Polar ejection forces, which push chromosomes away from spindle poles, modulate kinetochore–microtubule attachment stability.
Abstract
Chromosome biorientation promotes congression and generates tension that stabilizes kinetochore–microtubule (kt-MT) interactions. Forces produced by molecular motors also contribute to chromosome alignment, but their impact on kt-MT attachment stability is unclear. A critical force that acts on chromosomes is the kinesin-10–dependent polar ejection force (PEF). PEFs are proposed to facilitate congression by pushing chromosomes away from spindle poles, although knowledge of the molecular mechanisms underpinning PEF generation is incomplete. Here, we describe a live-cell PEF assay in which tension was applied to chromosomes by manipulating levels of the chromokinesin NOD (no distributive disjunction; Drosophila melanogaster kinesin-10). NOD stabilized syntelic kt-MT attachments in a dose- and motor-dependent manner by overwhelming the ability of Aurora B to mediate error correction. NOD-coated chromatin stretched away from the pole via lateral and end-on interactions with microtubules, and NOD chimeras with either plus end–directed motility or tip-tracking activity produced PEFs. Thus, kt-MT attachment stability is modulated by PEFs, which can be generated by distinct force-producing interactions between chromosomes and dynamic spindle microtubules.
Introduction
Establishing bioriented chromosomes with sister kinetochores attached to microtubules from opposing spindle poles is essential for maintaining genomic integrity though cell division. Mitotic forces select for bioriented attachments through tension-dependent stabilization of kinetochore–microtubule (kt-MT) attachments (Nicklas and Koch, 1969; Li and Nicklas, 1995; King and Nicklas, 2000; Nicklas et al., 2001; Akiyoshi et al., 2010). Polar ejection forces (PEFs) have been implicated in chromosome alignment since their discovery (Rieder et al., 1986; Rieder and Salmon, 1994). PEFs are predominantly generated by kinesin-10 family members—chromokinesins that are proposed to walk chromosome arms away from poles and toward the plus ends of spindle microtubules. Perturbation of chromokinesin function in multiple model systems disrupts the proper and timely congression of chromosome arms (Zhang et al., 1990; Theurkauf and Hawley, 1992; Afshar et al., 1995a; Antonio et al., 2000; Funabiki and Murray, 2000; Levesque and Compton, 2001; Goshima and Vale, 2003; Powers et al., 2004; Tokai-Nishizumi et al., 2005; Wignall and Villeneuve, 2009; Magidson et al., 2011; Stumpff et al., 2012; Wandke et al., 2012) but the extent to which PEFs contribute to chromosome alignment remains unclear as inhibition of chromokinesins in several cell types results in subtle or even undetectable effects on congression (Dumont et al., 2010; Kitajima et al., 2011).
An underappreciated feature of chromosomal positioning by PEFs is the potential regulation of kinetochore function by kinesin-10 motors. PEFs are well-positioned to impact kt-MT interactions by producing forces along chromosome arms that are transmitted through the kinetochore and it has been hypothesized that PEFs could regulate motility of bioriented chromosomes by creating tension at kinetochores (Skibbens et al., 1993; Rieder and Salmon, 1994). Furthermore, misaligned chromosomes where one (monotelic) or both (syntelic) kinetochores are attached to a single pole could come under tension when kinetochore-dependent poleward pulling forces are opposed by PEFs (Cassimeris et al., 1994; Rieder et al., 1995). In fact, applying tension with microneedles to unipolar bivalents attached to the same spindle pole in spermatocytes stabilized this normally unstable orientation (Nicklas and Koch, 1969) to the point that the spindle assembly checkpoint was satisfied and the cells entered anaphase (Li and Nicklas, 1995). Despite the fact that PEFs are likely to influence the production of tension at kinetochores, the contribution of PEFs to kt-MT attachment stability has never been directly tested.
PEFs were initially proposed to be generated by two nonexclusive sources: chromosome-associated motor proteins and the polymerization of microtubules (Rieder et al., 1986; Rieder and Salmon, 1994). The chromokinesin Kid (kinesin-10) was later identified as the principal mediator of PEF generation in vertebrate cells (Antonio et al., 2000; Funabiki and Murray, 2000; Brouhard and Hunt, 2005). NOD (no distributive disjunction) is the Drosophila melanogaster kinesin-10 family member that, like Kid, localizes to chromosomes and is required for generating PEFs (Theurkauf and Hawley, 1992; Afshar et al., 1995a, b). However, NOD is classified as a nonmotile kinesin because it fails to exhibit activity in conventional microtubule gliding assays (Matthies et al., 2001), whereas Kid is a bona fide plus end–directed motor (Yajima et al., 2003; Brouhard and Hunt, 2005; Bieling et al., 2010a). NOD has been shown to preferentially bind microtubule plus ends in vitro (Cui et al., 2005) and it has been postulated, based on analyses of its catalytic domain, that NOD generates force by associating with the plus ends of polymerizing microtubules, a behavior termed end tracking (Cochran et al., 2009). Thus, although PEF production by kinesin-10 chromokinesins is evolutionarily conserved, the molecular mechanism by which kinesin-10 motors transmit force is thought to differ.
Whether derived from motility or end tracking, individual PEF-producing interactions are most likely weak so that the DNA is not damaged (Brouhard and Hunt, 2005). Consistent with this presumed constraint, the PEF has been measured as 0.5 pN per microtubule on mammalian chromosomes (Brouhard and Hunt, 2005) and ∼1 pN in Drosophila embryos (Marshall et al., 2001). In principle, either motility or end tracking could generate PEFs of this magnitude because both motile kinesins and polymerizing microtubules generate forces in the low pN range (Dogterom and Yurke, 1997; Visscher et al., 1999). Although it is thought that NOD is nonmotile and that it produces PEFs solely by end tracking on polymerizing microtubules, the molecular mechanism of PEF production by NOD is unclear.
Results
NOD overexpression stabilizes syntelic attachments in a dose-dependent manner
To experimentally manipulate PEFs in living cells, the Drosophila chromokinesin NOD (Zhang et al., 1990; Afshar et al., 1995a) was fused to mCherry and placed under the control of a copper-inducible promoter. NOD-mCherry localized exclusively to mitotic chromosomes over a broad range of expression levels that varied on a cell-by-cell basis. Using a custom-made polyclonal peptide antibody, induced NOD, but not endogenous NOD, was detectable on mitotic chromatin by immunofluorescence (IF) and was detectable by Western blot of cell extracts as an ∼105-kD protein band (Fig. S1, A–C). An inability to detect endogenous NOD by Western blot or IF has also been reported for another NOD antibody (Afshar et al., 1995a, b). Our antibody efficiently detected NOD-mCherry by IF in the highest-expressing cells (Fig. S1, D and E), but in cells expressing lower levels of NOD the antibody’s sensitivity rapidly diminished. These data suggest that endogenous NOD levels are low in mitotic cells. Despite its low abundance, NOD generates an away-from-the-pole force in mitotic Drosophila cells as NOD RNAi (Fig. S2, A and B) caused an inward movement of kinetochores and chromosome arms in monopolar spindles (Fig. S2, D and E), which was also observed after Kid inhibition in vertebrate cells (Levesque and Compton, 2001; Stumpff et al., 2012; Wandke et al., 2012). Thus, our results are consistent with previous ones showing that NOD regulates mitotic chromosome behavior (Zhang and Hawley, 1990; Rasooly et al., 1991; Goshima and Vale, 2003; Goshima et al., 2007).
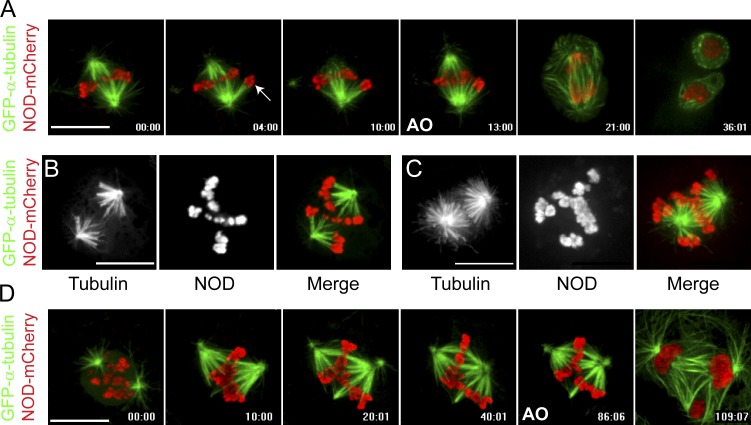
Aberrant spindle morphologies were evident in GFP–α-tubulin–expressing cells coexpressing NOD-mCherry. As observed in embryonic cells (Afshar et al., 1995b), monopolar spindles assembled in S2 cells expressing the highest levels of NOD. Cells expressing low or intermediate levels of NOD did not form monopoles but often assembled unusual spindles with robust kinetochore fibers (k-fibers) connecting chromosomes to the same spindle pole (Fig. 1 A and Video 1). At high-intermediate concentrations of NOD, spindles were often comprised of two fan-shaped half spindles, each with its own associated subset of chromosomes (Fig. 1 B and Video 2). In general, the intermediate range of NOD expression levels yielded spindles lacking normal metaphase plates (Fig. 1, C and D; and Video 3). Despite the absence of metaphase alignment, NOD-expressing cells were able to satisfy the spindle assembly checkpoint and enter anaphase (Fig. 1, A and D; and Videos 1 and 3).
Figure 1.
NOD-mCherry–expressing cells do not form a well-defined metaphase plate. (A–D) Two-color confocal imaging of GFP–α-tubulin (green)– and NOD-mCherry (red)–expressing S2 cells. (A) Selected frames from a time lapse of a cell with a pair of sister chromatids that are attached to the same pole (arrow). The aberrant attachment state persists and anaphase onset (AO) ensues without error correction. See Video 1. (B and C) Chromosomes move away from the poles but fail to align along a well-defined metaphase plate, particularly in cells expressing high levels of NOD-mCherry. See Video 2. (D) Selected micrographs from a time lapse of a NOD-mCherry–expressing cell as it progresses through mitosis. A mixture of attachment states are established within the first 10 min of nuclear envelope breakdown and persist until the cell enters anaphase with uncorrected syntelic attachments, resulting in chromosome mis-segregation and multiple nuclei. See Video 3. Bars, 10 µm.
The spindle morphologies in NOD-expressing cells indicated a prevalence of syntelic attachments, a nonbioriented attachment state where both sister kinetochores are attached to the same spindle pole. To confirm that syntelic attachments were being formed, NOD-mCherry was coexpressed with Ndc80-GFP to label kinetochores. Persistent syntelic attachments, as defined by clearly juxtaposed sister kinetochores facing the same spindle pole, were observed after induction of NOD (Fig. 2 A and Video 4). This phenotype was not caused by a dominant-negative effect of NOD expression as syntelic attachments still formed when NOD-mCherry was induced after targeting endogenous NOD by RNAi (Fig. S2 C). Thus, NOD overexpression yielded spindles with elevated levels of syntelic attachments.
