al., 2012). In the late pathway, cytoplasmic dynein is targeted to growing microtubule plus ends by the plus-end tracking protein Bik1 (bilateral karyogamy defect; Sheeman et al., 2003), which itself is targeted to plus ends by three partially redundant mechanisms. In the first mechanism, Bik1 is transported as cargo by the plus end–directed kinesin Kip2 (Carvalho et al., 2004). Cytoplasmic dynein is thus transported as cargo to the bud cortex by Bik1–Kip2 complexes (Fig. 3 D). In the absence of Kip2, Bik1 (and therefore dynein) can still track growing microtubule plus ends either through a second mechanism that requires the C-terminal tyrosine residue of α-tubulin or a third mechanism that requires the plus end–tracking protein Bim1 (Caudron et al., 2008). This is partially consistent with results of reconstitution experiments with purified proteins showing that the Bik1 homologue, CLIP170, tracks growing plus ends through a mechanism that involves binding to both the Bim1 homologue, EB1, and the C-terminal tyrosine-containing motif of α-tubulin (Bieling et al., 2008). When a microtubule plus end carrying dynein contacts the yeast-specific cortical protein Num1, Num1 apparently stimulates off-loading of the dynein tail onto the cortex so that the dynein motor domains engage the microtubule in a cortical pulling reaction (Fig. 3, E and F). Deletion of Num1 results in accumulation of inactive dynein at plus ends (Lee et al., 2003; Sheeman et al., 2003; Markus and Lee, 2011). In contrast with the GPR-dependent end-on cortical interactions observed in C. elegans, dynein-dependent cortical pulling is mediated by lateral sliding of astral microtubules along the yeast bud cortex (Fig. 3, E and F; Adames and Cooper, 2000). Also, unlike cortical GPR/LIN-5 in animal cells, cortical Num1 is distributed in patches throughout both mother and daughter cells (Heil-Chapdelaine et al., 2000b) and even participates in mitochondrial positioning and fission throughout the cell (Cerveny et al., 2007; Hammermeister et al., 2010). If there is no asymmetrically distributed cortical activator of dynein, how is dynein-mediated pulling directed specifically toward the bud cortex?
Why both spindle poles are not pulled to the same cortex in budding yeast.
The budding yeast spindle, rather than the cortex, is asymmetrical. At metaphase, Kar9 is asymmetrically localized on the spindle pole oriented toward the bud and on the plus ends of astral microtubules emanating from that bud-proximal spindle pole (Liakopoulos et al., 2003). This alone would explain why only one spindle pole moves toward the bud but then leaves the question of how Kar9 asymmetry is established. Cepeda-García et al. (2010) found that Kar9 at spindle poles became symmetrical and reduced in concentration after depolymerization of actin cables, depolymerization of microtubules, or disruption of the myosin V–Kar9 interaction. When microtubules were repolymerized, Kar9 was initially symmetrical on both spindle poles and quickly repolarized onto the first microtubule to make a functional cortical contact. These results suggested a positive feedback loop in which functional cortical pulling by Bim1–Kar9–Myo2 complexes causes loading of additional Kar9. Cytoplasmic dynein also accumulates preferentially on the plus-end tips that reach the bud neck first (Fig. 3, D and E) and on the bud-proximal spindle pole during metaphase. The asymmetrical accumulation of dynein on the bud-proximal microtubules and bud-proximal spindle pole requires kinases that are found at the bud neck. Thus, the asymmetry of dynein localization may be generated by a positive feedback loop, as suggested for Kar9 asymmetry. After the spindle is pulled into the bud neck, during anaphase, dynein becomes symmetrical on the plus ends emanating from both poles (Fig. 3 F; Grava et al., 2006). This regulation prevents both poles from moving toward the bud during metaphase and then prevents the spindle from being pulled all the way into the bud during anaphase. Asymmetric localization of dynein on spindle poles or microtubule plus ends has not yet been reported in animal cells.
Other spindle positioning mechanisms
In the examples of the C. elegans and budding yeast mitotic spindles, long astral microtubules are in contact with the cell cortex. In cells where spindles have no astral microtubules, other mechanisms must be at work. Female meiotic spindles are universally positioned with one spindle pole contacting the oocyte cortex so that one set of chromosomes can be eliminated in a polar body through an extremely asymmetrical division (Fabritius et al., 2011). Female meiotic spindles of at least three animal phyla (Chordata, Nematoda, and Arthropoda), however, have no centrioles in their spindle poles and no apparent astral microtubules. Work in C. elegans and mice suggests that different species have evolved different mechanisms for acentriolar meiotic spindle positioning.
Parallel metaphase meiotic spindles.
