Abstract
Intestinal mucin 2 (MUC2) encodes a heavily glycosylated, gel-forming mucin, which creates an important protective mucosal layer along the gastrointestinal tract in humans and other species. This first line of defense guards against attacks from microorganisms and is integral to the innate immune system. As a first step towards characterizing the innate immune response of MUC2 in different species, we report the cloning of a full-length, 11,359 bp chicken MUC2 cDNA, and describe the genomic organization and functional annotation of this complex, 74.5 kb locus. MUC2 contains 64 exons and demonstrates distinct spatiotemporal expression profiles throughout development in the gastrointestinal tract; expression increases with gestational age and from anterior to posterior along the gut. The chicken protein has a similar domain organization as the human orthologue, with a signal peptide and several von Willebrand domains in the N-terminus and the characteristic cystine knot at the C-terminus. The PTS domain of the chicken MUC2 protein spans ∼1600 amino acids and is interspersed with four CysD motifs. However, the PTS domain in the chicken diverges significantly from the human orthologue; although the chicken domain is shorter, the repetitive unit is 69 amino acids in length, which is three times longer than the human. The amino acid composition shows very little similarity to the human motif, which potentially contributes to differences in the innate immune response between species, as glycosylation across this rapidly evolving domain provides much of the musical barrier. Future studies of the function of MUC2 in the innate immune response system in chicken could provide an important model organism to increase our understanding of the biological significance of MUC2 in host defense and highlight the potential of the chicken for creating new immune-based therapies.
Introduction
The vast majority of the gastrointestinal tract is covered by a mucosal surface, which creates an important biological barrier that shields the epithelial lining. The top layer of the mucus gel surface, which is the first line of the innate immune defense, is composed primarily of a family of proteins called mucins (MUC). Mucin family members are broadly grouped into secretory and membrane-associated mucins. Membrane associated mucins are involved in signal transduction, oncogenic processes and/or gel formation [1]. Secretory gel-forming mucins (i.e. MUC2, MUC5AC, MUC5B, MUC6, MUC7 and MUC19) contain at least one repetitive domain rich in Pro, Thr and Ser (i.e. the PTS-domain), as well as von Willebrand domains (B, D or C), a cysteine rich domain (CysD), and a cystine knot (CT) [2], [3]. O-linked glycosylation occurs in the PTS domain, while the VWB, VWD, VWC, CysD and CT regions facilitate oligomerization and polymerization. In the small and large intestine, the primary gel-forming mucin is MUC2, although there are detectable levels of MUC5AC and MUC6 in the large intestine [4].
Human MUC2 is a large (5179 amino acid) heterologous glycoprotein that can be modified posttranslationally with more than 100 different oligosaccharides [5]. The oligosaccharides attach along the middle of the protein throughout the mucin domain [6]. The cystine knots at the C-terminus facilitate homodimerization in the endoplasmic reticulum [7], while trimerization occurs in the Golgi through the formation of disulfide bonds at the N-terminus [8]. This produces a 6-membered homopolymer that potentially oligomerizes into hexagonal sheets [4], [9], [10], [11]. Interactions between internal CysD sites create the 3D architecture of the mucosal gel surface [12]. In the intestinal lumen, the charged sugar chains efficiently trap water molecules, creating a stable continuous network that functions analogously to a protective semi-permeable membrane [13]. This protective structure is continually assaulted by physical shear stress due to luminal fluid flow, microbial forging and erosion from proteases or chemical degradation [4].
MUC2 is fundamental in maintaining the architecture of the gel layer on the intestinal surface and in preventing microorganisms from approaching the innermost mucus layer [6]. Alternate splicing of MUC2 and the heterologous nature of the attachment of the sugar molecules generate a highly heterogeneous mucin gel layer, which creates a broad innate defense mechanism within the gastrointestinal tract. Deficiency of or missense mutations in Muc2 causes the epithelial barrier to become permeable to bacteria, leading to colonic inflammation and spontaneous colitis in mice [14], [15], as well as increased susceptibility to infection by enteric nematodes [16]. In humans, rare short MUC2 exonic minisatellites comprised of sequences from the tandem repeat PTS cassettes, have been associated with the onset of gastric cancer [17].
Functional annotation of MUC2 in humans indicates the presence of two polymorphic PTS cassettes [18] and 11 alternatively spliced MUC2 transcripts (UniProtKB, Swiss-Prot) [19]. In addition, analysis of MUC2 in the LS174T derived HM7 colon cancer cell line led to the identification of a transcript variant that lacked the second PTS domain [20]. The presence of this highly polymorphic PTS VNTR (variable number of tandem repeats) has inhibited the resolution of the full-length mRNA, as well as the functional annotation of the complete DNA sequence in many species, including mouse and human [4], [9], [10]. Despite these efforts, the precise annotation of these alternatively spliced MUC2 transcripts remains incomplete, and the length of the PTS domain, which is predicted to span 55–110 cassettes, remains highly polymorphic within the human population [18]. Although the biological relevance of these alternatively spliced products in human is not fully understood, it is believed that they are associated with pathogenesis of intestinal diseases. Although functional studies in mice have indicated that Muc2 plays roles in the biology and health of the gut [15], [21], [22], [23], the function of the PTS domain in mice is less clear, due to the annotation of a relatively short and imprecise repetitive cassette [24].
Evolutionary studies predict that the gel forming mucins share a common ancestor with lower metazoa, as their domain structures are well conserved across a wide range of species from invertebrates to humans [3], [25]. However, relatively few MUC genes have been identified in avians and amphibians. The first Mucin gene cloned in chicken was ovomucin alpha-subunit [26], now annotated as MUC5B. In silico predictions [3], [27] and annotation of short mRNAs and expressed sequence tags (ESTs) have generated a putative partial MUC2 cDNA in chicken. However, these studies have provided very little functional annotation evidence of the genomic organization of the chicken MUC2 locus. To determine the structure, expression, biosynthesis and gene signatures of intestinal mucins from a functional and evolutionary perspective, we cloned the chicken MUC2 cDNA that encodes the MUC2 peptide backbone. We achieved this by analyzing and assembling more than 85 cDNA clones that were generated by overlapping RT-PCR products, rapid amplification of cDNA ends (RACE), sequencing of ESTs, and incorporating functional annotation data (i.e. mRNAs and ESTs) from the UCSC database [28] and NCBI [29]. We also compared our sequence to the predicted chicken cDNA (http://www.medkem.gu.se/mucinbiology/databases/). We found that the 11,359 bp chicken cDNA spans 74.5 kb of genomic DNA and is comprised of at least 64 exons. MUC2 is expressed in multiple regions of the gastrointestinal tract, and we detected transcripts as early as embryonic day 14.5. We found several alternatively spliced products, and characterized the splice junctions of one of these transcripts. We determined that the chicken MUC2 protein is remarkably similar to human and mouse outside of the central PTS domain, but is highly divergent within this central repetitive structure. In humans, this PTS domain is highly glycosylated by O-glycans in the Golgi, and it is predicted that these posttranslational modifications largely contribute to the innate immune response, as proteolytic cleavage of these sugar chains occurs in the outer mucus layer when these molecules come into contact with foreign pathogens [30]. It will be interesting to compare the posttranslational modifications in chicken with other species, especially given the high degree of divergence in this region.
Methods
Tissue biopsy, total RNA isolation
Ethics statement: This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was approved by the Purdue University Animal Care and Use Committee, protocol #03-095. Euthanasia was performed using CO2 inhalation, and all efforts were made to minimize suffering. Intestinal samples (50–100 mg) were taken from chicken embryos at embryonic day (E) 21.5, hatchlings and White Leghorn adult male birds. Tissues were stored in RNAlater, snap frozen in LN2 or processed immediately for RNA isolation. Fertile chicken eggs (n = 720) were obtained and incubated (Jamesway Incubator Company Inc., Cambridge, Ontario, Canada) for gene expression studies.
Since intestinal segments can be identified by E14.5, embryonic intestinal tracts (n = 5–8) from E14.5, E15.5, E16.5, E18.5, and E21.5 of incubation and post-hatch chickens (d 1, 3 and 7) were dissected as discussed previously [31]. Intestinal regions include: duodenum (from the ventriculus to the end of the pancreatic loop), jejunum (from the duodenum to the yolk sac), and ileum (from the jejunum to the ileal-cecal junction). Total RNA was isolated using TRIzol® (Invitrogen, Carlsbad, CA). For most studies, 5 µg of total RNA was reverse transcribed with M-MLV (Invitrogen, Carlsbad, CA) using random hexamers. To ensure transcripts of appropriate length, the reverse transcription reaction in studies involving qRT-PCR was performed using the iScript cDNA synthesis kit (Bio-Rad Life Science Research, Hercules, CA), which contains a mixture of random hexamers and oligo d(T). Alternately, some samples were reverse transcribed using oligo d(T) and SuperScript III (Invitrogen, Carlsbad, CA) or SMARTScribe™ (Clontech, Mountain view, CA). Each PCR was performed at least twice to ensure consistency.
RNA-ligase-mediated rapid amplification of cDNA ends (RLM-RACE)
Total RNA was purified using the DNA-free™ DNase Treatment and Removal Kit (Ambion Inc., Austin, TX) as described [32]. Integrity was verified by gel electrophoresis (1% agarose, 1× TAE). RLM-RACE was performed using the GeneRacer™ RLM-RACE kit (Invitrogen Inc., Carlsbad, CA) according to the manufacturer's protocol. Briefly, full-length capped mRNA was obtained by treating purified total RNA with calf intestinal phosphatase (CIP), which removes fragmented mRNA and non-mRNA. The protective 5′ cap structure from full-length mRNA was then dephosphorylated with tobacco acid pyrophosphatase to facilitate ligation of an RNA oligo to the 5′ end by T4 RNA ligase. Ligated mRNA (2 µg) was reverse transcribed using SuperScript™ III RT and GeneRacer™ Oligo d(T) primers.
