Abstract
The aim of this investigation was to compare the transcriptional expression of starch metabolism, involving genes and physiological characters, in seedlings of two contrasting salt-tolerant rice genotypes, in response to salt-stress. The soluble sugar content in rice seedlings of both salt-tolerant and salt-sensitive genotypes was enriched, relating to starch degradation, in plants subjected to 200 mM NaCl. In the salt-tolerant cultivar Pokkali, a major source of carbon may be that derived from the photosynthetic system and starch degradation. In starch degradation, only Pho and PWD genes in Pokkali were upregulated in plants subjected to salt stress. In contrast, the photosynthetic abilities of IR29 salt-susceptible cultivar dropped significantly, relating to growth reduction. The major source of carbohydrate in salt-stressed seedlings of the IR29 cultivar may be gained from starch metabolism, regulated by ADP-glucose pyrophosphorylase (AGP), starch synthase (SS), starch branching enzyme (SBE), starch debranching enzyme (ISA), glucan-water dikinase (GWD), dispropotionating enzyme (DPE), phospho glucan-water dikinase (PWD) and starch phosphorylase (Pho). Also, the major route of soluble sugar in salt-stressed Pokkali seedlings was derived from photosynthesis and starch metabolism. This was identified as novel information in the present study.
Keywords: Gene expression, Indica rice, Net photosynthetic rate, Salt tolerance, Soluble starch
Introduction
Salt-affected soil is a cause of abiotic stress in plants, and is one of the most serious factors to limit plant growth and development, especially in crop species which are classified as salt-susceptible (Qadir et al. 2008). In arid and semi-arid zones, large areas, including Australia, Asia Pacific, Latin America, Europe, North America, Africa and New East are identified as having salt affected soils. These soils have an excess of water–soluble salts, especially NaCl (Tanji 2002), which generate toxic ions, i.e. Na+ and Cl−, causing ionic and osmotic stress at the plant cellular level, especially in glycophyte species (Mansour and Salama 2004; Chinnusamy et al. 2005). In contrast, in halophyte species there are many defence mechanisms against salt, including ion homeostasis, osmoregulation, antioxidants and the hormonal regulation system (Hasegawa et al. 2000).
Carbohydrate is a candidate target of the salt defence mechanism and is controlled by the osmoticum in plant cells subjected to salt stress (Djanaguiraman et al. 2006; Chen et al. 2008; Rosa et al. 2009). Starch is the principle carbon reserve for energy preservation in higher plants. Also, starch may play a critical role as a Na+-starch binding granule, detoxifying toxic ions (Kanai et al. 2007). The chloroplast organelle, especially in the endosperm of cereals, is the origin of starch biosynthesis, forming complex polysaccharides, including linear and branched molecules of glucose polymer, amylose and amylopectin, by ADP-glucose pyrophosphorylase (ADP-AGPase), starch synthase (SS) and starch branching enzymes (SBE). In starch synthesis, ADP-glucose derived from photosynthesis is synthesised by the action of ADP-GPPase, which is the case in rice (Ballicora et al. 2004; Lee et al. 2007). The elongation of starch proceeds via SS activity that catalyses the transfer of glycosyl moiety of ADP-glucose to the non-reducing end of pre-existing α-D-1,4-glucan primer. Amylopectin formation requires the participation of starch branching enzyme (SBE), transferring the segment of an α-D-1,4-glucan to the sixth carbon of glycosyl moieties. The synthesis of starch depends not only on SS or SBE, but also on the participation of limit-dextrinase (pullulanase) and isoamylase. In starch degradation, the starch granule is subsequently cleaved into smaller molecules of glucan by a set of enzymes; glucan-water dikinase (GWD), phosphoglucan-water dikinase (PWD), debranching enzyme (DE), beta-amylase (BAM) and disproportionating enzyme (DPE), to provide a balanced energy reservoir. Relevant to this reaction is GWD, an enzyme that transfers the phosphate (Pi) of ATP to glycosyl moieties of amylopectin. PWD catalyses the reaction in a similar way to glucan-water dikinase. The formation of linear glucans results from the action of amylases and debranching enzymes on the branched starch. Several types of these enzymes have multiple isoforms, which are well established in different plant species (James et al. 2003; Tetlow 2006; Zeeman et al. 2007; Orzechowski 2008; Kötting et al. 2010; Zeeman et al. 2010). Soluble sugars derived from photosynthesis and starch degradation play acts as osmotic regulators in the salt defence mechanism, as is well established in many plant species (Balibrea et al. 2000; Kerepesi and Galiba 2000; Liu and van Staden 2001; Gupta and Kaur 2005; Brumós et al. 2009). The aim of this investigation was to compare the transcriptional expression of starch metabolism, involving genes and physiological characters, in seedlings of two contrasting rice cultivars, Pokkali (salt-tolerant) and IR29 (salt-susceptible) in response to salt-stress.
