Cathepsin L degrades 53BP1 to overcome genomic instability and growth arrest in BRCA1-deficient and triple-negative breast cancers.
Abstract
Loss of 53BP1 rescues BRCA1 deficiency and is associated with BRCA1-deficient and triple-negative breast cancers (TNBC) and with resistance to genotoxic drugs. The mechanisms responsible for decreased 53BP1 transcript and protein levels in tumors remain unknown. Here, we demonstrate that BRCA1 loss activates cathepsin L (CTSL)–mediated degradation of 53BP1. Activation of this pathway rescued homologous recombination repair and allowed BRCA1-deficient cells to bypass growth arrest. Importantly, depletion or inhibition of CTSL with vitamin D or specific inhibitors stabilized 53BP1 and increased genomic instability in response to radiation and poly(adenosine diphosphate–ribose) polymerase inhibitors, compromising proliferation. Analysis of human breast tumors identified nuclear CTSL as a positive biomarker for TNBC, which correlated inversely with 53BP1. Importantly, nuclear levels of CTSL, vitamin D receptor, and 53BP1 emerged as a novel triple biomarker signature for stratification of patients with BRCA1-mutated tumors and TNBC, with potential predictive value for drug response. We identify here a novel pathway with prospective relevance for diagnosis and customization of breast cancer therapy.
Introduction
BRCA1 is a well-established tumor suppressor, and women carrying germline mutations in BRCA1 have a high risk of developing breast and ovarian cancer (Neuhausen and Marshall, 1994; Wooster and Weber, 2003). Tumors that arise often lack expression of estrogen and progesterone receptors and Her2, being classified as triple-negative breast cancers (TNBC; Turner and Reis-Filho, 2006). BRCA1 participates in DNA double-strand break (DSB) repair, S and G2/M phase cell-cycle checkpoints after damage, control of centrosome numbers, maintenance of heterochromatin, and transcriptional regulation of several genes (Scully and Livingston, 2000; Mullan et al., 2006; Zhu et al., 2011). In addition, BRCA1 function is linked to epigenetic mechanisms such as DNA methylation and miRNA biogenesis (Shukla et al., 2010; Kawai and Amano, 2012; Tanic et al., 2012).
Recruitment of BRCA1 to DNA DSBs facilitates repair by homologous recombination (HR), and loss of BRCA1 results in genomic instability characterized by unrepaired DNA breaks and complex chromosomal rearrangements that compromise cell viability (Scully et al., 1997a; Moynahan et al., 1999; Snouwaert et al., 1999). As such, BRCA1 knockout mice and mice carrying a BRCA1 deletion mutant (BRCA1Δ11/Δ11) are embryonic lethal (Xu et al., 2001; Evers and Jonkers, 2006). Although lethality in BRCA1Δ11/Δ11 mice can be rescued by abrogation of ATM, Chk2, or p53, these mice ultimately develop tumors and premature aging (Cao et al., 2006). Recently, loss of the DNA repair factor 53BP1 was shown to rescue embryonic lethality in BRCA1-deficient mice while maintaining a low incidence of tumorigenesis and normal aging (Cao et al., 2009). This is in contrast to 53BP1 knockout mice, which are cancer prone (Ward et al., 2003), suggesting that 53BP1 contributes to the developmental defects of BRCA1-deficient mice and that 53BP1 loss has different consequences for cancer and aging in the context of BRCA1 proficiency or deficiency.
Loss of 53BP1 promotes viability of BRCA1-deficient cells by rescuing HR function (Cao et al., 2009; Bouwman et al., 2010; Bunting et al., 2010). Importantly, down-regulation of 53BP1 was observed in human BRCA1-related breast cancer and TNBC and was suggested to allow these tumors to overcome the genomic instability caused by HR defects (Bouwman et al., 2010). 53BP1 facilitates DNA DSB repair by nonhomologous end joining (NHEJ; Schultz et al., 2000; Fernandez-Capetillo et al., 2002; Wang et al., 2002; Xie et al., 2007) and also affects HR via inhibition of BRCA1-mediated DSB end-resection (Bunting et al., 2010). The current model is that BRCA1 deficiency hinders end-resection of DSBs by CtIP and the Mre11–Rad50–Nbs1 complex, an essential event in HR. Accumulation of 53BP1 in this context promotes indiscriminate NHEJ and chromosomal instability that ultimately causes proliferation arrest or cell death. Conversely, in cells double deficient in BRCA1 and 53BP1, end-resection is allowed, rescuing HR (Bunting et al., 2010). Consistent with this model, 53BP1 loss reduces the sensitivity of BRCA1-deficient cells to genotoxic agents such as cisplatin and mitomycin C (Bouwman et al., 2010) and to poly(ADP-ribose) polymerase inhibitors (PARPi; Farmer et al., 2005; Bunting et al., 2010), compounds at the forefront for breast cancer therapy (Gartner et al., 2010). Thus, BRCA1-deficient cells are thought to down-regulate 53BP1 as a means to ensure proliferation/viability.
Up-regulation of 53BP1 levels represents a promising strategy for treatment of breast tumors with the poorest prognosis and for improving their response to PARPi and other DNA-damaging strategies. However, we lack knowledge about how 53BP1 mRNA and protein levels are down-regulated in cancer cells. We previously identified a pathway regulating 53BP1 protein levels (Gonzalez-Suarez et al., 2011; Redwood et al., 2011a,b). Up-regulation of the cysteine protease cathepsin L (CTSL) leads to accumulation of the protease in the nucleus, degradation of 53BP1 protein, and defects in NHEJ. Importantly, inhibition of CTSL activity by treatment with vitamin D or specific inhibitors stabilizes 53BP1 protein levels and rescues NHEJ defects (Gonzalez-Suarez et al., 2011).
Here, we demonstrate that BRCA1-deficient cells activate CTSL-mediated degradation of 53BP1 as a means to overcome genomic instability and growth arrest. In addition, depletion or inhibition of CTSL in these cells increases genomic instability in response to ionizing radiation (IR) or PARPi. Lastly, we identify high levels of nuclear CTSL and low levels of 53BP1 and vitamin D receptor (VDR) as a novel signature in subsets of breast cancer patients. We envision that the status of nuclear CTSL, VDR, and 53BP1 could be used for customization of breast cancer therapy.
Results
BRCA1-deficient cells activate CTSL-mediated degradation of 53BP1 to bypass growth arrest
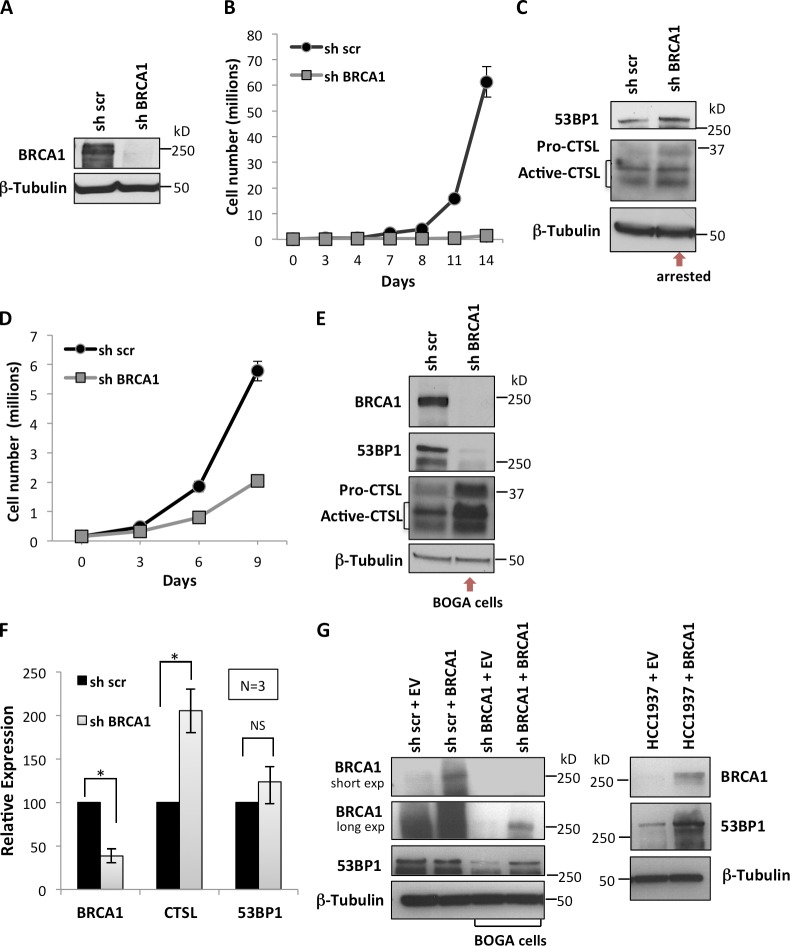
Previous studies demonstrated that loss of 53BP1 rescues the BRCA1-deficient phenotype (Cao et al., 2009; Bouwman et al., 2010; Bunting et al., 2010). We also showed that CTSL regulates the stability of 53BP1 (Gonzalez-Suarez et al., 2011; Redwood et al., 2011a,b). Here, we investigated whether breast tumor cells are able to down-regulate 53BP1 upon loss of BRCA1 to restore proliferation/viability and if CTSL is one of the factors responsible for the depletion of 53BP1. The human breast cancer cell line MCF7, which is BRCA1 and 53BP1 proficient, was depleted of BRCA1 via lentiviral transduction with shRNAs (Fig. 1 A and Fig. S1 A). As previously shown in human fibroblasts (Tu et al., 2011), depletion of BRCA1 in MCF7 cells induces growth arrest (Fig. 1 B). BRCA1-deficient cells did not show differences in the levels of 53BP1 or CTSL proteins immediately after growth arrest (Fig. 1 C). Interestingly, after approximately two weeks in culture, BRCA1-deficient cells resumed proliferation, albeit at a slower rate than control cells (Fig. 1 D). Importantly, BRCA1-deficient cells that overcome growth arrest (herein referred to as BOGA cells, for clarity) exhibit increased CTSL and decreased 53BP1 protein levels (Fig. 1 E). This signature was also observed in MDA-MB-231 breast cancer cells that overcome the growth arrest induced by depletion of BRCA1 (Fig. S1 B). Similar changes in CTSL and 53BP1 protein levels were observed with different shRNAs for depletion of BRCA1 (Fig. S1 C). Monitoring transcripts levels by quantitative RT-PCR (qRT-PCR) revealed transcriptional up-regulation of CTSL in BOGA cells without significant changes in 53BP1 transcript levels, indicating a decrease in 53BP1 protein stability (Fig. 1 F). To confirm a role for BRCA1 in the regulation of 53BP1 levels, we reconstituted BRCA1 via transient transfection in BOGA cells and the BRCA1-deficient breast cancer cell line HCC1937. Ectopic expression of BRCA1 resulted in stabilization of 53BP1 in both cell lines (Fig. 1 G), revealing a novel function of BRCA1 in the stabilization of 53BP1 protein.
Figure 1.
Bypass of growth arrest after BRCA1 loss is associated with CTSL up-regulation and 53BP1 degradation. (A) MCF7 cells were lentivirally transduced with an shRNA for depletion of BRCA1 (shBRCA1) or a control shRNA scrambled (sh scr) and BRCA1 levels were assessed by Western blot immediately after selection. (B) Proliferation rate shows that BRCA1 depletion induces growth arrest. The mean ± SD of three independent experiments is shown (SD too small to show at some time points). (C) Western blots show 53BP1 and CTSL levels in growth-arrested cells. (D) Proliferation rate of BRCA1-deficient cells that overcome growth arrest. The mean ± SD of three independent experiments is shown (SD too small to show at some time points). (E) Western blots show levels of BRCA1, 53BP1, and CTSL in control and BOGA cells (representative experiment of 25 biological repeats). (F) Relative expression of BRCA1, CTSL, and 53BP1 in control and BOGA cells as determined by qRT-PCR. The mean ± SD of three independent experiments is shown. Asterisk shows p-value of statistical significance (*, P ≤ 0.05). NS, not statistically significant differences. (G) Western blots show BRCA1 and 53BP1 levels upon reconstitution of BRCA1 by transient transfection into BRCA1-deficient cell lines—BOGA cells (left) and HCC1937 (right). An empty vector (EV) was used as control. β-Tubulin was the loading control.
