Abstract
Bipolar disorder (BD) is a severe psychiatric disorder that affects 1% of the population. Recently, there have been many attempts to identify specific genes that are involved in BD; however, the task of finding susceptibility genes is not easy due to the complexity of the disorder. Since lithium (Li) has been used for over 40 years now as an effective prophylactic agent and response to Li treatment seems to be, at least in part, genetically determined, classification according to Li response is a manner through which more homogeneous populations can be obtained for investigation. It has previously been suggested that Li exerts an effect on signal transduction pathways, such as the cyclic adenosine monophosphate (cAMP) pathway. We carried out an association study of BD with CREB1, CREB2 and CREB3 genes, located at ch 2q32.3-q34, 22q13.1 and 9pter-p22.1, respectively. A total of three promoter single nucleotide polymorphisms (SNP), 14 SNPs in the UTR, 6 exonic and 15 intronic SNPs were investigated for their frequency and haplotype distribution in a BD sample of 180 lithium responders and 69 nonresponders and 127 controls using a SNaPshot multiplex reaction from Applied Biosystems, a modified fluorescent single base pair extension procedure. Following correction for multiple testing, our results suggest that the CREB1-1H SNP (G/A change, p < 0.002) and the CREB1-7H SNP (T/C change, p < 0.002) may be associated with BD and/or lithium response.
Introduction
Bipolar disorder (BD) affects approximately 1% of the general population and is characterized by manic and depressive episodes (Akiskal et al., 2000);(APA, 1994). Family, twin and adoption studies support the hypothesis that genetic factors play a role in the aetiology of BD (Craddock and Jones, 2001). An interesting family of genes to investigate is that comprising genes coding for CREB (cyclic-AMP responsive element binding) proteins.
The cAMP signal transduction pathway is activated through ligand binding to G-protein coupled receptors and culminates with the phosphorylation of the CREB protein allowing it to bind the cAMP responsive element (CRE) recognition sequence of cAMP sensitive genes and regulate transcription. There have been a number of studies looking at the effects of lithium, an effective prophylactic treatment for BD, on CREB DNA binding, as well as its effects on gene expression. They all concur in that lithium decreases CREB phosphorylation leading to inefficient DNA binding resulting in altered expression of cAMP responsive genes (Bezchlibnyk and Young, 2002). Thus it is possible that genetic variation in genes coding for CREB proteins may help determine response to lithium prophylaxis. The aim of our study was to investigate single nucleotide polymorphisms (SNPs) in three genes that are part of the CREB family (CREB1, CREB2 and CREB3) in a sample prospectively characterized for lithium response.
Materials and Methods
Subjects
The sample used for this study was composed of 258 patients with bipolar disorder. There were one hundred fifty-two females (mean age: 48.1 ± 13.7 years) and 106 males (mean age: 49.9 ± 13.3 years); the average age of disease onset was 26.0 ± 9.7 years. The majority of probands included in this study were selected based on prospectively characterized response to lithium treatment. Of these, 180 were responders and 69 were non-responders to long term lithium treatment. The remaining patients could not be classified as they either never received lithium, or were treated with lithium only briefly. (As clear-cut response or non-response to lithium treatment was an ascertainment criterion for a majority of subjects, the proportions of responders and non-responders do not necessarily correspond to response rates in bipolar disorder rin general).
All subjects included in this study were Caucasian of European (non-Mediterranean) origin that were recruited in centers that are part of the International Group for the Study of Lithium (IGSLi), a collaborative group of specialized clinics in Canada, Austria, Czech Republic, Denmark, Germany, and Sweden.