Materials and methods
Plant materials and salt stress treatments
Seeds of two rice cultivars, Pokkali (salt-tolerant) and IR29 (salt-susceptible), were obtained from the germplasm bank, Rice Research Center, Thailand. The seeds were manually dehusked, sterilised once in 5 % (v/v) Clorox® (0.05 % sodium hypochlorite, active ingredient) overnight, once in 25 % (v/v) Clorox® for 30 min, and then rinsed three times with sterile distilled-water. Surface-sterilised seeds were germinated on 0.25 % Phytagel®-solidified MS media (Murashige and Skoog 1962) with 3 % sucrose (photomixotrophic condition) in a 250 mL glass vessel. The media were adjusted to pH 5.7 before autoclaving. Rice seedlings were cultured in vitro under conditions of 25 ± 2 °C ambient temperature, 60 ± 5 % relative humidity (RH) and 60 ± 5 μmol m-2 s−1 photosynthetic photon flux density (PPFD) provided by fluorescent lamps with a 16 h d−1 photoperiod. Fourteen-day-old seedlings were aseptically transferred to MS-liquid sugar-free media (photoautotrophic conditions) using vermiculite as supporting material for 2 weeks. Air-exchange in the glass vessels was adjusted to 2.32 μmol CO2 h−1 by punching a hole in the plastic cap (Ø 1 cm) and covering the hole with gas-permeable microporous polypropylene film (0.22 mm pore size, Nihon Millipore Ltd., Japan). After acclimatising the seedlings for 1 week, the sodium chloride (NaCl) concentration in the culture medium was adjusted to 0 (control) or 200 mM (salt stress). The experiment was arranged as 2 × 2 factorials in Completely Randomised Block Design (CRBD) with four replicates (n = 4). The mean values obtained were compared using Duncan’s New Multiple Range Test (DMRT) and analyzed with SPSS software.
RNA extraction and cDNA preparation
Whole leaf blade from the rice seedlings were collected at 0, 6, 12, 18, 24, 30, 36, 42 and 48 h after NaCl treatment and immediately frozen at −80 °C, prior to the extraction of total RNA. Total RNA was pooled from rice seedlings and extracted by the guanidine hydrochloride method (Sambrook et al. 1989). Ground leaves of rice seedlings were homogenised in guanidinium thiocyanate solution (0.75 M NaCitrate at pH 7.0, 10 % Sarcosyl and 2 M 2-β-mercaptoethanol), NaAcetate (pH 4.0) and phenol-chloroform solution. After chilling on ice for 15 min, the homogenate was centrifuged at 10,000 g for 20 min at 4 °C. The aqueous phase was separated and mixed with 1× vol isopropanol, then kept at −20 °C for 1 h before centrifuging at 10,000 g for 15 min at 4 °C. The pellet was completely dissolved in 0.3 mL guanidinium thiocyanate solution and precipitated with ethanol. Contaminant DNA in the RNA preparations was then treated with RQ1 RNase-Free DNase (Promega) and total RNA was purified by phenol-chloroform extraction. First-stranded DNA was synthesised with 3 μg total RNA per sample, using ImPromp-II ™ Reverse Transcriptase (Promega) and oligo-dT15 primer.
Gene expression analysis by semi-quantitative PCR
The PCR reaction was performed using a Veriti ® Thermal Cycler (Applied Biosystems, CA, USA). All primer sequences and their annealing temperature are demonstrated in Table 1. The PCR reaction was performed with 70–100 ng total RNA, 10 pM primer and EmeraldAmp® GT PCR Master Mix (Takara, Japan). The PCR profiles were as follows, using the conditions: 94 °C for 3 min, 18–37 cycles of 94 °C for 30 s, 56–67 °C for 30 s, 72 °C for 30 s and 72 °C for 5 min. The conditions and cycle numbers were determined in order to avoid saturation of DNA amplification. The DNA obtained was subjected to agarose gel electrophoresis and stained with ethidium bromide. The signal intensity of the stained bands was photographed with a Gel Doc image analysis system (Bio-Rad, Hercules) and the data were analysed using GeneTools™ (Syngene, Cambridge, UK) analysis software.
Table 1.
Primers used for analysis of genes expression profile
| Enzyme | Acc. no. | Gene name | Primer sequence | Product size (bp) | Annealing temp (°C) |
|---|---|---|---|---|---|
| ADP-glucose pyrophosphorylase small subunit 2b | AK103906 | OsAGPS2b | (F)CTCTGGGTGCCAACTACAGG | 528 | 64 |
| (R)CTTTTGCCCTCACGTCGTC | |||||
| ADP-glucose pyrophosphorylase large subunit 1 | D50317 | OsAGPL1 | (F)ATGCTGGCCCAGACACTCT | 550 | 67 |
| (R)GCTCGACTCTCTCCAACAGG | |||||
| ADP-glucose pyrophosphorylase large subunit 3 | AK069296 | OsAGPL3 | (F)ACCACAGGATGAAGCAAAGG | 498 | 63 |
| (R)TCAGCCAGCAGTTCTCCTCT | |||||
| Starch synthase I | D16202 | OsSSI | (F)GCCATTCCAGATCTCATGC | 500 | 60 |
| (R)TGAAGGCCCATTCGAAGA | |||||
| Starch synthase IIb | AF395537 | OsSSIIb | (F)CTAGAGGCCCCTCCCTTTC | 508 | 65 |
| (R)CCTGCAGCTTCCAGTCGT | |||||
| Starch synthase IIc | AF383878 | OsSSIIc | (F)CAGGGCAGGATATGGAGGTA | 508 | 63 |
| (R)TTGAGCTCCCATGCATAACC | |||||
| Starch synthase IIIa | AY100469 | OsSSIIIa | (F)TCGGAAGAAGGTGGAATCTATG | 618 | 61 |
| (R)TTGTGGCTTTGTCGCAGTATG | |||||
| Starch synthase IIIb | AF432915 | OsSSIIIb | (F)AGCCTCCACTCCGAAACCTA | 516 | 67 |
| (R)CCACTACTTGGCTGGCTTTG | |||||
| Starch synthase IVa | GQ151036 | OsSSIVa | (F)CTTGCAACCAAAGGGAACAT | 550 | 64 |