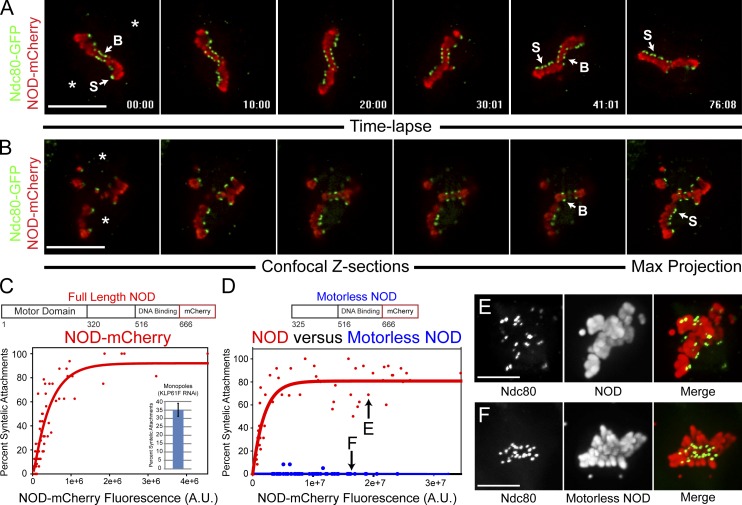
Figure 2.
NOD-mCherry expression stabilizes syntelic attachments. (A, B, E, and F) Two-color confocal imaging of Ndc80-GFP (green)– and NOD-mCherry (red)–expressing S2 cells. (A) Selected frames from a confocal time lapse of a cell with both bioriented (B) and syntelic (S) attachments (approximate pole positions are marked with asterisks). Note that the syntelic attachments persist for the duration of the time lapse. See Video 4. (B) Selected confocal Z-sections showing a combination of syntelic and bioriented kinetochore pairs in the same cell. The mCherry fluorescence intensity for each cell was quantified from the maximum intensity projection of the Z-sections (rightmost panel). (C) Plotting the percentage of syntelic attachments versus NOD-mCherry fluorescence reveals that NOD-mCherry stabilizes syntelic attachments in a dose-dependent fashion (n = 60 cells). Inset shows the mean percentage of syntelic attachments found in monopolar spindles assembled in the absence of Klp61F. (D) Syntelic stabilization by NOD-mCherry requires the motor domain of NOD (NOD, n = 57 cells; motorless NOD, n = 70 cells). (E and F) Maximum intensity projections of representative NOD-mCherry– and motorless NOD-mCherry–expressing cells with comparable expression levels (highlighted in D). Error bar represents the SEM. Bars, 10 µm.
The phenotypic variability exhibited by NOD overexpressing cells led to the development of a quantitative image-based assay (PEF assay) that was applied to further characterize the effects of NOD overexpression. In brief, two-color spinning disk confocal Z sections were acquired for individual cells coexpressing NOD-mCherry and Ndc80-GFP. NOD levels for a given cell were measured by quantifying the total integrated fluorescence intensity from the mCherry fluorophore, and each chromosome in that cell was individually scored as bioriented, syntelic, or other by examining Ndc80-GFP–labeled kinetochore pairs (Fig. 2 B and Fig. S1 F). Plotting the percentage of syntelic attachments in a given cell against the NOD-mCherry fluorescence intensity for that cell and repeating that analysis on a cell-by-cell basis over a range of expression levels revealed that NOD stabilized syntelic attachments in a dose-dependent manner (Fig. 2 C).
NOD-mediated stabilization of syntelic attachments is specific and motor dependent
The prevalence of syntelic attachments in NOD-expressing cells could not be attributed to monopolar spindle assembly because cells expressing the highest levels of NOD—those that assembled monopoles—were excluded from analysis. Furthermore, monopoles assembled after depletion of Klp61F (kinesin-5) contained a mean of ∼35% syntelic attachments, less than the mean percentage of syntelic attachments seen in the high NOD-expressing cells included in the analyses (Fig. 2 C). To address the possibility that NOD overexpression stabilized syntelic attachments by disrupting chromosome structure or by mislocalizing other chromosomal components, a NOD mutant lacking the N-terminal motor domain was expressed in S2 cells. Motorless NOD localized to mitotic chromosomes as efficiently as full-length NOD but did not increase the percentage of syntelic attachments (Fig. 2, D–F), demonstrating that the motor domain of NOD is required to stabilize syntelic attachments.
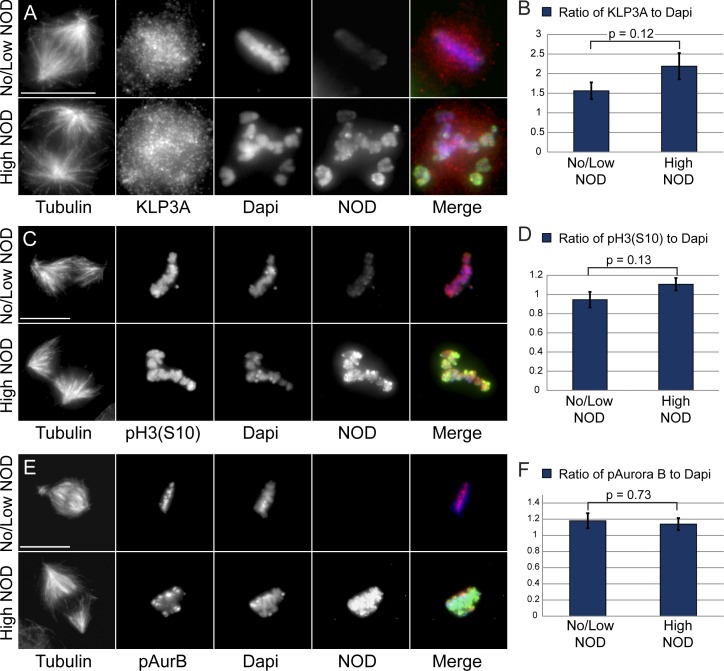
KLP3A (kinesin-4), the other major chromokinesin in Drosophila, regulates spindle pole separation in prometaphase and anaphase (Kwon et al., 2004), and its vertebrate homologue regulates chromosome oscillations and midzone assembly by suppressing microtubule plus end dynamics (Bringmann et al., 2004; Bieling et al., 2010b; Hu et al., 2011; Stumpff et al., 2012; Wandke et al., 2012). Like its vertebrate counterpart, KLP3A may act as an anti-PEF by inhibiting the polymerization of microtubules that come into contact with chromatin. We next examined whether NOD overexpression indirectly elevated PEFs by mislocalizing the anti-PEF motor KLP3A. As previously reported (Kwon et al., 2004), KLP3A localized to interphase nuclei as well as on and around mitotic chromosomes and along midzone and midbody microtubules. KLP3A localization and chromosomal association were unaffected by elevated NOD expression (Fig. 3, A and B).
Figure 3.
KLP3A localization and Aurora B localization and activity are not compromised in NOD-expressing cells. (A, C, and E) Representative maximum projections of tubulin, DAPI (blue), KLP3A (red; A), phospho–histone H3–serine 10 (red; C) or phospho–Aurora B (red; E), and NOD (green) for high and low/no NOD-expressing cells. (B, D, and F) Quantification of KLP3A (n = 102 cells; B), phospho–histone H3 (Serine 10; n = 31 cells; D), or phospho–Aurora B (n = 73 cells; F) signals relative to DAPI intensities for high and low/no NOD-expressing cells. There was not a statistically significant difference in levels of KLP3A, phospho–Aurora B, or phospho-H3 (Ser10) between high and no/low NOD-expressing cells. Two-tailed p-values are shown. Error bars represent the SEM. Bars, 10 µm.
Inhibition of Aurora B kinase, which destabilizes erroneous kt-MT attachments (for review see Maresca and Salmon, 2010), results in numerous syntelic attachments near spindle poles (Ditchfield et al., 2003; Hauf et al., 2003; Lampson et al., 2004). Despite the fact that chromosomes in NOD overexpressing cells were often pushed away from the poles and the NOD overexpression phenotype was distinct from the effects of Aurora B depletion in S2 cells (Adams et al., 2001; Giet and Glover, 2001), the striking abundance of stable syntelic attachments warranted a careful investigation of Aurora B localization and activity in NOD-expressing cells. We found that NOD overexpression did not affect the localization or activity of Aurora B as neither the phosphorylation of histone H3 at S10 (Fig. 3, C and D) nor the levels of active chromatin-associated and phosphorylated Aurora B (Fig. 3, E and F) was altered by NOD expression. The lack of syntelic attachments in motorless NOD overexpressing cells (Fig. 2 D) along with the fact that KLP3A localization and Aurora B localization/activity are unaffected by NOD overexpression strongly support the conclusion that NOD-dependent stabilization of syntelic attachments is a direct consequence of overexpressing full-length NOD and requires force generation by the motor.
Elevated PEFs produce cold-stable syntelic kt-MT attachments with reduced Mad1 levels
Syntelic attachments are typically repaired before cells enter anaphase. However, syntelic attachments assembled in NOD-expressing cells persisted until anaphase. We found that the GFP–α-tubulin fluorescence intensity of k-fibers was comparable for syntelic and amphitelic (connected to opposing spindle poles) attachments (Fig. 4, A and B). FRAP analysis of GFP–α-tubulin was performed near the spindle equator in control and NOD-expressing cells to determine whether excess NOD stabilizes microtubules in general. Similar to previous observations (Goshima et al., 2008), a t1/2 = 31 ± 2 s (n = 8 cells) was measured in control cells (Fig. S3, A and B). NOD overexpression did not significantly alter GFP–α-tubulin turnover as a t1/2 = 38 ± 3 s (n = 8 cells) was measured in cells expressing high levels of NOD-mCherry (Fig. S3, C and D). Thus, turnover of nonkinetochore microtubules is largely unaffected by NOD overexpression, indicating that the prevalence of syntelics was caused, not by stabilization of microtubules in general but, rather, by stabilization of kinetochore microtubules in particular. This finding is in agreement with the observation that overexpression of the motor domain alone, which binds exclusively to microtubules, does not stabilize syntelic attachments (unpublished data; Afshar et al., 1995b).
