Abstract
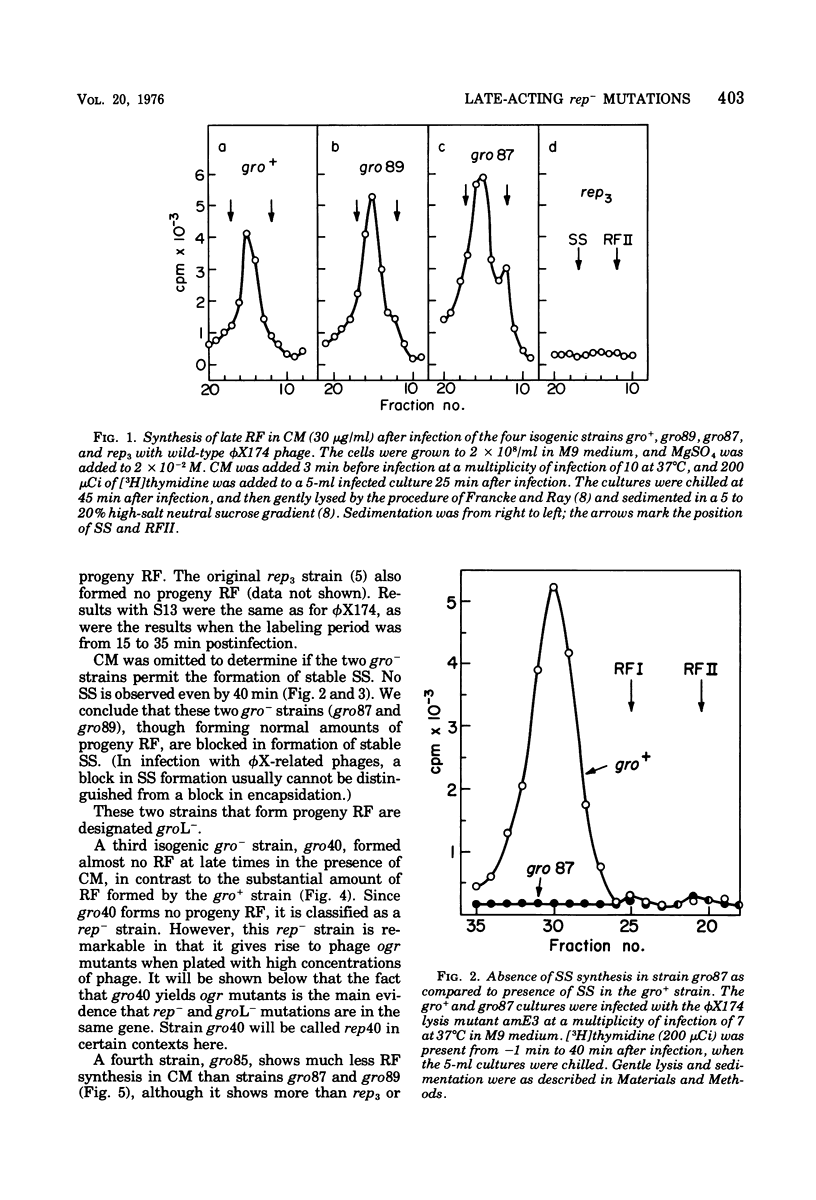
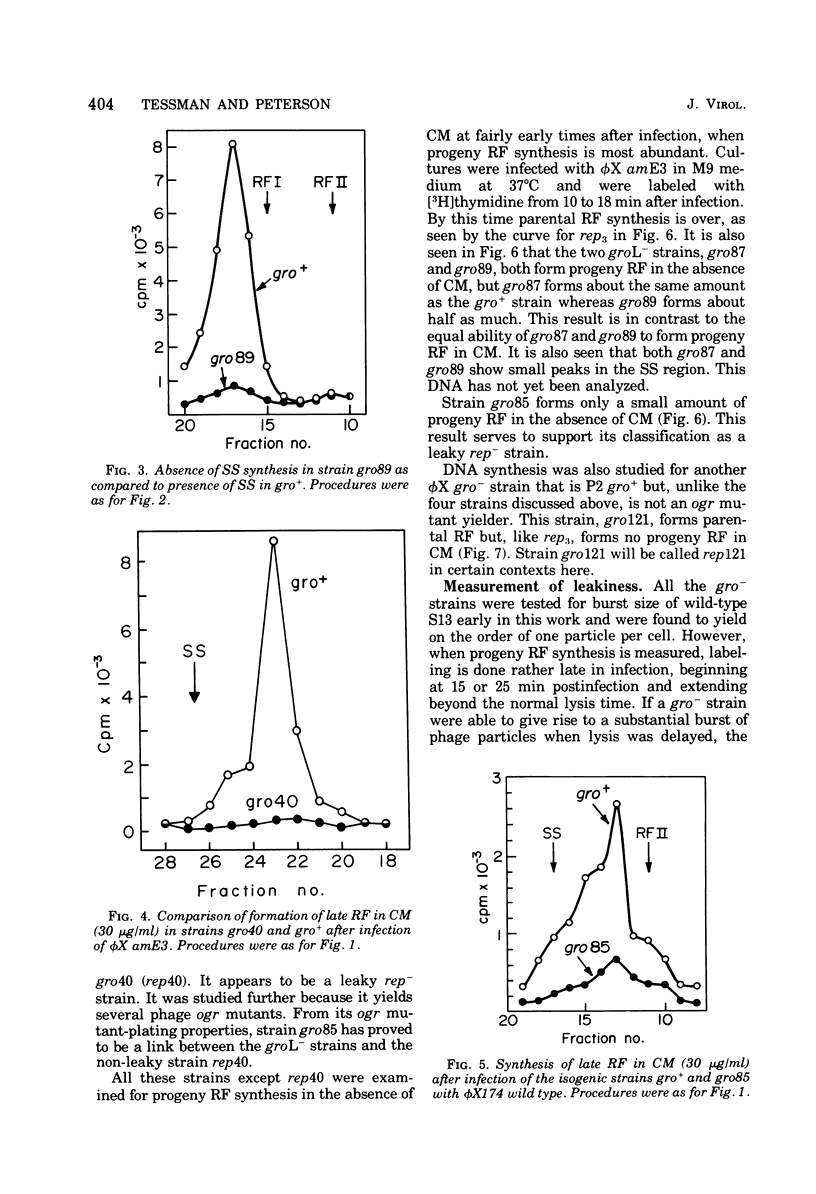
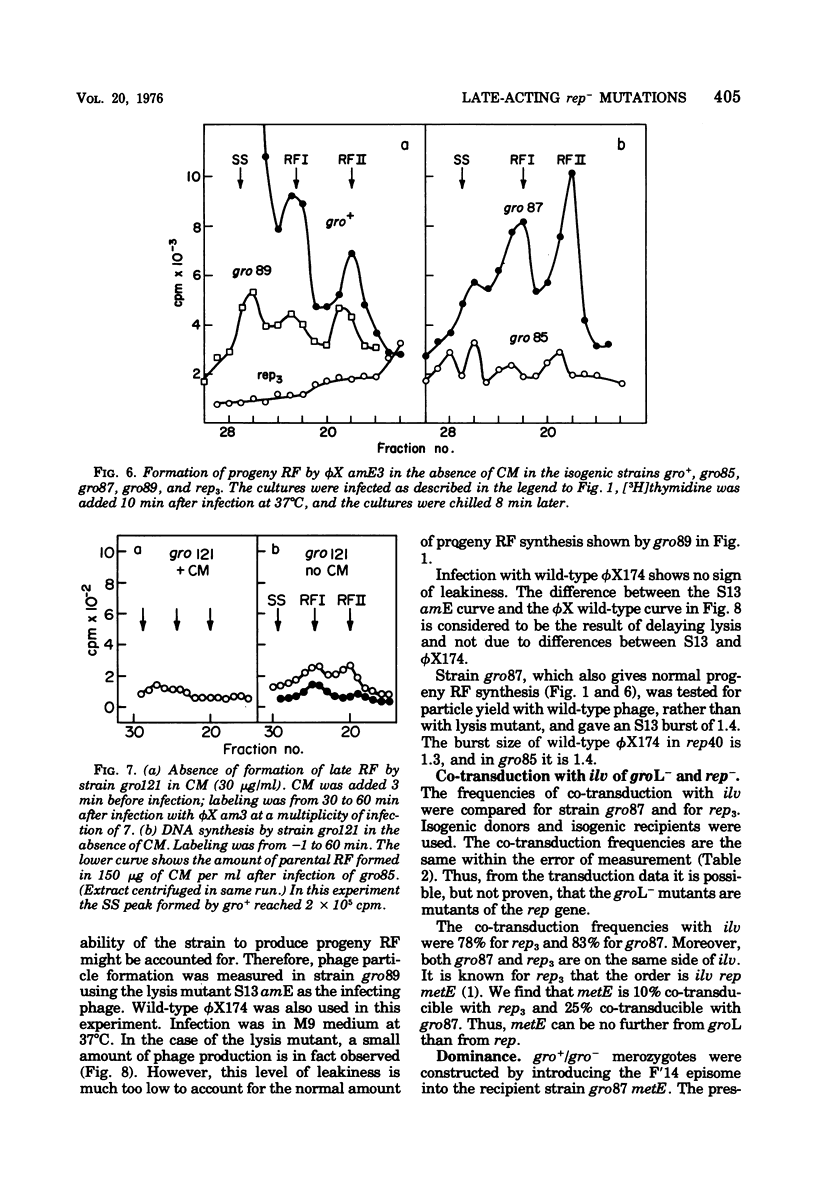
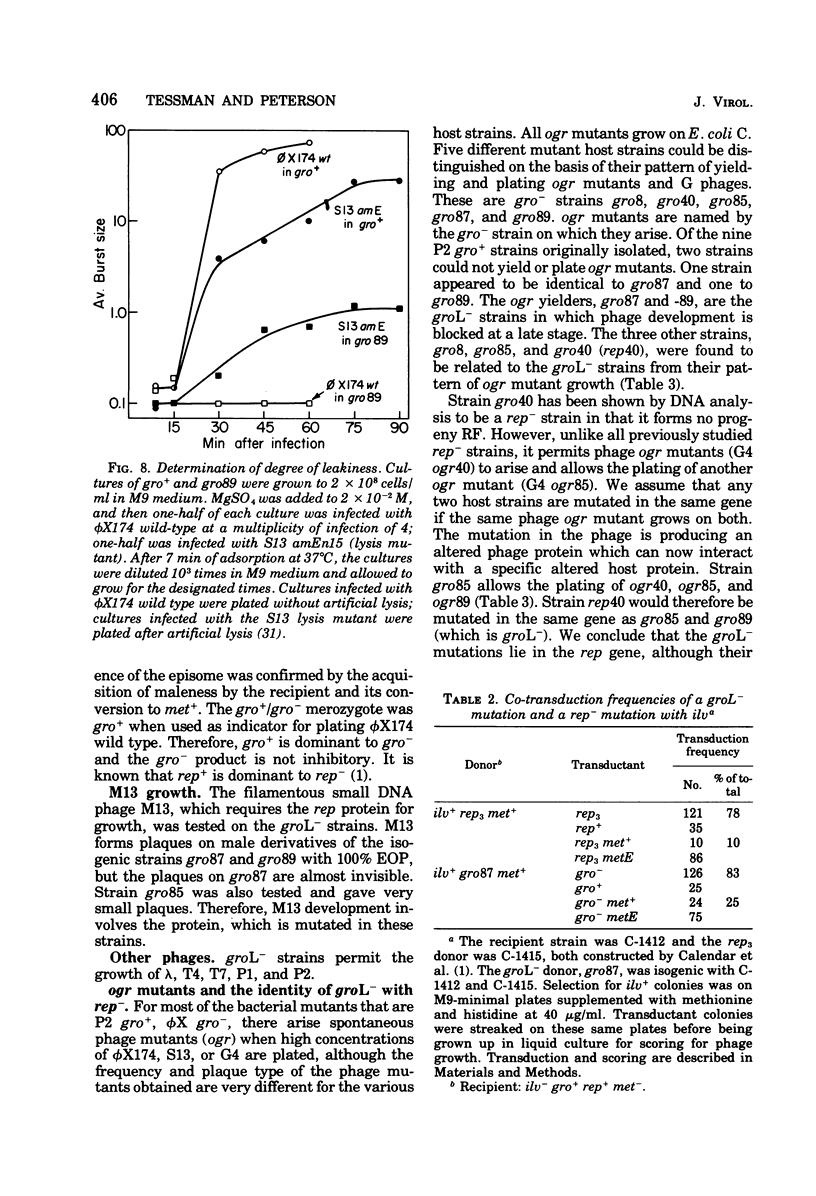
Several related mutants of Escherichia coli C have been isolated that block the growth of the small icosahedral DNA phages phiX174 and S13 late in infection. Phage G6 is also blocked, at a stage not yet known. Growth of the filamentous phage M13, though not blocked, is affected in these strains. These host mutations co-transduce with ilv at high frequency, as do rep- mutations. However, the new mutants, designated groL-, differ from previously studied rep- mutants in that they permit synthesis of progeny replicative-form DNA. The groL- mutants are blocked in synthesis of stable single-stranded DNA of phiX174 and related phages. They are gro+ for P2. Evidence that groL- mutations and rep- mutations are in the same gene is presented. Spontaneous mutants (ogr) of phiX174, S13, and the G phages can grow on groL- strains. The ogr mutations are located in the phage's major capsid gene, F, as determined by complementation tests. There are numerous sites for mutation to ogr. Some mutations in genes A and F interfere with the ogr property when combined with an ogr mutation on the same genome. The ogr mutations are cis acting in a groL- cell; i.e., an ogr mutant gives very poor rescue of a non-ogr mutant. The wild-type form of each G phage appears to be naturally in the ogr mutant state for one or more groL- strains. It is suggested that a complex between F and rep proteins is involved in phage maturation. The A protein appears to interact with this complex.
