Abstract
Identification of ethylene-regulated and ripening-related genes from banana (Musa acuminata Var. Harichaal) fruits using DDRT-PCR led to the isolation of differentially expressed partial cDNA of pectin methylesterase inhibitor (MaPMEI) gene. Its full-length cDNA sequence consisted of a 567 bp ORF, encoding a protein of 189 aa with deduced molecular mass 19.6 kDa. Expression pattern of MaPMEI gene revealed that upon ethylene treatment, this gene is up-regulated initially giving maximum expression in post-climacteric stage then decreases slightly in later stages of ripening. 1-MCP, a known ethylene perception inhibitor, inhibits both fruit ripening as well as the transcript level of this gene. Also, the transcripts of MaPMEI gene were not detected during the short time ethylene treatment suggesting this gene appears to be not directly induced by ethylene. Interestingly, MaPMEI gene showed fruit specific expression that indicates its possible role in the regulations of PMEs in fruits. In silico analysis revealed a predicted signal peptide sequence necessary for localization of MaPMEI in the cell wall. Furthermore, the four Cys residues involved in disulfide bridges are conserved in MaPMEI similar to other PMEIs and invertase inhibitors. Phylogenetic analysis further suggests that the MaPMEI identified in this study is more closely related to PMEIs than to invertase inhibitors.
Keywords: Banana, Fruit ripening, Pectin methylesterase inhibitor, Ethylene responsive expression, DDRT-PCR
Introduction
Ripening in banana is a highly programmed developmental event during which fruit undergoes through various physiological and biochemical changes that include conversion of starch to sugars and changes in color, flavor, aroma, texture and many more like in other climacteric fruits (Gupta et al. 2006; Srivastava and Gupta 2008). Among these, softening is a very important physiological change that attributes economic importance to the fruit as excessive softening renders the fruit inedible. Furthermore, once ripening process has been initiated in banana fruits it could not be controlled efficiently by modern storage facility and chemical sprays. Therefore, molecular genetic approach could be a better and attainable approach to control the rate of ripening and increase the shelf-life of banana fruits (Gupta et al. 2008).
Fruit softening is regulated by several carbohydrate-modifying enzymes like Endo-(1→4)β-D-glucanse, Xyloglucan endotransglucosylase hydrolase (XET), Expansin, Pectin Methyl Esterase (PME), Pectate Lyase (PL) and Polygalacturonase (PG) in plants (Lohani et al. 2004). Among these, PME (EC 3.1.1.11) plays a very significant role in the early stages of softening and is key control point for the assembly and disassembly of the pectic network of the fruit (Carpita and Gibeaut 1993). It de-esterifies polyuronides of the polygalacturonic acid (PGA) and makes the polyuronide susceptible for further degradation by other cell wall hydrolases like PG (Lohani et al. 2004). Activity of PME is regulated by mechanisms such as differential expression of isoforms at transcriptional level and formation of complex with inhibitory proteins at post-translational level (Balestrieri et al. 1990; Camardella et al. 2000; Jolie et al. 2010; Vandevenne et al. 2011). Among these, PME/Invertase inhibitor has been shown to regulate the activity of PME at post-translational level by binding to these proteins in a non-covalent 1:1 manner (Giovane et al. 2004; Jolie et al. 2009). The inhibitors of PME and Invertase are unique as they share very similar sequence and topology. These protein contain five Cys residues, four of which are conserved and connected by disulfide bridges, first to second and third to fourth. A cDNA encoding putative PME/Invertase inhibitor was identified from banana while isolation of differentially expressing genes during ripening by mRNA Differential Display Reverse Transcription-Polymerase Chain Reaction (DDRT-PCR) technology (Gupta et al. 2006). Based on the sequence and phylogenetic analysis, the putative PME/Invertase inhibitor obtained in the present study appears to be a pectin methylesterase inhibitor (PMEI) and thus it was christened as MaPMEI. The PMEIs and their inhibitory activity have also been reported from different plants (Di Matteo et al. 2005; Hao et al. 2008; Hong et al. 2010; Irifune et al. 2004; Wu et al. 2002). The production of transgenic crops containing PMEI genes is also an attractive possibility for inactivating endogenous PME activity (Irifune et al. 2004; Jiang et al. 2002). In addition, the application of PMEI protein to food technology is potentially interesting because PME causes many commercially deleterious effects such as phase separation during juice production and changes in tissue firmness during food processing (Castaldo et al. 1991). Apart from having its importance in food industry, biotechnological manipulation of PMEI in planta is likely to address the issue of over-softening owing to its ability to control the level of softening promoting PMEs. We report here the isolation and characterization of the first PMEI cDNA from banana.