| (R)GGCTTTTTGCTGCTTTCAAC | |||||
| Starch synthase IVb | GQ151051 | OsSSIVb | (F)CTATGGTTCTGGGACGCAAT | 480 | 63 |
| (R)ACCTAGTCCCTGGCAATGTG | |||||
| Granule-bound starch synthase II | AK067654 | OsGBSSII | (F)CCCTTTCTGTGCGGATTAAC | 492 | 56 |
| (R)CGAGGCATAAAAGGCAGAAC | |||||
| Starch branching enzyme I | EF122471 | OsBE1 | (F)CGCCTGCTGCACAAGAAG | 526 | 64 |
| (R)CCTGATCTGCTGCTGACTGC | |||||
| Starch branching enzyme IIa | AB023498 | OsBEIIa | (F)AAGATGGTGGCTCGAGGAG | 507 | 65 |
| (R)CTAGGTGTTGCCGGTCTGTC | |||||
| Starch branching enzyme IIb | D16201 | OsBEIIb | (F)AACCACGAGTTATCCCACCA | 452 | 64 |
| (R)TTCACCTGGAGCCTGCAC | |||||
| Starch debranching enzyme: Isomerase I | AF142590 | OsISAI | (F)CTTCAACTGTAATCATCCTGTGG | 460 | 60 |
| (R)GATACTGTGCCAAGGTTTCCT | |||||
| Starch debranching enzyme: Isomerase II | AC132483 | OsISAIIa | (F)CACCAGGACCTCTCACAAGG | 553 | 61 |
| (R)GAGAAGCTGGTGGTGTACCG | |||||
| Starch debranching enzyme: Isomerase III | AK101554 | OsISAIIb | (F)AACTGCAACCATCCTGTTGTC | 439 | 61 |
| (R)GGTCGCATAAAGTAAATCCATA | |||||
| Starch phosphorylase H | EF576564 | OsPHOH | (F)GCTGGGCTGAGGAAGGATAG | 483 | 64 |
| (R)CTCGTTGCTGGGTTCTCC | |||||
| Starch phosphorylase L | AF327055 | OsPHOL | (F)GTGTTGTTGGTGGGCATTC | 491 | 62 |
| (R)GCATACCCTTGGAACAAAGC | |||||
| Disproportionating enzyme I | AP004306 | OsDPE1 | (F)ATTCAGGACAGGATGCAAAC | 582 | 62 |
| (R)CATCAGGTGGAACACCACTAA | |||||
| Disproportionating enzyme II | NM_001067082 | OsDPE2 | (F)TGGACAAAATTGGGGTTTTC | 457 | 56 |
| (R)ACGCCGAATACTGTCTTCTTTC | |||||
| Glucan-water dikinase | AK103463 | OsGWD | (F)AGTGGCACCAGAAACTGCAC | 479 | 67 |
| (R)AGGAGAGCCAAGGAGCAAAG | |||||
| Phospho glucan-water dikinase | FN179404 | OsPWD | (F)GTCCCTTCTGGTGCTGTGAT | 525 | 67 |
| (R)GACCTCAGCCTGGACAACC | |||||
| β-amylase | L10345 | OsBAM | (F)CGACGATGCTGGAGAGTACA | 469 | 61 |
| (R)TGCATCATATCGACCGAGTG | |||||
| 18s rRNA | AK105009 | 18s | (F)GTGCAACAAACCCCGACT | 406 | 62 |
| (R)GCTGCTGGCACCAGACTT |
Starch and sugars determination
Starch content in the leaf blade tissues was determined with an EnzyChrom assay kit (BioAssay Systems, Hayward, CA), using an enzymatic colorimetric method, which was a 2-phase method to quantify the concentration of both soluble and resistant starch (McCleary and Monaghan 2002). A leaf sample of one hundred-milligrams was ground and the sugars extracted with 1 mL 90 % ethanol at 60 °C for 5 min with triple repeats. After ethanol evaporation, soluble and insoluble starch was dissolved in 0.5 mL water at 60 °C for 5 min and then mixed with 0.2 mL DMSO at 60 °C for 5 min. Concentrations of soluble and insoluble starch were determined by colourimetric measurement of glucose residue after digestion using amyloglucosidase and α-amylase.
Glucose and fructose in leaf blade tissues were analysed according to the modified method of Karkacier et al. (2003). In a pre-cooled mortar, 100 mg fresh weight tissue was ground with liquid nitrogen, extracted with 1 mL nanopure water, vigorously shaken for 15 s, sonicated for 15 min and then centrifuged at 12,000 rpm for 15 min. The supernatant was filtered through a 0.45 μm membrane filter and stored at −20 °C, prior to the measurement of sugar content (glucose and fructose) using high performance liquid chromatography (HPLC). A volume of 50 μL crude extracts was automatically injected into the HPLC system with a Waters 600 pump. Online detection was performed using a Waters 410 differential refractrometer detector and the data analysed by Empower software. The analytical column was a MetaCarb 87 °C equipped with a guard column. Deionised water was used as the mobile phase with a 0.5 mL min−1 flow rate. Glucose and fructose were used as standards.
Physiological characters
Chlorophyll fluorescence emission from the adaxial surface of the leaf was measured using a fluorescence monitoring system (FMS 2; Hansatech Instruments Ltd., Norfolk, UK) in the pulse amplitude modulation mode, as previously described by Loggini et al. (1999). A leaf, adapted to dark conditions for 30 min using leaf-clips, was initially exposed to the modulated measuring beam of far-red light (LED source with typical peak at wavelength 735 nm). Original (F0) and maximum (Fm) fluorescence yields were measured under weak modulated red light (<0.5 μmol m−2 s−1) with 1.6 s pulses of saturating light (>6.8 μmol m−2 s−1 PAR) and calculated using FMS software for Windows®. The variable fluorescence yield (Fv) was calculated by the equation of Fm–F0. The ratio of variable to maximum fluorescence (Fv/Fm) was calculated as maximum quantum yield of PSII photochemistry. The photon yield of PSII (ΦPSII) in the light was calculated by 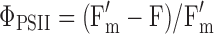 after 45 s illumination when steady state was achieved. In addition, photochemical quenching (qP) was calculated as described earlier (Maxwell and Johnson 2000).
after 45 s illumination when steady state was achieved. In addition, photochemical quenching (qP) was calculated as described earlier (Maxwell and Johnson 2000).