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Calendar R., Lindqvist B., Sironi G., Clark A. J. Characterization of REP- mutants and their interaction with P2 phage. Virology. 1970 Jan;40(1):72–83. doi: 10.1016/0042-6822(70)90380-6. [DOI] [PubMed] [Google Scholar]
- Casjens S., King J. Virus assembly. Annu Rev Biochem. 1975;44:555–611. doi: 10.1146/annurev.bi.44.070175.003011. [DOI] [PubMed] [Google Scholar]
- Chamberlin M. Isolation and characterization of prototrophic mutants of Escherichia coli unable to support the intracellular growth of T7. J Virol. 1974 Sep;14(3):509–516. doi: 10.1128/jvi.14.3.509-516.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denhardt D. T., Dressler D. H., Hathaway A. THE ABORTIVE REPLICATION OF PhiX174 DNA IN A RECOMBINATION-DEFICIENT MUTANT OF Escherichia coli. Proc Natl Acad Sci U S A. 1967 Mar;57(3):813–820. doi: 10.1073/pnas.57.3.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denhardt D. T. The single-stranded DNA phages. CRC Crit Rev Microbiol. 1975 Dec;4(2):161–223. doi: 10.3109/10408417509111575. [DOI] [PubMed] [Google Scholar]
- Dowell C. E., Sinsheimer R. L. The process of infection with bacteriophage phi-X174. IX. Studies on the physiology of three phi-X174 temperature-sensitive mutants. J Mol Biol. 1966 Apr;16(2):374–386. doi: 10.1016/s0022-2836(66)80180-8. [DOI] [PubMed] [Google Scholar]