Materials and methods
Plant material, treatments and northern blot preparation
Mature green bananas (Musa acuminata var. Harichaal, genome AAA type) were purchased from a local farm and treated with ethylene (100 μl/L) for 24 h and 1-methyl cyclopropene (1-MCP) (30 μl/L) for 12 h and tissues were harvested and stored at −70 °C as described in our earlier reports (Gupta et al. 2006). RNA was isolated from different fruit samples as described by Asif et al. (2000). Northern blots were prepared as described by Sambrook et al. (1989). DNA fragments were labeled by random priming using α-32PdCTP as the radiolabel and hybridizations were performed at 42 °C in a formamide based hybridization buffer as described by Sambrook et al. (1989). Blots were exposed to Kodak XOMAT X-ray film and stored at −70 °C for 1 to 5 days depending on signal intensity.
Isolation, sequencing and extension of MaPMEI
Differential display reverse transcription PCR (DDRT-PCR) was carried out as described by Liang and Pardee (1992) with slight modification as described by Gupta et al. (2006) using single base anchor primers AAGC(T)11A, AAGC(T)11 C and AAGC(T)11 G in combination with Gene Hunter primers (Gene Hunter Inc, USA). The cDNA fragment of PMEI gene obtained by DDRT-PCR was reamplified and cloned in pBluescriptII SK + (Stratagene, USA). Cloned DNA fragments were sequenced on an automated DNA sequencer (ABI 373A, Applied Biosystems Inc., USA) using the dye terminator cycle sequencing kit (ABI). Based on the sequences obtained, gene specific reverse primers for 5’RACE were designed. The cDNA for 5’RACE was generated using RNA from 2 day ethylene treated samples using the SMART 5’RACE kit (Clontech, USA). Amplified fragment was cloned in pTZ57R using the InsT/Aclone kit from MBI Fermentas and sequenced. Based on the sequence of 5’RACE fragment and fragment obtained by DDRT-PCR, respective extreme forward (MaPMEIFor: 5′ ATGGGAAGATTCACTTCCGTCTTCCTC 3′) and extreme reverse primers (MaPMEIRev: 5′ TCAACCCAGTCGAACGGCGATGGC 3′) were designed to isolate full-length cDNA of PMEI gene using banana cDNA library (ripe fruit) by PCR. Sequence analysis was carried out on NCBI database by using BLAST-x,-n, and -p algorithms. The phylogenetic analysis of sequences was performed using the maximum likelihood (ML), maximum parsimony (MP) and Neighbour-Joining (NJ) methods. Validation of contigs grouping in tree, calculations of the distance matrix and construction of phylogenetic trees with bootstrap tests was performed with MEGA 5 software (Tamura et al. 2011).