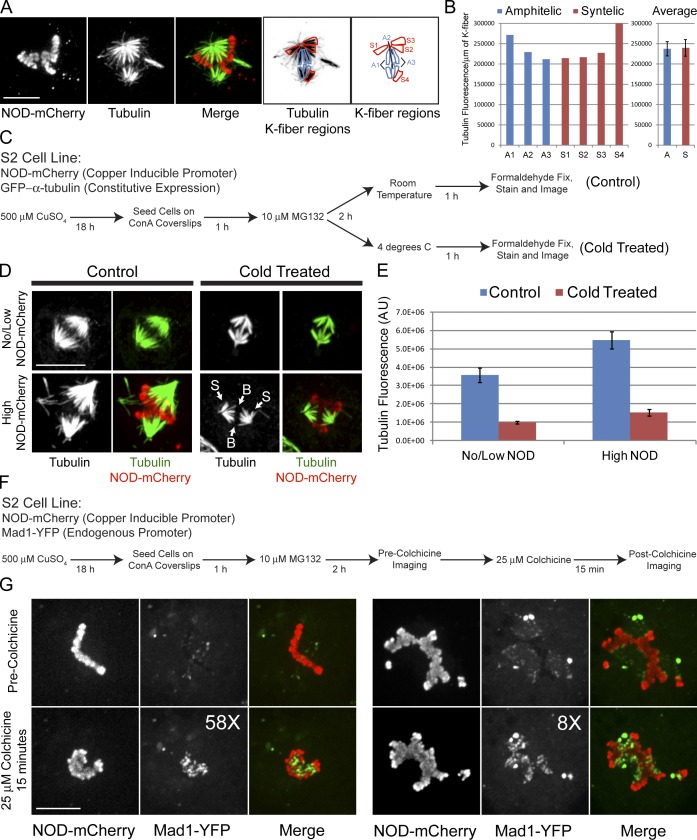
Figure 4.
NOD-mCherry–induced syntelic kt-MT attachments are stable and exhibit reduced levels of Mad1. (A) Maximum projection of two-color confocal Z-sections from a NOD-mCherry–expressing cell with syntelic attachments (S1–S4) and bioriented chromosomes with amphitelic attachments (A1–A3). (B) Quantification of k-fiber fluorescence intensity of the amphitelic (A1–A3) and syntelic attachments (S1–S4) highlighted in A. The tubulin fluorescence presented in the bar graph is the integrated fluorescence intensity per micrometer of k-fiber. (C) Flow chart outlining the cold stability assay used to probe syntelic k-fiber stability. (D) Micrographs of no/low NOD-mCherry– and high NOD-mCherry–expressing control and cold-treated cells. Note that both syntelic (S) and bioriented (B) chromosomes retain their k-fibers equally after cold treatment. (E) Quantification of tubulin fluorescence intensity for no/low NOD-mCherry– and high NOD-mCherry–expressing cells for control and cold-treated cells. (F) Flow chart outlining the protocol used to examine Mad1 reduction at syntelic attachments. (G) Spinning disk confocal imaging of NOD-mCherry– and Mad1-YFP–expressing cells before and after a 15-min colchicine treatment to depolymerize the spindle microtubules. The fold increase (58× and 8×) in Mad1 levels after the colchicine treatment is shown for each cell. Error bars represent the SEM. Bars, 10 µm.
We further probed the stability of NOD-induced syntelic attachments using a cold-stability assay, which selectively preserves microtubules that are stably bound to kinetochores. Induced NOD-mCherry, GFP–α-tubulin–expressing cells were treated with the proteasome inhibitor MG132 to arrest mitotic cells and were kept at room temperature or placed at 4°C for 1 h before fixing and staining the microtubules by IF (Fig. 4 C). Spindle fluorescence intensity and NOD-mCherry signals were quantified for control and cold-treated cells. Spindle fluorescence was 1.5× brighter in cells expressing high levels of NOD compared with cells with low or undetectable NOD-mCherry for both control and cold-treated cells. Only k-fibers remained after 1 h at 4°C, as reflected by a 3.7-fold reduction in the fluorescence intensity of spindle microtubules in both high and no/low NOD-expressing cells (Fig. 4, D and E). The stability of syntelic attachments was further indicated by the fact that both bioriented and syntelic attachments persisted within the same cold-treated spindles (Fig. 4 D).
Levels of the checkpoint protein Mad1 are low at stably attached kinetochores and high at unattached kinetochores (Chen et al., 1998; Shah et al., 2004). To investigate whether Mad1 levels were reduced at the syntelic attachments produced by NOD expression, we generated and imaged a stable cell line coexpressing inducible NOD-mCherry and Mad1-YFP under the control of its endogenous promoter. Induced cells were arrested in mitosis with MG132 and the same cell was imaged by spinning disk confocal microscopy both before and after depolymerizing microtubules with colchicine (Fig. 4 F). Syntelic and bioriented kinetochores had low levels of Mad1-YFP in MG132-treated cells before colchicine treatment (Fig. 4 G). Depolymerizing microtubules with a 15-min colchicine treatment resulted in an ∼22-fold mean increase in kinetochore-associated Mad1 in NOD-expressing cells. The observed reduction of Mad1 at kinetochores is consistent with the observation of anaphase onset in cells with NOD-stabilized syntelic attachments (Videos 1 and 3). A more detailed examination of the kinetics of mitotic progression and checkpoint protein depletion from syntelic attachments will be the focus of future work. Collectively, our findings support the conclusion that the stability of syntelic attachments is comparable to the stability of bioriented attachments in NOD-expressing cells.
Two types of chromatin stretch events occur in NOD overexpressing cells via distinct microtubule–chromatin interactions
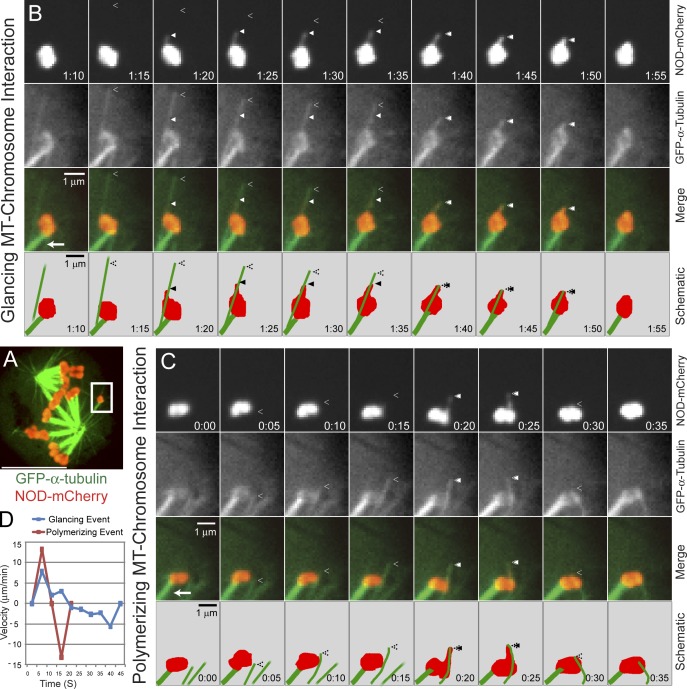
Elevated away-from-the-pole force production was indicated by frequent chromatin stretching events in which NOD-mCherry–coated chromatin extended away from chromosome arms. Stretch events were only observed in NOD-expressing cells and required microtubules (unpublished data). Therefore, we used near simultaneous two-color spinning disk confocal imaging of cells expressing NOD-mCherry and GFP–α-tubulin to more closely examine the relationship between chromatin stretches and dynamic microtubules (Fig. 5 A). Two distinct categories of microtubule–chromosome interactions were observed: glancing and polymerizing. In glancing interactions, chromatin laterally interacted with microtubules that extended beyond the chromosome. Polymerizing interactions, in contrast, occurred when growing microtubules collided with the chromatin in an end-on orientation. In one clear example of a glancing interaction, chromatin stretched toward the plus end of a microtubule after making lateral contact with the microtubule; the stretch persisted for ∼15 s until the chromatin recoiled when the microtubule depolymerized (Fig. 5, B and D; and Video 5). The same chromosome also underwent two definitive polymerizing interactions in which stretch events coincided with polymerizing microtubules (Video 5). During one of the polymerizing interactions, the growing microtubule visibly buckled as the chromatin reached maximum stretch and began to recoil (Fig. 5, C and D).
Figure 5.
NOD-dependent chromatin stretching events are associated with two different types of microtubule–chromatin interactions. (A) A whole cell two-color confocal image of a GFP–α-tubulin (green–) and NOD-mCherry (red)–expressing S2 cell with the chromosome shown in B and C highlighted (white box). (B and C) Selected frames from confocal time-lapse imaging of chromatin stretching events. The chromosome is attached to the pole through kinetochore microtubules (bottom left corner, arrow). (B) An example of a chromatin stretching event extending along a microtubule that makes a glancing interaction with the chromosome. The chromatin is stretched toward the plus end before it is retracted coincident with the depolymerizing microtubule. (C) A chromatin stretching event that is associated with a polymerizing microtubule–chromosome interaction. Note that the chromatin stretches along with the polymerizing microtubule before pausing and then rapidly retracting, causing the microtubule to buckle. The separation between the plus end of the microtubule and the stretched chromatin at t = 15 s is a consequence of sequential imaging. (D) A plot of velocity versus time for the two stretch events shown in B and C. The positive values represent extension velocities and the negative values reflect recoil velocities. Closed arrowheads denote the leading edges of stretched chromatin and open arrowheads mark the microtubule plus ends. See Video 5. Bars: (A) 10 µm; (B and C) 1 µm.