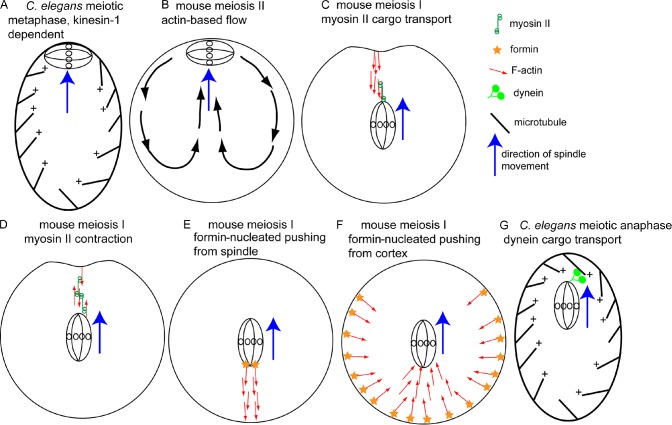
The metaphase I and metaphase II spindles of C. elegans (Fig. 4 A) and the metaphase II spindle of mouse (Fig. 4 B) are positioned at the cortex in a parallel orientation, with both poles equidistant from the cortex. In C. elegans, this parallel cortical positioning requires microtubules, kinesin-1, and a worm-specific kinesin-1–binding partner, KCA-1 (Yang et al., 2003, 2005), but is independent of F-actin (Yang et al., 2003). Kinesin-1 and microtubules are also required to move the nucleus to the cortex before germinal vesicle breakdown and to drive transport of yolk granules inward from the cortex, which results in a circular streaming pattern. It has been suggested that kinesin-1 may only move the germinal vesicle to the cortex and that an additional, unidentified pathway moves the spindle over the remaining distance to the cortex and establishes the parallel orientation (McNally et al., 2010). The mouse metaphase II spindle is maintained in a similar parallel orientation at the cortex, but this positioning requires the actin nucleator ARP2/3. ARP2/3 also drives streaming of actin filaments and cytoplasm in a pattern that has been proposed to push the spindle into the cortex to maintain parallel cortical position (Fig. 4 B; Yi et al., 2011).
Figure 4.
A plethora of nonastral spindle positioning mechanisms. (A) Metaphase C. elegans meiotic spindles are positioned in a parallel orientation at the cortex by microtubules and kinesin-1. (B) The mouse metaphase II spindle may be positioned by actin-dependent cytoplasmic streaming. Pole-first migration of the mouse meiosis I spindle to the cortex may be mediated by cargo transport on parallel actin filaments by spindle pole–bound myosin II (C), myosin II–based contraction of anti-parallel actin filaments (D), or pushing forces generated by polymerizing actin filaments nucleated by formin molecules on the spindle (E) or nucleated by formin molecules on the cortex (F). Red arrows indicate the pointed ends of actin filaments. (G) One spindle pole of the early anaphase C. elegans meiotic spindle may be transported to the cortex as cargo by dynein on polarized cytoplasmic microtubules.
Pole first migration of the mouse meiosis I spindle.
Unlike the C. elegans germinal vesicle, the mouse germinal vesicle is centered in the egg at germinal vesicle breakdown. Thus, the meiosis I spindle assembles near the center of the egg then migrates in a pole-first orientation to the nearest cortex so that it never adopts a parallel orientation (Fig. 4 C). This migration requires F-actin (Verlhac et al., 2000) and the actin nucleators Formin 2 (Dumont et al., 2007), Spire 1 and Spire 2 (Pfender et al., 2011), and ARP2/3 (Sun et al., 2011), but the mechanism of movement remains unclear. Schuh and Ellenberg (2008) demonstrated the existence of actin bundles extending between the spindle and an invagination of the cortex during spindle migration. The invagination indicated a pulling mechanism, and Schuh and Ellenberg (2008) suggested that myosin II on the spindle poles might walk on a discontinuous actin network with barbed ends oriented toward the cortex (Fig. 4 C). In support of this model, the Rho kinase inhibitor ML7 eliminated spindle pole staining by an antibody specific for phosphorylated myosin regulatory light chain and blocked spindle migration (Schuh and Ellenberg, 2008). However, Li et al. (2008) found that the myosin ATPase inhibitor, blebbistatin, had no effect on spindle migration even though it completely blocked polar body extrusion (cytokinesis). Because myosin II typically acts by forming bipolar thick filaments that exert contractile force on antiparallel actin filaments, a myosin II–based model would make more sense if myosin II was concentrated on antiparallel actin bundles extending between the spindle and cortex (Fig. 4 D). Myosin V is more typically associated with the transport of cargo on uniformly oriented actin filaments, and a recent study has shown that myosin V drives transport of secretory vesicles outward toward the cortex in germinal vesicle stage oocytes (Schuh, 2011). This study at least suggests that the cytoplasmic actin meshwork has a net polarity with barbed ends toward the cortex, a prerequisite for a myosin cargo transport model (Fig. 4 C).