Results and discussion
Isolation and sequence analysis of full-length MaPMEI gene
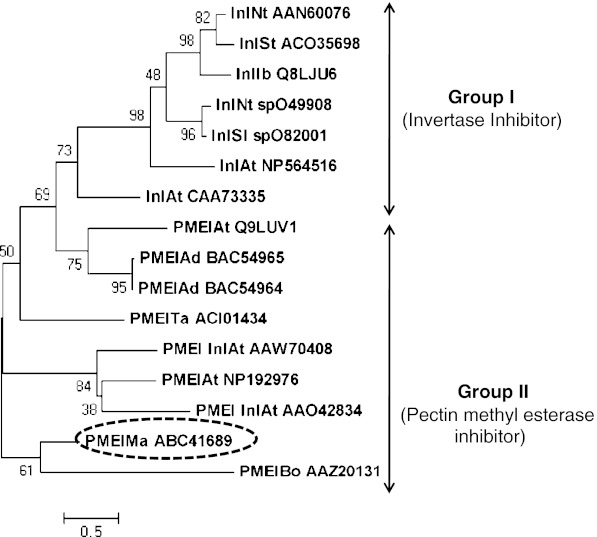
A cDNA fragment (250 bp) was obtained from DDRT-PCR, which showed homology to PME/invertase inhibitor proteins towards their 3′ region (Gupta et al. 2006). Gene specific reverse primers were designed to isolate the 5′ upstream region of putative MaPMEI and the 5′RACE yielded an amplicon of 550 bp. Assembly of these two sequences revealed that the cDNA of MaPMEI is comprised of 567 bp ORF region, 45 and 191 bp of 5′ and 3′ UTR regions, respectively. MaPMEI encodes a protein (ABC41689) of 189 aa with deduced molecular mass 19.6 kDa. The assembled putative full-length PME/Invertase inhibitor was validated to be part of one cDNA by isolating and sequencing the cDNA obtained by PCR using MaPMEIFor and MaPMEIRev primers. Analysis using neural networks and hidden Markov models hosted at SignalP server suggested that the ORF of MaPMEI comprised of an N-terminal 26 amino acid residues of a signal peptide. This region corresponds with the positions in other PMEIs, which is required for the extracellular targeting of the protein at the site of its function (Giovane et al. 2004). Protein sequence alignment and subsequent in silico analyses revealed the presence of all the four conserved cystine residues corresponding to other PMEIs and invertase inhibitors from other sources (Fig. 1). Based on the sequence analysis, the putative inhibitor obtained in the present study appears to be a PMEI and thus it was christened as MaPMEI. In order to see the relationship of MaPMEI (ABC41689) with other reported Invertase--and PME inhibitors, phylogenetic analysis was performed. The sequence of MaPMEI was compared with functionally characterized different members of Invertase--and PME inhibitors from other plants and molecular phylogenetic trees was constructed by using maximum parsimony, distance and maximum likelihood method (data not shown). All trees obtained by different methods showed more or less similar topology. Grouping of MaPMEI with other related proteins in Neighbour Joining (NJ) tree were consistent with similarity in sequences obtained by BLAST analysis. MaPMEI was grouped mainly with other PMEIs from different sources (Fig. 2). The analysis indicates that MaPMEI was likely to be the enzyme playing a role in regulating the activity of PMEs in banana during ripening. However, the functionality of the enzyme needs to be verified by characterizing the protein biochemically.
Fig. 1.

Alignment of the deduced amino acid sequences of MaPMEI (ABC41689) with other reported PMEIs and invertase inhibitors. PMEIs from M. acuminata (ABC41689), T. aestivum (ACI01434), B. oleracea (AAZ20131), A. deliciosa (BAC54965) and invertase inhibitors from Z. mays (NP001151081), N. tabacum (AAN60076), S. tuberosum (ACO35698), I. batatas (spQ8LJU6), S. lycopersicum (spO82001) and A. thaliana (NP564516) were taken for alignment using ClustalW program. Universally conserved Cys residues within the members of PME/Invertase inhibitors are highlighted in grey. Region from residue 1 up to black arrow shows putative signal peptide
Fig. 2.