To obtain the 3′ end of the MUC2 transcript, first strand cDNA was amplified using the provided 3′ anchor primer and a forward, gene specific 3′ primer (GSP). Hot-start Taq mixed with Pfu polymerase (Advantage© 2 system, Clontech Laboratories, Inc., Mountain View, CA) was used for the 3′ long-range PCR reaction. Amplification was performed under the following conditions: denaturation at 95°C for 1 min, followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 1 min, and extension at 68°C for 3 min. To amplify the 5′ end of MUC2, a reverse complement 5′-GSP and the 5′ anchor primer from the kit were used for a touchdown PCR with a long DNA polymerase (BIO-X-ACTTM Long Mix, Bioline, Tauton, MA). The conditions for the 5′ touch-down PCR reaction were: 2 min at 94°C for initial denaturation; 5 cycles of 30 s at 94°C followed by 90 s at 72°C; 5 cycles of 30 s at 94°C followed by 90 s at 70°C; 25 cycles of 94°C followed by 30 s at 68°C and 90 s at 70°C; and 7 min at 72°C for the final extension. To obtain the 5′ and 3′ ends, we performed nested PCR on 1 µl of the first round amplification reaction using internal MUC2-specific primers for both ends of the transcript and the corresponding anchor primers provided by the kit. RACE products were resolved on 1.2% agarose gels, purified with a gel recovery kit (Zymo Research Corp., Irvine, CA) and cloned using the TOPO TA cloning system (Invitrogen Inc., Carlsbad, CA). Internal primers were designed from either in silico sequences or RACE amplified reads. PCR conditions include initial denaturing at 95°C for 5 min followed by 33–34 cycles of denaturation at 94°C for 30 s, annealing at 58 to 63°C for 20 s, and extension at 72°C for 90 to 120 s, and extension at 72°C for 5 min.
Cloning and sequencing
RT-PCR products were inserted into a pCR-4 TOPO vector and chemically transformed into TOP10 E. coli cells (Invitrogen Inc., Carlsbad, CA) as previously described [32]. Long amplicons from RACE-PCR (>2 kb) were cloned into the T vector and chemically transformed into JM109 Competent cells (Promega, Madison, WI). Plasmids from each clone were prepared and purified using a Quicklyse Miniprep kit (Qiagen Inc., Valencia, CA) and digested with EcoRI. Digested fragments were resolved by gel electrophoresis on 1.5% agarose, 0.5× TBE gels. Three to ten subclones from each clone were sequenced bidirectionally using BigDye 3.1 on an ABI3730XL apparatus (ABI, Life Technologies). Resulting sequences were aligned using Sequencher™ Software (Gene Codes Corp., Ann Arbor, MI). Additionally, two overlapping EST clones (Accession #s BU287205 and BU368530) downstream to the annotated MUC2 transcript were purchased (ARK-Genomics, the Roslin Institute, UK) [33] and sequenced as described.
Genomic DNA was isolated from spleen from four independent chicken samples following proteinase K digestion and phenol/chloroform extraction. High molecular weight DNA was collected by spooling and diluted to a concentration of 50 ng/µl for PCR amplification. Following amplification and purification using the DNA Clean & Concentrator™-5 Kit (Zymo Research Corp., Irvine, CA) to remove free nucleotides and excess primers, the amplicons were sequenced using a ¼ BigDye 3.1 reaction. In a 10 µl reaction volume, this corresponds to 2 µl of 5× sequencing buffer, which ensures that the correct concentrations of reagents are included in the sequencing reaction, 5 µM primer, 2 µl of each amplicon, 1 µl of BigDye 3.1 and 5 µl of H2O. Sequencing reactions were purified using the ZR DNA Sequencing Clean-up Kit™ (Zymo Research Corp., Irvine, CA) and were sequenced as described above. ABI files were uploaded, aligned and analyzed using Sequencher™ Software (Gene Codes Corp., Ann Arbor, MI).
Northern blot hybridization
Total RNA prepared from chick intestine was denatured in 50% formamide (v/v), 5% formaldehyde (v/v) and 20 mM MOPS, pH 7.0, at 65°C for 10 min; electrophoresed in 1.2 to 1.3% agarose gels containing 5% formaldehyde (v/v); and transferred to Hybond N+ nylon membranes overnight. RNA was fixed by cross-linking under UV for 125 s. Membranes were prehybridized in ULTRAhyb® buffer (Ambion) for 1 h at 42°C. Hybridization was carried out at 42°C overnight in ULTRAhyb® buffer containing 32P-labeled probes and 0.1 mg/ml denatured salmon sperm. Probes for chicken MUC2 were prepared by asymmetric PCR or PCR in the presence of [γ-32P]dCTP using gel recovered RT-PCR products as the template. The RNA ladder was radioactively labeled using reverse transcription with random primers. Membranes were washed at 65°C in 2× SSC; 0.1× SDS; 1× SSC; 0.1× SDS, and subsequently 0.1× SSC; 0.1× SDS and exposed to Kodak XAR (Eastman Kodak, Rochester, NY) autoradiography film.
Quantitative RT-PCR
MUC2 expression was analyzed by quantitative RT-PCR (qRT-PCR) in embryonic and post-hatch tissues of chicks as described [32]. Primer pairs (Table 1, P34 to P37) for qRT-PCR analysis were optimized, and PCR products were cloned (into the pCR-4TOPO vector) and confirmed by sequencing. Assays were conducted in 15 µL reactions using iQ SYBR Green Supermix (Bio-Rad Life Science Research, Hercules, CA) with diluted first-strand cDNA. qRT-PCR programs for MUC2 and 18S RNA were: 5 min at 95°C, 40 cycles of 95°C for 15 sec, 56°C or 57°C for 15 sec, 72°C for 15 sec and 82°C or 83°C for 15 sec data collection, followed by 80 cycles for melting curve analysis. All cDNA samples calculated from 100 ng of total RNA per reaction were assayed in duplicate. Quantification standards were comprised of four 100-fold dilutions of purified plasmid DNA (containing from 108 to 102 molecules or 107 to 101 molecules) and assayed in triplicate with R square values of 0.99 or above. Standards were used to calculate a linear regression model for threshold cycle (Ct) relative to transcript abundance in each sample. The log value of MUC2 transcript starting abundance was calculated from the Ct values corrected by a factor calculated from 18S RNA as previously described [31].
Table 1. List of Primers.
| No | Product ID | Type | Oligo sequence (5′-3′) | |
| P1 | MUC2F1 | RT | F | TTT ATG CTC TGG CTG GCT CTT T |
| P2 | RT | R | GGA GTC CTC ATT TCC TTT ACA TGC | |
| P3 | MUC2F2 | RT | F | ATT GTC ACT CAC GCC TTA ATC TG |
| P4 | RT | R | TTT GTC ATC TAC TAA CAA CAC AAC AGT C | |
| P5 | MUC2F3 | RT | F | ATG TGG TGG TTT TCA GAT CAG ATG |
| P6 | RT | R | AGG TTC CAG ATA TGA CCC CTT GTA | |
| P7 | MUC2F4 | RT | F | TGT GGC TGC CCA GAT AAT ACA TAC |
| P8 | RT | R | CCA TTC CTG CTT GTA AAG TCA TTG | |
| P9 | MUC2F5 | RT | F | TGA TGT GCA TTA CCA GAA CAA GAC |
| P10 | RT | R | ATA TGT CGC CAT CCT TTA TTG TTG | |
| P11 | MUC2F6 | RT | F | TGG TGG AGA AAT ACC AAC AGA AGA |
| P12 | RT | R | TAT TGG TGG TAG GAC TGT GCT TGT | |
| P13 | MUC2F7 | RT | F | CCA CCA CAA GCC AGT CTC CA |
| P14 | RT | R | GCA GTA TGA AAC ATG GCC GTT G | |
| P15 | MUC2F8 | RT | F | TGA AGA ATT AGG GCA GAA GGT TGA |
| P16 | RT | R | GGG CAC TGC TAC TTG ACA CAG TC | |
| P17 | MUC2F9 | RT | F | AAC ACG TAC GAC TGT GTC AAG TAG C |
| P18 | RT | R | GCT GTT GTG CAC TCT GGA CTT AAT | |
| P19 | MUC2F10 | RT | F | ATT AAG TCC AGA GTG CAC AAC AGC |
| P20 | RT | R | GCA CTG CTA CTT GAC ACA GTC GT | |
| P21 | MUC2F11 | RT | F | ACA TTC CTA TAG AAG ATC TAG GGC AGA A |
| P22 | RT | R | CTC CCT CAA CAG GGG AAC AC | |
| P23 | MUC2F12 | RT | F | ACG ACT GTG TCA AGT AGC AGT GC |
| P24 | RT | R | TAC TTG ACA CAC TTG GAC TCG ACA | |
| P25 | MUC2F13 | RT | F | ACC ACC AGT ACA ACA GTG TCG AGT |
| P26 | RT | R | TTC CAC TTT CTG CCC TAG ATC TTC | |
| P27 | MUC2F14 | LR | F | ACA CAG TTC ACC CAC CTT AGC C |
| P4 | LR | R | TTT GTC ATC TAC TAA CAA CAC AAC AGT C | |
| P28 | MUC2F15 | LR | F | AAC GGC AAC TGA AAT AGT CTG CAC CTT C |
| P29 | LR | R | AAT GTG CTT TTA ATC ATT CAG AGA AAA TAA GTT GAT T | |
| P30 | MUC2F16 | LR | F | CGG GCC AAC ACC TAC CAC CTC |
| P29 | LR | R | AAT GTG CTT TTA ATC ATT CAG AGA AAA TAA GTT GAT T | |
| P31 | MUC2F17 | LR | F | TAA CTC AAA CCC ACT CTC CTC CAC CTT C |
| P29 | LR | R | AAT GTG CTT TTA ATC ATT CAG AGA AAA TAA GTT GAT T | |
| P19 | MUC2F18 | LR | F | ATT AAG TCC AGA GTG CAC AAC AGC |
| P22 | LR | R | CTC CCT CAA CAG GGG AAC AC | |
| P32 | MUC2F19 | 5′RACE | R | CTA ACA CAT GGA AAG CTC AGC CCA CC |
| P33 | MUC2F20 | 3′RACE | F | CGG GCC AAC ACC TAC CAC CTC |
| P34 | MUC2RQ | qRT | F | ATT GTG GTA ACA CCA ACA TTC ATC |
| P35 | qRT | R | CTT TAT AAT GTC AGC ACC AAC TTC TC | |
| P36 | 18S | qRT | F | GCC ACC CGA GAT TGA GCA ATA ACA |
| P37 | qRT | R | TAG ACA CAA GCT GAG CCA GTC AGT | |
| P38 | HPRT | RT-PCR | F | ATG ACC ACT GTC CAT GCC ATC |
| P39 | RT-PCR | R | AGG GAT GAC TTT CCC TAC AGC CTT | |
| P40 | ARK clone | Seq | F | GGA GAG AGT TGT CCT GAC TGA ATG |
| P41 | ARK clone | Seq | R | CAC AAG AGA AGA GCC ATC AG |
| P42 | MUC2F15,16,17 | Seq | R | TCC AGG TCT AAG TCG GGA AGT G |
| P43 | MUC2F15,16,17 | Seq | F | CAC CTC CTA AAC CCA CCT GCT |
| P44 | MUC2F15,16,17 | Seq | R | CCG CAG CTT TCC ACA TAC AC |
| P45 | MUC2F15,16,17 | Seq | F | GTG TTT GAG AAG TGC CGT GAA G |
| P46 | MUC2F15,17 | Seq | F | ACA CTC AAC CAC TAC AAC CAT |
| P47 | MUC2F15,17 | Seq | R | AAG GTA ATT GTC TGG CCG TGG TG |
| P48 | MUC2F15 | Seq | F | CCT GTT AAC ACA CAG TCT ACA GGA G |
| P49 | MUC2F15 | Seq | F | GCT CTT CAA CAG CTT CAG TTT |
| P51 | MUC2F15 | Seq | R | GGC TCA CAG ATT ACT GGA ACG A |
| P52 | MUC2F15 | Seq | R | ATT GGA GCA GGT GGG TTT AGG |
| P53 | E45F | DNA | F | AGA GCT CTC AGA CAC AGT GGT TGT |
| P54 | E46R | DNA | R | CAT TTT CCA TGA GCT CCC TTA CTT |
RT-PCR (RT); long-range PCR (LR); quantitative RT-PCR (qRT); sequencing (Seq); Forward (F); Reverse (R).