Li et al. (2008) proposed a completely different mechanism in which F-actin nucleated near the chromosomes generates a cloud of F-actin that pushes the spindle toward the cortex (Fig. 4 E) in a manner analogous to Listeria monocytogenes motility (Lambrechts et al., 2008). Support for this pushing model comes from imaging of an actin cloud behind the migrating spindle and localization of Formin 2 around the spindle (Li et al., 2008). In addition, Formin 2 overexpression causes invaginations in the nuclear envelope, which is consistent with inward pushing from the cortex, rather than protrusions that would be consistent with pulling forces from the cortex (Azoury et al., 2011). Formin 2 is symmetrical around the cortex during prophase but clears from the cortex in front of the migrating spindle, which is consistent with nucleation-based pushing from behind (Fig. 4 F; Azoury et al., 2011). As previously mentioned, cortical stiffness is a prerequisite for cortical pulling mechanisms because pulling on an unsupported plasma membrane should cause the membrane to invaginate inward instead of the spindle moving outward. Strikingly, cortical stiffness of the mouse oocyte decreases sixfold during spindle migration (Larson et al., 2010). Clearly, more work is required to resolve the mechanism of pole-first spindle migration in the mouse oocyte.
Rotation of the parallel metaphase spindle to a perpendicular anaphase spindle.
Activation of the anaphase-promoting complex results in rotation of the parallel metaphase meiotic spindle to a perpendicular orientation in both meiotic divisions of C. elegans, whereas fertilization induces rotation during meiosis II in mouse. In kinesin-1–depleted C. elegans embryos, the metaphase I spindle is far from the cortex and initiates pole-first migration to the cortex at the same time that wild-type rotation initiates (Yang et al., 2005). Both spindle rotation and late spindle migration in a kinesin mutant require cytoplasmic dynein (Ellefson and McNally, 2009, 2011). These results indicate that spindle rotation is simply migration of one spindle pole toward the cortex (analogous to spindle migration during mouse meiosis I). However, unlike dynein-dependent migration of one spindle pole toward the cortex during C. elegans mitosis or HeLa cell mitosis, C. elegans meiotic spindle rotation does not require GPR-1,2 (van der Voet et al., 2009). One possible model for C. elegans meiotic spindle rotation is that cytoplasmic dynein, which accumulates on both spindle poles just before and during rotation, transports one spindle pole as cargo on cytoplasmic microtubules with minus ends anchored at the cortex (Fig. 4 G). The orientation of these microtubules is inferred from the direction of kinesin-dependent yolk granule movement (McNally et al., 2010) and hook decoration in Xenopus laevis oocytes (Pfeiffer and Gard, 1999). This model is essentially the same as the myosin cargo transport model proposed for mouse meiosis I (Fig. 4 C). Rotation of the mouse meiosis II spindle is actin dependent (Maro et al., 1984) and myosin II dependent (Matson et al., 2006; Wang et al., 2011), but the mechanism is unknown.
Unifying themes and future directions
Work in HeLa cells and budding yeast suggests that negative feedback loops might generally lead to spindle centering and that positive feedback loops might generally lead to asymmetrical spindle positioning. Whereas the recent x-ray crystal structures of cytoplasmic dynein (Carter et al., 2011; Kon et al., 2012) have revealed great insights into how the motor walks, they have revealed little about how the motor is locally activated and anchored at the cortex or how GPR-1/LIN-5 switches the motor from a side-on motor to an end-on motor. More attention needs to be focused on the distinction between cortical stiffness and cortical anchoring required in any cortical pulling mechanism. The nonastral spindle positioning mechanisms acting in oocytes of mouse and C. elegans will require a much more detailed understanding of the polarity of cytoplasmic actin filaments and cytoplasmic microtubules.
Acknowledgments
Thanks to Lesilee Rose, Marina Ellefson Crowder, Amy Fabritius, Karen McNally, and the anonymous reviewers for critical comments.
F.J. McNally is supported by grant 2R01GM079421-05A1 from the National Institute of General Medical Sciences.