Phylogram of functionally characterized members of PMEIs and Invertase inhibitors including the putative MaPMEI (ABC41689) from banana. PMEIs from M. acuminata (ABC41689), T. aestivum (ACI01434), B. oleracea (AAZ20131), A. deliciosa (BAC54964), A. deliciosa (BAC54965), A thaliana (NP192976, Q9LUV1, AA042834, AAW70408) and Invertase inhibitors from N. tabacum (spO49908, AAN60076), S. lycopersicum (spO82001), I. batatas (spQ8LJU6), S. tuberosum (ACO35698) and A. thaliana (NP564516, CAA73335) were taken to construct phylogenetic tree. MaPMEI was grouped with PMEIs, which is shown in dotted circle. Phylogenetic analyses were conducted in MEGA 5 software (Tamura et al. 2011)
Expression analysis of MaPMEI during ripening
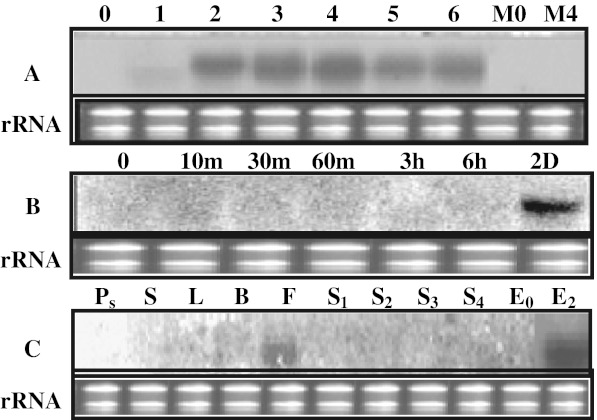
As mentioned earlier, partial cDNA of MaPMEI was obtained from ripe stage of banana, we undertook the expression analysis using northern blot analysis in order to establish that the MaPMEI gene is ripening related. The expression patterns were monitored during the entire course of ripening as well as in fruit in which ripening was blocked by 1-MCP (an ethylene perception inhibitor). Expression pattern of this gene revealed that upon ethylene treatment, this gene is up regulated initially giving maximum expression in post climacteric stage then decreases slightly in later stage of ripening. As shown in Fig. 3a, transcripts of PMEI gene starts accumulating right from day 1, reaches maximum on 4th day and maintained until day 6. The PMEI inhibitor protein has also been reported to be up-regulated during ripening in kiwifruit (Balestrieri et al. 1990; Jolie et al 2009; Vandevenne et al. 2011). Apart from the transcriptional control, the PME-PMEI complex has also been shown at structural level (Di Matteo et al. 2005; Hao et al. 2008; Hothorn et al. 2004) for the regulation of PMEs. Furthermore, PMEIs have been shown to have a broad specificity towards different PME isoforms (Wu et al. 2002). These PME isoforms are encoded by a family of genes in dicot plants (Giovane et al. 1994), which may be expressed in constitutive or in differential manner depending upon the developmental stage and specific tissue (Gaffe et al.1997). Furthermore, ripe fruits are more susceptible to phytopathogenic fungi and bacteria that also produce PME isoforms (Giovane et al. 2004), higher expression of MaPMEI even at later stages of ripening, may also be playing a role in controlling the non-host PMEs. Although, increase in PME activity during ripening in banana (Lohani et al. 2004) and high level of continued expression of MaPMEI specifically in fruit strongly suggests the regulation of PMEs also occurs at post-translational level, however, the exact mechanism and level of specificity for PME isoforms needs to be demonstrated using the biochemical and structural biology approach. In 1-MCP (ethylene perception inhibitor) treated fruit, transcripts of MaPMEI were strongly repressed indicating that this gene is ripening related and the expression was either directly or indirectly dependent on ethylene, as the 1-MCP treated fruit did not get ripe (Fig. 3a). In order to distinguish between direct and indirect ethylene regulation we treated mature green fruit with ethylene for a short duration (10 min–6 h) and RNA from these samples was tested for gene expression. Since ripening has not progressed much in these samples, in such short duration, this treatment allowed us to study any expression that is governed primarily by ethylene and not by secondary ripening related changes. As shown in Fig. 3b, the transcripts of MaPMEI gene were not detected upto 6 h which, strongly suggested that MaPMEI appears not to be directly induced by ethylene. In order to study the tissue specific expression of the gene, northern analyses were performed in different developing stages of the unripe fruit and in different tissues such as pseudo stem (Ps), stem (S), leaves (L), flowers (F) and bract (B). RNA from 2 days ethylene treated banana samples (E2) was used as control. As shown in Fig. 3c, MaPMEI gene was strongly expressed in ripe fruit and to a lower extent in flower, which indicates that MaPMEI is a ripening related gene having mainly fruit specific expression.
Fig. 3.