Results
Cloning the chicken MUC2 cDNA
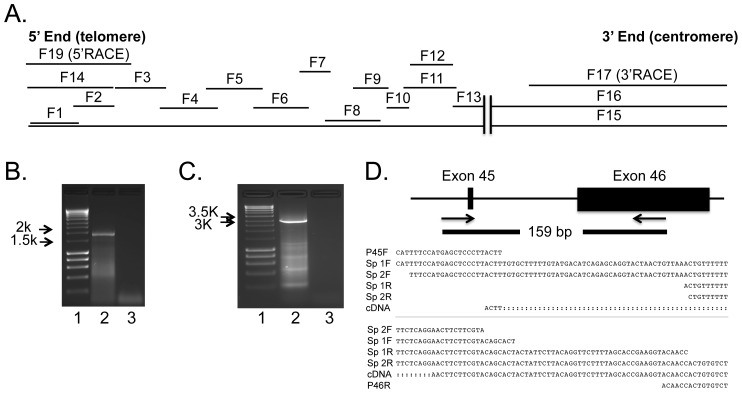
In our aim to clone the full-length chicken MUC2 gene, we amplified, cloned and sequenced 16 overlapping MUC2 RT-PCR products (F1–F14), two expressed sequence tags (ESTs) from the 3′ end of the MUC2 gene (not shown), and products from 5′ and 3′ RACE (F17, F19) (Figure 1A). We sequenced the 1.5 kb 3′-RACE and 3.3 kb 5′-RACE clones in their entirety using multiple internal primers (Figure 1B and 1C). RT-PCR clones derived from internal primers were sequenced to confirm the exon-intron junctions of the 5′ RACE product (F14, Figure 1A). Long-range RT-PCR was performed to determine the sequence of the central and 3′ terminal exons of MUC2, resulting in amplification of two fragments close to 3.7 kb in size (F15 and F16, Figure 1A). We sequenced two overlapping EST clones (Accession #s BU287205 and BU368530) [33] located at the 3′ end of the cDNA in an attempt to close the gap (Figure 1A) produced by the highly polymorphic PTS domain, however this was not successful..
Figure 1. MUC2 cDNA contig.
A. MUC2 cDNA contigs. Sixteen overlapping RT-PCR amplicons and two RACE products are depicted. The double vertical lines represent the cDNA gap in the PTS domain. EST sequences are not shown. B. 5′ RACE. Representative image from 5′ RACE products size fractionated on a 1.2% 1× TAE gel. Molecular weight marker (Lane 1); 5′ RACE produce (Lane 2); Water control (Lane 3). C. 3′ RACE. Representative image from 3′ RACE products size fractionated on a 1.5% 1× TAE gel. Molecular weight marker (Lane 1); 3′ RACE produce (Lane 2); Water control (Lane 3). D. Genomic PCR. We amplified a 159 bp fragment that spanned the gap between exons 45 and 46. Exons are depicted to scale. The forward and reverse sequences from two spleen (Sp 1 and Sp2) samples are shown, along with the cDNA sequence for exons 45 and 46 and the primers. The last 9 bp from the reverse primer are not shown.
To close the gap, we used BLAT alignment [34] to map the cDNA to the UCSC reference sequence (Nov. 2011 (ICGSC Gallus_gallus–4.0/galGal4)) [28]. Based on the genomic alignment, the UCSC database predicted that there was no gap in the cDNA, as the two exons spanning the gap (Table 2, exons 45 and 46) were located within a 136 bp sequence with a short intron. To confirm the genomic organization, we designed primers that flanked the putative gap in the genome. PCR amplification and sequence analysis confirmed that the UCSC annotation was correct, and that we had spanned the gap between the two cDNA contigs (Figure 1D).
Table 2. Genomic Structure of Chicken MUC2.
| Exon | cDNA | Exon | UCSC Alignment1 | Intron | Splice Junctions | |||
| # | Start | Stop | length | Start | Stop | length | 5′ Splice acceptor | 3′ Splice donor |
| 1a | 1 | 94 | 94 | 14368918 | 14368825 | 1490 | cagtggtattCACAGTTCAC | ATAAGAAAAGgtaagctcta |
| 2 | 95 | 359 | 265 | 14367335 | 14367071 | 2588 | aacttaacagGAAGGACAAG | ATGGGCGGATgtaagtacat |
| 3 | 360 | 474 | 115 | 14364483 | 14364369 | 698 | ttcttcctagTGTCAAGACA | TGCACTGATGgtatgtaaaa |
| 4 | 475 | 565 | 91 | 14363671 | 14363581 | 411 | cattacctagGTGGAGCTGG | ATTAGTGGAGgtgagtcata |
| 5 | 566 | 675 | 110 | 14363170 | 14363061 | 237 | tttctcacagTTGCAAGCTA | TAATGAGCATgtgagtaagc |
| 5b | 566 | 744 | 179 | 14363060 | 14362992 | 168 | taatgagcatGTGAGTAAGC | TGAGATGTTAgtgtgtgaag |
| 6 | 745 | 961 | 217 | 14362824 | 14362608 | 450 | tccttcccagCGTGAGGAAT | AACTTTTGCTgtaagcagct |
| 7 | 962 | 1087 | 126 | 14362158 | 14362033 | 260 | tttttttcagACAAAACATG | TGCCCCGAAGgcaagtgtat |
| 8 | 1088 | 1205 | 118 | 14361773 | 14361656 | 318 | tgctttacagGGACTGTGTA | GTGAAGAATGgtaaggatca |
| 9 | 1206 | 1344 | 139 | 14361338 | 14361200 | 663 | ttgtttatagCACCTGTGAT | GTTGGCCAAGgtactgtata |
| 10 | 1345 | 1455 | 111 | 14360537 | 14360427 | 1040 | gtgattgtagAGTGCTGTGA | CAAAAAAAATgtaagtgttg |
| 11 | 1456 | 1525 | 70 | 14359387 | 14359318 | 1334 | ttttatacagGTGGTGGTTT | CATGTGTCAGgtaaggacca |
| 12 | 1526 | 1660 | 135 | 14357984 | 14357850 | 527 | tgtttttcagCTAGCTTCTC | GACATTCAAGgcaagtgtcc |
| 12b | 1526 | 1735 | 210 | 14357849 | 14357774 | 451 | acattcaaggCAAGTGTCCA | ACATGGGTGGtaattcatag |
| 13 | 1736 | 1900 | 165 | 14357323 | 14357159 | 608 | tttttgatagGTCTTTGTGG | ATTGAAAGTGgtaagcttgc |
| 14 | 1901 | 1995 | 95 | 14356551 | 14356457 | 623 | ttttttgcagCAAATTATGC | ATACTATAAGgtatggtaat |
| 15 | 1996 | 2122 | 127 | 14355834 | 14355708 | 578 | gctttttcagAGATGTAAAT | AGTGTCTGCTgtaagtaatg |
| 16 | 2123 | 2378 | 256 | 14355130 | 14354875 | 854 | tcccctccagCTGATGAAGT | GAGAACGTTGgtatgtgtta |
| 17 | 2379 | 2431 | 53 | 14354021 | 14353969 | 289 | tctttcctagTGTTTGCCGA | AGAATGAAAAgtaagtgaca |
| 18 | 2432 | 2541 | 110 | 14353680 | 14353571 | 435 | taactttcagGTATAACAGA | AACTGATCTTgtatgtattc |
| 19 | 2542 | 2693 | 152 | 14353136 | 14352985 | 433 | tgctccttagTATCAAGGTG | GCAACACCTGgtatgctggt |
| 20 | 2694 | 2832 | 139 | 14352552 | 14352414 | 543 | tctcttctagTACCTGCCAG | GGCTACTCAGgtaatgctca |
| 21 | 2833 | 2943 | 111 | 14351871 | 14351761 | 342 | ttatccacagGATTATTGCG | GTTTATAGGGgtgagtaatg |
| 22 | 2944 | 3123 | 180 | 14351419 | 14351240 | 995 | taccatttagAAAACTGAAC | TGACTATAAAgtaagttaga |
| 23 | 3124 | 3363 | 240 | 14350245 | 14350006 | 1149 | ccttttgcagGGGAAAGTGT | TCATTCCAAGgtttgtagat |
| 24 | 3364 | 3520 | 157 | 14348857 | 14348701 | 1075 | tttccaacagGTGAACCCAT | GATATATGCCgtgagtaaca |
| 25 | 3521 | 3649 | 129 | 14347626 | 14347498 | 1713 | ttctcatcagCAATATTCTG | TACCTGGAAGgtaataaatt |
| 26 | 3650 | 3797 | 148 | 14345785 | 14345638 | 911 | gcatcaatagGTTGCTACCC | GTACAAAATGgtatgtaaaa |
| 27 | 3798 | 3841 | 44 | 14344727 | 14344684 | 473 | ttgcttttagTATCTGTCGC | CCAATTCCAGgtaaatagtg |
| 28 | 3842 | 4039 | 198 | 14344211 | 14344014 | 354 | tttcttgcagGATGTCCTTG | ACCACCATAGgtaagttttc |
| 29 | 4040 | 4066 | 27 | 14343660 | 14343634 | 350 | tctatttcagTTACCACAAG | GTACCTACAAgtaagttttg |
| 30 | 4067 | 4543 | 477 | 14343284 | 14342808 | 852 | tttcttgcagCTCCATGCCT | GGCTCCACAGgtatttagca |