Net photosynthetic rate (Pn; μmol m−2 s−1), stomatal conductance (gs; mmol m−2 s−1), transpiration rate (E; mmol m−2 s−1) and water use efficiency (WUE; %) were measured using a Portable Photosynthesis System (Model LI 6400, LI-COR®Inc, Lincoln, Nebraska, USA) with an Infra-red Gas Analyser following Cha-um et al. (2007). WUE was calculated according to the equation: 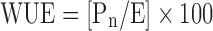 .
.
Growth performance
Fresh weight (FW), dry weight (DW), shoot height (SH) and root length (RL) of rice seedlings were measured, as described by Cha-um et al. (2006). Seedlings were dried at 80 °C in a hot-air oven for 2 days, and then incubated in desiccators before the measurement of dry weight.
Results and discussion
Physiological and growth characters
Maximum quantum yield of PSII (Fv/Fm) in the salt-stressed leaves of IR29 was significantly decreased by 52.6 %, whereas that in Pokkali was maintained (only 0.5 % diminution) when subjected to 200 mM NaCl (Table 2). Photon yield of PSII (ΦPSII) and photochemical quenching (qP) in both IR29 genotypes decreased when seedlings were exposed to 200 mM NaCl, by 63.6 % and 55.3 %, respectively. Conversely, the same parameters in the Pokkali variety decreased by only 19.6 % and 18.1 %, respectively. Water use efficiency (WUE), net photosynthetic rate (Pn), and stomatal conductance (gs) in both IR29 and Pokkali dropped significantly, but not transpiration rate (E) which increased (Table 3). WUE, Pn and gs in salt-stressed seedlings of IR29 (95.4 %, 74.4 % and 92.5 % reduction respectively) were lower than in seedlings of the Pokkali genotype (80.3 %, 44.4 % and 68.1 % reduction). The reduction in physiological character parameters directly affected overall growth, indicated by fresh weight (FW), dry weight (DW), shoot height (SH) and root length (RL). FW, DW, SH and RL values in Pokkali (13.3 %, 6.2 %, 6.6 % and 11.3 % reduction respectively) were alleviated significantly and were better than those of IR29 (26.2 %, 24.1 %, 19.6 % and 37.1 % reduction) in plants subjected to 200 mM NaCl (Table 4).
Table 2.
Maximum quantum yield of PSII (Fv/Fm), photon yield of PSII (ΦPSII) and photochemical quenching (qP) in the leaf tissues of IR29 and Pokkali seedlings grown under 0 mM or 200 mM NaCl treatment for 8 days
| Rice | NaCl (mM) | Fv/Fm | ΦPSII | qP |
|---|---|---|---|---|
| IR29 | 0 | 0.858a | 0.634a | 0.742a |
| 200 | 0.407b | 0.231c | 0.332c | |
| Pokkali | 0 | 0.866a | 0.567a | 0.678a |
| 200 | 0.862a | 0.456b | 0.555b |
Different letters in each column show significant difference at p ≤ 0.01 by Duncan’s New Multiple Range Test (DMRT)
Table 3.
Water use efficiency (WUE), net-photosynthetic rate (Pn), transpiration rate (E) and stomatal conductance (gs) in the leaf tissues of IR29 and Pokkali seedlings grown under 0 mM or 200 mM NaCl treatment for 8 days
| Rice | NaCl (mM) | WUE (%) | Pn (μmol m−2 s−1) | E (mmol m−2 s−1) | gs (mmol H2O m−2 s−1) |
|---|---|---|---|---|---|
| IR29 | 0 | 8.11a | 1.48a | 0.18b | 30.50ab |
| 200 | 0.37c | 0.35c | 1.05a | 2.28c | |
| Pokkali | 0 | 5.63b | 1.35a | 0.25b | 38.45a |
| 200 | 1.11c | 0.75b | 0.71b | 12.26bc |
Different letters in each column show significant difference at p ≤ 0.01 by Duncan’s New Multiple Range Test (DMRT)
Table 4.
Fresh weight (FW), dry weight (DW), shoot height (SH) and root length (RL) in IR29 and Pokkali seedlings grown under 0 mM or 200 mM NaCl treatment for 8 days
| Rice | NaCl (mM) | FW (mg) | DW (mg) | SH (cm) | RL (cm) |
|---|---|---|---|---|---|
| IR29 | 0 | 246.6b | 39.9ab | 29.1b | 7.0a |
| 200 | 182.1c | 30.3b | 23.4c | 4.4c | |
| Pokkali | 0 | 371.7a | 48.3a | 36.3a | 6.2b |
| 200 | 322.4a | 45.3a | 33.9a | 5.5b |
Different letters in each column show significant difference at p ≤ 0.01 by Duncan’s New Multiple Range Test (DMRT)
In the present study, photosynthetic abilities, chlorophyll fluorescence and net photosynthetic rate in the Pokkali salt-tolerant cultivar were better than in IR29, salt-susceptible, when subjected to 200 mM NaCl, leading to maintenance of growth efficiency. Similar results are well established in the case of physiological and morphological characters in response to salt stress of salt-tolerant [Pokkali and FL478 (Pokkali × IR29 isogenic line)] and salt-sensitive (IR24, Peta, Nonabokra, BRRI Dhan29 and IR29) rice cultivars (Walia et al. 2005; Senadheera et al. 2009). In general, sodium ions (Na+) are taken up into the root tissues and translocated to leaf tissues, especially in salt-sensitive cultivars (Golldack et al. 2003; Kader and Lindberg 2005; Malagoli et al. 2008). For example, Na+ in IR29 rice seedlings (leaf and culm tissues) was enriched significantly in plants subjected to 50 mM NaCl for 2 days and was greater than that in FL478 (Walia et al. 2005; Senadheera et al. 2009). Also, the photosynthetic pigments and activities were the final targets to be damaged by Na+, which was identified by pigment degradation (chlorophyll), chlorophyll fluorescence diminution (PSI and PSII) and net photosynthetic rate (Pn) reduction. In present study, Fv/Fm and ΦPSII in salt stressed seedlings of IR29 were sharply dropped for 52.6 % and 63.6 %, respectively. Similar result has been well established in PT1 rice cultivar exposed to 342 mM NaCl (Cha-um et al. 2009). The photosynthetic abilities of salt-tolerant rice varieties could be maintained and were better than those of the salt-sensitive variety, leading to maintained growth when subjected to salt stress (Tiwari et al. 1997; Lee et al. 2003; Ferdose et al. 2009; Wang et al. 2009; Cha-um et al. 2010; Ghosh et al. 2011).