All psychiatric assessments were carried out by experienced clinicians using the SADS-L structured interviews, and the RDC and DSM-IV criteria. Diagnostic and treatment information was reviewed blindly by a panel of experienced psychiatrists. To ensure diagnostic uniformity across centres all subjects were evaluated by the same senior clinician (PG). Excellent lithium response was established as described previously and summarized in Table 1 (Grof et al., 1994). The number of pretreatment episodes (± standard deviation) was 8.0 ± 5.8 and the patients remained episode-free on lithium monotherapy for 13.3 ± 7.3 years on average. To be considered non-responders, patients had to experience at least two recurrences during lithium treatment with confirmed therapeutic levels of lithium (Grof et al. 1994). Controls included in this study were 118 psychiatrically healthy individuals (mean age 48.6 ± 10.7) assessed using the same criteria as the patients.
Table 1.
Criteria used to diagnose bipolar patients as excellent lithium responders
| Each patient must meet criteria A, B and C | |
| A. | Diagnosis of primary episodic bipolar disorder based on the SADS-L (lifetime version) interview and Research Diagnostic Criteria (RDC). |
| B. | High Recurrence Risk |
| either | B1: five or more episodes prior to lithium treatment |
| or | B2: four episodes prior to lithium; of these two or more during the 2 years preceding lithium treatment |
| or | B3: three episodes prior to lithium, plus one more within 12 months after lithium discontinuation |
| C. | Unequivocal Lithium Response |
| C1: No recurrence requiring additional biological intervention (ECT, antidepressants, neuroleptics) during the entire observation time on lithium monotherapy | |
| and | C2: minimum period of observation of 3 years |
| and | C3: average plasma lithium concentration over 0.6 mEq L−1 |
DNA Analysis
Genomic DNA was extracted using a standardized method (Sambrook et al., 1989) from venous blood samples. Three CREB genes were investigated; CREB1, CREB2 and CREB3. A total of 40 single nucleotide polymorphisms (SNP); three promoter SNPs, 14 SNPS in the UTR, six exonic and 15 intronic SNPs, spaced out along each CREB gene were investigated using the ABI SNaPshot multiplex reaction and the ABI 3100 automated genetic analyzer from Applied Biosystems (Foster City, CA, USA). SNP selection was based on location with the gene, availability of heterozygosity information and validation status on dbSNP. Polymerase chain reaction (PCR) was carried out in a total volume of 10 μl which contained 30 ng genomic DNA, 0.2μM of each primer, 0.25mM of each dATP, dNTP, dTTP and dGTP (Invitrogen Canada, Burlington, Ontario), 0.5 units of Taq DNA polymerase, 1.0 μl of 10x buffer containing 25 mM MgCl2 (Qiagen Inc. Canada, Mississauga, Ontario). The samples were put through between 30 to 40 cycles of denaturation at 94°C, annealing at specific primer temperatures, elongation at 72°C and a final extension at 72°C. PCR product amplification was verified by running 5 μl of product on a 2% agarose gel. The remaining product was then processed as per the ABI SNaPshot protocol, using primers designed for fluorescent dideoxy nucleotide termination. SNP analysis was carried out on the ABI 3100 genetic analyzer. Genotypes were determined automatically using the Genemapper software.
Allele-Specific PCR
Allele-specific PCR was carried out for six of the aforementioned SNPs, which provided unclear results with the SNaPshot reaction. PCR conditions were similar to those described above, apart from the use of two antisense primers, each with one of the possible alleles at the 3′ end (Newton et al., 1989).
Statistical Analysis
Hardy-Weinberg calculations were done using QuickChi. The SNPs found to be in Hardy-Weinberg disequilibrium in both cases and controls were removed prior to further analyses. Allelic and genotypic frequency distributions were compared between cases and controls, responders versus controls, nonresponders versus controls and finally responders versus nonresponders using a χ2 test and the computer program SPSS (Version 11.5) for each CREB gene. Haplotype analysis was carried out using the Haploview program (Barrett et al., 2005) on those SNPs found to be polymorphic. Haplotype blocks were defined as those assessed by the “Strong LD Spine” algorithm using a D′ cut-off of 0.65.