| 31 | 4544 | 4735 | 192 | 14341956 | 14341765 | 1072 | tctcttccagTATCAACCAC | TCGCAACCAGgtgattaatt |
| 32 | 4736 | 5338 | 603 | 14340693 | 14340091 | 1025 | tgtctctcagTAGGAAATTG | TCGACAGAAGgtaattgtct |
| 33 | 5339 | 5577 | 239 | 14339066 | 14338828 | ttttccacagGTCCCACTCC | GACAACTGAAggtaatgtct | |
| 34c | 5578 | 5775 | 198 | nnnnnnnnnnATAGTCTGCA | AGAAGATCTAnnnnnnnnnn | |||
| 35 | 5776 | 5786 | 11 | 14338698 | 14338688 | tgtttgtcttGGGCA | GAAAGTcccttagtc | |
| 36d | 5787 | 6209 | 423 | nnnnnnnnnnGGAATGCGAT | CGTCGACAGAnnnnnnnnnn | |||
| 37 | 6210 | 6453 | 244 | 14336621 | 14336378 | gtgttatcccAGGTCCCACT | GACAGAAGGTaattgtctgg | |
| 38c | 6454 | 6497 | 44 | nnnnnnnnnnCCCACTTCCC | CGAGTCCAAGnnnnnnnnnn | |||
| 39e | 6498 | 6675 | 178 | 14335163 | 14334986 | cacgtacgacTGTGTCAAGT | ACCCACCACCnnnnnnnnnn | |
| 40d | 6676 | 6704 | 29 | nnnnnnnnnnTCCGTAACAC | CGTCGACAGAnnnnnnnnnn | |||
| 41 | 6705 | 6946 | 242 | 14313408 | 14313167 | 1173 | gtgttatcccAGGTCCCACT | TCGACAGAAGgtaattgtct |
| 42 | 6947 | 7203 | 257 | 14311994 | 14311738 | gttttcccagGTCCCACTTC | GACAGAAGGTaattgtctgg | |
| 43c | 7204 | 8072 | 869 | nnnnnnnnnnCCCACTCCTG | CGTCGACAGAnnnnnnnnnn | |||
| 44 | 8073 | 8359 | 287 | 14310552 | 14310266 | 1463 | gtgttatcccAGGTCCCACT | TTGAGGGAGAggtgaaggcc |
| 45 | 8360 | 8363 | 4 | 14308803 | 14308800 | gagctcccttAC | TTtgtgcttttt | |
| 46 | 8364 | 8823 | 460 | 14308740 | 14308281 | 1002 | ttttctcaggAACTTCTTCG | AACTCATGCGgttagtgaat |
| 47 | 8824 | 8998 | 175 | 14307279 | 14307105 | 566 | ccaaatacagCCGGGTGAGT | GAGTGTGATTgtaagtatat |
| 48 | 8999 | 9246 | 248 | 14306539 | 14306292 | 888 | gtttttgcagGCTACTGCAC | GGAAGTAGAGgtattggaga |
| 49 | 9247 | 9430 | 184 | 14305404 | 14305221 | 134 | tatttactagGTGACTGTAA | GGACAGTGTGgtaaggctta |
| 50 | 9431 | 9635 | 205 | 14305087 | 14304883 | 644 | cccattttagGCATTTGCAA | TTTGGGGAAGgtaggcatgc |
| 51 | 9636 | 9814 | 179 | 14304239 | 14304061 | 176 | ctgtttttagTGTGTTTGAG | GGTGTTTGCTgtaagtattt |
| 52f | 9815 | 9817 | 3 | 14303885 | 14303883 | 1879 | ccgctctctgGC | Tgtgtgtgtgg |
| 53 | 9818 | 9887 | 70 | 14302004 | 14301935 | 315 | cccattccagCTTATGAATG | GCAAGTCAAGgtaaagaatt |
| 54b , f | 9888 | 9988 | 101 | 14301620 | 14301520 | 157 | tctcatacagTCCACAAAA | AATACCTGTGgtgagtttta |
| 55 | 9989 | 10020 | 32 | 14301363 | 14301332 | 157 | attttcacagGCTGTGTTGG | ACCAAGAGAGgtacgctgcc |
| 56 | 10021 | 10198 | 178 | 14301175 | 14300998 | 707 | ttaatgacagTTTGGAGAAA | GTCACTTGCAgtgagttaat |
| 57 | 10199 | 10303 | 105 | 14300291 | 14300187 | 576 | tttttttcagAATGCAACAC | TACAAGTGTGgtaagtcctt |
| 58 | 10304 | 10344 | 41 | 14299611 | 14299571 | 644 | ttgttaacagTTCCCAAGAA | TGAGTTCTTGgtaagttaat |
| 59 | 10345 | 10467 | 123 | 14298927 | 14298805 | 452 | aaatttatagCCTAATTCCT | TTGTGAACCAgtaagtgatg |
| 60 | 10468 | 10566 | 99 | 14298353 | 14298255 | 625 | taatatctagGGATATGAAC | CATACTGAATgtaagtatgc |
| 61 | 10567 | 10695 | 129 | 14297630 | 14297502 | 665 | tcacttgcagCCTGGAGAGT | CTGCAAACCTgtaagtagat |
| 62 | 10696 | 10735 | 40 | 14296837 | 14296798 | 289 | gttcttttagGGAACTGTTA | TGCAAAACCTgtaagtatgt |
| 63 | 10736 | 10865 | 130 | 14296509 | 14296380 | 1513 | ttccctttagGTATACCTCT | CTTTCTCACTgtaagtaaac |
| 64g | 10866 | 11359 | 494 | 14294867 | 14294374 | atcttaacagGTACTCTGTT | ATTAAAAAAAaaaacatggg | |
Nov 2011 Build (ICGSC Gallus_gallus-4.0/galGal4).
Translational start site at nucleotide 25.
Predicted alternate splicing events (exons 5b & 12b) or exons (exon 55) from reference [3] that were not cloned in this study.
Assembly error—Exon is missing from the current assembly.
Assembly error—Exon located in Gap.
Assembly error—Exon ends in Gap.
Sequence was found in cerebrum and in primordial germ cells in the embyronic gondal, but not cloned in intestine in this study.
Translational stop site at nucleotide 11117.
We next assembled all of the cDNA clones, as well as the predicted cDNA and annotated mRNAs and ESTs from the UCSC and NCBI databases into an 11,359 bp chicken MUC2 cDNA sequence (Figure S1), which has been deposited into GenBank (Accession # JX284122). Translation of the cDNA indicates that we identified a 3697 amino acid protein (Figure S2), which is 1482 amino acids shorter than the predicted human orthologue [5] and 1017 amino acids longer than the annotated mouse protein [24], [35].
MUC2 genomic organization and protein structure
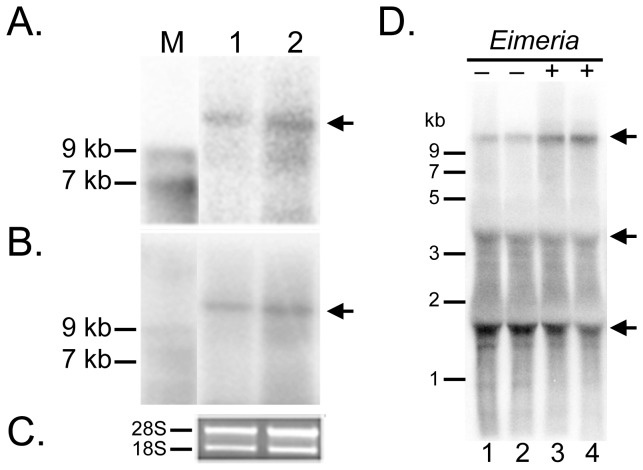
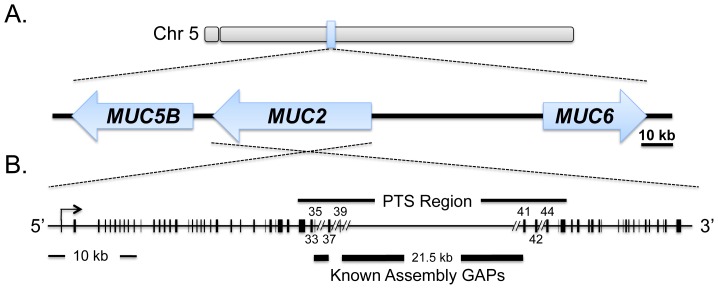
Using northern blot analysis, we estimated the size of the full-length MUC2 transcript to be approximately 12 kb using probes targeting the 3′ and 5′ termini (Figure 2A and 2B). MUC2 is expressed from the (–) strand and spans 74.5 kb of genomic DNA (nucleotides 14368918 to 14294373) on chicken chromosome 5 (Table 2; Figure 3A, 3B). Alignment of our MUC2 cDNA with the Nov 2011 Build (ICGSC Gallus_gallus–4.0/galGal4) of the chicken reference genome [28] indicates that MUC2 spans at least 64 exons (Table 2; Figure 3B). The translational start site occurs within exon 1 at nucleotide 25, while the translational stop site is found at position 11,117 in exon 64.
Figure 2. Northern blot analysis of intestinal MUC2.
A. Transcripts detected with a 5′ probe. B. Transcripts detected with a 3′ probe. Arrows show that both probes detected the same band. Membranes were stripped and demonstrated to show no signal prior to rehybridization. Lane M, ssRNA ladder (kilobase); Lane 1 and 2, 20 µg total RNA per lane. C. RNA integrity. Ethidium bromide-stained rRNAs of the same RNA samples electrophoresed on a 1.0% TAE gel are shown as an internal control. D. MUC2 expression following Eimeria infection. MUC2 transcripts detected in total RNA (30 µg) from cecal tonsils of chickens infected with Eimeria protozoa (+, lanes 3 and 4) or not (−, lane 1 and 2) were identified by probe 3 (1329-bp), which was synthesized by asymmetric PCR. Each lane represents pooled samples from 4 to 5 chickens. Arrow head: MUC2 mRNA.
Figure 3. Chicken MUC2 physical map and genomic organization.