Expression levels of starch involving genes under salt stress
The transcriptional level of starch biosynthesis and degradation genes in IR29, salt-susceptible, and Pokkali, salt-tolerant, were chosen as the target of salt responsive genes. In the case of starch biosynthesis, the expression level of AGPL1 in salt-stressed seedlings of Pokkali, treated with 200 mM NaCl (salt-stress) for 18–48 h, increased obviously when compared with the control seedlings (0 mM NaCl) and was higher than in IR29 by 15 times (Fig. 3a). The expression level of AGPL3 was unaffected, in either NaCl-treated or non-treated seedlings of both IR29 and Pokkali (Fig. 1b). In contrast, the expression of cytosolic AGPS2b was enriched in salt-stressed seedlings of IR29 and was higher than that of the non-treated sample by 2 times for 18–48 h (Fig. 1c). For starch synthase (SS) isoforms, from 6–48 h, expression levels of SSI gene in salt-stressed seedlings were increased in IR29 relative to increasing salt exposure time, whereas it was down-regulated in Pokkali (Fig. 2a). The level of SSIIb expression was down-regulated in both IR29 and Pokkali genotypes when seedlings were subjected to salt stress for 24–42 h (Fig. 2b). Relative expression of SSIIc in salt-stressed seedlings was unchanged (Fig. 2c). In Pokkali, the expression of SSIII class, either isoform SSIIIa or isoform SSIIIb, was stabilised in rice seedlings with or without NaCl treatment, whereas those were down-regulated when IR29 was treated with NaCl (Fig. 2d and e). The expression profiles of SSIVa and SSIVb in salt-stressed seedlings of Pokkali and IR29 were down-regulated when the seedlings were exposed to 200 mM NaCl for 36–48 h (Fig. 2f and g). Similarly, the relative level of GBSSII was down-regulated in salt-stressed Pokkali at 42 h and 48 h after exposure to NaCl (Fig. 2h). Three isoforms of starch branching enzyme (SBE), SBEI, SBEIII and SBEIV were chosen as targets for gene expression. In IR29, the expression levels of SBEI and SBEIV were up-regulated when plants were subjected to 200 mM NaCl for 42–48 h (Fig. 3a and c), while SBEIII was down-regulated (Fig. 3b). The expression level of SBEIII in salt-stressed Pokkali was down-regulated at 36–42 h of treatment (Fig. 3b). Starch debranching genes i.e. ISAI, ISAIIa and ISAIIb were also determined. The expression level of ISAI in salt-stressed Pokkali was down-regulated, whereas in IR29 it was unchanged (Fig. 3d). In contrast, relative expressions of ISAIIa and ISAIIb in IR29 were up-regulated when rice seedlings were subjected to 200 mM NaCl for 42–48 h (Fig. 3e and f).
Fig. 3.
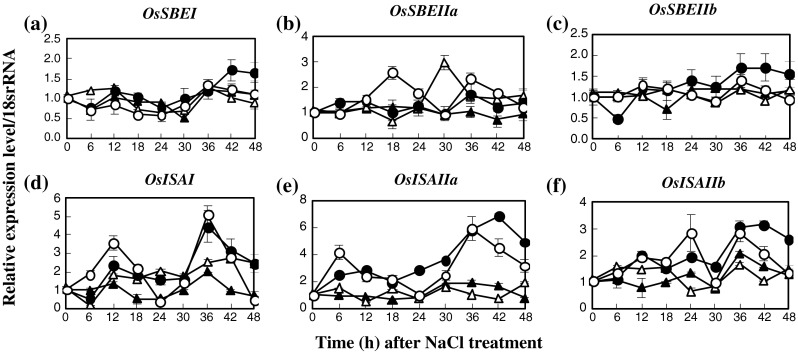
Expression patterns of SBE (a–c) and SDE (d–f) in leaf tissues of IR29 (circle) and Pokkali (triangle) seedlings grown under 0 (opened symbol) or 200 mM NaCl treatment (closed symbol) for 48 h
Fig. 1.
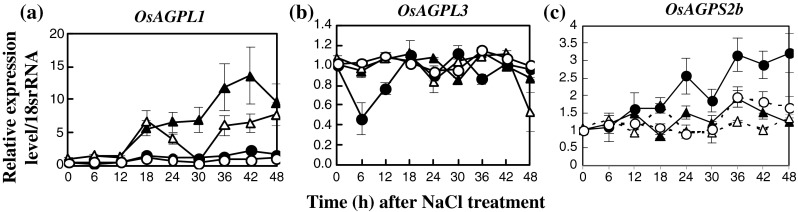
Expression patterns of ADP-glucose pyrophosphorylase (AGPase) large subunit (L; a–b) and small subunit (S; c) in leaf tissues of IR29 (circle) and Pokkali (triangle) seedlings grown under 0 (opened symbol) or 200 mM NaCl treatment (closed symbol) for 48 h
Fig. 2.