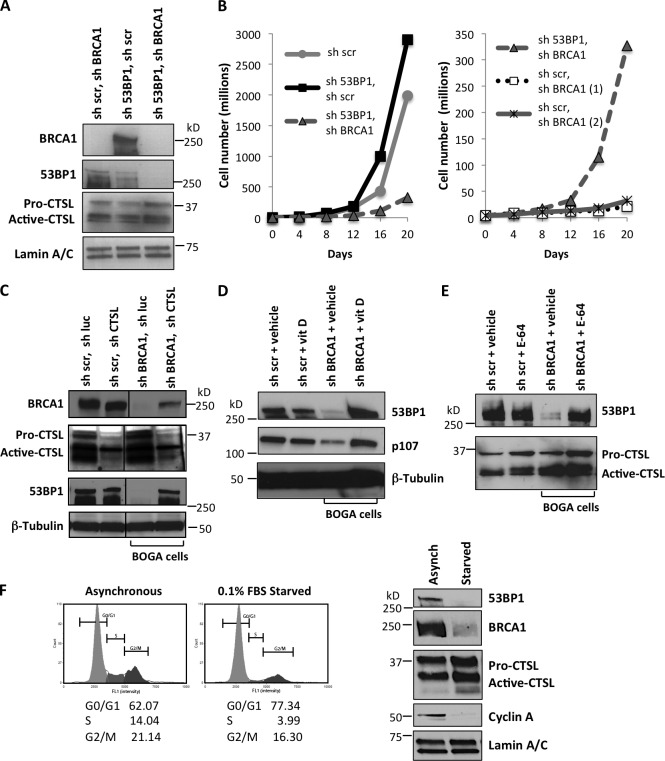
To determine if 53BP1 loss is responsible for the bypass of growth arrest in BRCA1-deficient cells, we depleted 53BP1 before BRCA1 depletion. As shown in Fig. 2 A, we achieved a marked reduction of both 53BP1 and BRCA1 proteins. Importantly, previous depletion of 53BP1 prevented the characteristic growth arrest that follows BRCA1 depletion (Fig. 2 B). These data indicate that loss of 53BP1 allows BRCA1-deficient cells to bypass growth arrest. Interestingly, cells that were depleted of 53BP1 before depletion of BRCA1 also up-regulated CTSL (Fig. S2 A), suggesting that up-regulation of CTSL is independent of 53BP1 status.
Figure 2.
CTSL is responsible for the degradation of 53BP1 that allows bypass of growth arrest in BOGA cells. (A) MCF7 cells were transduced with sh53BP1, shBRCA1, or both. After selection, BRCA1, 53BP1, and CTSL levels were monitored by Western blot. (B) Graphs compare proliferation rates between control cells and 53BP1/BRCA1-depleted cells (left) and between 53BP1/BRCA1-depleted cells and BRCA1-depleted cells (right). Depletion of 53BP1 prevents growth arrest upon BRCA1 loss (representative experiment of three biological repeats). (C) Control and BOGA cells were transduced with shCTSL or control shRNA (sh luc) and the levels of CTSL, BRCA1, and 53BP1 were monitored by Western blot. Vertical lines show where gel pieces were juxtaposed. (D) Control or BOGA cells were treated with vitamin D (10−7 M) or vehicle (BGS) for 24 h and the levels of 53BP1 and p107, a known target of CTSL degradation, were monitored by Western blot. (E) Control or BOGA cells were incubated with the broad cathepsin inhibitor E-64 (10 µM) or with vehicle (H2O) for 24 h, and the levels of 53BP1 and CTSL were monitored by Western blot. Note how cathepsin inhibition stabilizes 53BP1. (F) MCF7 cells growing asynchronously or growth arrested for 48 h by serum deprivation were analyzed for cell cycle profile (left) and for levels of CTSL, 53BP1, and BRCA1 by Western blot (right). Representative experiment of three biological repeats. Either Lamin A/C or β-tubulin was used as loading control.
Next, we determined whether CTSL is responsible for the degradation of 53BP1 in BRCA1-deficient cells by depleting CTSL in control and BOGA cells (Fig. 2 C and Fig. S2 B). Depletion of CTSL stabilized 53BP1 protein levels in BOGA cells, mirroring those of control cells, whereas transcript levels of BRCA1 were not affected by depletion of CTSL (Fig. S2 B). Intriguingly, we observed a slight increase in BRCA1 protein in BOGA cells depleted of CTSL, suggesting a possible feedback mechanism of CTSL on BRCA1 protein levels, a notion that remains to be tested.
We previously demonstrated that vitamin D inhibits CTSL activity and stabilizes 53BP1 protein in mouse embryonic fibroblasts (MEFs; Gonzalez-Suarez et al., 2011). Here, we show that treatment of BOGA cells with vitamin D (1α,25-dihydroxyvitamin D3) stabilized the levels of 53BP1 (Fig. 2 D). Similarly, treatment with the cathepsin inhibitor E-64 led to increased levels of 53BP1 protein (Fig. 2 E). These data demonstrate that cells growth arrested after BRCA1 loss activate CTSL-mediated degradation of 53BP1 to bypass the growth arrest imposed by BRCA1 deficiency. In addition, depletion or inhibition of CTSL can increase 53BP1 levels in the context of BRCA1 deficiency with potential therapeutic effects. Interestingly, growth arrest induced by serum deprivation in MCF7 cells also led to decreased levels of BRCA1, up-regulation of CTSL, and down-regulation of 53BP1 (Fig. 2 F). These data suggest that BRCA1 might regulate CTSL-mediated degradation of 53BP1 during the cell cycle and that the growth arrest induced by BRCA1 depletion itself could contribute to the activation of CTSL-mediated degradation of 53BP1.
CTSL-mediated degradation of 53BP1 rescues HR defects in BRCA1-deficient cells
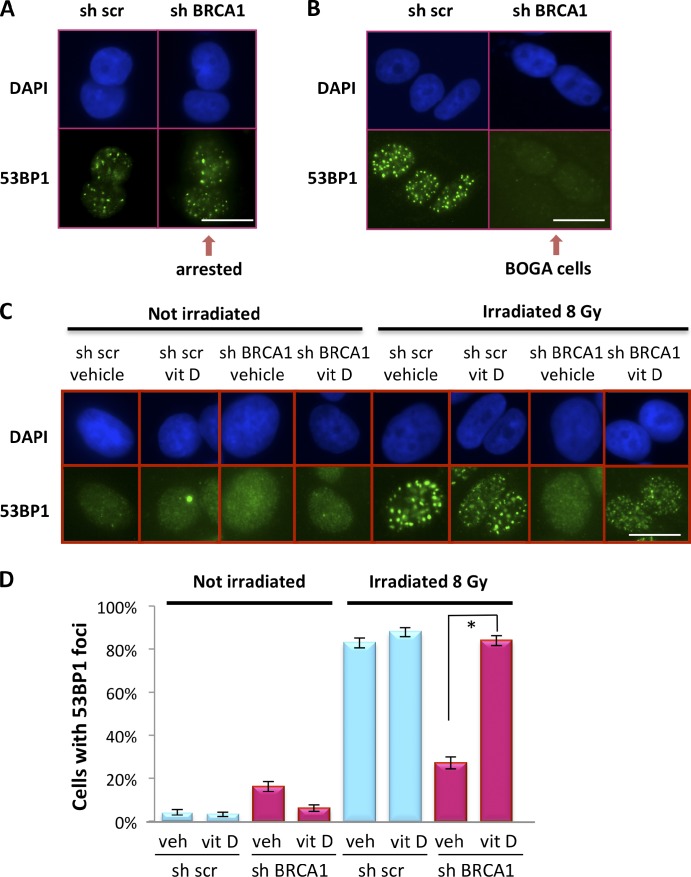
BRCA1 deficiency impairs DNA end-resection at DSBs and formation of RAD51-coated filaments that facilitate subsequent HR steps (Scully et al., 1997a,b; Moynahan et al., 1999; Snouwaert et al., 1999; Bhattacharyya et al., 2000; Sung et al., 2003; Schlegel et al., 2006). Interestingly, loss of 53BP1 in BRCA1-deficient cells partially rescues HR and accumulation of RAD51 at IR-induced foci (IRIF; Bunting et al., 2010). Here, we determined how CTSL-mediated degradation of 53BP1 impacts 53BP1 and RAD51 recruitment to DNA DSBs. We show that growth-arrested MCF7 cells immediately after BRCA1 depletion retained their ability to recruit 53BP1 protein to IRIF (Fig. 3 A), which is consistent with normal 53BP1 levels. In contrast, BOGA cells were unable to form 53BP1 IRIF (Fig. 3 B), which is consistent with their decreased 53BP1 levels. Next, we determined if the deficiency in 53BP1 foci formation could be rescued by inhibiting CTSL. BOGA cells treated with vehicle were defective in the formation of 53BP1 IRIF, whereas treatment with vitamin D rescued 53BP1 IRIF (Fig. 3, C and D; and Fig. S3 A).
Figure 3.
Regulation of 53BP1 IRIF in BOGA cells by vitamin D. (A) Immunofluorescence to detect 53BP1 IRIF in control (sh scr) and growth-arrested BRCA1-depleted cells (shBRCA1) 1 h after 8 Gy of IR. (B) The same assay was performed as in A but after bypass of growth arrest (BOGA cells). (C) Immunofluorescence performed in control and BOGA cells treated with vitamin D or vehicle 24 h before IR. Note how treatment with vitamin D restores 53BP1 IRIF in BOGA cells. (D) Graph shows the percentage of cells that form 53BP1 IRIF in the presence of vitamin D or vehicle. Cells with >10 53BP1 IRIF were considered positive. At least 1,000 cells were analyzed per condition in one experiment. Error bars were calculated using the exact binomial test. Asterisk shows p-value of statistical significance (*, P ≤ 0.05). Bars, 10 µm.
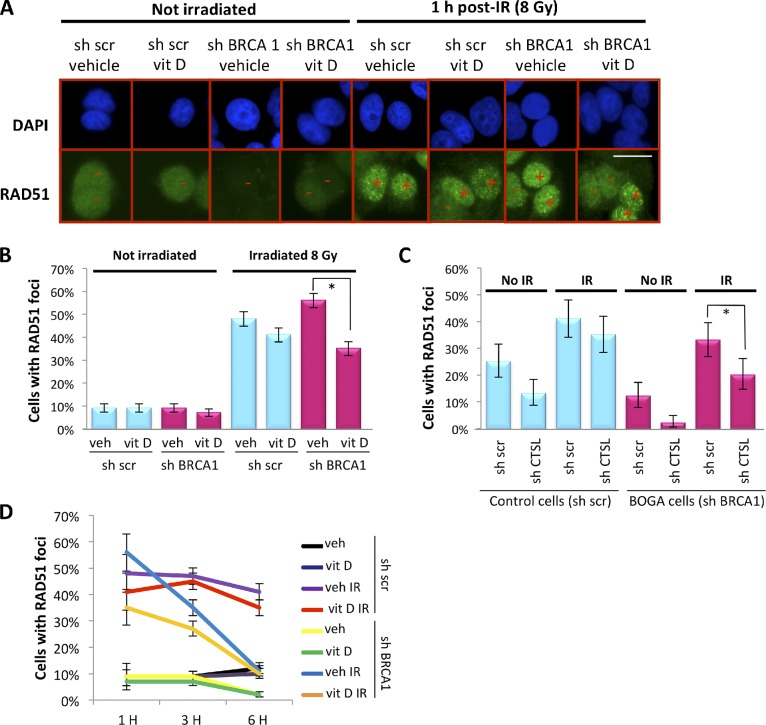
In contrast to 53BP1, RAD51 recruitment to IRIF was inhibited shortly after BRCA1 depletion in growth-arrested cells (Fig. S3 B). This defect was rescued in BOGA cells at 1 h after IR (Fig. 4, A and B). The rescue was not because of an increase in BRCA1 levels, as BOGA cells were unable to form BRCA1-labeled IRIF (Fig. S3 C). We tested whether activation of CTSL-mediated degradation of 53BP1 is behind the rescued recruitment of RAD51 to DSBs in BOGA cells. First, we show that stabilization of 53BP1 by vitamin D treatment reduced the extent of RAD51 IRIF (Fig. 4, A and B), revealing an unprecedented role for vitamin D in modulating HR. Similarly, depletion of CTSL reduced the ability of BOGA cells to recruit RAD51 to DSBs (Fig. 4 C). Intriguingly, although control cells formed RAD51 IRIF for up to 6 h, the percentage of BOGA cells positive for RAD51 IRIF decreased significantly over time (Fig. 4 D and Fig. S4), indicating that 53BP1 deficiency in BOGA cells does not completely compensate for the absence of BRCA1. These data demonstrate that by activating CTSL-mediated degradation of 53BP1, BRCA1-deficient cells can rescue to a certain extent the HR defects to promote survival and that this pathway can be disrupted by inhibiting CTSL activity. These findings provide a novel strategy to modulate HR efficiency in BRCA1-deficient cells.
Figure 4.
Regulation of RAD51 IRIF in BOGA cells by CTSL and vitamin D. (A) Immunofluorescence to detect RAD51 IRIF after 8 Gy of IR in control and BOGA cells after treatment with vitamin D or vehicle. Plus denotes positive cells (>10 IRIF) and minus negative cells. (B) Graph shows the percentage of cells that form RAD51 IRIF 1 h after IR in the presence of vitamin D or vehicle. Approximately 1,000 cells were analyzed per condition. Error bars were calculated using the exact binomial test. Asterisk shows p-value of statistical significance (*, P ≤ 0.05). (C) Quantitation of RAD51 IRIF in control or BOGA cells proficient or deficient in CTSL showing reduced number of positive cells in CTSL-depleted BOGA cells. At least 200 cells were analyzed per condition. Error bars were calculated using the exact binomial test. Asterisk shows p-value of statistical significance (*, P ≤ 0.05). (D) Quantitation of percentage of cells positive for RAD51 IRIF at different times after IR and upon treatment with vitamin D or vehicle. Note how BOGA cells exhibit defects in RAD51 IRIF 3 and 6 h after IR. Bar, 10 µm.