Results
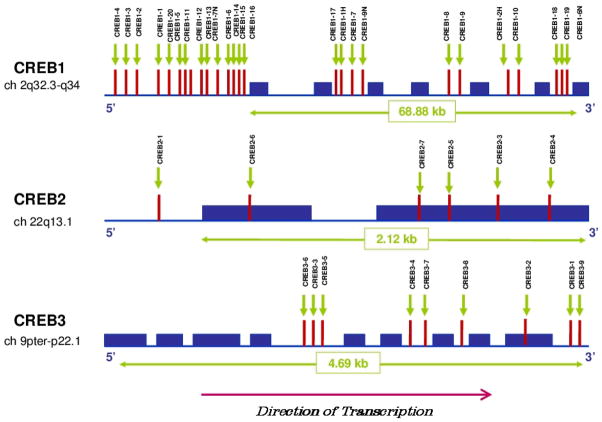
Information on the location of all SNPs investigated in each gene is detailed in Figure 1 and Table 2. Five SNPs were significantly different either between cases and controls or when each response group (LiR, LiNR) were compared to controls (CREB1-4, CREB1-1H, CREB1-2H, CREB1-7H and CREB2-5). Following Bonferroni correction for multiple testing (with 20 comparisons) most of these differences disappeared. However, some results remained significant, specifically the association of the CREB1-1H GA genotype (p < 0.002) and the association of the CREB1-7H T allele and TT genotype (p < 0.002). Table 3 outlines the genotypic and allelic distributions and the respective χ2 values for the SNPs that remained significant following correction.
Figure 1.
This figure depicts each of the genes investigated; CREB1, CREB2 and CREB3, along with the location of each single nucleotide polymorphism (SNP) investigated.
Table 2.
Summary of all SNPs investigated, the allelic change and their location
| Gene | SNP | Alleles | Location | dbSNP # | AA CHNG |
|---|---|---|---|---|---|
| CREB1 | CREB1-1 | A/G | UTR | 1050641 | |
| CREB1-2 | T/G | PMT | 2464976 | ||
| CREB1-3 | A/G | PMT | 2551638 | ||
| CREB1-4 | A/T | PMT | 2709377 | ||
| CREB1-5 | C/T | UTR | 889895 | ||
| CREB1-6 | C/T | UTR | 2709356 | ||
| CREB1-7 | C/T | INTRON | 2551919 | ||
| CREB1-8 | C/T | INTRON | 2551921 | ||
| CREB1-9 | C/T | INTRON | 3770704 | ||
| CREB1-10 | A/G | INTRON | 2709387 | ||
| CREB1-11 | C/T | UTR | 13422714 | ||
| CREB1-12 | C/T | UTR | 13422727 | ||
| CREB1-13 | A/T | UTR | 12466352 | ||
| CREB1-14 | A/G | UTR | 2709357 | ||
| CREB1-15 | A/G | UTR | 2709359 | ||
| CREB1-16 | C/T | UTR | 2709360 | ||
| CREB1-17 | A/T | INTRON | 2059336 | ||
| CREB1-18 | A/C | INTRON | 2254137 | ||
| CREB1-19 | C/T | INTRON | 7369949 | ||
| CREB1-20 | A/G | UTR | 2551639 | ||
| CREB1-6N | A/G | INTRON | 2464978 | ||
| CREB1-7H | A/G | UTR | 2551640 | ||
| CREB1-9N | C/G | INTRON | 2551920 | ||
| CREB1-1H | A/G | INTRON | 208254857 | ||
| CREB1-2H | A/G | INTRON | 208267282 | ||
| CREB2 | CREB2-1 | C/T | UTR | 3171687 | |
| CREB2-2 | A/G | UTR | 1060142 | ||
| CREB2-3 | G/C | NS-CHNG | 1803323 | PRO/ALA | |
| CREB2-4 | C/G | NS-CHNG | 1803324 | GLU/ASP | |
| CREB2-5 | A/G | NS-CHNG | 2228181 | ILE/VAL | |
| CREB2-6 | A/C | NS-CHNG | 4894 | PRO/GLN | |
| CREB2-7 | G/A | NS-CHNG | 11556099 | ASN/ASP | |
| CREB3 | CREB3-1 | A/C | UTR | 1058797 | |
| CREB3-2 | C/T | NS-CHNG | 11996 | GLN/Stop | |
| CREB3-3 | G/T | INTRON | 2181028 | ||
| CREB3-4 | C/T | INTRON | 2381399 | ||
| CREB3-5 | C/T | INTRON | 10814274 | ||
| CREB3-6 | A/C | INTRON | 12340601 | ||
| CREB3-7 | C/T | INTRON | 12352355 | ||
| CREB3-8 | C/T | INTRON | 12339916 | ||
| CREB3-9 | T/G | 3′ UTR | 1058796 |
UTR: untranslated region; PMT: promoter; NS-CHNG: non-synonymous change
Table 3.