References
- Adames N.R., Cooper J.A. 2000. Microtubule interactions with the cell cortex causing nuclear movements in Saccharomyces cerevisiae. J. Cell Biol. 149:863–874 10.1083/jcb.149.4.863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azoury J., Lee K.W., Georget V., Hikal P., Verlhac M.H. 2011. Symmetry breaking in mouse oocytes requires transient F-actin meshwork destabilization. Development. 138:2903–2908 10.1242/dev.060269 [DOI] [PubMed] [Google Scholar]
- Bellanger J.M., Carter J.C., Phillips J.B., Canard C., Bowerman B., Gönczy P. 2007. ZYG-9, TAC-1 and ZYG-8 together ensure correct microtubule function throughout the cell cycle of C. elegans embryos. J. Cell Sci. 120:2963–2973 10.1242/jcs.004812 [DOI] [PubMed] [Google Scholar]
- Bergman Z.J., Xia X., Amaro I.A., Huffaker T.C. 2012. Constitutive dynein activity in She1 mutants reveals differences in microtubule attachment at the yeast spindle pole body. Mol. Biol. Cell. 23:2319–2326 10.1091/mbc.E12-03-0223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieling P., Kandels-Lewis S., Telley I.A., van Dijk J., Janke C., Surrey T. 2008. CLIP-170 tracks growing microtubule ends by dynamically recognizing composite EB1/tubulin-binding sites. J. Cell Biol. 183:1223–1233 10.1083/jcb.200809190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter A.P., Cho C., Jin L., Vale R.D. 2011. Crystal structure of the dynein motor domain. Science. 331:1159–1165 10.1126/science.1202393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho P., Gupta M.L., Jr, Hoyt M.A., Pellman D. 2004. Cell cycle control of kinesin-mediated transport of Bik1 (CLIP-170) regulates microtubule stability and dynein activation. Dev. Cell. 6:815–829 10.1016/j.devcel.2004.05.001 [DOI] [PubMed] [Google Scholar]
- Caudron F., Andrieux A., Job D., Boscheron C. 2008. A new role for kinesin-directed transport of Bik1p (CLIP-170) in Saccharomyces cerevisiae. J. Cell Sci. 121:1506–1513 10.1242/jcs.023374 [DOI] [PubMed] [Google Scholar]
- Caydasi A.K., Ibrahim B., Pereira G. 2010. Monitoring spindle orientation: Spindle position checkpoint in charge. Cell Div. 5:28 10.1186/1747-1028-5-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cepeda-García C., Delgehyr N., Juanes Ortiz M.A., ten Hoopen R., Zhiteneva A., Segal M. 2010. Actin-mediated delivery of astral microtubules instructs Kar9p asymmetric loading to the bud-ward spindle pole. Mol. Biol. Cell. 21:2685–2695 10.1091/mbc.E10-03-0197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerveny K.L., Studer S.L., Jensen R.E., Sesaki H. 2007. Yeast mitochondrial division and distribution require the cortical num1 protein. Dev. Cell. 12:363–375 10.1016/j.devcel.2007.01.017 [DOI] [PubMed] [Google Scholar]
- Collins E.S., Balchand S.K., Faraci J.L., Wadsworth P., Lee W.L. 2012. Cell cycle-regulated cortical dynein/dynactin promotes symmetric cell division by differential pole motion in anaphase. Mol. Biol. Cell. 23:3380–3390 10.1091/mbc.E12-02-0109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couwenbergs C., Labbé J.C., Goulding M., Marty T., Bowerman B., Gotta M. 2007. Heterotrimeric G protein signaling functions with dynein to promote spindle positioning in C. elegans. J. Cell Biol. 179:15–22 10.1083/jcb.200707085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogterom M., Yurke B. 1997. Measurement of the force-velocity relation for growing microtubules. Science. 278:856–860 10.1126/science.278.5339.856 [DOI] [PubMed] [Google Scholar]
- Dumont J., Million K., Sunderland K., Rassinier P., Lim H., Leader B., Verlhac M.H. 2007. Formin-2 is required for spindle migration and for the late steps of cytokinesis in mouse oocytes. Dev. Biol. 301:254–265 10.1016/j.ydbio.2006.08.044 [DOI] [PubMed] [Google Scholar]
- Ellefson M.L., McNally F.J. 2009. Kinesin-1 and cytoplasmic dynein act sequentially to move the meiotic spindle to the oocyte cortex in Caenorhabditis elegans. Mol. Biol. Cell. 20:2722–2730 10.1091/mbc.E08-12-1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellefson M.L., McNally F.J. 2011. CDK-1 inhibits meiotic spindle shortening and dynein-dependent spindle rotation in C. elegans. J. Cell Biol. 193:1229–1244 10.1083/jcb.201104008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espiritu E.B., Krueger L.E., Ye A., Rose L.S. 2012. CLASPs function redundantly to regulate astral microtubules in the C. elegans embryo. Dev. Biol. 368:242–254 10.1016/j.ydbio.2012.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabritius A.S., Ellefson M.L., McNally F.J. 2011. Nuclear and spindle positioning during oocyte meiosis. Curr. Opin. Cell Biol. 23:78–84 10.1016/j.ceb.2010.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gönczy P., Bellanger J.M., Kirkham M., Pozniakowski A., Baumer K., Phillips J.B., Hyman A.A. 2001. zyg-8, a gene required for spindle positioning in C. elegans, encodes a doublecortin-related kinase that promotes microtubule assembly. Dev. Cell. 1:363–375 10.1016/S1534-5807(01)00046-6 [DOI] [PubMed] [Google Scholar]