A. Physical Map. The MUC2 locus lies on chromosome 5 between MUC5B and MUC6, and spans 74.5 kb of genomic DNA. B. Gene Structure. We identified an 11,359 bp MUC2 cDNA. Black vertical lines represent the location and relative size of the exons. There are two known assembly gaps in the chicken genome, based on the UCSC Nov 2011 Build (ICGSC Gallus_gallus-4.0/galGal4). Exon 36 (423 bp) is located in the smaller gap, and therefore not included in this figure, as it is unknown where it lies in relationship to its flanking exons. Exon 39 ends blindly in the large (∼21 kb) assembly gap. Exon 40 (29 bp, also not drawn in this map) is located within the gap, and exon 41 is intact on the other side of the gap. The location of these gaps are denoted by large black bars under the gene structure, and gaps are denoted in the genomic structure, when possible (the space between exons 33 and 35 is too small to insert a gap). Exons 34 (198 bp), 38 (44 bp) and 43 (869 bp), which are also not included in this figure) do not fall within the annotated gaps, but are missing in the genomic sequence, indicating errors in the assembly.
By comparing the positions of known chicken mRNAs, ESTs and predicted transcripts, as well as cross-species comparison of human, turkey and helmeted guineafowl mRNAs with our cloned cDNA, we demonstrate strong evidence for our annotation of the genomic structure of MUC2 in chicken (Table 2, Figure 4). Three partial chicken MUC2 mRNAs share significant overlap with our gene. HQ739084 (derived from spleen) and JN639849 share perfect homology with exons 9–11, while CR386462 overlaps with exons 42, 44, 50, 51, 53, 54–57. However, exons 54–56 are annotated as one exon in cDNA CR386462, and exon 57 is smaller than the sequence we cloned. Several chicken ESTs map to our MUC2 exons and add two additional exons. BU296220 overlaps with exons 23–27, while CD753801 maps to exons 32 and 33. BU288276 aligns with exons 42, 44, 50, 51, 53, 54 and DR410193 shares sequence identity with exons 48–51, 53–56. DN928031 maps to exons 50, 51, 53–56; BU368530 overlaps with exons 51–53, 55–60; and BU124202 has significant overlap with exons 57–64. BU371904 and BU287205 share exons 60–64; BU302198 lies within exons 62–64; BU122782 is located within exons 63 and 64; and CF250458 and CD738616 map to exon 64. The human MUC2 transcript overlaps with exons 2, 4, 6–10, 12–17, 19–25, 33, 47–49, 54, 56, 61, 62 and 64, while the helmeted guineafowl (HQ829292) and the turkey (JN942583) transcripts share homology with exons 9–11.
Figure 4. Functional Annotation of MUC2.
The compiled MUC2 cDNA is indicated by exon number (1–64). Black cells represent cDNAs, ESTs or predicted sequences that show the same exon structure as the reported cDNA; Grey boxes represent sequences that are shorter or longer than the compiled cDNA, or that contain significant numbers of mismatches compared to the intestinal MUC2 cDNA; blank cells indicate that this sequence is not present in the corresponding sequence. 1 G. gallus intestinal MUC2 cDNA from this report; 2Helmeted guineafowl; 3Turkey.
Four predicted transcripts provide additional support for our MUC2 gene structure (Figure 4). The most complete predicted chicken sequence [3] overlaps with exons 1–6, 8–20, 22–37, 43, 46–51, 53–64. However, exons 1, 6, 31, 32, 37, 43, 46 and 64 are shorter than the cloned intestinal sequences. The GeneScan predicted gene, Chr5.385, overlaps exons 1–5, 11, 16, 21, 23, 24, 28, 30, 48–51, 58, 63 and 64 of our cDNA. Significant overlap occurs with two additional predicted MUC2 chicken cDNAs (XM_421035 and XM_001234581) that have been annotated in GenBank (www.ncbi.nlm.nih.gov/nucleotide/) [36]. Although XM_421035 has been removed, BLAT analysis of the sequence aligns perfectly with exons 2–31 of our cloned MUC2 cDNA, while XM_001234581 aligns directly with exons 51, 53, and 55–64 of our annotated MUC2 cDNA.
The alignment of our cDNA, along with the chicken mRNAs, ESTs and putative transcripts, to the annotated genome matches very well between exons 1–33 and 46–64. However, there are several inconsistencies between exons 34 and 46 (Table 2; Figure 3B). Exons 34, 38 and 43 are completely missing from the assembly, while exon 36 is located within the small gap and exon 39 ends abruptly in the large 21.5 kb gap (Figure 3B). In an attempt to close these gaps, we designed primers that spanned exons 33–35. The predicted amplicon from this region is 2,340 bp. Despite repeated efforts, we were unsuccessful in generating the correct amplicon, due to the fact that the genomic DNA flanking both of these exons contains several elements that are repeated between exons 32 through 39 and exons 41 through 44. In addition, the presence of multiple poly T and poly A tracts within these regions hampered amplification and/or sequencing efforts due to slippage of the polymerase. Similar challenges occurred when we tried to design primers to amplify the region between exons 37 and 39 and between exons 42 and 44.
The chicken MUC2 locus contains a 21,496 bp gap in the assembly. When we align the compiled cDNA to the genomic locus, we were surprised to discover that the only exon that falls within this large gap is exon 40 (29 bp). Since exon 40 lies within the highly repetitive PTS domain, attempts at cloning the intervening sequences by PCR of genomic DNA have been unsuccessful. Similar challenges occur in the human and mouse genes, and it is likely that additional exons in this region could be identified when the technology becomes available to sequence long DNA or cDNA molecules, as assembling DNA or cDNA that contains multiple repeated cassettes is a major challenge with the current Sanger sequencing and next generation sequencing technologies.
Expression analyses of MUC2
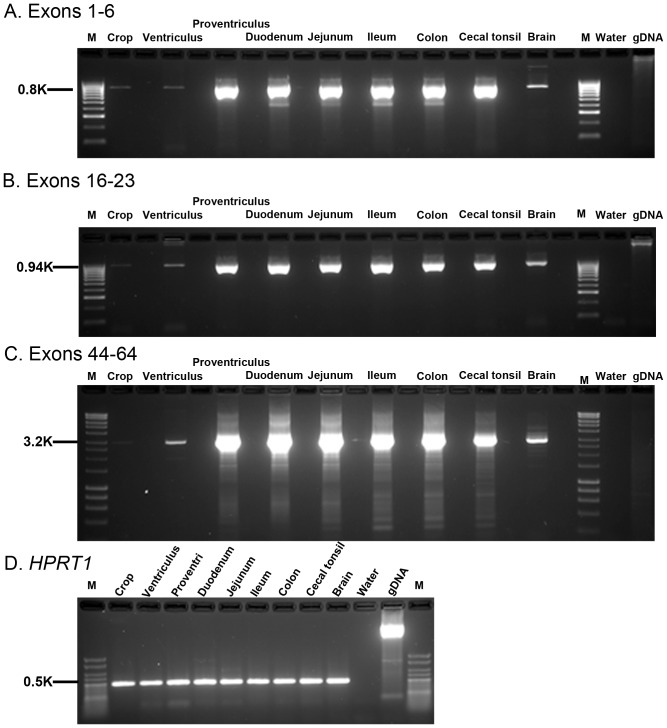
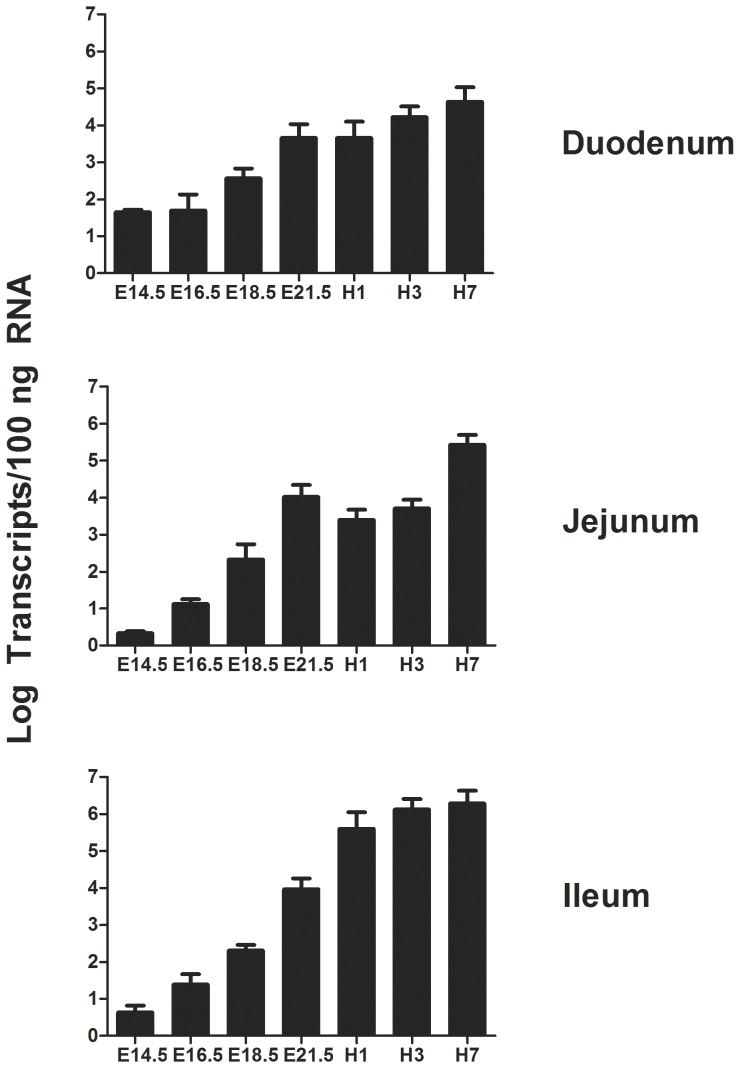
We investigated spatial expression of MUC2 throughout the gastrointestinal tract by RT-PCR and temporal expression in the small intestine at embryonic (E) days 14.5, 16.5, 18.5, 21.5 of incubation and 1, 3, 5 days post-hatch by qRT-PCR (Figures 5 and 6). We used amplicons that spanned three distinct regions of the gene (Exons 1–6; 16–23 and 44–64). MUC2 is highly expressed throughout the gastrointestinal tract, with weak signals in the crop and brain (Figure 5A, 5B and 5C). We observed no alternative splicing using any of these primer pairs. Quantitative RT-PCR analysis of intestinal MUC2 (Exons 25–26; primers P34 and P35) during embryogenesis indicates that expression initiates during late embryogenesis, increasing as development progresses (Figure 6). In the duodenum, jejunum and ileum, MUC2 mRNA levels are relatively low at E14.5, and steadily increase through E21.5. Expression of MUC2 at E14.5 was further confirmed by gel electrophoresis (data not shown). At day of hatch (E21.5), relative MUC2 mRNA levels show a spike (1 to 2 logs) in duodenal and ileal tissues, followed by a steady increase throughout the post-hatch time points. In the jejunum, MUC2 mRNA levels surge to an approximate 2-log increase at H1 followed by a decrease from 1 to 3 d post-hatch, and remain high at 7 d post-hatch.