- Farber M. B. Purification and properties of bacteriophage phi X 174 gene D product. J Virol. 1976 Mar;17(3):1027–1037. doi: 10.1128/jvi.17.3.1027-1037.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francke B., Ray D. S. Cis-limited action of the gene-A product of bacteriophage phiX174 and the essential bacterial site (E. coli-electron microscopy-cis-acting protein-specifically-nicked RF). Proc Natl Acad Sci U S A. 1972 Feb;69(2):475–479. doi: 10.1073/pnas.69.2.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francke B., Ray D. S. Formation of the parental replicative form DNA of bacteriophage phi-X174 and initial events in its replication. J Mol Biol. 1971 Nov 14;61(3):565–586. doi: 10.1016/0022-2836(71)90065-9. [DOI] [PubMed] [Google Scholar]
- Georgopoulos C. P. Bacterial mutants in which the gene N function of bacteriophage lambda is blocked have an altered RNA polymerase. Proc Natl Acad Sci U S A. 1971 Dec;68(12):2977–2981. doi: 10.1073/pnas.68.12.2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgopoulos C. P., Hendrix R. W., Kaiser A. D., Wood W. B. Role of the host cell in bacteriophage morphogenesis: effects of a bacterial mutation on T4 head assembly. Nat New Biol. 1972 Sep 13;239(89):38–41. doi: 10.1038/newbio239038a0. [DOI] [PubMed] [Google Scholar]
- Ghysen A., Pironio M. Relationship between the N function of bacteriophage lambda and host RNA polymerase. J Mol Biol. 1972 Mar 28;65(2):259–272. doi: 10.1016/0022-2836(72)90281-1. [DOI] [PubMed] [Google Scholar]
- Godson G. N. Evolution of phi-chi 174. Isolation of four new phi-chi-like phages and comparison with phi-chi 174. Virology. 1974 Mar;58(1):272–289. doi: 10.1016/0042-6822(74)90161-5. [DOI] [PubMed] [Google Scholar]