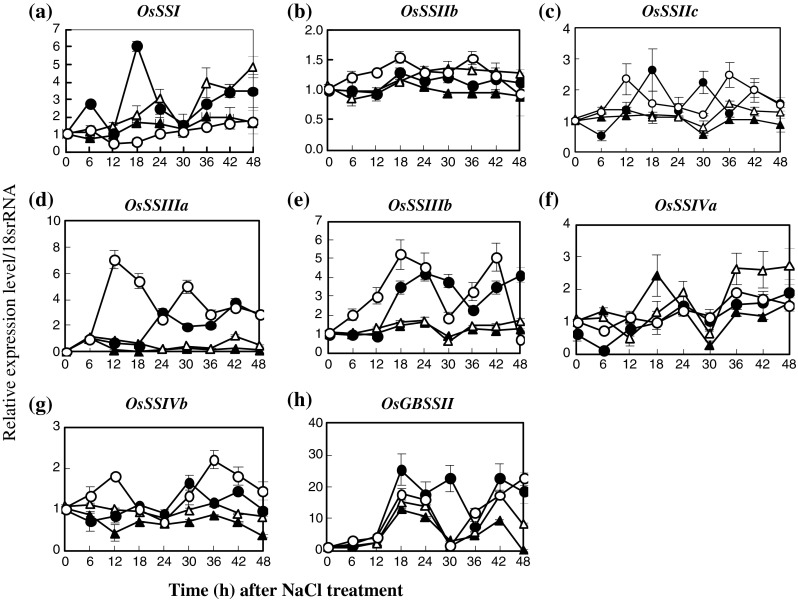
Expression patterns of starch synthase (SS; a–g) and granule-bound starch synthase (GBSS; h) in leaf tissues of IR29 (circle) and Pokkali (triangle) seedlings grown under 0 (opened symbol) or 200 mM NaCl treatment (closed symbol) for 48 h
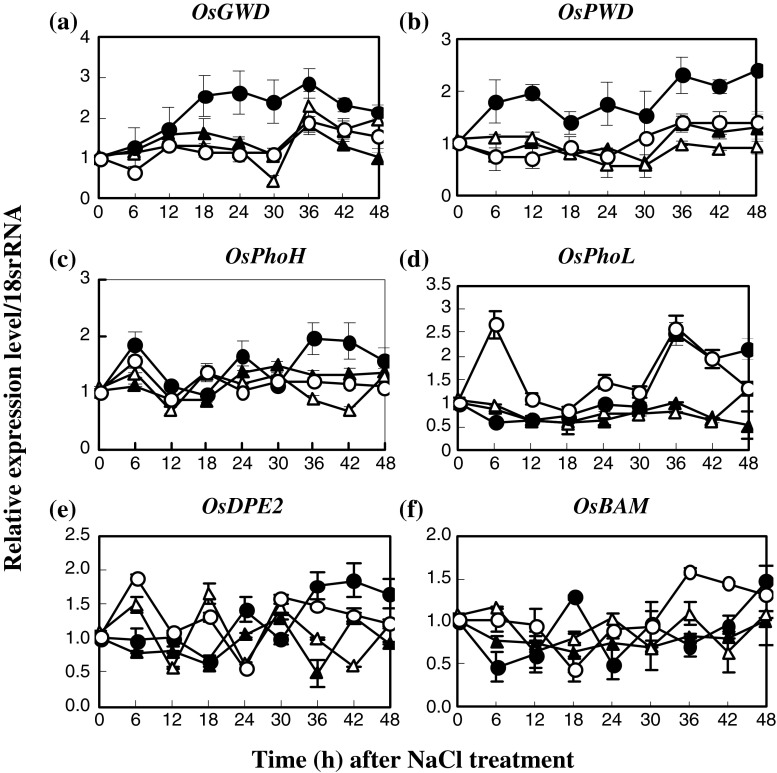
As for starch degradation, glucose-water dikinase (GWD) gene expression in salt-stressed IR29 was up-regulated in plants subjected to NaCl for 18–48 h, while it was down regulated in salt-stressed Pokkali (42–48 h) (Fig. 4a). Similarly, the expression level of phosphogluco-water dikinase (PWD) in salt-stressed IR29 was up-regulated when subjected to NaCl (Fig. 4b). The relative expressions of two starch phosphorelase (Pho) genes (isoforms; PhoL and PhoH) in salt-stressed IR29 were down-regulated when exposed to 200 mM NaCl from 0–30 h (Fig. 4c and d). The expression level of disproportionating enzyme (DPE2) in salt-stressed IR29 was increased in plants exposed to 200 mM NaCl for 36–48 h (Fig. 4e), while β-amylase (BAM) was decreased (36-42 h salt exposure) (Fig. 4f). The summary of gene regulation in starch metabolism is shown in Table 5. It should be identified as two groups, with early response (<6 h salt exposure) and late response (>36 h salt exposure). Also, a large number of genes were up-regulated in IR29 (10 genes), more than in Pokkali (only 3 genes). In early salt stress, starch biosynthesis in IR29 was up-regulated, while starch degradation in Pokkali was enhanced (Table 5).
Fig. 4.
Expression profiles of glucan-water dikinase (GWD; a), phosphogluco-water dikinase (PWD; b), starch phosphorylase H (PhoH; c), starch phosphorylase H (PhoL; d), disproportionating enzyme (DPE; e) and β-amylase (BAM; f) in leaf tissues of IR29 (circle) and Pokkali (triangle) seedlings grown under 0 (opened symbol) or 200 mM NaCl treatment (closed symbol) for 48 h
Table 5.