Consequences of CTSL-mediated degradation of 53BP1 for DNA repair and genomic stability
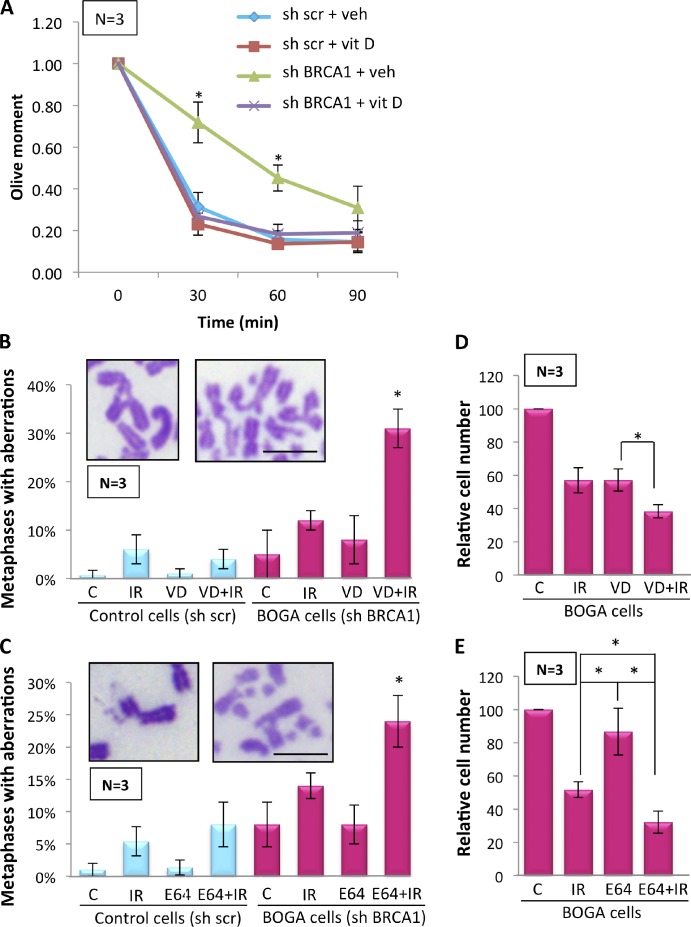
To determine the functional consequences of CTSL-mediated degradation of 53BP1 in BRCA1-deficient cells, we evaluated the kinetics of DNA DSB repair by performing comet assays under neutral, nondenaturing conditions (Olive et al., 1990). Fig. 5 A shows that BOGA cells exhibited defects in the fast phase of repair corresponding to classical NHEJ (Iliakis et al., 2004), which is consistent with our previous finding that up-regulation of CTSL in MEFs leads to defects in the fast phase of repair through degradation of 53BP1 (Gonzalez-Suarez et al., 2011). Furthermore, inhibition of CTSL with vitamin D rescued the kinetics of DNA DSB repair, mirroring control cells (Fig. 5 A). These results suggest that CTSL-mediated degradation of 53BP1 hinders NHEJ in BRCA1-deficient cells. However, these cells are still able to repair DSBs although at a lower rate, suggesting that repair by HR or alternative NHEJ might remain relatively intact, which is consistent with RAD51 foci being able to form early after IR.
Figure 5.
Effect of CTSL inhibition on DNA repair and genomic stability in BOGA cells. (A) Neutral comet assays after 8 Gy of IR show higher olive moments, and thus defects in the fast phase of DNA repair in BOGA cells. Inhibition of CTSL activity by treatment with vitamin D (10−7 M) 24 h before IR rescues defects in DNA DSB repair. (B) Control and BOGA cells were incubated with vehicle control (C) or vitamin D (VD) for 24 h before IR with 2 Gy (IR). Cells collected 24 h after IR were analyzed for genomic instability by quantitating the percentage of metaphases presenting with chromosomal aberrations, as shown in the images. N, number of independent experiments (50 metaphases analyzed per condition in each experiment). (C) Control and BOGA cells were incubated with the cathepsin inhibitor (E-64) or vehicle control (C) for 24 h before IR, and the extent of genomic instability was assessed as in B. (D) Graph shows relative numbers of BOGA cells 4 d after treatment with IR, vitamin D, or a combination of both. Treatment protocol as in B. (E) Graph shows relative numbers of BOGA cells 4 d after treatment with IR, E-64, or a combination of both. Treatment performed as in C. All values expressed as mean ± SD. N, number of independent experiments. Asterisk denotes a p-value of statistical significance (*, P ≤ 0.05). Bars, 10 µm.
Cells deficient in HR become dependent on alternative pathways of DNA DSB repair, which often form complex chromosomal aberrations that trigger cell cycle arrest or death. Loss of 53BP1 is sufficient to reduce the extent of aberrant chromosome structures in BRCA1-deficient cells (Bunting et al., 2010). Here, we determined whether stabilization of 53BP1 in BRCA1-deficient cells exacerbates the extent of genomic instability after IR by analyzing chromosomal aberrations in metaphase spreads. We did not find a profound increase in chromosome aberrations after IR in BOGA cells (Fig. 5 B), in agreement with the deficiency in BRCA1 and 53BP1. However, stabilization of 53BP1 in this context by vitamin D treatment significantly increased the percentage of metaphases with aberrant chromosomes after IR. Similarly, stabilization of 53BP1 in BOGA cells by treatment with the cathepsin inhibitor E-64 markedly increased genomic instability after IR (Fig. 5 C). Consistent with the increase in genomic instability, treatment of BOGA cells with vitamin D or E-64 significantly reduced their recovery from IR (Fig. 5, D and E). Thus, CTSL inhibition could represent a novel strategy to induce radiosensitivity in specific types of breast tumors.
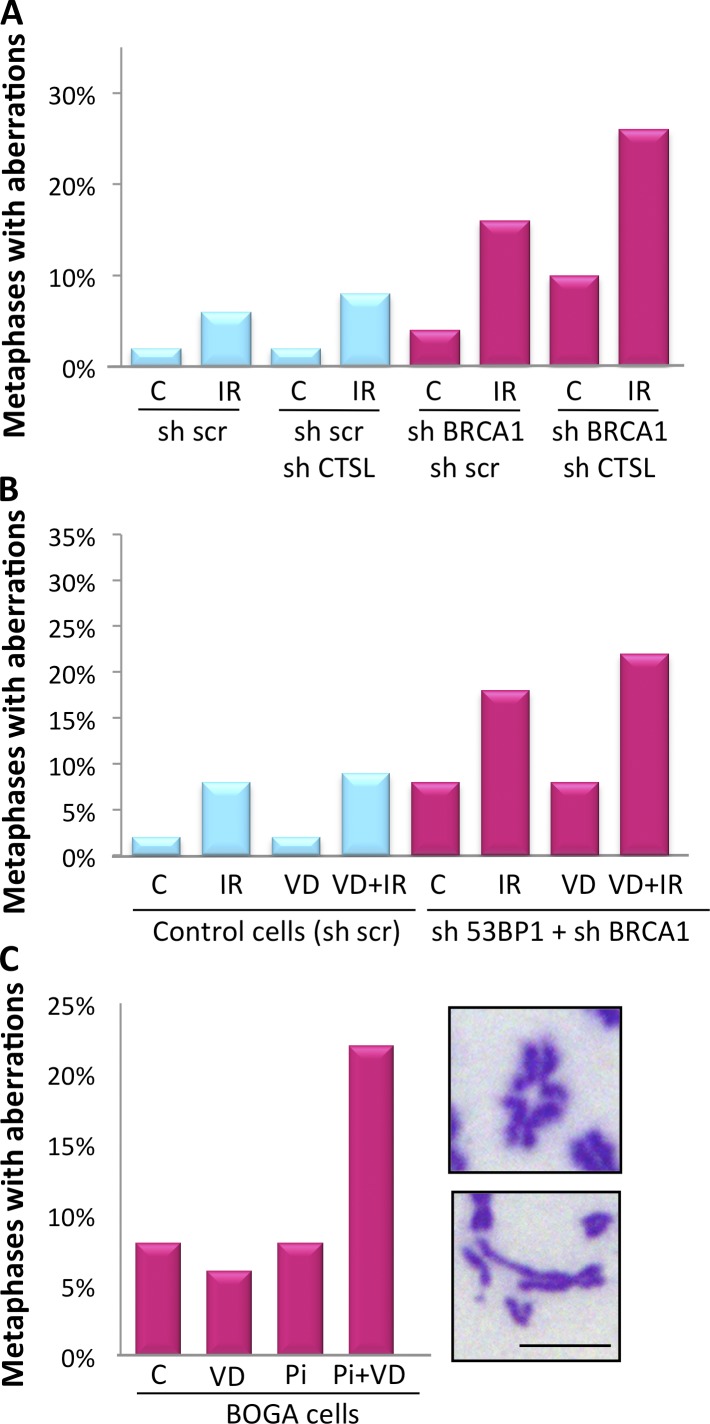
To confirm that inhibition of CTSL-mediated degradation of 53BP1 is responsible for the increase in chromosomal aberrations after IR, we analyzed genomic instability in BOGA cells depleted of CTSL. These cells showed a marked increase in chromosomal aberrations after IR, when compared with cells deficient in either BRCA1 or CTSL alone (Fig. 6 A). Furthermore, we determined if the effect of vitamin D increasing genomic instability after IR in BOGA cells is mediated by 53BP1 by monitoring chromosomal aberrations in cells doubly depleted of 53BP1 and BRCA1. In these cells, the combination of vitamin D and IR did not result in the profound increase in genomic instability (Fig. 6 B) that we observed in BOGA cells (Fig. 5 B). These results demonstrate that vitamin D exerts its effect in part by stabilizing 53BP1 levels and that the extent of CTSL-mediated degradation of 53BP1 is a key determinant of the ability of BRCA1-deficient cells to deal with the DNA damage generated by IR and putatively other genotoxic agents.
Figure 6.
Chromosomal aberrations in response to IR and PARPi. (A) BOGA cells transduced with an shRNA control (sh scr) or an shRNA for CTSL were irradiated with 2 Gy and the percentage of cells with aberrant metaphases was quantified 24 h after IR. A total of 400 metaphases were analyzed in one experiment (50 per condition). (B) MCF7 cells depleted of 53BP1 and BRCA1 (sh53BP1 + shBRCA1) were treated with vitamin D or vehicle 24 h before IR, and the percentage of metaphases with aberrant chromosomes was quantitated. A total of 800 metaphases were analyzed in one experiment (100 per condition). (C) BOGA cells were treated with vitamin D (10−7 M) 24 h before treatment with the PARPi EB-47 (Pi, 1.2 µg/ml) for an additional 48 h. Graph shows the percentage of metaphases with chromosomal aberrations. Images show the types of chromosomal aberrations observed. A total of 400 metaphases were analyzed in one experiment (100 per condition). Bar, 10 µm.
BRCA1-deficient cells are exquisitely sensitive to PARPi (Bryant et al., 2005; Drew et al., 2011). Importantly, loss of 53BP1 reduces the sensitivity of these cells to PARPi (Bunting et al., 2010; Aly and Ganesan, 2011). We assessed whether stabilization of 53BP1 in BOGA cells would increase the extent of genomic instability induced by PARPi. As shown in Fig. 6 C, treatment of BOGA cells with PARPi did not result in profound genomic instability, in agreement with the resistance of cells double deficient in 53BP1 and BRCA1 to this treatment. Interestingly, stabilization of 53BP1 by vitamin D increased the extent of chromosomal aberrations in response to PARPi, suggesting that inhibition of CTSL-mediated degradation of 53BP1 could induce sensitivity to PARPi.
Increased levels of nuclear CTSL in TNBC and tumors from patients with BRCA1 germline mutations
Although CTSL is one of the most abundant proteases in the endosomal/lysosomal compartment, it has also been identified in the nucleus (Goulet et al., 2004; Duncan et al., 2008). We previously showed that up-regulation of CTSL leads to accumulation of the protease in the nucleus and degradation of 53BP1 (Gonzalez-Suarez et al., 2011). Recent studies demonstrated that loss of 53BP1 is more frequent in TNBC and BRCA1-mutated human breast cancer (Bouwman et al., 2010). Here, we determined whether up-regulation of CTSL occurs in human breast cancers and if it correlates with decreased levels of 53BP1. In addition, given the inhibitory effect of vitamin D on this pathway, we monitored the levels of VDR, which mediates most of vitamin D’s cellular effects. We performed immunohistochemical analyses of multitumor tissue microarrays (TMAs) constructed with tissue from 249 patients with sporadic breast cancer (Fig. 7 and Table 1) classified into four molecular subtypes: luminal A, luminal B, Her2, and triple negative.
Figure 7.