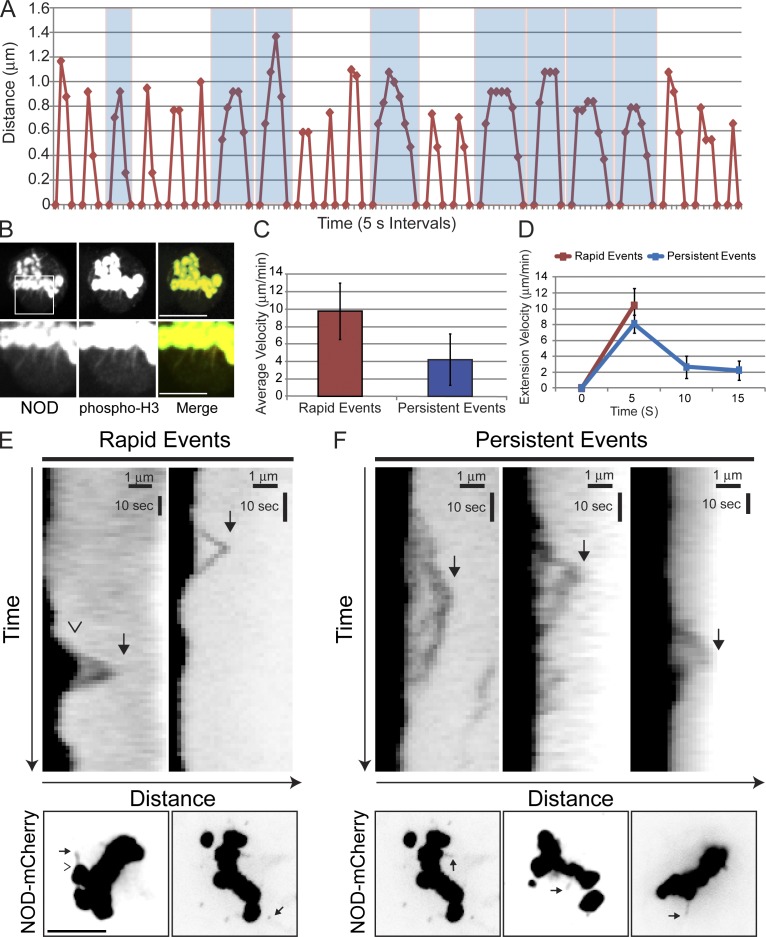
We reasoned that careful analysis of chromatin stretching events would provide insight into the molecular mechanism of force production by NOD. Stretched chromatin extended a mean distance of 0.94 µm (0.6–1.4-µm range) from the chromosome before recoiling (Fig. 6 A). Although stretch distances were largely uniform, two distinct types of stretch dynamics were observed: rapid and persistent. Stretch events were classified as rapid if complete extension and retraction onset occurred within 10 s. Events with an extension phase lasting 10 s or longer were deemed persistent (Fig. 6 A). Polymerizing microtubule–chromosome interactions coincided with rapid stretch events, whereas glancing microtubule–chromosome interactions were associated with persistent events (Fig. 5, B and C). The stretched NOD-mCherry signals were reporting on chromatin dynamics, as the stretches contained phospho–histone H3 (Fig. 6 B) and always recoiled (Fig. 6, E and F; and Fig. S4). The mean extension velocity for a rapid event was 10.4 ± 2.2 µm/min, which was more than twice the mean velocity of 4.2 ± 2.9 µm/min for persistent events (Fig. 6 C). However, rapid and persistent events were not differentiated solely by their extension velocities. Rapid events were simple: maximum extension of the chromatin was achieved within 10 s at a mean rate of ∼10.4 ± 2.2 µm/min (Fig. 6 D) before the chromatin completely recoiled within the next 5–10 s. Persistent stretch events were more complicated. The extension phase of a persistent stretch typically lasted 10–20 s and the extension velocity varied over time, starting at 8.1 ± 1.1 µm/min early and decreasing over time to ∼2.3 ± 1.2 µm/min (Fig. 6 D). Because the time scale of stretching was similar to our initial imaging frequency (5 s), stretch events were examined with higher temporal resolution by acquiring images at 1.5- to 2-s intervals. The additional data thereby obtained for individual stretch events afforded a more detailed view of their dynamic properties (Fig. 6, E and F). It is apparent from kymographs of chromatin stretching that rapid events are uniform in their extension and recoil phases with comparable velocities, whereas persistent events are complex and vary in both their extension and recoil velocities. Thus, NOD-mediated chromatin stretching events are characterized either by rapid ∼10-µm/min bursts that are followed by rapid recoiling or more complicated gradual extensions and recoils that change velocity over time.
Figure 6.
Two types of chromatin stretch events occur in NOD-mCherry–expressing cells. (A) Plot of distance versus time (5-s intervals) for 21 separate chromatin stretching events. Persistent stretch events are highlighted in blue. (B) NOD-mCherry stretches contain phospho-H3 (Serine 10)–positive chromatin. NOD is red and phospho-H3 is green in the merged image. (C) The mean extension velocities of rapid and persistent stretch events. (D) Chromatin in rapid events extends at ∼10 µm/min, whereas extension in persistent events starts at ∼8 µm/min and slows over time to ∼2 µm/min. (E and F) Kymographs of rapid and persistent stretch events imaged by spinning disk confocal microscopy with high temporal resolution (1.5–2-s intervals). Whole cell images are shown below each kymograph highlighting the stretch event that is represented in the kymograph. In the first image, the open arrowhead marks the periphery of the chromosome and the solid arrow marks the edge of the stretch event. Stretches are highlighted in subsequent images with solid arrows. (E) Rapid events exhibit rapid and uniform extension and recoil velocities. (F) Persistent stretch events exhibit variable extension and recoil velocities over time. Bars show standard deviation. Bars: (B, top; and E and F, bottom) 5 µm; (B, bottom) 2.5 µm; (E and F, top) 1 µm (horizontal) and 10 s (vertical).
NOD associates with the plus ends of polymerizing microtubules
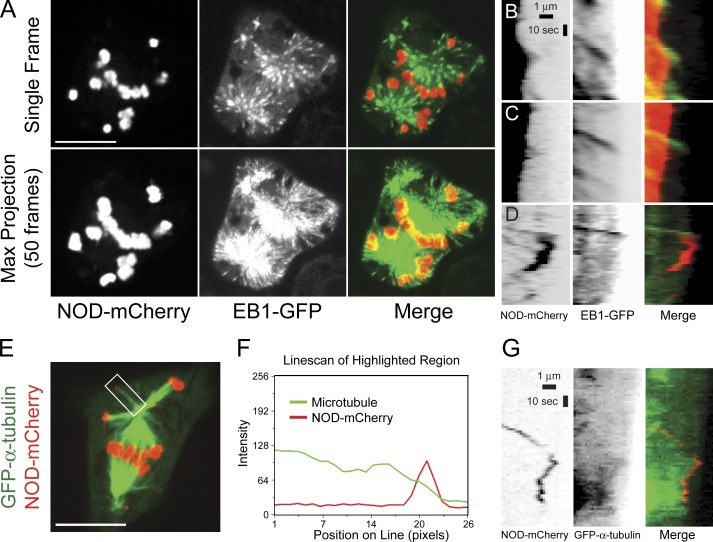
The mean extension velocity of rapid stretch events closely approximated the reported microtubule growth rate of 10.8 µm/min in mitotic S2 cells (Li et al., 2011). Furthermore, physical interactions must exist between the growing plus end of a microtubule and NOD-mCherry–coated chromatin for microtubule buckling to occur (Fig. 5 C and Video 5). Thus, the characteristics of NOD-dependent rapid stretch events are in agreement with the hypothesis that NOD generates PEFs by end tracking on polymerizing microtubules (Cui et al., 2005; Cochran et al., 2009). To further test if rapid stretch events associated with growing microtubule plus ends, a stable cell line coexpressing end binding 1 (EB1)–GFP, which labels the plus ends of polymerizing microtubules, and inducible NOD-mCherry was created (Fig. 7 and Video 6). Rapid chromatin stretch events colocalized with EB1-GFP comets (Fig. 7, B and C; and Video 7), and disappearance of the EB1 comets coincided with maximal chromatin stretch, indicating that the microtubule with which the chromatin was interacting had ceased polymerizing. Small pieces of NOD-coated chromatin were sometimes torn from chromosome arms and transported through the cytoplasm. NOD-positive fragments were significantly smaller than the smallest chromosome (number 4) in Drosophila. A subset of motile NOD-mCherry fragments colocalized with EB1-GFP comets (Fig. 7 D and Video 8). A closer look at motile NOD-mCherry fragments in cells coexpressing GFP–α-tubulin also revealed that they tracked the plus ends of microtubules (Fig. 7, E–G; and Video 9). Thus, our data support the hypothesis that NOD is capable of generating PEFs through association with polymerizing microtubule plus ends.
Figure 7.
High spatial and temporal resolution imaging reveals that NOD-mCherry associates with the ends of polymerizing microtubules. (A) Selected frame (top) and maximum projection of 50 frames (bottom) from a confocal time lapse of an EB1-GFP (green)– and NOD-mCherry (red)–expressing S2 cell. See Video 6. (B–D) Kymographs of NOD-mCherry and EB1-GFP. (B and C) The extension phases of two rapid stretch events that colocalize with EB1-GFP comets are shown. The kymograph in C is from Video 7. (D) A NOD-mCherry spot that is propelled through the cytoplasm colocalizes with an EB1-GFP comet. See Video 8. (E–G) NOD-mCherry tracks on dynamic microtubules. See Video 9. (E) Selected frame from a confocal time lapse of a GFP–α-tubulin (green)– and NOD-mCherry (red)–expressing mitotic S2 cell showing a NOD-mCherry fragment localized at the microtubule plus end. The fragment is considerably smaller than the syntelically attached chromosome 4 located below it. (F) Line scan of the highlighted region shown in E. (G) Kymographs of NOD-mCherry and GFP–α-tubulin during a tracking event. NOD-mCherry (red) tracks the growing, shortening, and paused plus end of the microtubule (green). Bars: (A and E) 10 µm; (B–D and G) 1 µm (horizontal) and 10 s (vertical).
NOD chimeras with either plus end–directed motility or tip tracking activity produce PEFs
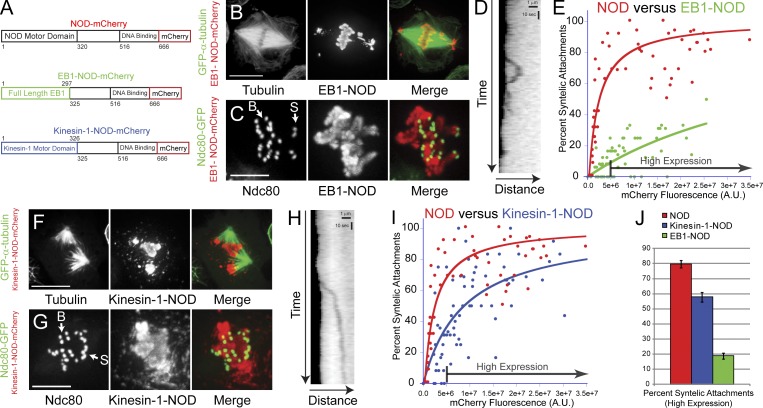
The observation of two types of chromatin stretch events associated with different microtubule–chromatin interactions suggested that PEFs could be produced by two distinct force-producing activities. To further investigate this possibility, two NOD chimeras were created to specifically isolate the contributions of plus end–directed motility and microtubule tip tracking to kt-MT stabilization in the PEF assay (Fig. 8 A). In one chimera, the motor domain of NOD was replaced with EB1 (Lee et al., 2000; Rogers et al., 2002; Tirnauer et al., 2002; Bieling et al., 2007). The EB1-NOD-mCherry chimera exhibited plus end tip tracking in interphase and became highly enriched on chromosomes during mitosis (Fig. 8, B and C; and Fig. S5). EB1-NOD–dependent stretch events were observed with lower frequency than in NOD-expressing cells, although, like NOD-dependent stretching, EB1-NOD stretches extended 1–2 µm in length (Fig. 8 D). The dynamics of EB1-NOD–dependent stretches were similar to those of rapid NOD-dependent stretch events, as maximum extension in the EB1-NOD cells was completed within less than 10 s (mean = 7.6 s). The mean extension velocity of EB1-NOD stretches was 7.6 ± 2.1 µm/min, which was slower than the rapid stretches observed in NOD overexpressing cells. Furthermore, pause events were more frequent during EB1-NOD stretches compared with rapid NOD-dependent stretching. Nonetheless, EB1-NOD-mCherry expression stabilized syntelic attachments in a dose-dependent manner, albeit with significantly lower efficiency than wild-type NOD-mCherry (Fig. 8, C, E, and J).