- Gotta M., Dong Y., Peterson Y.K., Lanier S.M., Ahringer J. 2003. Asymmetrically distributed C. elegans homologs of AGS3/PINS control spindle position in the early embryo. Curr. Biol. 13:1029–1037 10.1016/S0960-9822(03)00371-3 [DOI] [PubMed] [Google Scholar]
- Grava S., Schaerer F., Faty M., Philippsen P., Barral Y. 2006. Asymmetric recruitment of dynein to spindle poles and microtubules promotes proper spindle orientation in yeast. Dev. Cell. 10:425–439 10.1016/j.devcel.2006.02.018 [DOI] [PubMed] [Google Scholar]
- Green R.A., Paluch E., Oegema K. 2012. Cytokinesis in animal cells. Annu. Rev. Cell Dev. Biol. 28:29–58 10.1146/annurev-cellbio-101011-155718 [DOI] [PubMed] [Google Scholar]
- Grill S.W., Hyman A.A. 2005. Spindle positioning by cortical pulling forces. Dev. Cell. 8:461–465 10.1016/j.devcel.2005.03.014 [DOI] [PubMed] [Google Scholar]
- Grill S.W., Gönczy P., Stelzer E.H., Hyman A.A. 2001. Polarity controls forces governing asymmetric spindle positioning in the Caenorhabditis elegans embryo. Nature. 409:630–633 10.1038/35054572 [DOI] [PubMed] [Google Scholar]
- Grill S.W., Howard J., Schäffer E., Stelzer E.H., Hyman A.A. 2003. The distribution of active force generators controls mitotic spindle position. Science. 301:518–521 10.1126/science.1086560 [DOI] [PubMed] [Google Scholar]
- Grishchuk E.L., Molodtsov M.I., Ataullakhanov F.I., McIntosh J.R. 2005. Force production by disassembling microtubules. Nature. 438:384–388 10.1038/nature04132 [DOI] [PubMed] [Google Scholar]
- Grissom P.M., Fiedler T., Grishchuk E.L., Nicastro D., West R.R., McIntosh J.R. 2009. Kinesin-8 from fission yeast: a heterodimeric, plus-end-directed motor that can couple microtubule depolymerization to cargo movement. Mol. Biol. Cell. 20:963–972 10.1091/mbc.E08-09-0979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta M.L., Jr, Carvalho P., Roof D.M., Pellman D. 2006. Plus end-specific depolymerase activity of Kip3, a kinesin-8 protein, explains its role in positioning the yeast mitotic spindle. Nat. Cell Biol. 8:913–923 10.1038/ncb1457 [DOI] [PubMed] [Google Scholar]
- Gusnowski E.M., Srayko M. 2011. Visualization of dynein-dependent microtubule gliding at the cell cortex: implications for spindle positioning. J. Cell Biol. 194:377–386 10.1083/jcb.201103128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamaguchi M.S., Hiramoto Y. 1986. Analysis of the role of astral rays in pronuclear migration in sand dollar eggs by the colcemid-UV method. Dev. Growth Differ. 28:143–156 10.1111/j.1440-169X.1986.00143.x [DOI] [PubMed] [Google Scholar]
- Hammermeister M., Schödel K., Westermann B. 2010. Mdm36 is a mitochondrial fission-promoting protein in Saccharomyces cerevisiae. Mol. Biol. Cell. 21:2443–2452 10.1091/mbc.E10-02-0096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heil-Chapdelaine R.A., Tran N.K., Cooper J.A. 2000a. Dynein-dependent movements of the mitotic spindle in Saccharomyces cerevisiae do not require filamentous actin. Mol. Biol. Cell. 11:863–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heil-Chapdelaine R.A., Oberle J.R., Cooper J.A. 2000b. The cortical protein Num1p is essential for dynein-dependent interactions of microtubules with the cortex. J. Cell Biol. 151:1337–1344 10.1083/jcb.151.6.1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holy J., Schatten G. 1991. Differential behavior of centrosomes in unequally dividing blastomeres during fourth cleavage of sea urchin embryos. J. Cell Sci. 98:423–431 [DOI] [PubMed] [Google Scholar]
- Hwang E., Kusch J., Barral Y., Huffaker T.C. 2003. Spindle orientation in Saccharomyces cerevisiae depends on the transport of microtubule ends along polarized actin cables. J. Cell Biol. 161:483–488 10.1083/jcb.200302030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura K., Kimura A. 2011. Intracellular organelles mediate cytoplasmic pulling force for centrosome centration in the Caenorhabditis elegans early embryo. Proc. Natl. Acad. Sci. USA. 108:137–142 10.1073/pnas.1013275108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyomitsu T., Cheeseman I.M. 2012. Chromosome- and spindle-pole-derived signals generate an intrinsic code for spindle position and orientation. Nat. Cell Biol. 14:311–317 10.1038/ncb2440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kon T., Oyama T., Shimo-Kon R., Imamula K., Shima T., Sutoh K., Kurisu G. 2012. The 2.8 Å crystal structure of the dynein motor domain. Nature. 484:345–350 10.1038/nature10955 [DOI] [PubMed] [Google Scholar]