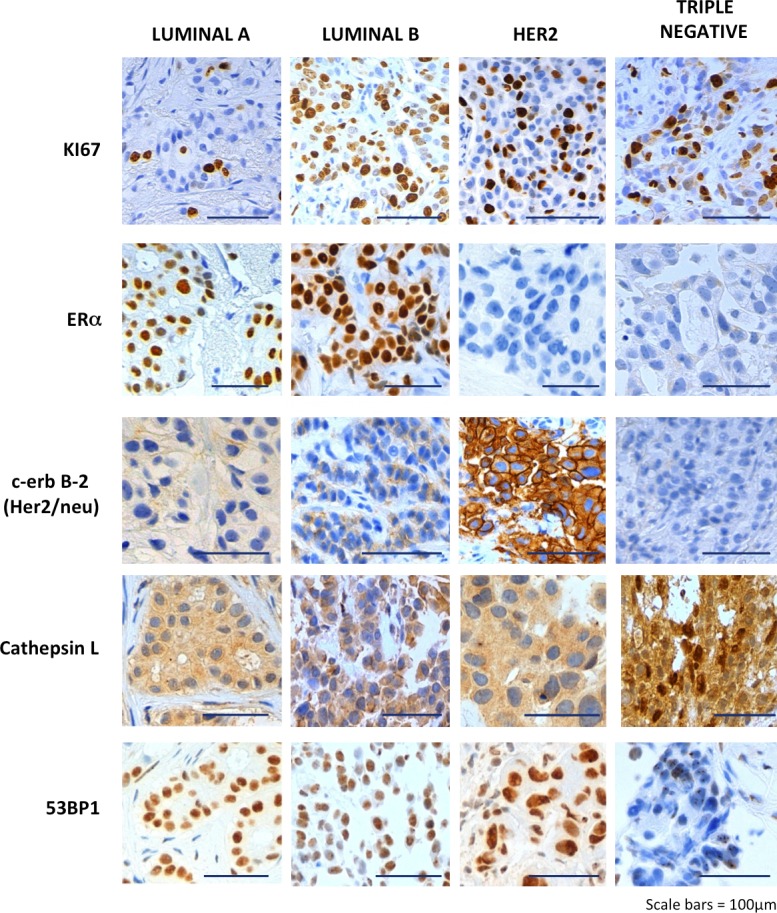
A new signature for subsets of TNBC patients. Immunohistochemical analysis was performed in breast tumor TMAs from 249 patients, which included four molecular subtypes: luminal A, luminal B, Her2, and triple negative. Representative images of immunohistochemical labeling with Ki67, ERα, Her2, CTSL, and 53BP1 are shown. Note that although cytoplasmic CTSL is observed in all tumor subtypes, nuclear CTSL is markedly up-regulated in a subset of TNBC. In addition, TNBC tumors exhibit a marked decrease in 53BP1. Bars, 100 µm.
Table 1.
Immunohistochemical analysis of CTSL, 53BP1, and VDR expression in sporadic human breast cancer
| Proteins | Molecular type | H score | Kruskal-Wallis test p-value | ||
| Mean (SD) | Median [P25, P75] | Min–Max | |||
| Cytoplasmic CTSL | Luminal A | 137 (36.6) | 135 [110,160] | 50–205 | 0.50 |
| Luminal B | 134 (35.2) | 130 [110,155] | 75–-230 | ||
| Erbb2 (−1) | 141 (43.1) | 135 [110,171] | 50–230 | ||
| Triple negative (−1) | 145 (35.2) | 140 [120,175] | 80–200 | ||
| Nuclear CTSL | Luminal A | 8 (18.0) | 0 [0, 0] | 0–90 | <0.0001 |
| Luminal B | 8 (14.9) | 0 [0, 15] | 0–75 | ||
| Erbb2 (−1) | 9 (18.3) | 0 [0, 5] | 0–75 | ||
| Triple negative (−1) | 42 (44.3) | 30 [0, 83] | 0–125 | ||
| 53BP1 | Luminal A | 155 (56.5) | 160 [120, 200] | 0–270 | 0.0002 |
| Luminal B | 150 (45.8) | 150 [120, 170] | 40–280 | ||
| Erbb2 (−4) | 154 (58.1) | 150 [110, 200] | 20–300 | ||
| Triple negative | 112 (44.4) | 105 [80, 143] | 0–190 | ||
| Cytoplasmic VDR | Luminal A (−4) | 50 (50.9) | 50 [0, 90] | 0–185 | 0.22 |
| Luminal B (−4) | 52 (50.8) | 50 [0, 80] | 0–190 | ||
| Erbb2 (−2) | 53 (48.8) | 50 [0, 95] | 0–160 | ||
| Triple negative (−3) | 69 (50.6) | 50 [20, 110] | 0–170 | ||
| Nuclear VDR | Luminal A (−4) | 121 (68.2) | 110 [100, 170] | 0–300 | 0.78 |
| Luminal B (−4) | 125 (65.2) | 130 [90, 160] | 0–300 | ||
| Erbb2 (−2) | 122 (73.3) | 125 [95, 160] | 0–300 | ||
| Triple negative (−3) | 114 (62.3) | 110 [80, 150] | 0–270 | ||
Values are mean and median Hscores for nuclear and cytoplasmic CTSL, 53BP1, and VDR per tumor type. Min–Max denotes minimal and maximal values within the tumor subtype. Dispersion is assessed by SD and percentiles 25 and 75 ([P25, P75]); (−X) represents X missing values per molecular type as a result of insufficient specimen. Bolded Hscore values highlight the molecular subtype responsible for the statistically significant difference identified with the Kruskal-Wallis test that compares all molecular types (bold p-value if significant difference).
Immunohistochemical scores (Hscores; ranging from 0 to 300) for Ki67, ER, CTSL, 53BP1, and VDR provided a semi-quantitative measurement of their expression for each tumor subtype (Pallares et al., 2009). As shown in Fig. 7, staining of CTSL was both cytoplasmic and nuclear, whereas 53BP1 staining was only nuclear. Table 1 summarizes the immunohistochemistry results. Whereas cytoplasmic CTSL Hscores were similar in all tumor subtypes, nuclear CTSL Hscores were markedly enhanced in triple-negative tumors. In agreement with the in vitro findings, these high nuclear CTSL Hscores concur with lower 53BP1 Hscores in TNBC compared with all other tumor types. Furthermore, using the median nuclear Hscores for CTSL and 53BP1 of 0 and 150, respectively, as cut-off points with identical statistical power, we confirmed statistically significant differences in CTSL and 53BP1 expression among molecular tumor types, with TNBC emerging as a remarkably different tumor subtype. Table 2 shows that 60% of triple-negative tumors elicited Hscores for nuclear CTSL >0, a frequency more than twofold higher than for any other molecular type (P = 0.0013). Also, 75% of triple-negative tumors expressed 53BP1 Hscores below 150 compared with 39–49% of all other tumor types (P = 0.0049), clearly showing that high expression of nuclear CTSL and low expression of 53BP1 is significantly more associated with TNBC than any other molecular type. Thus, we identified nuclear CTSL as a novel biomarker for subsets of TNBC patients (Fig. S5). Importantly, this new signature (high nuclear CTSL and low 53BP1) could serve to stratify TNBC patients.
Table 2.
Frequency of CTSL, 53BP1, and VDR expression within molecular subtype relative to the median values in sporadic human breast cancer
| Proteins | Molecular type | n (%) | Fisher exact test p-value |
| Nuclear CTSL >0 | Luminal A | 23 (23.2) | 0.0013 |
| Luminal B | 22 (31.9) | ||
| Erbb2 (−1) | 12 (27.3) | ||
| Triple negative (−1) | 21 (60.0) | ||
| 53BP1 <150 | Luminal A | 59 (59.6) | 0.0049 |
| Luminal B | 35 (50.7) | ||
| Erbb2 (−4) | 21 (51.2) | ||
| Triple negative | 9 (25.0) | ||
| Nuclear VDR <120 | Luminal A | 46 (48.4) | 0.34 |
| Luminal B | 40 (61.5) | ||
| Erbb2 (−4) | 25 (58.1) | ||
| Triple negative | 16(48.5) |
Values denote the absolute (n) and the relative (%) frequencies of tumors with Hscore values above (nuclear CTSL) or below (nuclear 53BP1 and VDR) the median Hscore values for each protein in the overall population of sporadic breast cancer. Bolded frequency values highlight the molecular subtype responsible for the statistically significant difference identified with the Fisher exact test that compares all molecular types (bold p-value if significant difference).
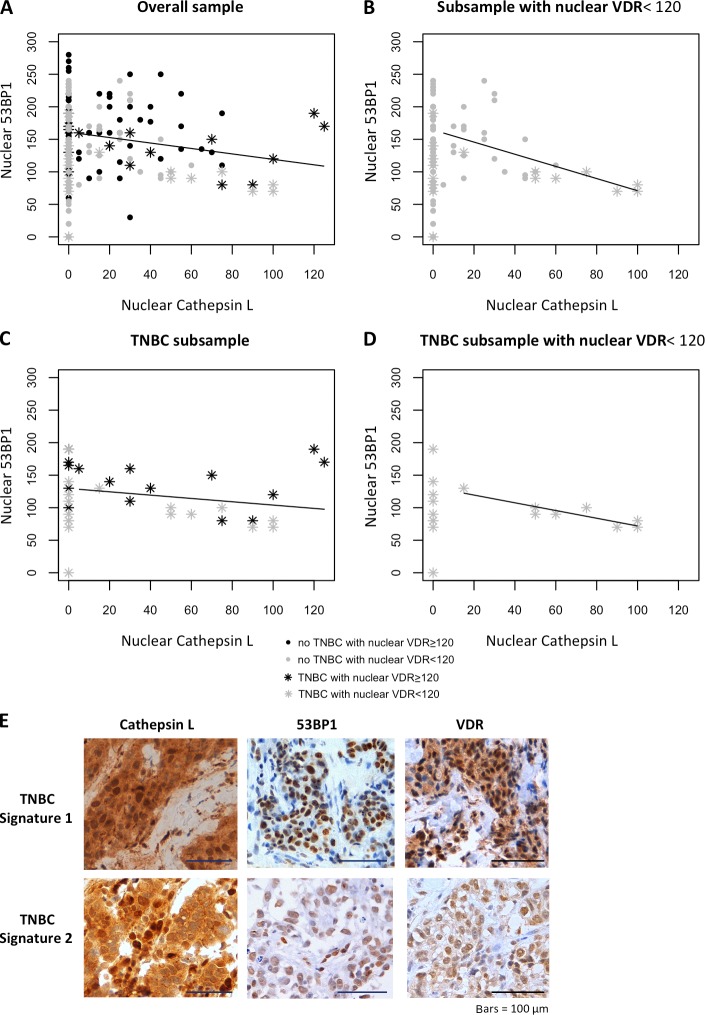
Next, we analyzed breast tumors from patients with germline mutations in BRCA1 (n = 18) or BRCA2 (n = 14) by immunohistochemistry (Tables 3 and 4). In comparison with sporadic TNBC, tumors from patients with BRCA1 germline mutations elicited the same high Hscores for nuclear CTSL (P = 0.95) and low Hscores for 53BP1 (P = 1). In contrast, tumors from patients with BRCA2 germline mutations had nuclear CTSL Hscores significantly lower than BRCA1-related tumors. Accordingly, 53BP1 Hscores were higher in tumors from patients with BRCA2 germline mutations than in BRCA1-related tumors or all molecular subtypes of sporadic tumors. These results support our in vitro data for a role of CTSL in the degradation of 53BP1 in BRCA1-deficient cells. Importantly, Fig. 8 A shows a statistically significant inverse linear correlation between Hscores for nuclear 53BP1 and CTSL in all tumor subtypes with positive nuclear CTSL expression. However, a coefficient of determination of only 6.6% indicates that 93.4% of the variability in 53BP1 Hscores cannot be accounted for by increases in nuclear CTSL. Thus, additional factors might contribute to CTSL-mediated degradation of 53BP1 in these tumors. Identifying these factors could help to discriminate subsets of patients in which this pathway is activated.
Table 3.
Immunohistochemical analysis of CTSL, 53BP1, and VDR expression in tumors from patients with BRCA1 or BRCA2 germline mutations
| Proteins | Mutation type | H score | M-W test p-value (vs. sporadics) | M-W test p-value (BRCA1 vs. BRCA2) | ||
| Mean (SD) | Median [P25, P75] | Min–Max | ||||
| Cytoplasmic CTSL | BRCA1 mutation (−4) | 119 (34.2) | 110 [100, 138] | 60–190 | 0.0563 | 0.88 |
| BRCA2 mutation (−1) | 118 (46.2) | 120 [100, 120] | 100–150 | 0.0437 | ||
| Nuclear CTSL | BRCA1 mutation (−4) | 38 (45.2) | 30 [15, 45] | 0–180 | 0.0001 | 0.0494 |
| BRCA2 mutation (−1) | 15 (22.7) | 4 [0, 15] | 0–75 | 0.19 | ||
| 53BP1 | BRCA1 mutation (−1) | 111 (28.4) | 115 [100, 125] | 70–175 | 0.0016 | 0.0001 |
| BRCA2 mutation (−1) | 198 (43.6) | 210 [185, 220] | 110–270 | 0.0008 | ||
| Cytoplasmic VDR | BRCA1 mutation | 86 (38.5) | 100 [50, 100] | 0–150 | 0.0048 | 0.0011 |
| BRCA2 mutation (−3) | 145 (36.7) | 150 [115, 165] | 100–200 | <0.0001 | ||
| Nuclear VDR | BRCA1 mutation | 66 (52.9) | 53 [27, 100] | 0–180 | 0.0010 | 0.0001 |
| BRCA2 mutation (−3) | 175 (57.8) | 170 [135, 193] | 110–300 | 0.0074 | ||
Values are mean and median Hscores for nuclear and cytoplasmic CTSL, 53BP1, and VDR in BRCA1- or BRCA2-related tumors. Min–Max denotes minimal and maximal values within the tumor. Dispersion is assessed by SD and percentiles ([P25, P75]); (−X) represents X missing values. Bolded p-values highlight the statistical significance of differences measured by Mann-Whitney test (M-W) between each tumor mutation subtype versus the overall population of sporadic breast cancer or between tumors with BRCA1 versus BRCA2 germline mutations.