- Goldberg R. B., Bender R. A., Streicher S. L. Direct selection for P1-sensitive mutants of enteric bacteria. J Bacteriol. 1974 Jun;118(3):810–814. doi: 10.1128/jb.118.3.810-814.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwaya M., Denhardt D. T. The mechanism of replication of phi X174 single-stranded DNA. II. The role of viral proteins. J Mol Biol. 1971 Apr 28;57(2):159–175. doi: 10.1016/0022-2836(71)90339-1. [DOI] [PubMed] [Google Scholar]
- Jazwinski S. M., Lindberg A. A., Kornberg A. The gene H spike protein of bacteriophages phiX174 and S13. I. Functions in phage-receptor recognition and in transfection. Virology. 1975 Jul;66(1):283–293. doi: 10.1016/0042-6822(75)90198-1. [DOI] [PubMed] [Google Scholar]
- Jazwinski S. M., Marco R., Kornberg A. The gene H spike protein of bacteriophages phiX174 and S13. II. Relation to synthesis of the parenteral replicative form. Virology. 1975 Jul;66(1):294–305. doi: 10.1016/0042-6822(75)90199-3. [DOI] [PubMed] [Google Scholar]
- Jeng Y., Gelfand D., Hayashi M., Shleser R., Tessman E. S. The eight genes of bacteriophages phi X174 and S13 and comparison of the phage-specified proteins. J Mol Biol. 1970 Apr 28;49(2):521–526. doi: 10.1016/0022-2836(70)90262-7. [DOI] [PubMed] [Google Scholar]
- Lane H. E., Denhardt D. T. The rep mutation. III. Altered structure of the replicating Escherichia coli chromosome. J Bacteriol. 1974 Nov;120(2):805–814. doi: 10.1128/jb.120.2.805-814.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane H. E., Denhardt D. T. The rep mutation. IV. Slower movement of replication forks in Escherichia coli rep strains. J Mol Biol. 1975 Sep 5;97(1):99–112. doi: 10.1016/s0022-2836(75)80025-8. [DOI] [PubMed] [Google Scholar]