Figure 8.
Syntelic attachments are stabilized by NOD chimeras that possess either plus end–directed motility or tip-tracking activity. (A) Schematic diagrams of full-length NOD-mCherry and EB1- and kinesin-1-NOD-mCherry. (B) Maximum intensity projection of a GFP–α-tubulin (green)– and EB1-NOD-mCherry (red)–expressing S2 cell showing enrichment of EB1-NOD on chromosomes and the presence of misaligned chromosomes. (C) Maximum intensity projection of a cell expressing Ndc80-GFP (green) and EB1-NOD-mCherry (red) with syntelic (S) and bioriented (B) attachments. (D) Kymograph of a rapid EB1-NOD–mediated chromatin stretch event. (E) Plot of percentage of syntelic attachments versus mCherry fluorescence for EB1-NOD cells. EB1-NOD-mCherry overexpression stabilizes syntelic attachments at a significantly lower frequency than wild-type NOD-mCherry (NOD, n = 57 cells; EB1-NOD, n = 71 cells). (F) Maximum intensity projection of a GFP–α-tubulin (green)– and kinesin-1-NOD-mCherry (red)–expressing cell showing aberrant spindle morphology. (G) Maximum intensity projection of an Ndc80-GFP (green)– and kinesin-1-NOD-mCherry (red)–expressing S2 cell with a mixture of syntelic (S) and bioriented (B) attachments. (H) Kymograph of a persistent kinesin-1-NOD–mediated chromatin stretch event. (I) Plot of percentage of syntelic attachments versus mCherry fluorescence for kinesin-1-NOD cells. Kinesin-1-NOD-mCherry overexpression induces a dose-dependent increase in the percentage of syntelic attachments that rises more slowly and plateaus at a lower percentage of syntelics than the corresponding increase seen for wild-type NOD-mCherry (NOD, n = 57 cells; kinesin-1-NOD, n = 72 cells). (J) The percentage of syntelic attachments in high-expressing cells (defined as >5.0e6 A.U) for NOD-, kinesin-1-NOD-, and EB1-NOD-mCherry–expressing cells. Two-tailed p-values are <0.0005. Error bars are the SEM. Curves were fit with a hyperbolic function. R values are 0.66 (EB1-NOD), 0.7 (kinesin-1-NOD), and 0.86 (NOD). Bars: (B and F) 10 µm; (C and G) 5 µm; (D and H) 1 µm (horizontal) and 10 s (vertical). See Video 10.
In the second NOD chimera, the motor domain of NOD was replaced with the motor domain of human kinesin-1, a highly processive plus end–directed motor (Howard et al., 1989; Block et al., 1990; Hackney, 1995). Kinesin-1-NOD-mCherry stabilized syntelic attachments in a dose-dependent manner with greater potency than EB1-NOD but with lower efficiency than wild-type NOD-mCherry (Fig. 8, G, I, and J). Expression of kinesin-1-NOD-mCherry resulted in chromatin being extensively stretched from chromosome arms (Fig. 8, F–H; and Video 10). Interestingly, neither of the chimeras, each of which possesses a unique but singular force-producing activity, was capable of stabilizing syntelics as efficiently as wild-type NOD (Fig. 8, E, I, and J).
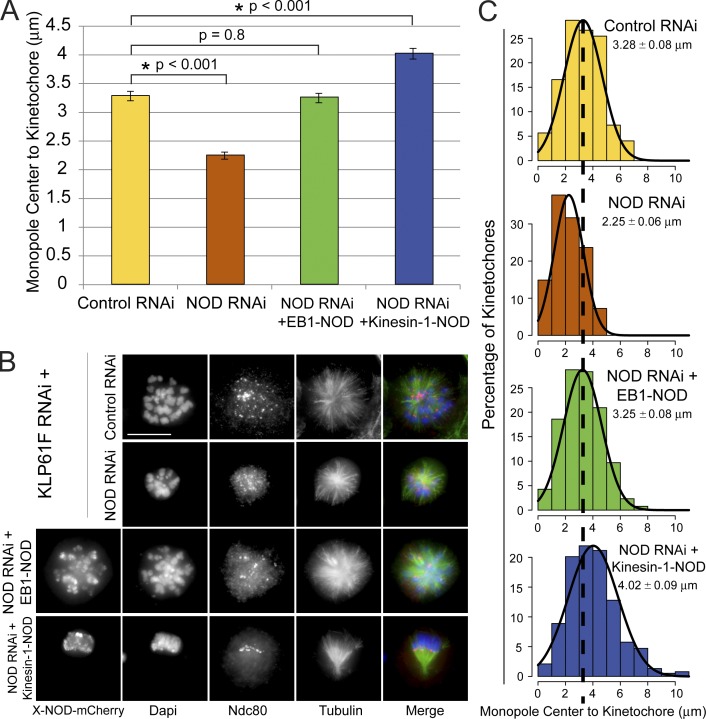
Chromosomes are positioned at the periphery of monopolar spindles by PEFs (Fig. S2, D and E) (Levesque and Compton, 2001; Stumpff et al., 2012; Wandke et al., 2012). To further test if the chimeras produced PEFs we tested how expression of the NOD chimeras affected chromosome positioning in monopoles. NOD depletion resulted in the inward movement of kinetochores and chromosome arms within monopoles (Fig. 9). Notably, the effects of NOD depletion could be rescued by overexpressing either EB1-NOD-mCherry or kinesin-1-NOD-mCherry (Fig. 9, A–C). Collectively, results from the PEF and monopolar spindle assays demonstrate that a chromosome-associated protein with either plus end–directed motility or tip-tracking activity can generate PEFs.
Figure 9.
NOD chimeras with either plus end–directed motility or tip tracking activity produce PEFs. (A) Distance between the monopole center and Ndc80-stained kinetochores. The distance between kinetochores and monopole centers decreased by 30% in the absence of NOD and was rescued in NOD-depleted cells by inducing either EB1-NOD-mCherry or kinesin-1-NOD-mCherry (control, n = 283 kinetochore pairs; NOD RNAi, n = 302; NOD RNAi + EB1-NOD-mCherry, n = 293; NOD RNAi + kinesin-1-NOD-mCherry, n = 383). Error bars are SEM. (B) Representative maximum projection images for each condition. Monopoles were generated in each condition by depleting Klp61F. The chimeric NOD-mCherry protein is shown only in black and white. In the merged images DNA is blue, Ndc80 is red, and microtubules are green. (C) Histograms of the distribution of pole–kinetochore measurements for the four experimental conditions each fit with a Gaussian function (black lines). The dashed line extending through the histograms marks the mean pole–kinetochore distance in control RNAi-treated cells. Two-tailed p-values are shown. Bar, 10 µm.
Discussion
A live-cell assay for studying tension-dependent kt-MT stabilization
Over 40 years ago, Nicklas and Koch (1969) stabilized erroneous kt-MT attachments in grasshopper spermatocytes by artificially creating tension with microneedles. We propose that NOD overexpression is the molecular equivalent of Nicklas’ microneedles and that elevated PEFs produced by NOD overexpression stabilize syntelic attachments by introducing tension at kinetochores. The pioneering spermatocyte studies provided the first direct evidence that tension regulates interactions between chromosomes and the spindle. However, the use of microneedles is technically challenging, requires significant time investment per cell/experiment, and is restricted to a small number of manipulatable cell types that are not genetically tractable. The PEF assay developed here overcomes these previous limitations because (a) force application simply requires the addition of CuSO4 to the growth media, (b) proteins of interest in Drosophila S2 cells can be readily manipulated by RNAi, overexpression, and molecular engineering, and (c) the assay is scalable because many cells can be examined in one experiment. Consequently, we envision that the PEF assay will provide a powerful tool for studying tension-dependent regulation of kt-MT attachment stability in living cells.
PEFs are well-positioned to regulate chromosome oscillations and error correction
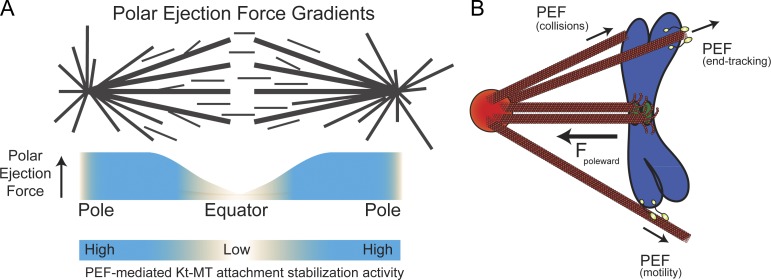
Since its discovery, the PEF has been implicated in chromosome positioning via regulation of both chromosome oscillation and congression. During chromosome oscillations, movement is driven by the poleward moving or leading kinetochore. The poleward moving kinetochore remains attached to its depolymerizing k-fiber and pulls the lagging sister kinetochore, which must elongate its k-fiber by microtubule polymerization (Khodjakov and Rieder, 1996). A change in direction has been hypothesized to be triggered by the introduction of tension at the leading kinetochore as it approaches the pole and experiences increasing levels of opposing PEFs (Fig. 10 A; Rieder et al., 1986; Skibbens et al., 1993; Rieder and Salmon, 1994; Ke et al., 2009). Our observations support this model and are in agreement with recent cell-based examinations of the contribution of PEFs to chromosome behavior (Stumpff et al., 2012; Wandke et al., 2012) as well as the finding that the application of tension to MT-associated kinetochore particles inhibited catastrophes and promoted rescues (Akiyoshi et al., 2010). Thus, emerging evidence supports chromosome oscillation models where the introduction of tension by PEFs at the leading kinetochore promotes a directional switch by rescuing depolymerizing kt-MTs.
Figure 10.
Models for PEF-based modulation of error correction around spindle poles and sources of PEF production. (A) A model of PEF gradients across the metaphase spindle predicts that as a chromosome approaches a spindle pole it will experience progressively higher levels of PEF-mediated kt-MT stabilization because of elevated tension at kinetochores. (B) Model for generation of PEFs that oppose kinetochore-mediated pulling forces to create tension at kinetochores. We propose that chromosome-associated proteins with either plus end–directed motility or end tracking activity can generate PEFs.
The fact that ∼80% of the attachments in high NOD-expressing cells are syntelic suggests that most chromosomes establish improper attachments before becoming bioriented. This mirrors a recent characterization of chromosome biorientation in meiosis I mouse oocytes, where ∼90% of chromosomes experienced at least one round of Aurora kinase–dependent error correction before biorientation (Kitajima et al., 2011). Thus, transient formation of incorrect attachments is commonplace during cell division. Interestingly, improperly attached chromosomes often move to the spindle poles where they remain until error correction occurs (Lampson et al., 2004). Misoriented chromosomes must experience increasingly higher levels of PEFs as they move poleward (Fig. 10 A). Hence, the fact that elevated PEFs counteract error correction presents a conundrum: the spindle pole, where error correction often takes place, is also where PEFs are highest. Over time, baseline error correction mechanisms may win out over the stabilizing effects of the PEFs. Alternatively, other kt-MT attachment destabilizing activities may exist to counter the stabilizing effects of PEFs.