Table 4.
Frequency of CTSL, 53BP1, and VDR expression within BRCA1- and BRCA2-related tumors
| Proteins | Mutation type | n (%) | Fisher exact test p-value (vs. sporadics) | Fisher exact test p-value (BRCA1 vs. BRCA2) |
| Nuclear CTSL >0 | BRCA1 mutation (−4) | 12 (85.7) | 0.0001 | 0.10 |
| BRCA2 mutation (−1) | 7 (53.8) | 0.13 | ||
| 53BP1 <150 | BRCA1 mutation (−1) | 2 (11.8) | 0.0019 | 0.0001 |
| BRCA2 mutation (−1) | 11 (84.6) | 0.0210 | ||
| Nuclear VDR <120 | BRCA1 mutation (−1) | 3 (16.7) | 0.0027 | 0.0051 |
| BRCA2 mutation (−1) | 8 (72.7) | 0.35 |
Values denote the absolute (n) and the relative (%) frequencies of BRCA1- or BRCA2-related tumors with Hscore values above (nuclear CTSL) or below (nuclear 53BP1 and VDR) the median Hscore values for each protein in sporadic breast tumors. Bolded p-values highlight the statistically significant difference measured by Fisher exact test comparing a molecular subtype with either the overall population of sporadic breast cancer or between tumors with BRCA1 versus BRCA2 germline mutations.
Figure 8.
Nuclear 53BP1 expression correlates inversely with nuclear CTSL expression in sporadic human breast cancer. (A–D) Linear regression analysis between Hscores for nuclear 53BP1 and CTSL in sporadic breast cancer samples with Hscores for nuclear CTSL >0. Note the high variability in nuclear 53BP1 Hscores in tumors without nuclear CTSL. The linear regression analysis of the association between nuclear 53BP1 and CTSL depicted in A (linear regression coefficient r= −0.42; P = 0.02; coefficient of determination r2 = 6.6%) and B (r = −0.93; P = 0.0025; r2 = 29.2%) include Hscores from both TNBC and no TNBC tumors, whereas C (r = −0.255; P = 0.294; r2 = 5.8) and D (r = −0.89; P = 0.0027; r2 = 80.2%) depict TNBC only. Note the marked increase in percentage of the variability in nuclear 53BP1 levels that can be explained by changes in nuclear CTSL parameters of B and D when only tumors with nuclear VDR Hscores <120 are included in the regression analysis. (E) Images of immunohistochemical analysis results in TNBC patients. Two different signatures were observed in these patients (as well as in patients with BRCA1 germline mutations). Top panels show the signature of a few TNBC patients: high nuclear CTSL, 53BP1, and VDR. Bottom panels show the most common signature in TNBC patients: high nuclear CTSL, low 53BP1, and low nuclear VDR. Bars, 100 µm.
Previous studies in human colon cancer cells showed a correlation between expression of VDR and cystatin D, an inhibitor of several cathepsins including CTSL, and up-regulation of cystatin D by vitamin D (Alvarez-Díaz et al., 2009). In addition, our in vitro data show that vitamin D inhibits CTSL-mediated degradation of 53BP1. Vitamin D actions require a functional nuclear VDR (Dusso et al., 2005). Because VDR levels are reduced in several human cancers and BRCA1 loss causes defective VDR translocation to the nucleus (Deng et al., 2009), we hypothesized that there might be a threshold for nuclear VDR that inhibits CTSL-mediated degradation of 53BP1. High levels of nuclear VDR could explain the signature of tumors with high Hscores for both nuclear CTSL and 53BP1. Our hypothesis is supported by the findings of a direct linear correlation between nuclear levels of VDR and 53BP1 for all 249 tumor types (Pearson correlation r = 0.238; P = 0.0002). By analyzing the linear relationship for those tumors with nuclear VDR expression below the median Hscore of 120 obtained in the 249 sporadic tumors (Fig. 8 B), we found an increase in the slope of the linear regression of nuclear 53BP1 and CTSL Hscores as well as increased coefficient of determination (from 6.6 to 29.2%). Furthermore, when the correlation between 53BP1 and CTSL was examined exclusively in TNBC, the nonsignificant correlation depicted in Fig. 8 C becomes significant when only tumors with nuclear VDR <120 are examined (P < 0.0027), as seen in Fig. 8 D, with a coefficient of determination of 80.2%. Indeed, the outliers in Fig. 8 C correspond to patients with VDR Hscores far above 120, which maintain high Hscores for nuclear 53BP1 despite high Hscores for nuclear CTSL (Fig. 8 E, top). The bottom panels of Fig. 8 E show the most common biomarker signature found in TNBC and BRCA1-related tumors. Furthermore, mean nuclear Hscores for VDR are lower in tumors from patients with BRCA1 mutations compared with TNBC (Tables 1 and 3), suggesting that loss of BRCA1 may also impair VDR translocation to the nucleus. The VDR Hscores in BRCA2 germline mutations depicted in Table 3 suggest that only BRCA1 mutations impact VDR translocation to the nucleus, resulting in less inhibition of CTSL-mediated degradation of 53BP1.
In summary, this study reveals a new triple biomarker signature—nuclear VDR, CTSL, and 53BP1—for stratification of patients with TNBC (in which BRCA1 is frequently somatically altered) and tumors from patients with BRCA1 germline mutations (Tables 2 and 4). Based on our in vitro data, these signatures could potentially be used as predictors of the response of specific tumors to radiation, cross-linking reagents, and PARPi.
Discussion
Breast cancers classified as triple negative or BRCA1 deficient are among the most aggressive and difficult to treat. These tumors harbor similar DNA repair deficiencies and gene expression profiles (Foulkes et al., 2010). Of particular relevance is the loss of BRCA1 function and decrease in 53BP1 levels, two factors with a decisive role in the choice of DNA DSB repair mechanisms: HR or NHEJ (Bouwman et al., 2010). Recent landmark studies demonstrated that loss of 53BP1 allows survival of BRCA1-deficient cells and induces their resistance to DNA-damaging therapeutic strategies (Cao et al., 2009; Bothmer et al., 2010; Bouwman et al., 2010; Bunting et al., 2010). Thus, stabilization of 53BP1 levels represents a promising new strategy for the treatment of these cancers. However, before this study, no information was available about how the levels of 53BP1 mRNA and/or protein are down-regulated in breast tumor cells.
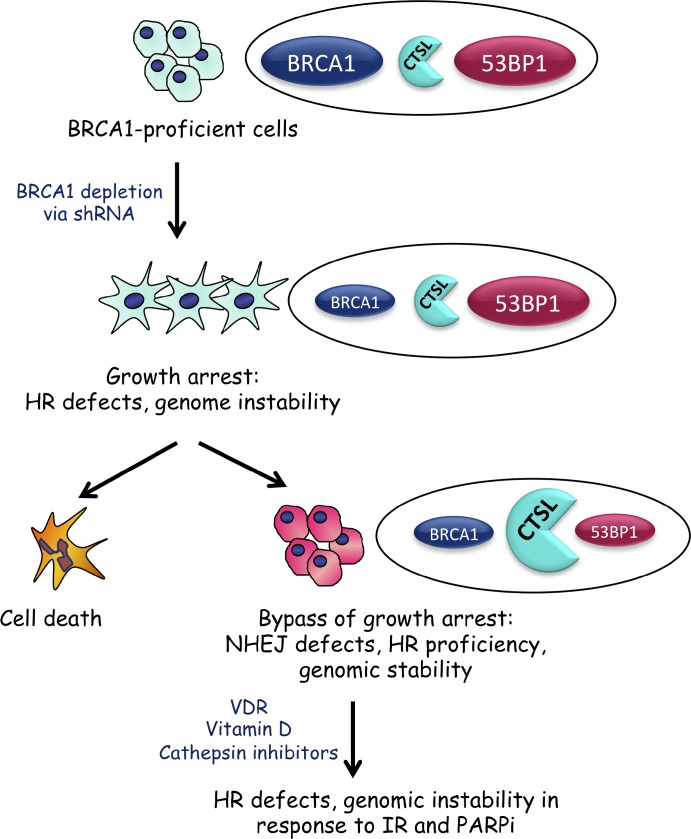
This study demonstrates that up-regulation of CTSL is a mechanism responsible for lowering 53BP1 protein levels in BRCA1-deficient cells, allowing bypass of the characteristic growth arrest upon loss of BRCA1 function (Fig. 9). It is possible that other mechanisms such as degradation of 53BP1 by the proteasome also contribute to 53BP1 reduction in BRCA1-deficient tumor cells. In addition, previous studies showed that subsets of BRCA1-mutated tumors also exhibit reduced 53BP1 mRNA levels (Bouwman et al., 2010), indicating that different mechanisms can be activated in BRCA1-deficient cells to lower 53BP1 levels and ensure survival. Importantly, depletion of CTSL or inhibition of its activity stabilizes 53BP1 protein levels and induces genomic instability in BRCA1-deficient cells after IR or treatment with PARPi.
Figure 9.
Activation of CTSL-mediated degradation of 53BP1 allows BRCA1-deficient cells to overcome genomic instability and growth arrest. Depletion of BRCA1 in breast cancer cells leads to defects in HR, genomic instability, and growth arrest. Over time, BRCA1-deficient cells activate CTSL-mediated degradation of 53BP1, which rescues HR defects while inhibiting NHEJ. This allows BRCA1-deficient cells to overcome growth arrest. Inhibition of CTSL activity via treatment with vitamin D or specific inhibitors stabilizes 53BP1 protein levels and induces genomic instability in response to IR and PARPi. Furthermore, up-regulation of CTSL-mediated degradation of 53BP1 could be regulated by nuclear VDR.
Furthermore, we show a significant negative correlation between 53BP1 and nuclear CTSL in human tumors. The highest median levels of nuclear CTSL concur with the lowest levels of 53BP1 in a subset of TNBC and tumors from patients with BRCA1 germline mutations and with low nuclear VDR levels. This study has revealed a new pathway, activated upon loss of BRCA1 function, which is anticipated to contribute to the progression of breast cancers with the poorest prognoses. Inhibition of this pathway by treatment with vitamin D or cathepsin inhibitors could provide a new therapeutic strategy for breast cancer. Importantly, the status of the pathway offers great potential as a predictive biomarker for response to therapy.
CTSL up-regulation after growth arrest by BRCA1 loss and serum deprivation
In a variety of cancers, up-regulation of CTSL has been associated with increased invasiveness, metastasis, and overall degree of malignancy (Jedeszko and Sloane, 2004; Skrzydlewska et al., 2005; Gocheva and Joyce, 2007). Thus, inhibition of CTSL activity could represent a strategy for cancer treatment (Lankelma et al., 2010). Our previous studies in MEFs revealed novel roles for CTSL in the degradation of nuclear factors with functions in cell cycle regulation (Rb family members) and DNA repair (53BP1; Gonzalez-Suarez et al., 2011). The present study shows that CTSL is up-regulated in breast cancer cells after BRCA1 depletion and that increased CTSL activity impacts mechanisms of DNA DSB repair. Several lines of evidence implicate BRCA1 in transcriptional regulation, chromatin remodeling (Bochar et al., 2000; Mullan et al., 2006), and maintenance of heterochromatin silencing (Zhu et al., 2011). Epigenetic mechanisms such as DNA methylation and expression of miRNAs have been linked to BRCA1 function (Shukla et al., 2010; Kawai and Amano, 2012; Tanic et al., 2012). Although the mechanism by which BRCA1-deficient cells activate CTSL remains unknown, the latency in the activation of this pathway indicates that CTSL is not a direct transcriptional target of BRCA1. Rather, we speculate that the loss of BRCA1 might result in alterations in chromatin structure that either make the CTSL gene more permissive to transcriptional activation over time and/or alter the stability of CTSL mRNAs. Those BRCA1-deficient cells that are able to activate CTSL-mediated degradation of 53BP1 would be poised to continue proliferation.