- Lindqvist B. H., Sinsheimer R. L. The process of infection with bacteriophage phi-X174. XV. Bacteriophage DNA synthesis in abortive infections with a set of conditional lethal mutants. J Mol Biol. 1967 Nov 28;30(1):69–80. doi: 10.1016/0022-2836(67)90244-6. [DOI] [PubMed] [Google Scholar]
- Luria S E. Mutations of Bacterial Viruses Affecting Their Host Range. Genetics. 1945 Jan;30(1):84–99. doi: 10.1093/genetics/30.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D. F., Godson G. N. Identification of a phiX174 coded protein involved in the shut-off of host DNA replication. Biochem Biophys Res Commun. 1975 Jul 8;65(1):323–330. doi: 10.1016/s0006-291x(75)80096-9. [DOI] [PubMed] [Google Scholar]
- Rosner J. L. Formation, induction, and curing of bacteriophage P1 lysogens. Virology. 1972 Jun;48(3):679–689. doi: 10.1016/0042-6822(72)90152-3. [DOI] [PubMed] [Google Scholar]
- SINSHEIMER R. L., STARMAN B., NAGLER C., GUTHRIE S. The process of infection with bacteriophage phi-XI74. I. Evidence for a "replicative form". J Mol Biol. 1962 Mar;4:142–160. doi: 10.1016/s0022-2836(62)80047-3. [DOI] [PubMed] [Google Scholar]
- Salstrom J. S., Pratt D. Role of coliphage M13 gene 5 in single-stranded DNA production. J Mol Biol. 1971 Nov 14;61(3):489–501. doi: 10.1016/0022-2836(71)90061-1. [DOI] [PubMed] [Google Scholar]
- Siden E. J., Hayashi M. Role of the gene beta-product in bacteriophage phi-X174 development. J Mol Biol. 1974 Oct 15;89(1):1–16. doi: 10.1016/0022-2836(74)90159-4. [DOI] [PubMed] [Google Scholar]