Our findings also bear upon the interplay between force-dependent stabilization of kt-MT attachments and Aurora B–mediated error correction. Application of force to reconstituted kinetochore particles stabilized kt-MT attachments in the absence of Aurora B (Akiyoshi et al., 2010). We have shown that the application of force to kinetochores in living cells stabilizes kt-MT attachments even in the presence of active Aurora B. Thus, kinetochore tension is capable of overpowering the ability of Aurora B to mediate error correction without compromising its activity.
How does NOD generate force?
Our data support the hypothesis that NOD end tracks on polymerizing microtubules. But what is the molecular basis of NOD end tracking? That NOD fragments associate with paused and depolymerizing microtubule plus ends, when EB1 is absent (Fig. 7 G and Video 9), suggests that NOD could track nonpolymerizing microtubule ends in an EB1-independent manner although tracking on polymerizing ends by NOD may require EB1. NOD end tracking has been envisioned as an EB1-independent phenomenon although it has never been directly demonstrated (Cochran et al., 2009). Thus, it will be important to determine whether NOD behaves like budding yeast dynein, which is targeted to microtubule plus ends independent of EB1 (Carvalho et al., 2004; Markus et al., 2011), or like MCAK (kinesin-13), which contains an S/TxIP motif and exhibits EB1-dependent tip tracking (Domnitz et al., 2012).
Because NOD has never been shown to possess plus end–directed motility in vitro, it is currently classified as a nonmotile kinesin. However, the observation of persistent chromatin stretching events that moved along the sides of microtubules toward the plus ends provides compelling evidence that NOD could exhibit plus end–directed motility in cells. We feel this work strongly supports the NOD end tracking hypothesis but does not rule out plus end–directed motility as another potential source of force production by NOD. It will be worthwhile to further test the hypothesis that NOD possesses two force-producing activities.
PEF generation through multiple molecular mechanisms
Microtubule polymerization and molecular motors have long been proposed as possible sources of the PEF (Rieder et al., 1986; Rieder and Salmon, 1994) and the focus has rightfully been placed on molecular motors since the discovery of chromokinesins. Here we report that PEFs can be generated not only by plus end–directed chromokinesins but also by chromosome-associated factors that associate with polymerizing plus ends (Fig. 10 B). Thus, it may be time to look beyond the motility of kinesin-10 motors and consider chromosome-based tip-tracking factors as potential mediators of PEF production.
Materials and methods
Cell culture
Drosophila S2 cells were cultured at 24°C in Schneider’s media (Life Technologies) supplemented with 10% heat-inactivated fetal bovine serum (Life Technologies) and 0.5× antibiotic-antimycotic cocktail (Life Technologies).
Generation of S2 cell lines
The Drosophila NOD gene (CG1763) was amplified from cDNA clone SD02282 with a 5′ KpnI site and 3′ SpeI site. The resulting product was inserted into the multiple cloning site of a pMT/V5 His-B vector (Invitrogen) containing the mCherry gene. The NOD-EGFP construct was generated by inserting the NOD gene between the 5′ KpnI and 3′ SpeI sites in the pMT/V5 His-B vector containing the EGFP gene, lacking a stop codon, inserted between the second XbaI and SacII sites. Motorless NOD was produced by PCR amplification of the portion of the NOD gene corresponding to aa 325–666. The Kin1-NOD and EB1-NOD cells were both produced by ligating the kinesin-1 and EB1 regions into the motorless NOD vector. For the Kin1-NOD cells, the motor domain (corresponding to aa 1–326) of human kinesin-1 (gift of J. Ross, University of Massachusetts, Amherst, MA) was amplified with 5′ XbaI and 3′ KpnI cut sites flanking the gene by PCR. To produce the EB1-NOD cell line, a full-length Drosophila EB1 isoform variant A (CG3265) was amplified from a cDNA clone with 5′ XbaI and 3′ KpnI sites and inserted upstream of motorless NOD-mCherry. See Table 1 for the primers used in cloning. All cell lines were generated by transfecting DNA constructs into S2 cells using the Effectene Transfection Reagent system (QIAGEN), according to product directions. The transfected cells were grown in Schneider’s media containing 10% fetal bovine serum. After 4 d, they were transferred to a 25-cm2 flask. Cells were then grown in media containing blasticidin at a concentration of 0.025 mg/ml until cell death ceased. At that point cells were maintained in media containing no blasticidin. Cell lines were induced by adding either 500 µM (high induction) or 25 μm (low induction) CuSO4 for 6–18 h.
Table 1.
The primers used in this study
| Primer | Sequence |
| FL-NOD cloning (forward) | 5′-GGGGTACCATGGAGGGCGCCAAATTAAGCGCA-3′ |
| Motorless NOD cloning (forward) | 5′-CGGGGTACCATGCAAGTGGCGCGCCAGAA-3′ |
| NOD cloning (reverse) | 5′-GGACTAGTAGTGTCGAGACAGTTAATTTGACAAATCGTTC-3′ |
| EGFP cloning (forward) | 5′-GGACTAGTATGGTGAGCAAGGGCGAGGAG-3′ |
| EGFP cloning (reverse no stop codon) | 5′-GGCCGCGGCTTGTACAGCTCGTCCATGCC-3′ |
| NOD RNAi (forward) | 5′-TAATACGACTCACTATAGGGATGGAGGGCGCCAAATTAAG-3′ |
| NOD RNAi (reverse) | 5′-TAATACGACTCACTATAGGGCGCGCCGCCACCATGGGCAT-3′ |
| KLP61F RNAi (forward) | 5′-TAATACGACTCACTATAGGGATGTCCAGCGAGGATCCCAG-3′ |
| KLP61F RNAi (reverse) | 5′-TAATACGACTCACTATAGGGTTGACCTCCCTGATGTCTAC-3′ |
| Kinesin1 aa 1–326 cloning (forward) | 5′-GCTCTAGAATGGCGGACCTGGCCGAGTG-3′ |
| Kinesin1 aa 1–326 cloning (reverse) | 5′-ATTGGTACCCTTAATTGTTTTGGCCCTTT-3′ |
| EB1 cloning (forward) | 5′-GCTCTAGAATGGCTGTAAACGTCTACTC-3′ |
| EB1 cloning (reverse) | 5′-ATTGGTACCATACTCCTCGTCCTCTGGTG-3′ |
The bases underlined in the RNAi primers represent the T7 promoter sequence. The bases underlined in the cloning primers are restriction sites.
Production of double-stranded RNAs (dsRNAs)
DNA templates for NOD (CG1763) and KLP61F (CG9191) were produced to contain ∼500 bp of complementary sequence flanked with the T7 promoter sequence. dsRNAs were synthesized overnight at 37°C from the DNA templates using the T7 RiboMax Express Large Scale RNA Production System (Promega). For RNAi experiments, media was aspirated off semi-adhered cells at 25% confluence and replaced with 1 ml of serum-free Schneider’s medium containing ∼20 µg dsRNA. After 1 h, 1 ml of fresh Schneider’s plus FBS was added to the wells and incubated for 2–4 d at 24°C. See Table 1 for target sequences.
Live-cell imaging
Cells were seeded onto concanavalin A (Sigma-Aldrich)–treated acid-washed coverslips (Corning) for 1 h. The coverslips were assembled into rose chambers containing Schneider’s media and imaged at room temperature. Cells were imaged on two different spinning disk confocal systems: (1) a TE300 microscope stand (Nikon) equipped with a CSU10 spinning disk confocal head (Yokogawa) attached to a cooled charge-coupled device Orca ER camera (Hamamatsu) using a 100× 1.4 NA Plan Apochromat (Apo) differential interference contrast objective, and (2) a TiE inverted microscope (Nikon) with a CSU-X1 spinning disk confocal head (Yokogawa) and an iXON EMCCD camera (Andor Technology) using a 100× 1.4 NA Plan Apo violet-corrected series differential interference contrast objective (Nikon). Metamorph software (Molecular Devices) was used to control the imaging systems. Images for all figures were processed in Photoshop (Adobe).
PEF assay
Ndc80-GFP and NOD-mCherry S2 cells were treated with 500 µM CuSO4 for 6–18 h to induce expression of NOD-mCherry. Two-color Z-series consisting of ∼30 planes at 0.2-µm intervals were then acquired for both the mCherry and Ndc80-GFP channels. A region of interest was drawn around the mCherry-positive chromosomes in a maximum intensity projection of the mCherry Z-series. After recording the integrated fluorescence intensity of the chromosomal area, the region of interest was moved to a nonchromosomal area and the background integrated fluorescence intensity was measured and then subtracted from the chromosomal mCherry signal to yield the corrected mCherry values, which are presented in the PEF assay graphs. The Z-series from the Ndc80-GFP channel was then carefully examined by eye and the percentage of syntelic attachments, defined as those with a pair of juxtaposed kinetochores facing the same pole, was recorded. Cells with evident monopolar chromosomal arrangements were not imaged.
Antibody production
A polyclonal peptide antibody (antibody 5444) was generated in a rabbit and affinity purified by GenScript. The peptide sequence to which the antibody was raised was EAPYRQFLGRREPSC, corresponding to aa 15–28 of the NOD protein. Recombinant GST-KLP3A tail domain (a gift from J. Scholey, University of California, Davis, Davis, CA) was expressed in BL21 cells with 1 mM IPTG and purified using glutathione sepharose 4B (GE Healthcare) following the manufacturer’s protocol. Cells were lysed in a buffer containing 50 mM Tris, 10 mM KCl, 1% Triton X-100, 10% glycerol, 1 mM MgCl2, 1 mM EGTA, 1 mM DTT, 0.2 mM PMSF, and protease inhibitors (Roche), and the protein was eluted in 10 mM of reduced l-glutathione. Anti-KLP3A was affinity purified against GST-KLP3A by immobilizing the purified protein on a nitrocellulose membrane (Bio-Rad Laboratories). The antibody was eluted using 0.2 M glycine, pH 2.3, and immediately neutralized with 1 M Tris, pH 8.0.