- Korinek W.S., Copeland M.J., Chaudhuri A., Chant J. 2000. Molecular linkage underlying microtubule orientation toward cortical sites in yeast. Science. 287:2257–2259 10.1126/science.287.5461.2257 [DOI] [PubMed] [Google Scholar]
- Kotak S., Busso C., Gönczy P. 2012. Cortical dynein is critical for proper spindle positioning in human cells. J. Cell Biol. 199:97–110 10.1083/jcb.201203166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlowski C., Srayko M., Nedelec F. 2007. Cortical microtubule contacts position the spindle in C. elegans embryos. Cell. 129:499–510 10.1016/j.cell.2007.03.027 [DOI] [PubMed] [Google Scholar]
- Krueger L.E., Wu J.C., Tsou M.F., Rose L.S. 2010. LET-99 inhibits lateral posterior pulling forces during asymmetric spindle elongation in C. elegans embryos. J. Cell Biol. 189:481–495 10.1083/jcb.201001115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laan L., Pavin N., Husson J., Romet-Lemonne G., van Duijn M., López M.P., Vale R.D., Jülicher F., Reck-Peterson S.L., Dogterom M. 2012. Cortical dynein controls microtubule dynamics to generate pulling forces that position microtubule asters. Cell. 148:502–514 10.1016/j.cell.2012.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbé J.C., Maddox P.S., Salmon E.D., Goldstein B. 2003. PAR proteins regulate microtubule dynamics at the cell cortex in C. elegans. Curr. Biol. 13:707–714 10.1016/S0960-9822(03)00251-3 [DOI] [PubMed] [Google Scholar]
- Lambrechts A., Gevaert K., Cossart P., Vandekerckhove J., Van Troys M. 2008. Listeria comet tails: the actin-based motility machinery at work. Trends Cell Biol. 18:220–227 10.1016/j.tcb.2008.03.001 [DOI] [PubMed] [Google Scholar]
- Larson S.M., Lee H.J., Hung P.H., Matthews L.M., Robinson D.N., Evans J.P. 2010. Cortical mechanics and meiosis II completion in mammalian oocytes are mediated by myosin-II and Ezrin-Radixin-Moesin (ERM) proteins. Mol. Biol. Cell. 21:3182–3192 10.1091/mbc.E10-01-0066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee L., Tirnauer J.S., Li J., Schuyler S.C., Liu J.Y., Pellman D. 2000. Positioning of the mitotic spindle by a cortical-microtubule capture mechanism. Science. 287:2260–2262 10.1126/science.287.5461.2260 [DOI] [PubMed] [Google Scholar]
- Lee W.L., Oberle J.R., Cooper J.A. 2003. The role of the lissencephaly protein Pac1 during nuclear migration in budding yeast. J. Cell Biol. 160:355–364 10.1083/jcb.200209022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Guo F., Rubinstein B., Li R. 2008. Actin-driven chromosomal motility leads to symmetry breaking in mammalian meiotic oocytes. Nat. Cell Biol. 10:1301–1308 10.1038/ncb1788 [DOI] [PubMed] [Google Scholar]
- Liakopoulos D., Kusch J., Grava S., Vogel J., Barral Y. 2003. Asymmetric loading of Kar9 onto spindle poles and microtubules ensures proper spindle alignment. Cell. 112:561–574 10.1016/S0092-8674(03)00119-3 [DOI] [PubMed] [Google Scholar]
- Lombillo V.A., Stewart R.J., McIntosh J.R. 1995. Minus-end-directed motion of kinesin-coated microspheres driven by microtubule depolymerization. Nature. 373:161–164 10.1038/373161a0 [DOI] [PubMed] [Google Scholar]
- Lutz D.A., Hamaguchi Y., Inoué S. 1988. Micromanipulation studies of the asymmetric positioning of the maturation spindle in Chaetopterus sp. oocytes: I. Anchorage of the spindle to the cortex and migration of a displaced spindle. Cell Motil. Cytoskeleton. 11:83–96 10.1002/cm.970110202 [DOI] [PubMed] [Google Scholar]
- Markus S.M., Lee W.L. 2011. Regulated offloading of cytoplasmic dynein from microtubule plus ends to the cortex. Dev. Cell. 20:639–651 10.1016/j.devcel.2011.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markus S.M., Kalutkiewicz K.A., Lee W.L. 2012. She1-mediated inhibition of dynein motility along astral microtubules promotes polarized spindle movements. Curr. Biol. 22:2221–2230 10.1016/j.cub.2012.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maro B., Johnson M.H., Pickering S.J., Flach G. 1984. Changes in actin distribution during fertilization of the mouse egg. J. Embryol. Exp. Morphol. 81:211–237 [PubMed] [Google Scholar]