Early (<6 h; normal text) and late (>36 h; bold text) responses of specific isoform of each gene family related to starch metabolism in leaf tissues of IR29 and Pokkali seedlings grown under 200 mM NaCl treatment
| Genotype | Up regulation | Down regulation |
|---|---|---|
| IR29 | OsAGPS2b, OsSSI, OsSBEI, OsSBEIV, OsISAIIa, OsISAIIb , OsGWD, OsPWD, OsPhoH, OsDPE2 | OsSSIIb, OsSSIIIa, OsSSIIIb, OsSSIVb, OsSBEIII |
| Pokkali | OsAGPL1, OsPWD, OsPhoH | OsSSI, OsSSIIb, OsSSII c, OsSSIVa, OsSSIVb, OsGBSSII, OsSBEIII, OsISAI, OsGWD |
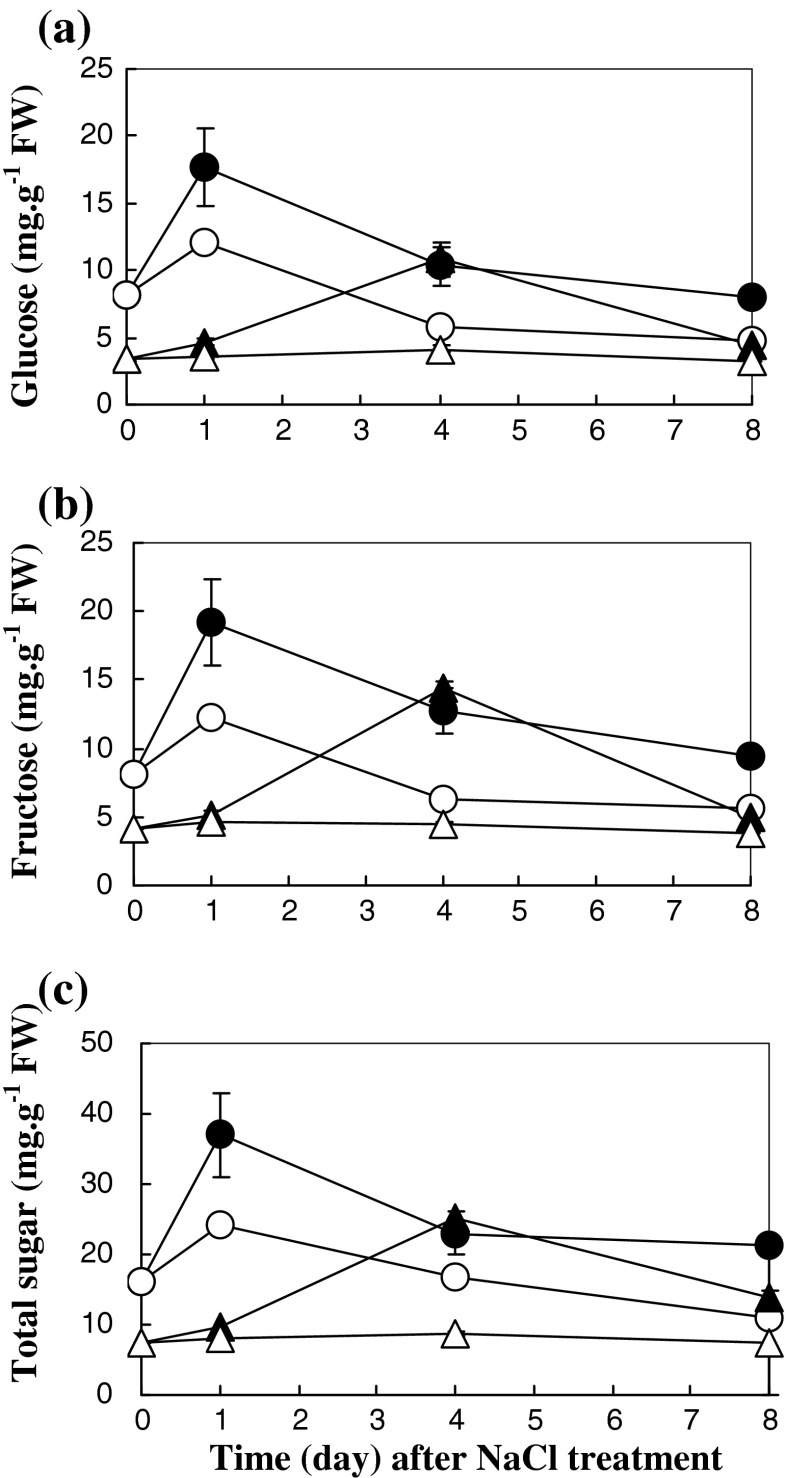
Starch and sugar profiles in the leaf tissues of rice seedlings were evaluated. Insoluble and soluble starch accumulated in salt-stressed Pokkali subjected to 200 mM NaCl for 1–8 days, whereas in IR29 the levels were unchanged (Fig. 5a and b). A positive correlation between soluble starch and soluble sugar accumulation in both IR29 (r2 = 0.50) and Pokkali (r2 = 0.98) was demonstrated (Fig. 5c and d). Glucose, fructose and total sugar content in salt-stressed Pokkali increased in plants subjected to NaCl for 4 days. In contrast, the sugar content in salt-stressed IR29 was enriched when seedlings were exposed to NaCl for 1–8 days (Fig. 6). In the present study, the regulation of starch metabolism in IR29, salt-susceptible, was identified as a single channel for sugar production when subjected to salt stress. In contrast, starch metabolism and photosynthesis in salt stressed Pokkali were played as major route for the sugar enrichment.