Figure 5. Expression of MUC2 in the gastrointestinal tract and brain.
RNAs were reverse transcribed using SMARTScribe™ (Clontech) with Oligo d(T) to generate long, full-length cDNA. We performed 33 cycles of RT-PCR amplification on 40 ng of cDNA with three sets of MUC2 primers. Alternating blank lanes lack reverse transcriptase. A. Exons 1–6. MUC2 is highly expressed in the proventriculus, duodenum, jejunum, ileum, colon, and cecal tonsil, with lower levels in the brain and minimal expression in the crop and ventriculus. Although these primers (P27 and P2) amplify genomic DNA, Genomic DNA controls demonstrate the lack of genomic contamination in all samples, indicating that observed expression is from cDNA B. Exons 16–23. MUC2 is highly expressed in the proventriculus, duodenum, jejunum, ileum, colon, and cecal tonsil, with lower levels in the brain and minimal expression in the crop and ventriculus. Although these primers (P7 and P8) amplify genomic DNA, Genomic DNA controls demonstrate the lack of genomic contamination in all samples, indicating that observed expression is from cDNA C Exons 44–65. A touchdown long-range PCR was used to amply the 3′ end of MUC2 using an internal primer and a primer targeting the exact end of the MUC2 cDNA (P30 and P29). This region demonstrates a very similar pattern of expression, with high levels detected in all tissues, except brain, which shows low-level expression,and ventriculus which has minimal expression. No expression is detected in the crop in this analysis. D. HPRT1 control gene. All samples express HPRT, and lack the presence of the genomic DNA band, indicating that the samples do not have genomic contamination. RT-PCR products were examined by electrophoresis through a 2.5% agarose gel in 0.5× TBE (A and B) or 1.2% TAE; water and genomic DNA were used as controls.
Figure 6. Temporal expression of MUC2 transcripts.
Quantification of MUC2 transcripts in embryonic (E) and post-hatch (H) ages in duodenum (top), jejunum (middle) and ileum (bottom) using qRT-PCR. MUC2 expression was normalized to 18S RNA. The MUC2 primers hybridize to exons 25 (P35) and 26 (P34) and generate a 135 bp product that spans intron 25 (1710 bp).
Alternative splicing of chicken MUC2
We investigated the presence of alternative splicing events of MUC2 by RT-PCR, long-range PCR and available ESTs. Several sets of primers spanning the entire cDNA were assayed in multiple tissue types. We identified and characterized one distinct splicing event (Figure 7); we detected one shorter fragment in cecal tonsil samples, which revealed that this transcript used internal splice acceptor/donor sites in exons 41 and 43, removing exon 42. This product is 495 bp shorter than the full-length transcript, but is predicted to result in an in-frame deletion of 165 amino acids within the central PTS domain. Moreover, to explore whether massive alternative splicing events of MUC2 gene would occur in infected versus normal intestine, MUC2 transcripts in Eimeria infected chicks were analyzed, as MUC2 has reported to be aberrantly expressed and critically involved in the pathogenesis of coccidiosis [20], a prevalent protozoal disease in the gastro-intestinal tract of the chicken. However, no detectable alternative splicing event(s) were observed at this the resolution (Figure 2D).
Figure 7. Alternate splicing.
We characterized one of the alternatively spliced products and determined that it was generated from an internal 3′ splice donor in exon 41. This transcript skipped exon 42, and used an alternate splice acceptor site in exon 43.
Predicted amino acid sequences and protein structure
We used a combination of protein analysis software (Interproscan; http://www.ebi.ac.uk/Tools/pfa/iprscan/) [37] and analysis of the domain structure of the predicted protein by The Mucin Biology Group (http://www.medkem.gu.se/mucinbiology/databases/) to determine the putative domain structure of chicken MUC2 (Figure 8A). The deduced amino acid sequence of MUC2 contains several elements common to gel-forming mucins, including: VWD and VWC regions; a central PTS domain interspersed with CysD motifs; and a characteristic cystine knot (CT) at the C terminus (Figure S2). Other prominent features include four cysteine-rich regions (C8) and two trypsin inhibitor-like, cysteine rich domains (TIL).
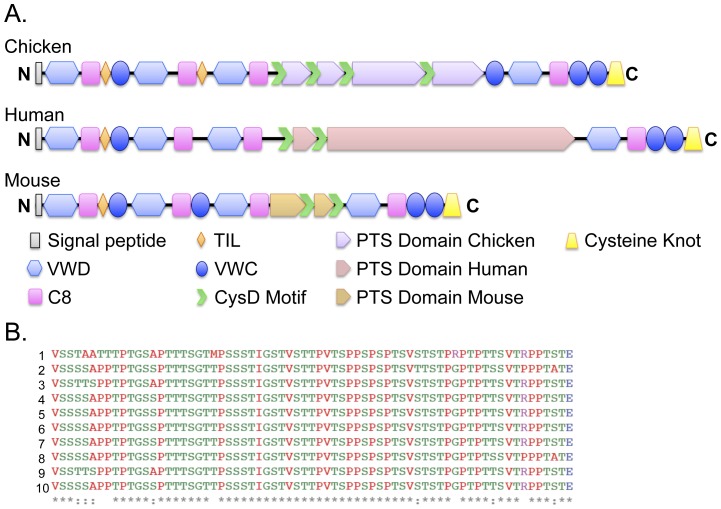
Figure 8. Cross-species comparison of the MUC2 protein structure.
A. Protein structure of chicken, human and mouse MUC2. InterProScan protein domain prediction analysis (www.ebi.ac.uk/Tools/pfa/iprscan/) [37] indicates that the full-length chicken intestinal MUC2 transcript encodes a 3697 amino acid (aa) protein with a short signal peptide at the N-terminus, multiple von Willebrand factor domain structures (VWD, VWC), several cysteine-rich domains (C8), two trypsin Inhibitor-like cysteine-rich domains (TIL), a 1614 amino acid central PTS domain that is interspersed with four CysD motifs and a C-terminal cystine knot (CT). The structure of the human (5179 aa) and mouse (2680 aa) proteins shows strong homology on both sides of the central PTS domain. The two exceptions are that humans and mice lack the second TIL domain, and mice have an additional VWC motif. Although the N-terminal and C-terminal sequences are highly conserved amongst species, the PTS domain is highly divergent, containing different types and varying numbers of repeat cassettes within the central domain. In chicken, this region stretches between aa 1308 and 2922. The different colors in the cartoons represent the finding that the PTS domains are highly divergent among the three species. B. Sequence comparison of the 10 repeats within the PTS domain. Amino acids 1702 through 2763 demarcate this highly repetitive element, which spans exons 32 through 45. RADAR analysis (http://www.ebi.ac.uk/Tools/Radar/) [38] indicates that these cassettes consist of three blocks of repetitive elements interspersed with two CysD domains. Each repeat is 69 amino acids in length and contains one of two short spacer motifs. Repeats 1 and 2 are located in block one, repeats 3–8 are located in block two and repeats 9 and 10 are located in block 3.
We used Rapid Automatic Detection and Alignment of Repeats (RADAR) profiling (http://www.ebi.ac.uk/Tools/Radar/) [38] to detect a core repetitive cassette within the PTS domain. There are 10 total cassettes within this region in chicken, which encompasses amino acids 1702 through 2763 (nt 5131 through 8313) and spans exons 32 through 44. These 10 cassettes are split into three regions containing varying numbers of a highly similar 69 amino acid repetitive element (Figure 8B): element one contains two repeats interspersed with a GPTPESTTRTT motif; element two contains 6 repeats interspersed with alternating GPTPESTTRTT and GPTSQSTTSTTVSSPS motifs; while element three contains two repetitive cassettes with a GPTPESTTRTT linker motif. These three regions are divided by two of the four CysD domains.
Although the N-terminus and the C-terminus share significant identity among human, mouse and chicken, the PTS domain is highly divergent amongst these three species. The human MUC2 protein contains two types of PTS motifs. The larger one contains 97 highly identical direct head to tail repeats of a 23 amino acid sequence (PTGTQTPTTTPITTTTTVTPTPT). The PTS domain in the mouse is separated in two clusters; cluster one contains nine imperfect duplications of an 8 amino acid repeat, while section two contains 15 imperfect duplications of a 10 amino acid cassette [24].
Discussion
It has been over two decades since the initial cloning of the first intestinal mucin gene in humans [39]. Although the physiological implications and disease associations of mucins on various mucosal surfaces have been well recognized, many questions remain as to how and why the gene architecture of this family contributes to diverse protein modifications that may display distinct functionalities. Different species demonstrate structural and sequence conservations as well as their own uniqueness. Chicken, the most-studied and characterized avian species, bridges the evolutionary gap between mammals and non-amniote vertebrates, providing an excellent model system for agricultural and biological research.
In the mucin family, the PTS-domains (or mucin domains) are highly polymorphic in both length and sequence in humans, which is primarily due to the presence of multiple alleles of various number of tandem repeats (VNTRs). However, the presence of the VNTR, as well as the cDNA sequence within the PTS domain, is not highly conserved evolutionarily [3], highlighting the distinct possibility that broad functional differences exist between species [40]. Our data indicate that the PTS domain of the chicken MUC2 protein contains a vastly different repeat structure than the human protein. Although the chicken PTS region is shorter, the central repeat motif is 69 amino acids in length (as opposed to a 23 amino acid cassette in humans) and shows very little identity with the human motif.
Recent in vitro studies using human intestinal cells demonstrated that the intestinal mucins isolated from chicken were detrimental to the proliferation of Campylobacter jejuni, an infectious bacteria causing acute gastroenteritis in humans but not in chicken [10], [40], [41]. In addition these studies demonstrated that the chicken mucins attenuated the invasiveness of Campylobacter jejuni, suggesting that differences in mucin protein sequence or structure between humans and chicken could account for the differences in susceptibility to infection. Alternatively, the functional differences between human and chicken may imply species-specific divergence in intestinal mucus composition and/or structures. This could also occur through differences in posttranslational modifications of the human and chicken proteins. Outside of the PTS domain, the human and chicken MUC2 proteins share large blocks of highly conserved sequences, strongly suggesting that this variable PTS region could account for the phenotypic differences. Plausibly, MUC2 is of utmost importance, as the functionality of intestinal mucus was proposed to rely primarily on MUC2 encoded mucins [10]. Therefore, the full understanding of the functional divergence and prognostic implications of chicken mucins compared to their mammalian orthologues necessitates identification and comparisons of the gene sequences across species.