IF
S2 cells were allowed to adhere to concanavalin A–coated coverslips before being quickly rinsed in BRB80 and then fixed in 10% paraformaldehyde. Cells were then permeabilized for 10 min in PBS plus 1% Triton X-100, washed three times for 5 min each in PBS plus 0.1% Triton X-100, and blocked in boiled donkey serum for 30–60 min. Primary antibodies were diluted into boiled donkey serum. Anti-KLP3A (a gift of J. Scholey) was used at a concentration of 1:100, anti-phospho-H3 Serine 10 (Abcam) at 1:20,000, phospho–Aurora A/B/C (Cell Signaling Technology) at 1:500, and anti-NOD (GenScript) at 1:1,000. All secondary antibodies (Jackson ImmunoResearch Laboratories, Inc.) were diluted 1:200 in boiled donkey serum. Cells were treated with DAPI (1:100) and sealed in mounting media containing 20 mM Tris, pH 8.0, 0.5% N-propyl gallate, and 90% glycerol. The ratio of the fluorescence intensities of KLP3A, phospho-H3, or phospho–Aurora B (as appropriate) to DAPI were done similarly to previous work (Maresca and Salmon, 2009); larger and smaller regions were drawn manually in MetaMorph around the chromosomes and the regions were transferred to the images to be quantified (KLP3A, phospho-H3, or phospho–Aurora B). The total intensity measurements were then normalized to the DAPI total intensity. The following equations were used: backgound signal = (integrated fluorescence intensity of big area − integrated fluorescence intensity of small area)/(big area − small area). Total intensity = integrated fluorescence intensity of small area × (background signal × small area).
FRAP analysis
An ∼2–3-µm2 area of spindle microtubules labeled with GFP–α-tubulin was bleached near the spindle equator with a 5–10-s laser pulse from a micropoint laser (Andor Technology) using the 481-nm dye cell. Images were then acquired every 10 s for 5 min. The fluorescence intensity of the bleached region was measured over time using Metamorph (Molecular Devices) software. The recovery data were fit with a hyperbolic function, y = (x*m2)/(m1 + x), to determine the t1/2.
Western blotting
A total of 10 µg of protein was loaded into a 10% SDS-PAGE gel, run out, and transferred to PVDF membrane (Bio-Rad Laboratories) in transfer buffer containing 10% methanol. All antibodies were diluted in TBS with 0.1% Tween and 5% milk. The membrane was first incubated with either anti-GFP serum (gift of M. Bezanilla, University of Massachusetts, Amherst, MA) at 1:50 or anti-NOD antibody (antibody 5444) at 1:500 and then probed with DM1α (anti–α-tubulin antibody; Sigma-Aldrich) at 1:1,000. Rabbit and mouse HRP secondary antibodies (Jackson ImmunoResearch Laboratories, Inc.) were used in conjunction with their respective primaries and imaged with a GBox system controlled by GeneSnap software (Syngene).
Online supplemental material
Fig. S1 shows the detection of fluorescently labeled versions of NOD by IF and Western blot using an anti-NOD antibody generated for this study and the percentage of syntelic kt-MT attachments observed in Drosophila S2 cells exhibiting a range of NOD-mCherry expression levels. Fig. S2 shows the effect of NOD RNAi on levels of the protein, formation of stable syntelic kt-MT attachments in mitotic cells, and pole-to-centromere distance in monopolar mitotic spindles formed in the absence of the kinesin-5 family motor Klp61F. Fig. S3 shows the effect of NOD overexpression on GFP–α-tubulin turnover rate as calculated by FRAP analysis. Fig. S4 illustrates the dynamic properties of recoil for mitotic chromatin after NOD-mediated stretching away from the chromosome arm. Fig. S5 shows the localization of EB1-NOD-mCherry to the plus ends of polymerizing microtubules in interphase cells. Videos 1–3 show mitotic progression in S2 cells expressing GFP–α-tubulin and NOD-mCherry. Video 4 shows the persistence of syntelic kt-MT attachments in mitotic S2 cells expressing Ndc80-GFP and NOD-mCherry. Video 5 shows two rapid and one persistent NOD-mediated chromatin stretch event in an S2 cell expressing GFP–α-tubulin and NOD-mCherry. Videos 6–9 show mitotic S2 cells expressing EB1-GFP and NOD-mCherry. Video 10 shows S2 cells expressing GFP–α-tubulin and kinesin-1-NOD-mCherry. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201211119/DC1.
Supplementary Material
Acknowledgments
We would like to thank Ted Salmon and Rebecca Heald for sharing insightful discussions and for critically reading the manuscript. We acknowledge the University of Massachusetts Medical School Proteomics and Mass Spectrometry Facility. We also thank Magdalena Bezanilla for the gift of anti-GFP serum and Jonathan Scholey for generously sharing anti-KLP3A serum and a GST-KLP3a-tail expression vector. We are grateful to the Ross laboratory (University of Massachusetts [UMASS], Amherst) for sharing the human kinesin-1 construct. Thank you to Steven Markus and Wei-Lih Lee for help with R and KaleidaGraph and the Wadsworth laboratory (UMASS, Amherst) for sharing resources and expertise.
Footnotes
Abbreviations used in this paper:
- dsRNA
- double-stranded RNA
- EB1
- end binding 1
- IF
- immunofluorescence
- k-fiber
- kinetochore fiber
- kt-MT
- kinetochore–microtubule
- PEF
- polar ejection force
References
- Adams R.R., Maiato H., Earnshaw W.C., Carmena M. 2001. Essential roles of Drosophila inner centromere protein (INCENP) and aurora B in histone H3 phosphorylation, metaphase chromosome alignment, kinetochore disjunction, and chromosome segregation. J. Cell Biol. 153:865–880 10.1083/jcb.153.4.865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afshar K., Barton N.R., Hawley R.S., Goldstein L.S. 1995a. DNA binding and meiotic chromosomal localization of the Drosophila nod kinesin-like protein. Cell. 81:129–138 10.1016/0092-8674(95)90377-1 [DOI] [PubMed] [Google Scholar]
- Afshar K., Scholey J., Hawley R.S. 1995b. Identification of the chromosome localization domain of the Drosophila nod kinesin-like protein. J. Cell Biol. 131:833–843 10.1083/jcb.131.4.833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyoshi B., Sarangapani K.K., Powers A.F., Nelson C.R., Reichow S.L., Arellano-Santoyo H., Gonen T., Ranish J.A., Asbury C.L., Biggins S. 2010. Tension directly stabilizes reconstituted kinetochore-microtubule attachments. Nature. 468:576–579 10.1038/nature09594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonio C., Ferby I., Wilhelm H., Jones M., Karsenti E., Nebreda A.R., Vernos I. 2000. Xkid, a chromokinesin required for chromosome alignment on the metaphase plate. Cell. 102:425–435 10.1016/S0092-8674(00)00048-9 [DOI] [PubMed] [Google Scholar]
- Bieling P., Laan L., Schek H., Munteanu E.L., Sandblad L., Dogterom M., Brunner D., Surrey T. 2007. Reconstitution of a microtubule plus-end tracking system in vitro. Nature. 450:1100–1105 10.1038/nature06386 [DOI] [PubMed] [Google Scholar]
- Bieling P., Kronja I., Surrey T. 2010a. Microtubule motility on reconstituted meiotic chromatin. Curr. Biol. 20:763–769 10.1016/j.cub.2010.02.067 [DOI] [PubMed] [Google Scholar]
- Bieling P., Telley I.A., Surrey T. 2010b. A minimal midzone protein module controls formation and length of antiparallel microtubule overlaps. Cell. 142:420–432 10.1016/j.cell.2010.06.033 [DOI] [PubMed] [Google Scholar]
- Block S.M., Goldstein L.S., Schnapp B.J. 1990. Bead movement by single kinesin molecules studied with optical tweezers. Nature. 348:348–352 10.1038/348348a0 [DOI] [PubMed] [Google Scholar]
- Bringmann H., Skiniotis G., Spilker A., Kandels-Lewis S., Vernos I., Surrey T. 2004. A kinesin-like motor inhibits microtubule dynamic instability. Science. 303:1519–1522 10.1126/science.1094838 [DOI] [PubMed] [Google Scholar]
- Brouhard G.J., Hunt A.J. 2005. Microtubule movements on the arms of mitotic chromosomes: polar ejection forces quantified in vitro. Proc. Natl. Acad. Sci. USA. 102:13903–13908 10.1073/pnas.0506017102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho P., Gupta M.L., Jr, Hoyt M.A., Pellman D. 2004. Cell cycle control of kinesin-mediated transport of Bik1 (CLIP-170) regulates microtubule stability and dynein activation. Dev. Cell. 6:815–829 10.1016/j.devcel.2004.05.001 [DOI] [PubMed] [Google Scholar]
- Cassimeris L., Rieder C.L., Salmon E.D. 1994. Microtubule assembly and kinetochore directional instability in vertebrate monopolar spindles: implications for the mechanism of chromosome congression. J. Cell Sci. 107:285–297 [DOI] [PubMed] [Google Scholar]
- Chen R.H., Shevchenko A., Mann M., Murray A.W. 1998. Spindle checkpoint protein Xmad1 recruits Xmad2 to unattached kinetochores. J. Cell Biol. 143:283–295 10.1083/jcb.143.2.283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochran J.C., Sindelar C.V., Mulko N.K., Collins K.A., Kong S.E., Hawley R.S., Kull F.J. 2009. ATPase cycle of the nonmotile kinesin NOD allows microtubule end tracking and drives chromosome movement. Cell. 136:110–122 10.1016/j.cell.2008.11.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui W., Sproul L.R., Gustafson S.M., Matthies H.J., Gilbert S.P., Hawley R.S. 2005. Drosophila Nod protein binds preferentially to the plus ends of microtubules and promotes microtubule polymerization in vitro. Mol. Biol. Cell. 16:5400–5409 10.1091/mbc.E05-06-0582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditchfield C., Johnson V.L., Tighe A., Ellston R., Haworth C., Johnson T., Mortlock A., Keen N., Taylor S.S. 2003. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J. Cell Biol. 161:267–280 10.1083/jcb.200208091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogterom M., Yurke B. 1997. Measurement of the force-velocity relation for growing microtubules. Science. 278:856–860 10.1126/science.278.5339.856 [DOI] [PubMed] [Google Scholar]
- Domnitz S.B., Wagenbach M., Decarreau J., Wordeman L. 2012. MCAK activity at microtubule tips regulates spindle microtubule length to promote robust kinetochore attachment. J. Cell Biol. 197:231–237 10.1083/jcb.201108147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont J., Oegema K., Desai A. 2010. A kinetochore-independent mechanism drives anaphase chromosome separation during acentrosomal meiosis. Nat. Cell Biol. 12:894–901 10.1038/ncb2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funabiki H., Murray A.W. 2000. The Xenopus chromokinesin Xkid is essential for metaphase chromosome alignment and must be degraded to allow anaphase chromosome movement. Cell. 102:411–424 10.1016/S0092-8674(00)00047-7 [DOI] [PubMed] [Google Scholar]