Genotypic and Allelic Frequencies for selected CREB SNPs
| SNP | Allele | Genotype | Status | |||
|---|---|---|---|---|---|---|
|
| ||||||
| Cases | Controls | LiR | LiNR | |||
| CREB1-1H | A | 206 (0.493) | 74 (0.430) | 136 (0.5) | 64 (0.485) | |
| G | 212 (0.507) | 98 (0.570) | 136 (0.5) | 68 (0.515) | ||
| AA | 42 (0.201) | 5 (0.058) | 36 (0.265) | 5 (0.076) | ||
| AG | 122 (0.584) | 64 (0.744) | 64 (0.471) | 54 (0.818) | ||
| GG | 45 (0.215) | 17 (0.198) | 36 (0.265) | 7 (0.106) | ||
|
| ||||||
| CREB1-7H | T | 318 (0.869) | 131 (0.978) | 225 (0.907) | 89 (0.781) | |
| C | 48 (0.131) | 3 (0.022) | 23 (0.093) | 25 (0.219) | ||
| TT | 140 (0.765) | 64 (0.955) | 106 (0.855) | 32 (0.561) | ||
| TC | 38 (0.208) | 3 (0.045) | 13 (0.105) | 25 (0.439) | ||
| CC | 5 (0.027) | 0 (0) | 5 (0.040) | 0 (0) | ||
When performing haplotype analysis in Haploview using the “strong LD Spine” algorithm, with the D′ value set at greater than 0.65, we found three haplotype blocks. Block 1 consisted of markers CREB1-4 and CREB1-20; Block 2 included the CREB1-7H and CREB1-1H SNPs and lastly Block 3 spanned 3 markers; CREB1-2H, CREB1-10 and CREB1-19.
We did not find any association between sex or age of onset with genotypic or allelic presentation (data not shown).
Discussion
It is hypothesized that lithium’s chronic effect may occur through the alteration of transcription factors which are responsible for linking transient changes induced by ligand binding to G-protein coupled receptors to long-term cellular changes. Due to their role in gene expression, transcription factors are ideal candidates to be investigated in pharmacogenetics studies. The majority of studies investigating any of the CREB genes have focused on transcription factor activation or phosphorylation and specifically on genetic variation. Zubenko’s studies (Zubenko et al., 2002; Zubenko et al., 2003) of women with recurrent major depression (MD) were the first to investigate CREB1 genetic variation in the etiology of MD; whilst our study provides an initial look into variation in the CREB family of genes and their possible involvement in the etiology of BD and/or lithium response. Interestingly, although these two disorders do overlap to some extent, namely with respect to relatively early onset and highly recurrent course, there were no cases of BD among the probands or their relatives in the sample studied by Zubenko et al. (2002).