Furthermore, we show that cells growth arrested in G0/G1 by serum deprivation also exhibit low BRCA1, high CTSL, and low 53BP1 levels, suggesting a functional relationship between these proteins during the cell cycle. We envision a model where the decrease in BRCA1 levels in G0/G1 phases contributes to CTSL-mediated degradation of 53BP1. Up-regulation of CTSL could also trigger a feedback mechanism that lowers BRCA1 protein levels. This is supported by studies showing that BRCA1 is a target for degradation by cysteine proteases of the cathepsin family, although the specific cathepsin was not identified (Blagosklonny et al., 1999). Future studies need to determine if CTSL functions as a regulator of 53BP1 and BRCA1 protein stability during the cell cycle.
Vitamin D and cathepsin inhibitors can modulate DNA DSB repair choice
Our previous study in MEFs (Gonzalez-Suarez et al., 2011) and the present study in human breast cancer cells reveal an unprecedented role for vitamin D and cathepsin inhibitors in the regulation of DNA DSB repair choice. By stabilizing 53BP1 protein levels in the context of BRCA1 deficiency, CTSL inhibitors facilitate repair of DSBs by NHEJ while inhibiting HR. Importantly, our data indicate that both vitamin D and cathepsin inhibitors impact 53BP1 stability, especially in cells that up-regulate CTSL, i.e., BRCA1-deficient cells, showing a lesser effect in cells with normal CTSL expression. This is likely because up-regulation of CTSL leads to an increase in the levels of nuclear CTSL, which is low relative to other cellular compartments in normal cells (Gonzalez-Suarez et al., 2011). Although our attempts to localize nuclear CTSL in MCF7 BOGA cells by Western blot were unsuccessful, the immunohistochemical analysis of TMA clearly showed that a subset of TNBC and BRCA1-deficient tumors present with high levels of nuclear CTSL. Interestingly, these tumors are often also deficient in nuclear VDR and 53BP1.
The ability to impact the choice of DNA DSB repair pathway could have profound consequences for cancer therapy. In tumor cells that activate CTSL-mediated degradation of 53BP1 as a means to ensure survival, cathepsin inhibition could stabilize 53BP1, increase genomic instability, and induce growth arrest and/or cell death, especially in combination with IR or PARPi. Thus, treatment with vitamin D or CTSL inhibitors could represent a new therapeutic strategy for specific types of breast tumors, if they can be identified.
Nuclear levels of CTSL, 53BP1, and VDR: a new predictive biomarker signature for drug response?
The analysis of TMA indicates that levels of nuclear CTSL represent a new positive biomarker for subsets of tumors having abnormalities in BRCA1, including TNBC (currently defined solely by the absence of Her2/neu, estrogen and progesterone receptors, and frequently showing somatic alterations of BRCA1 function), and tumors arising in association with germline mutations in BRCA1. Similarly, we find that nuclear VDR levels correlate linearly with 53BP1 content in all tumor subtypes, and the lowest nuclear VDR levels correspond to TNBC and tumors from patients with BRCA1 germline mutations. The finding of the strong negative correlation between reductions in nuclear 53BP1 with increases in nuclear CTSL levels (80% coefficient of determination) in TNBC with nuclear VDR <120 is very interesting, as TNBC is associated with severe vitamin D deficiency (Peppone et al., 2011). Furthermore, a previous study showed a role for VDR in the up-regulation of cathepsin inhibitors (Alvarez-Díaz et al., 2009). Thus, it is tempting to speculate that vitamin D interventions could lead to VDR-induced expression of cystatins and the attenuation of CTSL-mediated degradation of 53BP1. Future studies testing this hypothesis might lead to new strategies of targeted therapy.
The combination of low nuclear VDR or high nuclear CTSL with low 53BP1 levels offers great potential for the stratification of BRCA1-deficient and TNBC patients into different subgroups and as a predictive biomarker for the response of these patients to current therapies. In particular, the use of PARPi as single agents or in combination with radiation or chemotherapy is a leading strategy for breast cancer management, especially for HR-deficient tumors (Farmer et al., 2005; Helleday et al., 2005; Fong et al., 2009; Tutt et al., 2010; Drew et al., 2011). However, a significant fraction of these cancers acquire resistance to PARPi. A recent study in cell culture and mouse models demonstrated that loss of 53BP1 reduces the sensitivity of BRCA1-deficient cells to PARPi (Bunting et al., 2010).
Our study suggests that BRCA1-deficient and TNBC patients that exhibit low nuclear VDR, high nuclear CTSL, and low 53BP1 levels are likely to be proficient in HR and resistant to PARPi. Therefore, these patients will not benefit from this specific treatment unless levels of 53BP1 are stabilized. For these patients, treatment with vitamin D or CTSL inhibitors to stabilize 53BP1 levels in combination with PARPi might result in the most effective therapy.
Materials and methods
Cells maintained in DMEM, 10% FBS, and antibiotics/antimycotics were transduced with shRNAs and selected in media containing 0.5 mg/ml Geneticin G418 or 2 µg/ml puromycin. Except for one shBRCA1 (5′-TCAGTACAATTAGGTGGGCTT-3′) that was constructed by J. Zhang, the rest were purchased from the Genome Institute at Washington University (shBRCA sequences: 1, 5′-GAGAATCCTAGAGATACTGAA-3′; 2, 5′-TATAGCTGTTGGAAGGACTAG-3′; 3, 5′-CCCTAAGTTTACTTCTCTAAA-3′; 4, 5′-GCCCACCTAATTGTACTGAAT-3′; 5, 5′-CCCACCTAATTGTACTGAATT-3′).
Lentiviral transductions were performed as previously described (Gonzalez-Suarez et al., 2009). In brief, 293T cells were transfected with viral packaging (pHR’8.2ΔR) and envelope (pCMV-VSV-G) plasmids along with the vector containing the shRNA of interest. After 48 h, viral media was collected and used to infect target cells in one 4–6-h infection. Cells were allowed to recover for 48 h and treated with the appropriate selection drug. Viral packaging and envelope plasmids were gifts from S. Stewart (Washington University, St. Louis, MO). HCC1937 cells were a gift from J. Zhang (Washington University, St. Louis, MO). HA-BRCA1 (Addgene) transient transfections were performed using the X-tremeGENE HP transfection reagent (Roche). For growth arrest by serum deprivation, MCF7 cells were incubated in DMEM, 0.1% FBS, and antibiotics/antimycotics for 48 h. DNA content was monitored after ethanol fixation and propidium iodide labeling (1 mg/ml in water).
Cell treatments
Vitamin D.
Cells were incubated with 10−7 M 1α,25-dihydroxyvitamin D3 (Sigma-Aldrich) for 24–48 h, as indicated. Aliquots of 1 nmol 1α,25-dihydroxyvitamin D3 were resuspended in 1 ml BGS and diluted in DMEM to a final concentration of 10% BGS. BGS was the vehicle control.
E-64.
Cells were incubated with the broad-spectrum cathepsin inhibitor E-64 (Sigma-Aldrich) diluted in water at a concentration of 10 µM for 24 h.
PARPi.
Cells were treated with the PARPi EB-47 (Sigma-Aldrich) diluted in water at a concentration of 1.2 µg/ml for 48 h.
IR.
For determining the extent of genomic instability, cells were irradiated with 2 Gy and analysis of metaphase spreads was performed 24 h after IR. For assaying formation of IRIF, cells were irradiated with 8 Gy and fixed and processed for immunofluorescence 1, 3, or 6 h after IR as indicated. For comet assays, cells were irradiated with 8 Gy.
Western blotting
Cells were lysed in RIPA buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, and 0.5% SDS) supplemented with PMSF, protease inhibitors (Sigma-Aldrich), and 20 mM DTT. Protein detection was performed using the following antibodies: BRCA1 (Santa Cruz Biotechnology, Inc.), 53BP1 (Novus Biologicals), CTSL (Santa Cruz Biotechnology, Inc.), β-Tubulin (Sigma-Aldrich), and p107 (Santa Cruz Biotechnology, Inc.).
Immunofluorescence
For immunofluorescence of RAD51 and 53BP1, cells were plated on coverslips and fixed in 3.7% formaldehyde and 0.2% Triton-X 100 in PBS for 10 min at room temperature. Coverslips were washed and blocked for 1 h at 37°C in 1% BSA and 0.1% Triton X-100 in PBS. Incubation with antibodies recognizing RAD51 (1:200; Santa Cruz Biotechnology, Inc.) or 53BP1 (1:1,000; Novus Biologicals) was performed for 1 h at 37°C, followed by washing in PBS and incubation with Alexa Fluor 488 goat anti–rabbit (1:1,000; Invitrogen) secondary antibody for 1 h at 37°C. After washing in PBS, coverslips were mounted using Vectashield with DAPI (Vector Laboratories).
BRCA1 immunofluorescence was performed following a protocol from the Fernandez-Capetillo laboratory (Centro Nacional de Investigaciones Oncológicas, Madrid, Spain). In brief, cells were washed twice with PBS and incubated in CSK I buffer (10 mM Pipes, pH 6.8, 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 1 mM EGTA, and 0.5% Triton X-100) for 5 min. Coverslips were washed five times with cold PBS and fixed in modified STF buffer (150 mM 2-Bromo-2-nitro-1,3-propanediol, 108 mM diazolidinyl urea, 10 mM sodium citrate, and 50 nM EDTA, pH 5.7) for 30 min at room temperature. Coverslips were then washed twice with cold PBS and permeabilized (PB buffer: 100 mM Tris-HCl, pH 7.4, 50 mM EDTA, pH 8.0, and 0.5% Triton X-100) for 15 min at room temperature followed by two washes in PBS. Blocking, antibody staining, and mounting were performed as previously described with BRCA1 antibody (Santa Cruz Biotechnology, Inc.) diluted at 1:200 and Alexa Fluor 594 goat anti–mouse secondary antibody (Invitrogen) diluted at 1:1,000.
Microscopy and photo capture was performed at room temperature on either an Eclipse 90i microscope (Nikon) using 60 or 100× oil objective lenses (NA 1.4 and 1.45, respectively) with a CoolSNAP ES2 digital camera (Photometrics) and MetaMorph (Version 7.1.2.0) or a DM5000 B microscope (Leica) using 63 or 100× oil objective lenses (NA 1.4 and 1.3, respectively) with a DFC350FX digital camera (Leica) and the Application Suite (Version 4.1.0; Leica).
Comet assay
Neutral comet assays were performed using CometSlide assay kits (Trevigen). Treated cells were irradiated with 8 Gy and incubated at 37°C for different periods of time to allow DNA to repair (0, 30, 60, and 90 min). Cells were embedded in agarose, lysed, and subjected to neutral gel electrophoresis. Cells were stained with ethidium bromide and visualized using a fluorescence microscope. Olive comet moments were calculated by multiplying the percentage of DNA in the tail by displacement between the means of the head and tail distributions as described previously (Olive et al., 1990). The program CometScore Version 1.5 (TriTek) was used to calculate olive moments. A total of 30 comets were analyzed per sample in each experiment.
Proliferation assay
To quantify growth upon depletion of BRCA1, cells were plated in triplicate at 150,000 cells/well and counted 96 h later. Cell proliferation was measured in each cell line four times within a 14-d period. To quantify the growth of the sh53BP1/shBRCA1 cells, 4 × 106 cells were plated in 15-cm dishes and counted every 96 h for 20 d. To extrapolate proliferation to the respective time periods, we used the equation N = N0ekt where N is the final number of cells, N0 is the starting number of cells, k is ln2/DT (doubling time), and t is time in days (Sherley et al., 1995). For each 96-h period we calculated the doubling time and used it to estimate the number of cells (N) that would result from initially plating 150,000 (N0) and cultured them for a given period of time. The doubling times for the control cells remained constant throughout the time period, whereas the doubling times for the shBRCA1 cells started increasing once the cells overcame the growth arrest.
qRT-PCR
RNA was isolated using the RNAqueous-4PCR kit (Ambion). cDNA was generated from 500 ng of total RNA using TaqMan Reverse Transcription Reagents (Applied Biosystems). BRCA1, 53BP1, CTSL, and 18S expression was determined using TaqMan Gene Expression Assays (Hs01556193_m1, Hs00996818_m1, Hs00377632_m1, and Hs99999901_s1; Applied Biosystems). All the reactions were performed in triplicate and the target gene and endogenous controls were amplified in the same plate. Relative quantitative measurements of target genes were determined by comparing the cycle thresholds.
Analysis of aberrant chromosomes
Cells were treated with vitamin D or E-64 for 24 h, irradiated, and allowed to recover for 24 h. Cells treated with PARPi were pretreated with vitamin D for 24 h followed by combined treatment with vitamin D and PARPi for 48 h. After all treatments, cells were arrested in mitosis by treatment with colcemid for 4 h and metaphase spreads were prepared by hypotonic swelling in 0.56% KCl, followed by fixation in 3:1 methanol/acetic acid. Cell suspensions were dropped onto slides and stained for 25 min in Wright-Giemsa Stain (Ricca Chemical Company) and then washed in water. Slides were allowed to dry and were mounted using Eukitt Mounting Reagent (Sigma-Aldrich) and analyzed at room temperature on a DM5000 B microscope using a 100× oil objective (NA 1.30). Images were acquired with the DFC350 FX digital camera using the Application Suite.