Expression studies of MaPMEI gene. a Northern blot analysis during different stages of ethylene induced ripening banana (0–6 days post ethylene treatment) and in fruits treated with 1-MCP. Numbers indicate days after ethylene treatment. M0—mature green unripe fruits treated with 30 μl/L 1-MCP for 12 h but no ethylene; M4—fruits samples collected 4 days after treatment with 1-MCP followed by 24 h ethylene treatment. b Northern blot analysis after a short ethylene treatment of mature green bananas. Fruits were treated with 100 ppm ethylene and samples collected 10 min, 30 min, 60 min, 3 h and 6 h after ethylene treatment. The 2 day ethylene treated fruit sample RNA was loaded as a control. c Northern blot analysis in different tissues such as pseudo stem (Ps), stem (S), bract (B), leaf (L) and flowers (F) and in different developing stages of unripe banana. Stages 1 to 4 (S1, S2, S3 and S4) represent developing fruits from a hand in order of increasing size. RNA from ethylene untreated (E0) and 2 days after ethylene treatment (E2) of bananas were loaded as controls of gene expression
To conclude, the present data strongly suggest that, ripening related MaPMEI gene might be regulating the activity of PMEs at post translational level during ripening in banana. However, the functionality of the gene needs to be verified by characterizing the protein biochemically. The MaPMEI over-expression studies might be an ideal approach not only to control the over fruit softening but also likely to extend banana shelf-life. In addition, characterization of MaPMEI cis-elements is likely to provide information on fruit specific expression that is distinct from fruit ripening related expression because expression profile suggests that its promoter might be endowed with elements that confers fruit specific and ripening related expression.
Acknowledgement
Authors are thankful to Department of Biotechnology (DBT), Government of India for funding the project and Council of Scientific and Industrial Research (CSIR), India for providing the Senior Research Fellowship to SS. Director, National Botanical Research Institute (NBRI) is also acknowledged for providing research facilities to carrying out the present work.
References
- Asif MH, Dhawan P, Nath P. A simple procedure for the isolation of high quality RNA from ripening banana fruit. Plant Mol Biol Rep. 2000;18:109–115. doi: 10.1007/BF02824018. [DOI] [Google Scholar]
- Balestrieri C, Castaldo D, Giovane A, Quagliuolo L, Servillo L. A glycoprotein inhibitor of pectin methylesterase in kiwi fruit. Eur J Biochem. 1990;193:183–187. doi: 10.1111/j.1432-1033.1990.tb19321.x. [DOI] [PubMed] [Google Scholar]
- Camardella L, Carratore V, Ciardiello MA, Servilo L, Balestrieri C, Giovane A. Kiwi protein inhibitor of pectin methylesterase. Eur J Biochem. 2000;267:4561–4565. doi: 10.1046/j.1432-1327.2000.01510.x. [DOI] [PubMed] [Google Scholar]
- Carpita NC, Gibeaut DM. Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. Plant J. 1993;3:1–30. doi: 10.1111/j.1365-313X.1993.tb00007.x. [DOI] [PubMed] [Google Scholar]
- Castaldo D, Loboi A, Quagliuolo L, Servillo L, Balestrieri C, Giovane A. Orange juices and concentrates stabilization by a protein inhibitor of pectin methylesterase. J Food Sci. 1991;56:1632–1634. doi: 10.1111/j.1365-2621.1991.tb08658.x. [DOI] [Google Scholar]
- Di Matteo A, Giovane A, Raiola A, Camardella L, Bonivento D, De Lorenzo G, Cervone F, Bellincampi D, Tsernoglou D. Structural basis for the interaction between pectin methyl esterase and a specific inhibitor protein. Plant Cell. 2005;17:849–858. doi: 10.1105/tpc.104.028886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffe J, Tiznado ME, Handa AK. Characterization and functional expression of a ubiquitously expressed tomato pectin methylesterase. Plant Physiol. 1997;114:1547–1556. doi: 10.1104/pp.114.4.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovane A, Servillo L, Balestrieri C, Quagliuolo L, La Ratta B, Loiudice R, Castaldo D. Purification and characterization of three forms of pectin methylesterase from tomato fruit. J Food Biochem. 1994;17:339–341. doi: 10.1111/j.1745-4514.1993.tb00478.x. [DOI] [Google Scholar]