Although identification of new MUC family members is ongoing, sequencing of most MUCIN genes is hampered due to the highly complex PTS cassettes clustered throughout the gene, and several gaps still remain in mouse and human family members [4], [9], [10]. In the case of the secretory mucins, this can largely be accounted for by the large, frequently repetitive PTS region. The presence of several different polymorphic elements in many of the MUCIN genes hinders annotation efforts at the gene and protein levels, and could even hamper the understanding of the biological significance and disease associations of the diverse family members. By using overlapping RT-PCR, long-range PCR and RACE techniques we have cloned an 11,359 bp chicken MUC2 cDNA. Previous annotations and predictive modeling validate our predicted gene structure. The cDNA that we cloned spans at least 64 exons on chicken 5q16. The central PTS region of the chicken MUC2 locus harbors four CysD motifs and contains 10 repeat cassettes. Although we have closed the gap across the PTS domain by sequencing overlapping cDNA clones derived primarily from chicken intestinal mRNA, it is likely that future studies using more sophisticated sequencing platforms will identify additional exons within the PTS domain. The highly complex nature of this motif indicates that obtaining the full-length MUC2 cDNA could be difficult in the absence of single molecule sequencing efforts. This problem is a common occurrence in the delineation of other mucin genes in mouse and human [4], [9], [10].
The 5′-end of the MUC2 mRNA contains two in-frame ATG codons. Comparing the surrounding sequences of the first ATG codon to the Kozak consensus sequence [42] indicates that the purine at −3 and the G at +6 of GCCGCCATGGGG are conserved within the optimal context for initiation of translation [43]. The sequences surrounding the second ATG codon (Met10; GCCTTTTTATGCTC) are non-consensus Kozak sequences with a T at position −3 and a C at +6. Additionally, analysis of human and mouse MUC2 proteins indicates that the first three amino acids are MGL, which strongly indicates that the first in-frame ATG codon is most likely the translational start site.
The initiating methionine residue is followed by a signal sequence of 18 amino acid residues (analyzed by Signal P3.0; HMM probability: 0.997) (http://www.cbs.dtu.dk/services/SignalP-3.0/) [44] that are rich in leucine but not isoleucine, and are plausibly cleaved to generate the mature mucin isoform during mucin biosynthesis. The amino-terminal region of MUC2, from its initiating methionine to the third C8 motif, spans 1,166 residues composed of multiple VWD and two TIL domains. TIL domains consist of 10 cysteines that are capable of forming disulfide bonds, indicating a high degree of secondary and tertiary structure is possible for these heterogeneous MUC2 protein isoforms. The carboxyl-terminus contains a terminal cystine knot (CT), as well as VWC, VWD domains. These domains are highly conserved throughout evolution [3].
In the endoplasmic reticulum, MUC2 forms disulfide-linked dimers via the VWD domains of the amino-terminus [7], [45], while the CT knot in the carboxy-terminus supports disulfide-linked trimerization in the trans-Golgi network [8]. CysD (C8) domains exert non-covalent cross-linkages in the MUC2 gel formation process, likely contributing to tertiary structure and determination of the pore size of the mucus network [12]. Chicken may plausibly carry more CysD domains than that of human, which may suggest that the polymeric net-like structure contains smaller pores in chickens than humans. This could account for differences in innate defense response to pathogens. The conservation of a cationic domain at the C-terminus observed in rodents was not found in chickens [46].
In human MUC2, two different PTS domains have been identified, both of which are located on the same large exon separated by ∼600 bp. One region consists of repeats that are interrupted in places by 21 to 24 bp segments. The other is composed of an uninterrupted array (of up to 115 repeated units) of a tandem 23-amino acid repeat cassette [18]. Due to the highly unpredictable but repetitive nature, the PTS regions are somewhat refractory to traditional cloning and sequencing technologies [24], [47]. In mice, partial cDNA sequences from the PTS domain suggest the presence of two repetitive PTS regions containing 8 or 10 repetitive units interspaced by a cysteine-rich domain [24]. These repeats are highly dissimilar from both the human and chicken PTS domain. The cDNA that we cloned is composed of 10 interspersed segmental duplications, with the following consensus sequence: VSSSSAPPTPTGSSPTTTSGTTP SSSTIGSTVSTTPVTSPPSPSPTSVSTSTPGPTPTTSVTRPPTSTE. The repetitive unit is rich in threonine (30%), proline (22%) and serine (29%), and is especially high in serine compared to human MUC2 (0% per repetitive unit in the human PTS-region 2). The significance of this is not clear, however, the PTS domains are highly modified posttranslationally by oligosaccharides in humans [30], and these differences could play a role in species-specific innate immune response.
The spatio-temporal expression of MUC2 transcripts follows a specific pattern in humans and rodents [24], [48], [49], [50]. Similarly, our data show that chicken intestinal MUC2 transcripts are expressed throughout the gastrointestinal tract and in embryos as early as E14.5. This is thereafter followed by a rapid increase that follows a developmental timeline. This pattern is seemingly disrupted during the developmental switch from E21.5 (hatch day) to post-hatch day 1 in the duodenal and jejunal tissue. These types of temporal trends in MUC2 expression patterns have been linked to a previous morphometric investigation of intestinal goblet cells, where a gradient increase in goblet cell density was observed along the gastrointestinal tract, and during the period from 3 d prior to and 7 d post hatch [51].
Conclusions
In summary, we have characterized the chicken MUC2 cDNA and identified several conserved structural features of the chicken gene, including VWC, VWD, TIL, C8 and CT domains, as well as large PTS tandem repeat region. Interestingly, although the VWC, VWD, TIL, C8 and CT domains are highly conserved amongst human, mouse and chicken, the PTS domain is quite divergent. Since MUC2 is highly glycosylated posttranslationally, this diversity could prove to be a valuable method for generating species-specific innate immune responses to different host pathogens. This is supported by the supposition that the different species could create mucin gel layers with vastly different pore sizes. This could hamper the ability of pathogens to invade different species and provide a mechanism for the different responses seen across species. Interestingly, known sequence variations in other species have elicited functional differences in cancer incidence, induction of virulence from pathogens, bacterial mucolysis, amongst others, suggesting that the heterogeneity of MUC2 plays an important role in many different biological processes. By defining the structure of mucin from an avian species, we provide important information pertaining to a deeper understanding of the evolutionary mechanisms by which genes contribute to innate barrier functions in the host amongst a wide variety of species. By understanding the role of MUC2 in innate host defense in chickens, we may be able to develop more effective therapies for creating enhanced defense mechanisms in humans.
Supporting Information
Chicken MUC2 cDNA. We identified an 11,359 bp cDNA for MUC2. We derived the cDNA from all available sources, including: in silico data, mRNAs, ESTs, RACE products and RT-PCR amplicons.
(DOCX)
Chicken MUC2 protein. Based on the cDNA sequence from Figure S1, we deduced that the MUC2 protein was 3697 amino acids. We used InterProScan (REF) to determine the different domains, and then compared this analysis to the protein structure provided (medkem). When necessary, we adjusted domains to corresponded with the medkem analysis. We used RADAR (REF) to identify repetitive elements within the protein. Domains are noted in bold, color-coded text, which corresponds to the color coding in Figure 8. Color coding is as noted: signal peptide (black), VWD domains (medium blue), C8 domains (fuschia), TIL motifs (orange), VWC domains (dark blue), CysD (green), PTS (purple), Cysteine knot (yellow). Shaded text indicates the location of the 69 bp repeat within the PTS motifs.
(DOCX)
Acknowledgments
The authors would like to thank Melissa Cramer, and Katherine Baumgarner for excellent technical support. We are grateful to Dr. Bidwell for his technical expertise and informative discussions regarding the northern studies and to Dr. Karcher for his help with embryo sample collection and RNA isolation.