- Matson S., Markoulaki S., Ducibella T. 2006. Antagonists of myosin light chain kinase and of myosin II inhibit specific events of egg activation in fertilized mouse eggs. Biol. Reprod. 74:169–176 10.1095/biolreprod.105.046409 [DOI] [PubMed] [Google Scholar]
- McIntosh J.R., Volkov V., Ataullakhanov F.I., Grishchuk E.L. 2010. Tubulin depolymerization may be an ancient biological motor. J. Cell Sci. 123:3425–3434 10.1242/jcs.067611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally K.L., Martin J.L., Ellefson M., McNally F.J. 2010. Kinesin-dependent transport results in polarized migration of the nucleus in oocytes and inward movement of yolk granules in meiotic embryos. Dev. Biol. 339:126–140 10.1016/j.ydbio.2009.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller R.K., Heller K.K., Frisèn L., Wallack D.L., Loayza D., Gammie A.E., Rose M.D. 1998. The kinesin-related proteins, Kip2p and Kip3p, function differently in nuclear migration in yeast. Mol. Biol. Cell. 9:2051–2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller R.K., Cheng S.C., Rose M.D. 2000. Bim1p/Yeb1p mediates the Kar9p-dependent cortical attachment of cytoplasmic microtubules. Mol. Biol. Cell. 11:2949–2959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison T., Wühr M., Nguyen P., Ishihara K., Groen A., Field C.M. 2012. Growth, interaction, and positioning of microtubule asters in extremely large vertebrate embryo cells. Cytoskeleton (Hoboken). 69:738–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin X., Bellaïche Y. 2011. Mitotic spindle orientation in asymmetric and symmetric cell divisions during animal development. Dev. Cell. 21:102–119 10.1016/j.devcel.2011.06.012 [DOI] [PubMed] [Google Scholar]
- Nguyen-Ngoc T., Afshar K., Gönczy P. 2007. Coupling of cortical dynein and G α proteins mediates spindle positioning in Caenorhabditis elegans. Nat. Cell Biol. 9:1294–1302 10.1038/ncb1649 [DOI] [PubMed] [Google Scholar]
- Olivera-Couto A., Graña M., Harispe L., Aguilar P.S. 2011. The eisosome core is composed of BAR domain proteins. Mol. Biol. Cell. 22:2360–2372 10.1091/mbc.E10-12-1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rourke S.M., Christensen S.N., Bowerman B. 2010. Caenorhabditis elegans EFA-6 limits microtubule growth at the cell cortex. Nat. Cell Biol. 12:1235–1241 10.1038/ncb2128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panbianco C., Weinkove D., Zanin E., Jones D., Divecha N., Gotta M., Ahringer J. 2008. A casein kinase 1 and PAR proteins regulate asymmetry of a PIP(2) synthesis enzyme for asymmetric spindle positioning. Dev. Cell. 15:198–208 10.1016/j.devcel.2008.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park D.H., Rose L.S. 2008. Dynamic localization of LIN-5 and GPR-1/2 to cortical force generation domains during spindle positioning. Dev. Biol. 315:42–54 10.1016/j.ydbio.2007.11.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschal B.M., Shpetner H.S., Vallee R.B. 1987. MAP 1C is a microtubule-activated ATPase which translocates microtubules in vitro and has dynein-like properties. J. Cell Biol. 105:1273–1282 10.1083/jcb.105.3.1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer D.C., Gard D.L. 1999. Microtubules in Xenopus oocytes are oriented with their minus-ends towards the cortex. Cell Motil. Cytoskeleton. 44:34–43 [DOI] [PubMed] [Google Scholar]
- Pfender S., Kuznetsov V., Pleiser S., Kerkhoff E., Schuh M. 2011. Spire-type actin nucleators cooperate with Formin-2 to drive asymmetric oocyte division. Curr. Biol. 21:955–960 10.1016/j.cub.2011.04.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pielak R.M., Gaysinskaya V.A., Cohen W.D. 2004. Formation and function of the polar body contractile ring in Spisula. Dev. Biol. 269:421–432 10.1016/j.ydbio.2004.01.033 [DOI] [PubMed] [Google Scholar]
- Rasmussen C.G., Humphries J.A., Smith L.G. 2011. Determination of symmetric and asymmetric division planes in plant cells. Annu. Rev. Plant Biol. 62:387–409 10.1146/annurev-arplant-042110-103802 [DOI] [PubMed] [Google Scholar]
- Redemann S., Pecreaux J., Goehring N.W., Khairy K., Stelzer E.H., Hyman A.A., Howard J. 2010. Membrane invaginations reveal cortical sites that pull on mitotic spindles in one-cell C. elegans embryos. PLoS ONE. 5:e12301 10.1371/journal.pone.0012301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuh M. 2011. An actin-dependent mechanism for long-range vesicle transport. Nat. Cell Biol. 13:1431–1436 10.1038/ncb2353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuh M., Ellenberg J. 2008. A new model for asymmetric spindle positioning in mouse oocytes. Curr. Biol. 18:1986–1992 10.1016/j.cub.2008.11.022 [DOI] [PubMed] [Google Scholar]