Studies investigating antidepressant response have shown a decreased level of phosphorylated cyclic AMP response element binding protein (pCREB). To activate the transcriptional process (pathway) CREB1 requires phosphorylation on serine 119 or 133, depending on the transcript (Daniel et al., 1998), thereby facilitating the binding of the CREB binding protein (CBP). Sulser et al. (2002) demonstrated that, antidepressants lead to a reduced expression of CREB1, as well as reduced efficacy of CREB1 to initiate transcription; this likely occurs through a decrease in DNA binding capability. Considering that lithium has been shown to produce a similar effect on CREB, decreased phosphorylation resulting in blunting of the cAMP mediated signal transduction pathway (Wang et al., 1999), CREB provides a likely first foray into elucidating the significance of the cAMP signal transduction pathway in lithium response.
The aim of the present study was to identify, among members of the CREB family, genetic variants associated with excellent response to lithium. The CREB family of proteins belong to the leucine zipper family of DNA-binding proteins able to recognize the cAMP responsive element (CRE). CREB1 is located on ch 2q32.3-q34 and exists in one of two isoforms (CREB347/CREB327) differing by a 14bp deletion (Daniel et al., 1998). CREB2 or ATF4 is located on ch 22q13.1 and is a repressor of CRE-dependent transcription (Karpinski et al., 1992). Lastly, there is the CREB3 gene, also referred to as LUMAN, located on ch 9pter-p22.1. Its function has not been completely elucidated, however, it appears that CREB3 behaves as a transcriptional activator similar to CREB1. In total, 40 SNPs were investigated spread over 3 CREB genes with the majority being on CREB1 and CREB3, for association to lithium response in bipolar disorder. Following Bonferroni correction for multiple testing, positive association still remained for the GA genotype (p<0.002) of the CREB1-1H SNP when LiR were compared to controls or LiNR. The TT genotype for CREB1-7H was significantly associated with cases and non-responders at a significance level of p<0.002, as was the case with the T allele as well. These SNPs are of interest due to their being HapMap SNPs as well as having been selected as tag SNPs for the CEPH population.
In recent months there have been several articles published looking at the applicability of HapMap tagSNPs to other populations, as well as to investigate common diseases. The results for tagSNP portability are promising and attest to the usefulness of the HapMap in association studies. In our investigation we looked at several HapMap validated SNPs, as well as tag SNPs in CREB1. However, we did not encounter such promising results due to markers having low heterozygosity and departures from HW disequilibrium. It is likely that the population differences can attest to the low transferability of HapMap SNPs. Even though our population is of European ancestry subjects may not have been assessed in similar fashion as the CEPH population.
When conducting case-control studies one must be wary of the possibility of the occurrence of false-positives resulting from the presence of different populations within the sample being studied. While we have collected individuals at different centres, our sample is of similar ethnic background, as all subjects were Caucasians of European decent. Furthermore, as is commonly the case in genetic studies of complex traits, independent replications are needed to validate our findings. In addition, more variants need to be investigated, particularly for CREB2 and CREB3, to draw more definite conclusions. Also, it would be interesting to genotype variants in genes at other levels of the cAMP signal transduction pathway. These can include protein kinase A (PKA), phosphatases and complementary transcription factors (eg. CREM and CBP).
Table 4.
Estimated Haplotype Frequencies for SNPs on CREB1
| Haplotype | P-Value | Empirical P-Value | Frequency | |
|---|---|---|---|---|
|
| ||||
| Cases | Controls | |||
| 122211112 | 0.00497 | 0.04 | 0.049838 | 0.007692 |
| 211112222 | 0.00006 | 0 | 0.067515 | 0 |
| 211211222 | 0 | 0 | 0.014181 | 0.092136 |
| 211212222 | 0.1495 | 0.13 | 0.121424 | 0.160445 |
| 211222212 | 0.22936 | 0.19 | 0.269107 | 0.312186 |
| All Rare | 0.38487 | 0.023438 | 0.003856 | 0.004914 |
Acknowledgments
Drs. Bernd Ahrens, Anne Berghöfer, Patrizia Cavazzoni, Anne Duffy, Marta Dvoráková, Eva Grof, Anita Holzinger, Eva Libigerová, Rasmus W. Licht, Bruno Müller-Oerlinghausen, Agnetta Nilsson, Helena Prochazka, Niels A. Rasmussen, Mogens Schou, Claudia Schumann, Kenneth Thau, Per Vestergaard, Milos Vojtechovský and Petr Zvolský contributed to the collection of clinical data. The study has been supported by the grant 64410 from the Canadian Institutes of Health Research to MA.