- Giovane A, Servillo L, Balestrieri C, Raiola A, D’Avino R, Tamburrini M, Ciardiello MA, Camardella L. Pectin methylesterase inhibitor. Biochim Biophys Acta. 2004;1696:245–252. doi: 10.1016/j.bbapap.2003.08.011. [DOI] [PubMed] [Google Scholar]
- Gupta SM, Srivastava S, Sane AP, Nath P. Differential expression of genes during banana fruit development, ripening and 1-MCP treatment: presence of distinct fruit specific, ethylene induced and repressed expression. Postharvest Biol Technol. 2006;42:16–22. doi: 10.1016/j.postharvbio.2006.05.002. [DOI] [Google Scholar]
- Gupta SM, Srivastava S, Gupta S, Ahmed Z. Genetic manipulation of fruit ripening: using Antisense mRNA strategies. J Appl Biosci. 2008;34:115–123. [Google Scholar]
- Hao Y, Huang X, Mei X, Li R, Zhai Z, Yin S, Huang Y, Luo Y. Expression, purification and characterization of pectin methylesterase inhibitor from kiwi fruit in Escherichia coli. Protein Expr Purif. 2008;60:221–224. doi: 10.1016/j.pep.2008.04.004. [DOI] [PubMed] [Google Scholar]
- Hong MJ, Kim DY, Lee TG, Jeon WB, Seo YW. Functional characterization of pectin methyl esterase inhibitor (PMEI) in wheat. Genes Genet Syst. 2010;85:97–106. doi: 10.1266/ggs.85.97. [DOI] [PubMed] [Google Scholar]
- Hothorn M, Wolf S, Aloy P, Greiner S, Scheffzek K. Structural insights into the target specificity of plant invertase and pectin methylesterase inhibitory proteins. Plant Cell. 2004;16:3437–3447. doi: 10.1105/tpc.104.025684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irifune K, Nishida T, Egawa H, Nagatani A. Pectin methylesterase inhibitor cDNA from kiwi fruit. Plant Cell Rep. 2004;23:333–338. doi: 10.1007/s00299-004-0835-6. [DOI] [PubMed] [Google Scholar]
- Jiang CM, Li CP, Chang JC, Chang HM. Characterization of pectinesterase inhibitor in jelly fig (Ficus awkeotsang Makino) achenes. J Agric Food Chem. 2002;50:4890–4894. doi: 10.1021/jf011568+. [DOI] [PubMed] [Google Scholar]
- Jolie RP, Duvetter T, Houben K, Clynen E, Sila DN, Van Loey AM, Hendrickx ME. Carrot pectin methylesterase and its inhibitor from kiwi fruit: Study of activity, stability and inhibition. Innov Food Sci Emerg Technol. 2009;10:601–609. doi: 10.1016/j.ifset.2009.02.003. [DOI] [Google Scholar]
- Jolie RP, Duvetter T, Van Loey AM, Hendrickx ME. Pectin methylesterase and its proteinaceous inhibitor: a review. Carbohydr Res. 2010;345:2583–2595. doi: 10.1016/j.carres.2010.10.002. [DOI] [PubMed] [Google Scholar]
- Liang P, Pardee AB. Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science. 1992;257:967–971. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- Lohani S, Trivedi PK, Nath P. Changes in activities of cell wall hydrolases during ethylene induced ripening in banana: effect of 1-MCP, ABA and IAA. Postharvest Biol Technol. 2004;31:119–126. doi: 10.1016/j.postharvbio.2003.08.001. [DOI] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. 2. New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Srivastava S, Gupta SM. Role of differentially expressed ripening related genes and promoters in banana fruit: identified by mRNA DDRT-PCR. Res Environ Life Sci. 2008;1:81–90. [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance and maximum parsimo-ny methods. [DOI] [PMC free article] [PubMed]
- Vandevenne E, Christiaens S, Buggenhout SV, Jolie RP, González-Vallinas M, Duvetter T, Declerck PJ, Hendrickx ME, Gils A, Loey AV. Advances in understanding pectin methylesterase inhibitor in kiwi fruit: an immunological approach. Planta. 2011;2:287–298. doi: 10.1007/s00425-010-1307-6. [DOI] [PubMed] [Google Scholar]
- Wu MC, Tseng KC, Huang TH, Chang HM. Pectinesterase Inhibitor in Rubbery Banana (Musa sapientum L.) J Food Sci. 2002;67:1337–1340. doi: 10.1111/j.1365-2621.2002.tb10284.x. [DOI] [Google Scholar]