- Sheeman B., Carvalho P., Sagot I., Geiser J., Kho D., Hoyt M.A., Pellman D. 2003. Determinants of S. cerevisiae dynein localization and activation: implications for the mechanism of spindle positioning. Curr. Biol. 13:364–372 10.1016/S0960-9822(03)00013-7 [DOI] [PubMed] [Google Scholar]
- Slaughter B.D., Smith S.E., Li R. 2009. Symmetry breaking in the life cycle of the budding yeast. Cold Spring Harb. Perspect. Biol. 1:a003384 10.1101/cshperspect.a003384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S.C., Wang Z.B., Xu Y.N., Lee S.E., Cui X.S., Kim N.H. 2011. Arp2/3 complex regulates asymmetric division and cytokinesis in mouse oocytes. PLoS ONE. 6:e18392 10.1371/journal.pone.0018392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X., Germain B.S., Lee W.L. 2012. A novel patch assembly domain in Num1 mediates dynein anchoring at the cortex during spindle positioning. J. Cell Biol. 196:743–756 10.1083/jcb.201112017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Hoopen R., Cepeda-García C., Fernández-Arruti R., Juanes M.A., Delgehyr N., Segal M. 2012. Mechanism for astral microtubule capture by cortical Bud6p priming spindle polarity in S. cerevisiae. Curr. Biol. 22:1075–1083 10.1016/j.cub.2012.04.059 [DOI] [PubMed] [Google Scholar]
- Theesfeld C.L., Irazoqui J.E., Bloom K., Lew D.J. 1999. The role of actin in spindle orientation changes during the Saccharomyces cerevisiae cell cycle. J. Cell Biol. 146:1019–1032 10.1083/jcb.146.5.1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallee R.B., Wall J.S., Paschal B.M., Shpetner H.S. 1988. Microtubule-associated protein 1C from brain is a two-headed cytosolic dynein. Nature. 332:561–563 10.1038/332561a0 [DOI] [PubMed] [Google Scholar]
- van der Voet M., Berends C.W., Perreault A., Nguyen-Ngoc T., Gönczy P., Vidal M., Boxem M., van den Heuvel S. 2009. NuMA-related LIN-5, ASPM-1, calmodulin and dynein promote meiotic spindle rotation independently of cortical LIN-5/GPR/Galpha. Nat. Cell Biol. 11:269–277 10.1038/ncb1834 [DOI] [PubMed] [Google Scholar]
- Varga V., Leduc C., Bormuth V., Diez S., Howard J. 2009. Kinesin-8 motors act cooperatively to mediate length-dependent microtubule depolymerization. Cell. 138:1174–1183 10.1016/j.cell.2009.07.032 [DOI] [PubMed] [Google Scholar]
- Verlhac M.H., Lefebvre C., Guillaud P., Rassinier P., Maro B. 2000. Asymmetric division in mouse oocytes: with or without Mos. Curr. Biol. 10:1303–1306 10.1016/S0960-9822(00)00753-3 [DOI] [PubMed] [Google Scholar]
- Walther T.C., Brickner J.H., Aguilar P.S., Bernales S., Pantoja C., Walter P. 2006. Eisosomes mark static sites of endocytosis. Nature. 439:998–1003 10.1038/nature04472 [DOI] [PubMed] [Google Scholar]
- Wang Q., Racowsky C., Deng M. 2011. Mechanism of the chromosome-induced polar body extrusion in mouse eggs. Cell Div. 6:17 10.1186/1747-1028-6-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff J.B., Drubin D.G., Barnes G. 2009. Dynein-driven mitotic spindle positioning restricted to anaphase by She1p inhibition of dynactin recruitment. Mol. Biol. Cell. 20:3003–3011 10.1091/mbc.E09-03-0186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H.Y., McNally K., McNally F.J. 2003. MEI-1/katanin is required for translocation of the meiosis I spindle to the oocyte cortex in C. elegans. Dev. Biol. 260:245–259 10.1016/S0012-1606(03)00216-1 [DOI] [PubMed] [Google Scholar]
- Yang H.Y., Mains P.E., McNally F.J. 2005. Kinesin-1 mediates translocation of the meiotic spindle to the oocyte cortex through KCA-1, a novel cargo adapter. J. Cell Biol. 169:447–457 10.1083/jcb.200411132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi K., Unruh J.R., Deng M., Slaughter B.D., Rubinstein B., Li R. 2011. Dynamic maintenance of asymmetric meiotic spindle position through Arp2/3-complex-driven cytoplasmic streaming in mouse oocytes. Nat. Cell Biol. 13:1252–1258 10.1038/ncb2320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H., Pruyne D., Huffaker T.C., Bretscher A. 2000. Myosin V orientates the mitotic spindle in yeast. Nature. 406:1013–1015 10.1038/35023024 [DOI] [PubMed] [Google Scholar]
- Zimniak T., Stengl K., Mechtler K., Westermann S. 2009. Phosphoregulation of the budding yeast EB1 homologue Bim1p by Aurora/Ipl1p. J. Cell Biol. 186:379–391 10.1083/jcb.200901036 [DOI] [PMC free article] [PubMed] [Google Scholar]