References
- Akiskal HS, Bourgeois ML, Angst J, Post R, Moller H, Hirschfeld R. Reevaluating the prevalence of and diagnostic composition within the broad clinical spectrum of bipolar disorders. J Affect Disord. 2000;59(Suppl 1):S5–S30. doi: 10.1016/s0165-0327(00)00203-2. [DOI] [PubMed] [Google Scholar]
- APA. Diagnostic and Statistical Manual of Mental Disorders (DSM-IV) 4. 1994. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Bezchlibnyk Y, Young LT. The neurobiology of bipolar disorder: focus on signal transduction pathways and the regulation of gene expression. Can J Psychiatry. 2002;47:135–148. doi: 10.1177/070674370204700203. [DOI] [PubMed] [Google Scholar]
- Craddock N, Jones I. Molecular genetics of bipolar disorder. Br J Psychiatry Suppl. 2001;41:s128–s133. [PubMed] [Google Scholar]
- Daniel PB, Walker WH, Habener JF. Cyclic AMP signaling and gene regulation. Annu Rev Nutr. 1998;18:353–383. doi: 10.1146/annurev.nutr.18.1.353. [DOI] [PubMed] [Google Scholar]
- Grof P, Alda M, Grof E, Zvolsky P, Walsh M. Lithium response and genetics of affective disorders. J Affect Disord. 1994;32:85–95. doi: 10.1016/0165-0327(94)90066-3. [DOI] [PubMed] [Google Scholar]
- Karpinski BA, Morle GD, Huggenvik J, Uhler MD, Leiden JM. Molecular cloning of human CREB-2: an ATF/CREB transcription factor that can negatively regulate transcription from the cAMP response element. Proc Natl Acad Sci U S A. 1992;89:4820–4824. doi: 10.1073/pnas.89.11.4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton CR, Graham A, Heptinstall LE, Powell SJ, Summers C, Kalsheker N, Smith JC, Markham AF. Analysis of any point mutation in DNA. The amplification refractory mutation system (ARMS) Nucleic Acids Res. 1989;17:2503–2516. doi: 10.1093/nar/17.7.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. A Laboratory Manual. 1989. Molecular Cloning. [Google Scholar]
- Wang JF, Asghari V, Rockel C, Young LT. Cyclic AMP responsive element binding protein phosphorylation and DNA binding is decreased by chronic lithium but not valproate treatment of SH-SY5Y neuroblastoma cells. Neuroscience. 1999;91:771–776. doi: 10.1016/s0306-4522(98)00627-7. [DOI] [PubMed] [Google Scholar]
- Zubenko GS, Hughes HB, III, Maher BS, Stiffler JS, Zubenko WN, Marazita ML. Genetic linkage of region containing the CREB1 gene to depressive disorders in women from families with recurrent, early-onset, major depression. Am J Med Genet. 2002;114:980–987. doi: 10.1002/ajmg.b.10933. [DOI] [PubMed] [Google Scholar]
- Zubenko GS, Hughes HB, III, Stiffler JS, Brechbiel A, Zubenko WN, Maher BS, Marazita ML. Sequence variations in CREB1 cosegregate with depressive disorders in women. Mol Psychiatry. 2003;8:611–618. doi: 10.1038/sj.mp.4001354. [DOI] [PubMed] [Google Scholar]