- Giet R., Glover D.M. 2001. Drosophila aurora B kinase is required for histone H3 phosphorylation and condensin recruitment during chromosome condensation and to organize the central spindle during cytokinesis. J. Cell Biol. 152:669–682 10.1083/jcb.152.4.669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., Vale R.D. 2003. The roles of microtubule-based motor proteins in mitosis: comprehensive RNAi analysis in the Drosophila S2 cell line. J. Cell Biol. 162:1003–1016 10.1083/jcb.200303022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., Wollman R., Goodwin S.S., Zhang N., Scholey J.M., Vale R.D., Stuurman N. 2007. Genes required for mitotic spindle assembly in Drosophila S2 cells. Science. 316:417–421 10.1126/science.1141314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., Mayer M., Zhang N., Stuurman N., Vale R.D. 2008. Augmin: a protein complex required for centrosome-independent microtubule generation within the spindle. J. Cell Biol. 181:421–429 10.1083/jcb.200711053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackney D.D. 1995. Highly processive microtubule-stimulated ATP hydrolysis by dimeric kinesin head domains. Nature. 377:448–450 10.1038/377448a0 [DOI] [PubMed] [Google Scholar]
- Hauf S., Cole R.W., LaTerra S., Zimmer C., Schnapp G., Walter R., Heckel A., van Meel J., Rieder C.L., Peters J.M. 2003. The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore–microtubule attachment and in maintaining the spindle assembly checkpoint. J. Cell Biol. 161:281–294 10.1083/jcb.200208092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard J., Hudspeth A.J., Vale R.D. 1989. Movement of microtubules by single kinesin molecules. Nature. 342:154–158 10.1038/342154a0 [DOI] [PubMed] [Google Scholar]
- Hu C.K., Coughlin M., Field C.M., Mitchison T.J. 2011. KIF4 regulates midzone length during cytokinesis. Curr. Biol. 21:815–824 10.1016/j.cub.2011.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke K., Cheng J., Hunt A.J. 2009. The distribution of polar ejection forces determines the amplitude of chromosome directional instability. Curr. Biol. 19:807–815 10.1016/j.cub.2009.04.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov A., Rieder C.L. 1996. Kinetochores moving away from their associated pole do not exert a significant pushing force on the chromosome. J. Cell Biol. 135:315–327 10.1083/jcb.135.2.315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King J.M., Nicklas R.B. 2000. Tension on chromosomes increases the number of kinetochore microtubules but only within limits. J. Cell. Sci. 113:3815–3823 [DOI] [PubMed] [Google Scholar]
- Kitajima T.S., Ohsugi M., Ellenberg J. 2011. Complete kinetochore tracking reveals error-prone homologous chromosome biorientation in mammalian oocytes. Cell. 146:568–581 10.1016/j.cell.2011.07.031 [DOI] [PubMed] [Google Scholar]
- Kwon M., Morales-Mulia S., Brust-Mascher I., Rogers G.C., Sharp D.J., Scholey J.M. 2004. The chromokinesin, KLP3A, dives mitotic spindle pole separation during prometaphase and anaphase and facilitates chromatid motility. Mol. Biol. Cell. 15:219–233 10.1091/mbc.E03-07-0489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampson M.A., Renduchitala K., Khodjakov A., Kapoor T.M. 2004. Correcting improper chromosome-spindle attachments during cell division. Nat. Cell Biol. 6:232–237 10.1038/ncb1102 [DOI] [PubMed] [Google Scholar]
- Lee L., Tirnauer J.S., Li J., Schuyler S.C., Liu J.Y., Pellman D. 2000. Positioning of the mitotic spindle by a cortical-microtubule capture mechanism. Science. 287:2260–2262 10.1126/science.287.5461.2260 [DOI] [PubMed] [Google Scholar]
- Levesque A.A., Compton D.A. 2001. The chromokinesin Kid is necessary for chromosome arm orientation and oscillation, but not congression, on mitotic spindles. J. Cell Biol. 154:1135–1146 10.1083/jcb.200106093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Nicklas R.B. 1995. Mitotic forces control a cell-cycle checkpoint. Nature. 373:630–632 10.1038/373630a0 [DOI] [PubMed] [Google Scholar]
- Li W., Miki T., Watanabe T., Kakeno M., Sugiyama I., Kaibuchi K., Goshima G. 2011. EB1 promotes microtubule dynamics by recruiting Sentin in Drosophila cells. J. Cell Biol. 193:973–983 10.1083/jcb.201101108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magidson V., O’Connell C.B., Lončarek J., Paul R., Mogilner A., Khodjakov A. 2011. The spatial arrangement of chromosomes during prometaphase facilitates spindle assembly. Cell. 146:555–567 10.1016/j.cell.2011.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maresca T.J., Salmon E.D. 2009. Intrakinetochore stretch is associated with changes in kinetochore phosphorylation and spindle assembly checkpoint activity. J. Cell Biol. 184:373–381 10.1083/jcb.200808130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maresca T.J., Salmon E.D. 2010. Welcome to a new kind of tension: translating kinetochore mechanics into a wait-anaphase signal. J. Cell Sci. 123:825–835 10.1242/jcs.064790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markus S.M., Plevock K.M., St Germain B.J., Punch J.J., Meaden C.W., Lee W.L. 2011. Quantitative analysis of Pac1/LIS1-mediated dynein targeting: implications for regulation of dynein activity in budding yeast. Cytoskeleton (Hoboken). 68:157–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall W.F., Marko J.F., Agard D.A., Sedat J.W. 2001. Chromosome elasticity and mitotic polar ejection force measured in living Drosophila embryos by four-dimensional microscopy-based motion analysis. Curr. Biol. 11:569–578 10.1016/S0960-9822(01)00180-4 [DOI] [PubMed] [Google Scholar]
- Matthies H.J., Baskin R.J., Hawley R.S. 2001. Orphan kinesin NOD lacks motile properties but does possess a microtubule-stimulated ATPase activity. Mol. Biol. Cell. 12:4000–4012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklas R.B., Koch C.A. 1969. Chromosome micromanipulation. III. Spindle fiber tension and the reorientation of mal-oriented chromosomes. J. Cell Biol. 43:40–50 10.1083/jcb.43.1.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklas R.B., Waters J.C., Salmon E.D., Ward S.C. 2001. Checkpoint signals in grasshopper meiosis are sensitive to microtubule attachment, but tension is still essential. J. Cell Sci. 114:4173–4183 [DOI] [PubMed] [Google Scholar]
- Powers J., Rose D.J., Saunders A., Dunkelbarger S., Strome S., Saxton W.M. 2004. Loss of KLP-19 polar ejection force causes misorientation and missegregation of holocentric chromosomes. J. Cell Biol. 166:991–1001 10.1083/jcb.200403036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasooly R.S., New C.M., Zhang P., Hawley R.S., Baker B.S. 1991. The lethal(1)TW-6cs mutation of Drosophila melanogaster is a dominant antimorphic allele of nod and is associated with a single base change in the putative ATP-binding domain. Genetics. 129:409–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder C.L., Salmon E.D. 1994. Motile kinetochores and polar ejection forces dictate chromosome position on the vertebrate mitotic spindle. J. Cell Biol. 124:223–233 10.1083/jcb.124.3.223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder C.L., Davison E.A., Jensen L.C., Cassimeris L., Salmon E.D. 1986. Oscillatory movements of monooriented chromosomes and their position relative to the spindle pole result from the ejection properties of the aster and half-spindle. J. Cell Biol. 103:581–591 10.1083/jcb.103.2.581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder C.L., Cole R.W., Khodjakov A., Sluder G. 1995. The checkpoint delaying anaphase in response to chromosome monoorientation is mediated by an inhibitory signal produced by unattached kinetochores. J. Cell Biol. 130:941–948 10.1083/jcb.130.4.941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers S.L., Rogers G.C., Sharp D.J., Vale R.D. 2002. Drosophila EB1 is important for proper assembly, dynamics, and positioning of the mitotic spindle. J. Cell Biol. 158:873–884 10.1083/jcb.200202032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah J.V., Botvinick E., Bonday Z., Furnari F., Berns M., Cleveland D.W. 2004. Dynamics of centromere and kinetochore proteins; implications for checkpoint signaling and silencing. Curr. Biol. 14:942–952 [DOI] [PubMed] [Google Scholar]
- Skibbens R.V., Skeen V.P., Salmon E.D. 1993. Directional instability of kinetochore motility during chromosome congression and segregation in mitotic newt lung cells: a push-pull mechanism. J. Cell Biol. 122:859–875 10.1083/jcb.122.4.859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpff J., Wagenbach M., Franck A., Asbury C.L., Wordeman L. 2012. Kif18A and chromokinesins confine centromere movements via microtubule growth suppression and spatial control of kinetochore tension. Dev. Cell. 22:1017–1029 10.1016/j.devcel.2012.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theurkauf W.E., Hawley R.S. 1992. Meiotic spindle assembly in Drosophila females: behavior of nonexchange chromosomes and the effects of mutations in the nod kinesin-like protein. J. Cell Biol. 116:1167–1180 10.1083/jcb.116.5.1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirnauer J.S., Grego S., Salmon E.D., Mitchison T.J. 2002. EB1-microtubule interactions in Xenopus egg extracts: role of EB1 in microtubule stabilization and mechanisms of targeting to microtubules. Mol. Biol. Cell. 13:3614–3626 10.1091/mbc.02-04-0210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokai-Nishizumi N., Ohsugi M., Suzuki E., Yamamoto T. 2005. The chromokinesin Kid is required for maintenance of proper metaphase spindle size. Mol. Biol. Cell. 16:5455–5463 10.1091/mbc.E05-03-0244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visscher K., Schnitzer M.J., Block S.M. 1999. Single kinesin molecules studied with a molecular force clamp. Nature. 400:184–189 10.1038/22146 [DOI] [PubMed] [Google Scholar]
- Wandke C., Barisic M., Sigl R., Rauch V., Wolf F., Amaro A.C., Tan C.H., Pereira A.J., Kutay U., Maiato H., et al. 2012. Human chromokinesins promote chromosome congression and spindle microtubule dynamics during mitosis. J. Cell Biol. 198:847–863 10.1083/jcb.201110060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wignall S.M., Villeneuve A.M. 2009. Lateral microtubule bundles promote chromosome alignment during acentrosomal oocyte meiosis. Nat. Cell Biol. 11:839–844 10.1038/ncb1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yajima J., Edamatsu M., Watai-Nishii J., Tokai-Nishizumi N., Yamamoto T., Toyoshima Y.Y. 2003. The human chromokinesin Kid is a plus end-directed microtubule-based motor. EMBO J. 22:1067–1074 10.1093/emboj/cdg102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Hawley R.S. 1990. The genetic analysis of distributive segregation in Drosophila melanogaster. II. Further genetic analysis of the nod locus. Genetics. 125:115–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Knowles B.A., Goldstein L.S., Hawley R.S. 1990. A kinesin-like protein required for distributive chromosome segregation in Drosophila. Cell. 62:1053–1062 10.1016/0092-8674(90)90383-P [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.