TMAs
A total of 249 tissue samples from patients with sporadic breast carcinoma were obtained at Hospital Universitari Arnau de Vilanova in Lleida, Lleida, Spain, from 1998 to 2012. An informed consent was obtained from each patient and the study was approved by the local Ethical Committee. The series of 249 tumor samples included formalin-fixed, paraffin-embedded blocks for all patients, 165 core biopsies, before the initiation of neoadjuvant treatment, and 84 surgical specimens, before the initiation of adjuvant treatment. Tumors were classified according to the expression of proteins Ki67, Erα, and Her2 into four molecular subtypes: luminal A (n = 99), luminal B (n = 69), Her2 (n = 45), and triple negative (n = 36). Luminal A tumors are steroid hormone receptor positive, are negative for Her2, contain less than 30% of Ki67 positive cells, and tend to have a good prognosis. Luminal B tumors are steroid hormone receptor–positive, negative for Her2, contain more than 30% Ki67-positive cells, and tend to have a worse prognosis than luminal A. In contrast, Her2 tumors are positive for Her2 and have been shown to have a poor prognosis. Triple-negative tumors are negative for steroid hormone receptors and Her2 and have the worst prognosis. Moreover, tissue samples were also obtained from 18 breast cancers from patients carrying a BRCA1 germline mutation (luminal A, n = 1; luminal B, n = 1; Her2, n = 2; triple negative, n = 15), and from 14 breast cancers from patients carrying a BRCA2 germ-line mutation (luminal A, n = 6; luminal B, n = 2; Her2, n = 3; triple negative, n = 3). These patients had been treated in Hospital Santa Creu I Sant Pau, Barcelona, Spain.
We used a tissue arrayer device (Beecher Instrument) to construct the TMAs. Representative areas of all samples were marked in the corresponding paraffin blocks. In each sample, we selected two cylinders (0.6 mm of largest diameter) for the immunohistochemical analysis.
Immunohistochemical analysis
TMA blocks were sectioned at a thickness of 3 µm, dried for 1 h at 65°C before being dewaxed in xylene and rehydrated through descending concentrations of ethanol, and washed with PBS. Ki67, ERα, and Her2 were used to determine molecular subtype. Comparative studies of CTSL, VDR, and 53BP1 expression were performed on sequential serial sections. Antigen retrieval for CTSL and ERα was achieved by heat treatment at 95°C for 20 min in a high pH solution (Dako). Heat-induced antigen retrieval for 53BP1 and Ki67 was performed in a low pH solution (Dako). Before staining the sections, endogenous peroxidase was blocked. Primary antibodies and incubation times were as follows: CTSL (1:50; Santa Cruz Biotechnology, Inc.; incubation overnight at 4°C); 53BP1 (1:2,500; Novus Biologicals; incubation 20 min at room temperature); VDR (1:2,000; Abcam; incubation 20 min at room temperature); Ki67 (Ready-to-use; Dako; incubation 20 min at room temperature); ERα (Ready-to-use; Dako; incubation 20 min at room temperature), and Her2 (Herceptest kit; Dako). The reaction was visualized with the Streptovidin-Biotin Complex (Dako) for CTSL and Envision Flex (Dako) for 53BP1, Ki67, and ERα. Sections were counterstained with hematoxylin. Appropriate positive and negative controls were also tested.
Hscores provide a semiquantitative measurement of protein expression per tumor by taking into consideration the percentage of positive cells and the intensity of their staining. An Hscore ranging from 0 (no immune reaction) to 300 (maximal immunoreactivity) was obtained with the formula Hscore = 1× (% light staining) + 2× (% moderate staining) + 3× (% strong staining). The reliability of such scores for the interpretation of immunohistochemical staining in TMAs has been reported previously (Pallares et al., 2009).
Her2 staining was evaluated according to a standard protocol (HercepTest; Dako) and scored as four intensities (i.e., negative = 0; weak = 1+; moderate = 2+; and strong = 3+), considering negative Her2 expression for intensity values of 0, 1+, and 2+ when there was no amplification by FISH and positive for intensity values of 3+ and 2+, when 2+ was amplified by FISH. For each marker, there were a variable number of nonassessable cases caused by technical problems including no representative tumor sample left in the cylinders, detachment, cylinders missed while constructing the array, necrosis, and absence of viable tumor cells in the TMA sections.
Statistical analysis
For the in vitro experiments, a two-tailed student’s t test was used to calculate statistical significance of the observed differences. Excel 2010 (Microsoft) was used for the calculations. In all cases, differences were considered statistically significant when P < 0.05. For some figures the 95% confidence interval based on an exact binomial distribution was calculated to determine significant differences among samples. For the TMA studies, a Kruskal-Wallis test was used to test the statistical significance of the observed differences in CTSL, VDR, and 53BP1 Hscores between molecular breast tumor subtypes. Tumors were partitioned according to cut-off nuclear Hscores for CTSL, 53BP1, and VDR selected by their median values as >0, <150, and <120, respectively. Once cut-off points were applied, Fisher exact test was used to assess the statistical significance of the differences in the distribution of the two categories of CTSL, 53BP1, and VDR Hscores above or below cut-off points for all breast tumor types. In the analysis of BRCA1 and BRCA2 samples, we used the Mann-Whitney test to analyze Hscore differences between them as well as differences of each of them with the sample of sporadic tumors, and we used the Fisher exact test to assess differences in the distributions of groups defined by the same cut-off points used for the sporadic tumors. The subsample of TNBC was also compared with the sample of germinal BRCA1 mutated cancers using the same statistical tests. The Pearson correlation coefficient together with linear regression models assessed the statistical significance of the relationship between nuclear CTSL and 53BP1 Hscores. R package was used to perform all TMA statistical tests. Differences were considered significant when P < 0.05.
Online supplemental material
Fig. S1 shows an entire BRCA1 Western blot as well as Western blots of activation of CTSL-mediated degradation of 53BP1 in MDA-MB-231 cells and by different BRCA1 shRNAs. Fig. S2 shows qRT-PCR results for the sh53BP1/shBRCA1 and the shBRCA1/shCTSL doubly depleted cells. Fig. S3 shows results from immunofluorescence studies of 53BP1, BRCA1, and RAD51 IRIF in the generated cell lines and upon different treatments. Fig. S4 shows RAD51 foci formation data at 3 and 6 h after IR. Fig. S5 shows differences in CTSL levels among different molecular subtypes of breast cancer. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201204053/DC1.
Supplementary Material
Acknowledgments
We thank Ray Kreienkamp for his contribution to the study during his rotation in the laboratory and Jesus Caceres for technical assistance. We thank D. Ferraro and J. Jaboin for helpful discussions and clinical advice.
Work performed in the laboratory of S. Gonzalo was supported by National Institute of General Medical Sciences grant RO1 GM094513-01, Department of Defense Breast Cancer Research Program Idea Award BC110089, and Research Development Award from Siteman Cancer Center. I. Gonzalez-Suarez was a recipient of a postdoctoral fellowship from the American Heart Association. Work in the laboratory of J. Zhang was supported by National Cancer Institute grant R01CA154625. A. Novell, M. Martinez-Alonso, X. Matias-Guiu, and A. Dusso were supported by grants 2009SGR794, RD06/0020/1034, Beca Marta Santamaria, and programa de intensificación de la investigación, Instituto Carlos III. A. Dusso was supported by a grant from Barnes Jewish Hospital Auxilary. Tumor samples were obtained with the support of Xarxa Catalana de Bancs de Tumors, the Tumor Banc Platform of Red Temática de Investigación Cooperativa en Cáncer and Red de Biobancos (RD09/0076/00059).
Authors have no conflicts of interest involving this manuscript.
Author contributions: D.A. Grotsky, I. Gonzalez-Suarez., A. Novell, and M.A. Neumann performed the majority of the experiments. D.A. Grotsky, A. Dusso, and X. Matias-Guiu contributed to the writing of the manuscript. S.C. Yaddanapudi, M. Croke, A.B. Redwood, and S. Ortega-Martinez contributed to some of the experiments. Z. Feng and J. Zhang provided essential reagents such as the shRNA specific for depletion of BRCA1 and HCC1937 transfected with BRCA1, as well as helpful discussions and advice. A. Novell, M. Martinez-Alonso., X. Matias-Guiu, and A. Dusso performed all the analysis of human TMA. E. Lerma and T. Ramon y Cajal provided breast tumor samples from patients with BRCA1/2 germline mutations. A. Dusso provided seminal advice and helpful discussions throughout the progression of the project, and her expertise in vitamin D biology and analysis of human tumors was instrumental for the study. S. Gonzalo supervised the research and the preparation of the manuscript.
Footnotes
Abbreviations used in this paper:
- CTSL
- cathepsin L
- DSB
- DNA double strand break
- HR
- homologous recombination
- Hscore
- immunohistochemical score
- IR
- ionizing radiation
- IRIF
- IR-induced foci
- MEF
- mouse embryonic fibroblast
- NHEJ
- nonhomologous end joining
- PARPi
- poly(ADP-ribose) polymerase inhibitor
- qRT-PCR
- quantitative RT-PCR
- TMA
- tissue microarray
- TNBC
- triple-negative breast cancer
- VDR
- vitamin D receptor
References
- Alvarez-Díaz S., Valle N., García J.M., Peña C., Freije J.M., Quesada V., Astudillo A., Bonilla F., López-Otín C., Muñoz A. 2009. Cystatin D is a candidate tumor suppressor gene induced by vitamin D in human colon cancer cells. J. Clin. Invest. 119:2343–2358 10.1172/JCI37205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aly A., Ganesan S. 2011. BRCA1, PARP, and 53BP1: conditional synthetic lethality and synthetic viability. J. Mol. Cell Biol. 3:66–74 10.1093/jmcb/mjq055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya A., Ear U.S., Koller B.H., Weichselbaum R.R., Bishop D.K. 2000. The breast cancer susceptibility gene BRCA1 is required for subnuclear assembly of Rad51 and survival following treatment with the DNA cross-linking agent cisplatin. J. Biol. Chem. 275:23899–23903 10.1074/jbc.C000276200 [DOI] [PubMed] [Google Scholar]
- Blagosklonny M.V., An W.G., Melillo G., Nguyen P., Trepel J.B., Neckers L.M. 1999. Regulation of BRCA1 by protein degradation. Oncogene. 18:6460–6468 10.1038/sj.onc.1203068 [DOI] [PubMed] [Google Scholar]
- Bochar D.A., Wang L., Beniya H., Kinev A., Xue Y., Lane W.S., Wang W., Kashanchi F., Shiekhattar R. 2000. BRCA1 is associated with a human SWI/SNF-related complex: linking chromatin remodeling to breast cancer. Cell. 102:257–265 10.1016/S0092-8674(00)00030-1 [DOI] [PubMed] [Google Scholar]
- Bothmer A., Robbiani D.F., Feldhahn N., Gazumyan A., Nussenzweig A., Nussenzweig M.C. 2010. 53BP1 regulates DNA resection and the choice between classical and alternative end joining during class switch recombination. J. Exp. Med. 207:855–865 10.1084/jem.20100244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwman P., Aly A., Escandell J.M., Pieterse M., Bartkova J., van der Gulden H., Hiddingh S., Thanasoula M., Kulkarni A., Yang Q., et al. 2010. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat. Struct. Mol. Biol. 17:688–695 10.1038/nsmb.1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant H.E., Schultz N., Thomas H.D., Parker K.M., Flower D., Lopez E., Kyle S., Meuth M., Curtin N.J., Helleday T. 2005. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 434:913–917 10.1038/nature03443 [DOI] [PubMed] [Google Scholar]