Funding Statement
This work was funded through state and federal funds appropriated to Purdue University, as well as through gift funds from Biomin Research Center, (Tulln, Austria). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Hollingsworth MA, Swanson BJ (2004) Mucins in cancer: protection and control of the cell surface. Nat Rev Cancer 4: 45–60. [DOI] [PubMed] [Google Scholar]
- 2. Hoorens PR, Rinaldi M, Li RW, Goddeeris B, Claerebout E, et al. (2011) Genome wide analysis of the bovine mucin genes and their gastrointestinal transcription profile. BMC Genomics 12: 140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lang T, Hansson GC, Samuelsson T (2007) Gel-forming mucins appeared early in metazoan evolution. Proc Natl Acad Sci U S A 104: 16209–16214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. McGuckin MA, Linden SK, Sutton P, Florin TH (2011) Mucin dynamics and enteric pathogens. Nat Rev Microbiology 9: 265–278. [DOI] [PubMed] [Google Scholar]
- 5. Larsson JM, Karlsson H, Sjovall H, Hansson GC (2009) A complex, but uniform O-glycosylation of the human MUC2 mucin from colonic biopsies analyzed by nanoLC/MSn. Glycobiology 19: 756–766. [DOI] [PubMed] [Google Scholar]
- 6. Johansson ME, Phillipson M, Petersson J, Velcich A, Holm L, et al. (2008) The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci U S A 105: 15064–15069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Asker N, Axelsson MA, Olofsson SO, Hansson GC (1998) Dimerization of the human MUC2 mucin in the endoplasmic reticulum is followed by a N-glycosylation-dependent transfer of the mono- and dimers to the Golgi apparatus. J Biol Chem 273: 18857–18863. [DOI] [PubMed] [Google Scholar]
- 8. Godl K, Johansson ME, Lidell ME, Morgelin M, Karlsson H, et al. (2002) The N terminus of the MUC2 mucin forms trimers that are held together within a trypsin-resistant core fragment. T J Biol Chem 277: 47248–47256. [DOI] [PubMed] [Google Scholar]
- 9. Hansson GC (2012) Role of mucus layers in gut infection and inflammation. Current Opin Microbiol 15: 57–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Johansson ME, Ambort D, Pelaseyed T, Schutte A, Gustafsson JK, et al. (2011) Composition and functional role of the mucus layers in the intestine. Cell Mol Life Sci 68: 3635–3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ambort D, Johansson ME, Gustafsson JK, Nilsson HE, Ermund A, et al. (2012) Calcium and pH-dependent packing and release of the gel-forming MUC2 mucin. Proc Natl Acad Sci U S A 109: 5645–5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ambort D, van der Post S, Johansson ME, Mackenzie J, Thomsson E, et al. (2011) Function of the CysD domain of the gel-forming MUC2 mucin. Biochem J 436: 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tam PY, Verdugo P (1981) Control of mucus hydration as a Donnan equilibrium process. Nature 292: 340–342. [DOI] [PubMed] [Google Scholar]
- 14. Heazlewood CK, Cook MC, Eri R, Price GR, Tauro SB, et al. (2008) Aberrant mucin assembly in mice causes endoplasmic reticulum stress and spontaneous inflammation resembling ulcerative colitis. PLoS Med 5: e54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Van der Sluis M, De Koning BA, De Bruijn AC, Velcich A, Meijerink JP, et al. (2006) Muc2-deficient mice spontaneously develop colitis, indicating that MUC2 is critical for colonic protection. Gastroenterology 131: 117–129. [DOI] [PubMed] [Google Scholar]
- 16. Hasnain SZ, Wang H, Ghia JE, Haq N, Deng Y, et al. (2010) Mucin gene deficiency in mice impairs host resistance to an enteric parasitic infection. Gastroenterology 138: 1763–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jeong YH, Kim MC, Ahn EK, Seol SY, Do EJ, et al. (2007) Rare exonic minisatellite alleles in MUC2 influence susceptibility to gastric carcinoma. PLoS One 2: e1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gum JR Jr, Hicks JW, Toribara NW, Siddiki B, Kim YS (1994) Molecular cloning of human intestinal mucin (MUC2) cDNA. Identification of the amino terminus and overall sequence similarity to prepro-von Willebrand factor. J Biol Chem 269: 2440–2446. [PubMed] [Google Scholar]
- 19. The UniProt Consortium (2012) Reorganizing the protein space at the Universal Protein Resource (UniProt). Nucleic Acids Res 40: D71–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sternberg LR, Byrd JC, Hansson GC, Liu KF, Bresalier RS (2004) Alternative splicing of the human MUC2 gene. Arch Biochem Biophys 421: 21–33. [DOI] [PubMed] [Google Scholar]
- 21. Burger-van Paassen N, van der Sluis M, Bouma J, Korteland-van Male AM, Lu P, et al. (2011) Colitis development during the suckling-weaning transition in mucin Muc2-deficient mice. Am J Physiol-Gastr L 301: G667–678. [DOI] [PubMed] [Google Scholar]
- 22. Lu P, Burger-van Paassen N, van der Sluis M, Witte-Bouma J, Kerckaert JP, et al. (2011) Colonic gene expression patterns of mucin Muc2 knockout mice reveal various phases in colitis development. Inflamm Bowel Dis 17: 2047–2057. [DOI] [PubMed] [Google Scholar]
- 23. Burger-van Paassen N, Loonen LM, Witte-Bouma J, Korteland-van Male AM, de Bruijn AC, et al. (2012) Mucin muc2 deficiency and weaning influences the expression of the innate defense genes reg3beta, reg3gamma and angiogenin-4. PLoS One 7: e38798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Escande F, Porchet N, Bernigaud A, Petitprez D, Aubert JP, et al. (2004) The mouse secreted gel-forming mucin gene cluster. Biochimi Biophys Acta 1676: 240–250. [DOI] [PubMed] [Google Scholar]
- 25. Wang P, Granados RR (1997) Molecular cloning and sequencing of a novel invertebrate intestinal mucin cDNA. J Biol Chem 272: 16663–16669. [DOI] [PubMed] [Google Scholar]
- 26. Watanabe K, Shimoyamada M, Onizuka T, Akiyama H, Niwa M, et al. (2004) Amino acid sequence of alpha-subunit in hen egg white ovomucin deduced from cloned cDNA. DNA sequence 15: 251–261. [DOI] [PubMed] [Google Scholar]
- 27. Lang T, Hansson GC, Samuelsson T (2006) An inventory of mucin genes in the chicken genome shows that the mucin domain of Muc13 is encoded by multiple exons and that ovomucin is part of a locus of related gel-forming mucins. BMC Genomics 7: 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dreszer TR, Karolchik D, Zweig AS, Hinrichs AS, Raney BJ, et al. (2012) The UCSC Genome Browser database: extensions and updates 2011. Nucleic Acids Res 40: D918–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pruitt KD, Tatusova T, Maglott DR (2007) NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res 35: D61–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Johansson ME, Larsson JM, Hansson GC (2011) The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proc Natl Acad Sci U S A 108 Suppl 1: 4659–4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Karcher DM, Fleming-Waddell JN, Applegate TJ (2009) Developmental changes in insulin-like growth factor (IGF)-I and -II mRNA abundance in extra-embryonic membranes and small intestine of avian embryos. Growth Horm IGF Res 19: 31–42. [DOI] [PubMed] [Google Scholar]
- 32. Jiang Z, Lossie AC, Applegate TJ (2011) Evolution of trefoil factor(s): genetic and spatio-temporal expression of trefoil factor 2 in the chicken (Gallus gallus domesticus). PLoS One 6: e22691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Boardman PE, Sanz-Ezquerro J, Overton IM, Burt DW, Bosch E, et al. (2002) A comprehensive collection of chicken cDNAs. Current biology 12: 1965–1969. [DOI] [PubMed] [Google Scholar]
- 34. Kent WJ (2002) BLAT–the BLAST-like alignment tool. Genome Res 12: 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Karlsson NG, Johansson ME, Asker N, Karlsson H, Gendler SJ, et al. (1996) Molecular characterization of the large heavily glycosylated domain glycopeptide from the rat small intestinal Muc2 mucin. Glycoconjugate J 13: 823–831. [DOI] [PubMed] [Google Scholar]
- 36. Benson DA, Karsch-Mizrachi I, Clark K, Lipman DJ, Ostell J, et al. (2012) GenBank. Nucleic Acids Res 40: D48–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hunter S, Jones P, Mitchell A, Apweiler R, Attwood TK, et al. (2012) InterPro in 2011: new developments in the family and domain prediction database. Nucleic Acids Res 40: D306–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Heger A, Holm L (2000) Rapid automatic detection and alignment of repeats in protein sequences. Proteins 41: 224–237. [DOI] [PubMed] [Google Scholar]
- 39. Gum JR, Byrd JC, Hicks JW, Toribara NW, Lamport DT, et al. (1989) Molecular cloning of human intestinal mucin cDNAs. Sequence analysis and evidence for genetic polymorphism. J Biol Chem 264: 6480–6487. [PubMed] [Google Scholar]
- 40. Byrne CM, Clyne M, Bourke B (2007) Campylobacter jejuni adhere to and invade chicken intestinal epithelial cells in vitro. Microbiology 153: 561–569. [DOI] [PubMed] [Google Scholar]
- 41. Collier CT, Hofacre CL, Payne AM, Anderson DB, Kaiser P, et al. (2008) Coccidia-induced mucogenesis promotes the onset of necrotic enteritis by supporting Clostridium perfringens growth. Veterinary Immunol Immunopath 122: 104–115. [DOI] [PubMed] [Google Scholar]
- 42. Galperin MY, Cochrane GR (2009) Nucleic Acids Research annual Database Issue and the NAR online Molecular Biology Database Collection in 2009. Nucleic Acids Res 37: D1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kozak M (1986) Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell 44: 283–292. [DOI] [PubMed] [Google Scholar]
- 44. Bendtsen JD, Nielsen H, von Heijne G, Brunak S (2004) Improved prediction of signal peptides: SignalP 3.0. Journal Mol Biol 340: 783–795. [DOI] [PubMed] [Google Scholar]
- 45. Lidell ME, Johansson ME, Morgelin M, Asker N, Gum JR Jr, et al. (2003) The recombinant C-terminus of the human MUC2 mucin forms dimers in Chinese-hamster ovary cells and heterodimers with full-length MUC2 in LS 174T cells. Biochem J 372: 335–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Xu G, Bell SL, McCool D, Forstner JF (2000) The cationic C-terminus of rat Muc2 facilitates dimer formation post translationally and is subsequently removed by furin. Eur J Biochem 267: 2998–3004. [DOI] [PubMed] [Google Scholar]
- 47. Desseyn JL, Tetaert D, Gouyer V (2008) Architecture of the large membrane-bound mucins. Gene 410: 215–222. [DOI] [PubMed] [Google Scholar]
- 48. Buisine MP, Devisme L, Savidge TC, Gespach C, Gosselin B, et al. (1998) Mucin gene expression in human embryonic and fetal intestine. Gut 43: 519–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chambers JA, Hollingsworth MA, Trezise AE, Harris A (1994) Developmental expression of mucin genes MUC1 and MUC2. J Cell Sci 107 (Pt 2) 413–424. [DOI] [PubMed] [Google Scholar]
- 50. Matsuoka Y, Pascall JC, Brown KD (1999) Quantitative analysis reveals differential expression of mucin (MUC2) and intestinal trefoil factor mRNAs along the longitudinal axis of rat intestine. Biochimi Biophys Acta 1489: 336–344. [DOI] [PubMed] [Google Scholar]
- 51. Uni Z, Smirnov A, Sklan D (2003) Pre- and posthatch development of goblet cells in the broiler small intestine: effect of delayed access to feed. Poultry Sci 82: 320–327. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Chicken MUC2 cDNA. We identified an 11,359 bp cDNA for MUC2. We derived the cDNA from all available sources, including: in silico data, mRNAs, ESTs, RACE products and RT-PCR amplicons.
(DOCX)
Chicken MUC2 protein. Based on the cDNA sequence from Figure S1, we deduced that the MUC2 protein was 3697 amino acids. We used InterProScan (REF) to determine the different domains, and then compared this analysis to the protein structure provided (medkem). When necessary, we adjusted domains to corresponded with the medkem analysis. We used RADAR (REF) to identify repetitive elements within the protein. Domains are noted in bold, color-coded text, which corresponds to the color coding in Figure 8. Color coding is as noted: signal peptide (black), VWD domains (medium blue), C8 domains (fuschia), TIL motifs (orange), VWC domains (dark blue), CysD (green), PTS (purple), Cysteine knot (yellow). Shaded text indicates the location of the 69 bp repeat within the PTS motifs.
(DOCX)