- Siegel J. E., Hayashi M. Phi-X-174 bacteriophage structural mutants which affect deoxyribonucleic acid synthesis. J Virol. 1969 Oct;4(4):400–407. doi: 10.1128/jvi.4.4.400-407.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunshine M. G., Sauer B. A bacterial mutation blocking P2 phage late gene expression. Proc Natl Acad Sci U S A. 1975 Jul;72(7):2770–2774. doi: 10.1073/pnas.72.7.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TESSMAN E. S. COMPLEMENTATION GROUPS IN PHAGE S13. Virology. 1965 Feb;25:303–321. doi: 10.1016/0042-6822(65)90208-4. [DOI] [PubMed] [Google Scholar]
- Takahashi H., Coppo A., Manzi A., Martire G., Pulitzer J. F. Design of a system of conditional lethal mutations (tab/k/com) affecting protein-protein interactions in bacteriophage T4-infected Escherichia coli. J Mol Biol. 1975 Aug 25;96(4):563–578. doi: 10.1016/0022-2836(75)90139-4. [DOI] [PubMed] [Google Scholar]
- Tessman E. S. Mutants of bacteriophage S13 blocked in infectious DNA synthesis. J Mol Biol. 1966 May;17(1):218–236. doi: 10.1016/s0022-2836(66)80104-3. [DOI] [PubMed] [Google Scholar]
- Tessman I., Ishiwa H., Kumar S., Baker R. Bacteriophage S13: a 7th gene. Science. 1967 May 12;156(3776):824–825. doi: 10.1126/science.156.3776.824. [DOI] [PubMed] [Google Scholar]
- Tonegawa S., Hayashi M. Intermediates in the assembly of phi X174. J Mol Biol. 1970 Mar 14;48(2):219–242. doi: 10.1016/0022-2836(70)90158-0. [DOI] [PubMed] [Google Scholar]
- Weisbeek P. J., Sinsheimer R. L. A DNA-protein complex involved in bacteriophage phi chi 174 particle formation. Proc Natl Acad Sci U S A. 1974 Aug;71(8):3054–3058. doi: 10.1073/pnas.71.8.3054. [DOI] [PMC free article] [PubMed] [Google Scholar]