- Bunting S.F., Callén E., Wong N., Chen H.T., Polato F., Gunn A., Bothmer A., Feldhahn N., Fernandez-Capetillo O., Cao L., et al. 2010. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 141:243–254 10.1016/j.cell.2010.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L., Kim S., Xiao C., Wang R.H., Coumoul X., Wang X., Li W.M., Xu X.L., De Soto J.A., Takai H., et al. 2006. ATM-Chk2-p53 activation prevents tumorigenesis at an expense of organ homeostasis upon Brca1 deficiency. EMBO J. 25:2167–2177 10.1038/sj.emboj.7601115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L., Xu X., Bunting S.F., Liu J., Wang R.H., Cao L.L., Wu J.J., Peng T.N., Chen J., Nussenzweig A., et al. 2009. A selective requirement for 53BP1 in the biological response to genomic instability induced by Brca1 deficiency. Mol. Cell. 35:534–541 10.1016/j.molcel.2009.06.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng C., Ueda E., Chen K.E., Bula C., Norman A.W., Luben R.A., Walker A.M. 2009. Prolactin blocks nuclear translocation of VDR by regulating its interaction with BRCA1 in osteosarcoma cells. Mol. Endocrinol. 23:226–236 10.1210/me.2008-0075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew Y., Mulligan E.A., Vong W.T., Thomas H.D., Kahn S., Kyle S., Mukhopadhyay A., Los G., Hostomsky Z., Plummer E.R., et al. 2011. Therapeutic potential of poly(ADP-ribose) polymerase inhibitor AG014699 in human cancers with mutated or methylated BRCA1 or BRCA2. J. Natl. Cancer Inst. 103:334–346 10.1093/jnci/djq509 [DOI] [PubMed] [Google Scholar]
- Duncan E.M., Muratore-Schroeder T.L., Cook R.G., Garcia B.A., Shabanowitz J., Hunt D.F., Allis C.D. 2008. Cathepsin L proteolytically processes histone H3 during mouse embryonic stem cell differentiation. Cell. 135:284–294 10.1016/j.cell.2008.09.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dusso A.S., Brown A.J., Slatopolsky E. 2005. Vitamin D. Am. J. Physiol. Renal Physiol. 289:F8–F28 10.1152/ajprenal.00336.2004 [DOI] [PubMed] [Google Scholar]
- Evers B., Jonkers J. 2006. Mouse models of BRCA1 and BRCA2 deficiency: past lessons, current understanding and future prospects. Oncogene. 25:5885–5897 10.1038/sj.onc.1209871 [DOI] [PubMed] [Google Scholar]
- Farmer H., McCabe N., Lord C.J., Tutt A.N., Johnson D.A., Richardson T.B., Santarosa M., Dillon K.J., Hickson I., Knights C., et al. 2005. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 434:917–921 10.1038/nature03445 [DOI] [PubMed] [Google Scholar]
- Fernandez-Capetillo O., Chen H.T., Celeste A., Ward I., Romanienko P.J., Morales J.C., Naka K., Xia Z., Camerini-Otero R.D., Motoyama N., et al. 2002. DNA damage-induced G2-M checkpoint activation by histone H2AX and 53BP1. Nat. Cell Biol. 4:993–997 10.1038/ncb884 [DOI] [PubMed] [Google Scholar]
- Fong P.C., Boss D.S., Yap T.A., Tutt A., Wu P., Mergui-Roelvink M., Mortimer P., Swaisland H., Lau A., O’Connor M.J., et al. 2009. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N. Engl. J. Med. 361:123–134 10.1056/NEJMoa0900212 [DOI] [PubMed] [Google Scholar]
- Foulkes W.D., Smith I.E., Reis-Filho J.S. 2010. Triple-negative breast cancer. N. Engl. J. Med. 363:1938–1948 10.1056/NEJMra1001389 [DOI] [PubMed] [Google Scholar]
- Gartner E.M., Burger A.M., Lorusso P.M. 2010. Poly(adp-ribose) polymerase inhibitors: a novel drug class with a promising future. Cancer J. 16:83–90 10.1097/PPO.0b013e3181d78223 [DOI] [PubMed] [Google Scholar]
- Gocheva V., Joyce J.A. 2007. Cysteine cathepsins and the cutting edge of cancer invasion. Cell Cycle. 6:60–64 10.4161/cc.6.1.3669 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Suarez I., Redwood A.B., Gonzalo S. 2009. Loss of A-type lamins and genomic instability. Cell Cycle. 8:3860–3865 10.4161/cc.8.23.10092 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Suarez I., Redwood A.B., Grotsky D.A., Neumann M.A., Cheng E.H., Stewart C.L., Dusso A., Gonzalo S. 2011. A new pathway that regulates 53BP1 stability implicates cathepsin L and vitamin D in DNA repair. EMBO J. 30:3383–3396 10.1038/emboj.2011.225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulet B., Baruch A., Moon N.S., Poirier M., Sansregret L.L., Erickson A., Bogyo M., Nepveu A. 2004. A cathepsin L isoform that is devoid of a signal peptide localizes to the nucleus in S phase and processes the CDP/Cux transcription factor. Mol. Cell. 14:207–219 10.1016/S1097-2765(04)00209-6 [DOI] [PubMed] [Google Scholar]
- Helleday T., Bryant H.E., Schultz N. 2005. Poly(ADP-ribose) polymerase (PARP-1) in homologous recombination and as a target for cancer therapy. Cell Cycle. 4:1176–1178 10.4161/cc.4.9.2031 [DOI] [PubMed] [Google Scholar]
- Iliakis G., Wang H., Perrault A.R., Boecker W., Rosidi B., Windhofer F., Wu W., Guan J., Terzoudi G., Pantelias G. 2004. Mechanisms of DNA double strand break repair and chromosome aberration formation. Cytogenet. Genome Res. 104:14–20 10.1159/000077461 [DOI] [PubMed] [Google Scholar]
- Jedeszko C., Sloane B.F. 2004. Cysteine cathepsins in human cancer. Biol. Chem. 385:1017–1027 10.1515/BC.2004.132 [DOI] [PubMed] [Google Scholar]
- Kawai S., Amano A. 2012. BRCA1 regulates microRNA biogenesis via the DROSHA microprocessor complex. J. Cell Biol. 197:201–208 10.1083/jcb.201110008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lankelma J.M., Voorend D.M., Barwari T., Koetsveld J., Van der Spek A.H., De Porto A.P., Van Rooijen G., Van Noorden C.J. 2010. Cathepsin L, target in cancer treatment? Life Sci. 86:225–233 10.1016/j.lfs.2009.11.016 [DOI] [PubMed] [Google Scholar]
- Moynahan M.E., Chiu J.W., Koller B.H., Jasin M. 1999. Brca1 controls homology-directed DNA repair. Mol. Cell. 4:511–518 10.1016/S1097-2765(00)80202-6 [DOI] [PubMed] [Google Scholar]
- Mullan P.B., Quinn J.E., Harkin D.P. 2006. The role of BRCA1 in transcriptional regulation and cell cycle control. Oncogene. 25:5854–5863 10.1038/sj.onc.1209872 [DOI] [PubMed] [Google Scholar]
- Neuhausen S.L., Marshall C.J. 1994. Loss of heterozygosity in familial tumors from three BRCA1-linked kindreds. Cancer Res. 54:6069–6072 [PubMed] [Google Scholar]
- Olive P.L., Banáth J.P., Durand R.E. 1990. Heterogeneity in radiation-induced DNA damage and repair in tumor and normal cells measured using the “comet” assay. Radiat. Res. 122:86–94 10.2307/3577587 [DOI] [PubMed] [Google Scholar]
- Pallares J., Santacana M., Puente S., Lopez S., Yeramian A., Eritja N., Sorolla A., Llobet D., Dolcet X., Matias-Guiu X. 2009. A review of the applications of tissue microarray technology in understanding the molecular features of endometrial carcinoma. Anal. Quant. Cytol. Histol. 31:217–226 [PubMed] [Google Scholar]
- Peppone L.J., Huston A.J., Reid M.E., Rosier R.N., Zakharia Y., Trump D.L., Mustian K.M., Janelsins M.C., Purnell J.Q., Morrow G.R. 2011. The effect of various vitamin D supplementation regimens in breast cancer patients. Breast Cancer Res. Treat. 127:171–177 10.1007/s10549-011-1415-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redwood A.B., Gonzalez-Suarez I., Gonzalo S. 2011a. Regulating the levels of key factors in cell cycle and DNA repair: new pathways revealed by lamins. Cell Cycle. 10:3652–3657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redwood A.B., Perkins S.M., Vanderwaal R.P., Feng Z., Biehl K.J., Gonzalez-Suarez I., Morgado-Palacin L., Shi W., Sage J., Roti-Roti J.L., et al. 2011b. A dual role for A-type lamins in DNA double-strand break repair. Cell Cycle. 10:2549–2560 10.4161/cc.10.15.16531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlegel B.P., Jodelka F.M., Nunez R. 2006. BRCA1 promotes induction of ssDNA by ionizing radiation. Cancer Res. 66:5181–5189 10.1158/0008-5472.CAN-05-3209 [DOI] [PubMed] [Google Scholar]
- Schultz L.B., Chehab N.H., Malikzay A., Halazonetis T.D. 2000. p53 binding protein 1 (53BP1) is an early participant in the cellular response to DNA double-strand breaks. J. Cell Biol. 151:1381–1390 10.1083/jcb.151.7.1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scully R., Livingston D.M. 2000. In search of the tumour-suppressor functions of BRCA1 and BRCA2. Nature. 408:429–432 10.1038/35044000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scully R., Chen J., Ochs R.L., Keegan K., Hoekstra M., Feunteun J., Livingston D.M. 1997a. Dynamic changes of BRCA1 subnuclear location and phosphorylation state are initiated by DNA damage. Cell. 90:425–435 10.1016/S0092-8674(00)80503-6 [DOI] [PubMed] [Google Scholar]
- Scully R., Chen J., Plug A., Xiao Y., Weaver D., Feunteun J., Ashley T., Livingston D.M. 1997b. Association of BRCA1 with Rad51 in mitotic and meiotic cells. Cell. 88:265–275 10.1016/S0092-8674(00)81847-4 [DOI] [PubMed] [Google Scholar]
- Sherley J.L., Stadler P.B., Stadler J.S. 1995. A quantitative method for the analysis of mammalian cell proliferation in culture in terms of dividing and non-dividing cells. Cell Prolif. 28:137–144 [DOI] [PubMed] [Google Scholar]
- Shukla V., Coumoul X., Lahusen T., Wang R.H., Xu X., Vassilopoulos A., Xiao C., Lee M.H., Man Y.G., Ouchi M., et al. 2010. BRCA1 affects global DNA methylation through regulation of DNMT1. Cell Res. 20:1201–1215 10.1038/cr.2010.128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skrzydlewska E., Sulkowska M., Koda M., Sulkowski S. 2005. Proteolytic-antiproteolytic balance and its regulation in carcinogenesis. World J. Gastroenterol. 11:1251–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snouwaert J.N., Gowen L.C., Latour A.M., Mohn A.R., Xiao A., DiBiase L., Koller B.H. 1999. BRCA1 deficient embryonic stem cells display a decreased homologous recombination frequency and an increased frequency of non-homologous recombination that is corrected by expression of a brca1 transgene. Oncogene. 18:7900–7907 10.1038/sj.onc.1203334 [DOI] [PubMed] [Google Scholar]
- Sung P., Krejci L., Van Komen S., Sehorn M.G. 2003. Rad51 recombinase and recombination mediators. J. Biol. Chem. 278:42729–42732 10.1074/jbc.R300027200 [DOI] [PubMed] [Google Scholar]
- Tanic M., Zajac M., Gómez-López G., Benítez J., Martínez-Delgado B. 2012. Integration of BRCA1-mediated miRNA and mRNA profiles reveals microRNA regulation of TRAF2 and NFκB pathway. Breast Cancer Res. Treat. 134:41–51 10.1007/s10549-011-1905-4 [DOI] [PubMed] [Google Scholar]
- Tu Z., Aird K.M., Bitler B.G., Nicodemus J.P., Beeharry N., Xia B., Yen T.J., Zhang R. 2011. Oncogenic RAS regulates BRIP1 expression to induce dissociation of BRCA1 from chromatin, inhibit DNA repair, and promote senescence. Dev. Cell. 21:1077–1091 10.1016/j.devcel.2011.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner N.C., Reis-Filho J.S. 2006. Basal-like breast cancer and the BRCA1 phenotype. Oncogene. 25:5846–5853 10.1038/sj.onc.1209876 [DOI] [PubMed] [Google Scholar]
- Tutt A., Robson M., Garber J.E., Domchek S.M., Audeh M.W., Weitzel J.N., Friedlander M., Arun B., Loman N., Schmutzler R.K., et al. 2010. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: a proof-of-concept trial. Lancet. 376:235–244 10.1016/S0140-6736(10)60892-6 [DOI] [PubMed] [Google Scholar]
- Wang B., Matsuoka S., Carpenter P.B., Elledge S.J. 2002. 53BP1, a mediator of the DNA damage checkpoint. Science. 298:1435–1438 [DOI] [PubMed] [Google Scholar]
- Ward I.M., Minn K., van Deursen J., Chen J. 2003. p53 Binding protein 53BP1 is required for DNA damage responses and tumor suppression in mice. Mol. Cell. Biol. 23:2556–2563 10.1128/MCB.23.7.2556-2563.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooster R., Weber B.L. 2003. Breast and ovarian cancer. N. Engl. J. Med. 348:2339–2347 10.1056/NEJMra012284 [DOI] [PubMed] [Google Scholar]
- Xie A., Hartlerode A., Stucki M., Odate S., Puget N., Kwok A., Nagaraju G., Yan C., Alt F.W., Chen J., et al. 2007. Distinct roles of chromatin-associated proteins MDC1 and 53BP1 in mammalian double-strand break repair. Mol. Cell. 28:1045–1057 10.1016/j.molcel.2007.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B., Kim St, andKastan M.B. 2001. Involvement of Brca1 in S-phase and G(2)-phase checkpoints after ionizing irradiation. Mol. Cell. Biol. 21:3445–3450 10.1128/MCB.21.10.3445-3450.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q., Pao G.M., Huynh A.M., Suh H., Tonnu N., Nederlof P.M., Gage F.H., Verma I.M. 2011. BRCA1 tumour suppression occurs via heterochromatin-mediated silencing. Nature. 477:179–184 10.1038/nature10371 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.