Fig. 5.
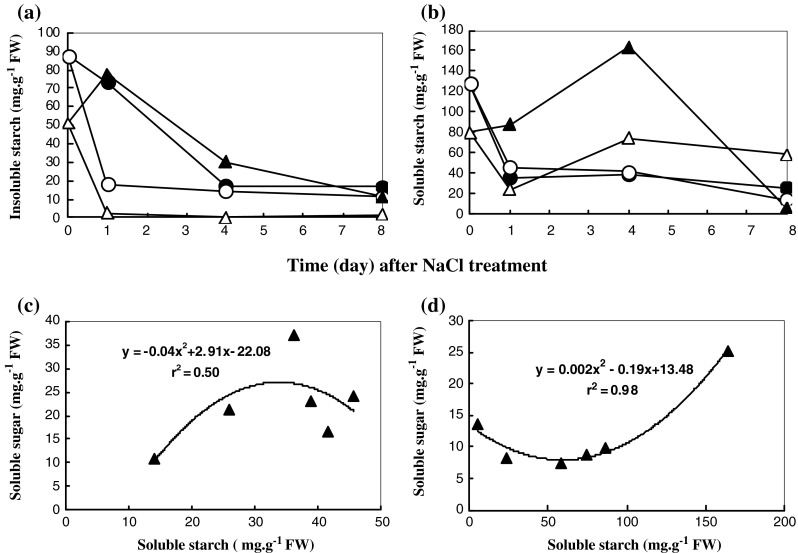
Insoluble (a) and soluble starch (b) contents in leaf tissues of IR29 (circle) and Pokkali (triangle) seedlings grown under 0 (opened symbol) or 200 mM NaCl treatment (closed symbol) for 8 days and relationship between soluble starch content and sugar content in leaf tissues of IR29 (c) and Pokkali (d)
Fig. 6.
Glucose (a), fructose (b) and total sugar contents (c) in leaf tissues of IR29 (circle) and Pokkali (triangle) seedlings grown under 0 (opened symbol) or 200 mM NaCl treatment (closed symbol) for 8 days
The gene expression of two isoforms of granule-bound starch synthase, GBSSI and GBSSII, in salt-stressed seedlings (200 mM NaCl) of the Tainung 67 rice cultivar has been reported as down regulation. Also, the activity of GBSS decreased significantly, leading to low starch content in seedlings exposed to 200 mM NaCl. In contrast, the activities of ADP-glucose pyrophosphorylase (AGPase), soluble starch synthase (SS) and starch branching enzyme (SBE) in starch biosynthesis were unchanged in plants subjected to salt stress (Chen et al. 2008). In the present study, the differential display of starch metabolism regulated genes, in both Pokkali, salt-tolerant and IR29, salt-susceptible, was demonstrated. An expression of AGPL1 gene in Pokkali rice seedlings was enriched when subjected to 200 mM NaCl for 24–48 h. In contrast, the expression of AGPS2b gene in salt stressed seedling of IR29 was up-regulated. The large subunit (L1) and small subunit (S1 and S2a) of AGPase protein in rice crop (cv. Dongjin) has been localized in plastid organelles, while S2b small subunit protein locates in cytosol (Lee et al. 2007). Also, the sensitivity and function of those subunits of AGPase protein relating to salt stress are still undiscovered. The regulation of those genes in starch biosynthesis i.e. AGP, GBSS, SS, SBE and starch debranching enzyme (ISA) and in starch degradation i.e. disproportionating enzyme (DPE), glucan-water dikinase (GWD), phospho glucan-water dikinase (PWD), β-amylase (BAM) and starch phosphorelase (Pho), depends on the degree of salt stress, the genotype and their interactions. In the salt-tolerant cultivar Pokkali, a major source of carbon is that derived from the photosynthetic system. Only Pho and PWD genes in Pokkali were upregulated for starch degradation in plants subjected to salt stress. In contrast, the photosynthetic abilities of IR29, salt-susceptible, dropped significantly. The major source of carbohydrate may flow from starch metabolism, regulated by AGP, SS, SBE, ISA, GWD, DPE, PWD and Pho. In the salt-sensitive rice cv. IR20, starch content decreased, relating to an increase in soluble sugar, including sucrose, when exposed to 50–100 mM NaCl (Djanaguiraman et al. 2006). Soluble sugar, sucrose, glucose and fructose content in the root and leaf tissues of two rice genotypes (HJ, salt-tolerant, and PT1, salt-susceptible) were enriched when plants were subjected to 342 mM NaCl for 7 d (Cha-um et al. 2009). In addition, soluble sugar content, fructose and glucose, in a chill-sensitive rice cultivar increased significantly, whereas it was unchanged in a chill-tolerant cultivar exposed to 100 mM NaCl for 4 d (Morsy et al. 2007). Similar results have been reported in the salt-sensitive tomato cv. Volgogradskij (Khelil et al. 2007), wheat cv. Ghods (Kafi et al. 2003), cashew nut (Voigt et al. 2009), cowpea (Praxedes et al. 2011) and quinoa (Rosa et al. 2009). From this evidence, it can be seen that the photosynthetic abilities in salt-sensitive rice dropped significantly when seedlings were subjected to salt stress. Soluble sugars are still enriched in the plant tissues, which is possibly the method of sugar enrichment. In the present study, soluble sugar enrichment may be contributed by starch metabolism, as identified by transcriptional expression of genes involved with starch, including AGP, SS, SBE, ISA, GWD, DPE, PWD and Pho. In quinoa (Chenopodium quinoa Willd.), AGP gene in the cotyledons (carbohydrate sink organs) was generally down-regulated, relating to low starch content, leading to increased soluble sugar in plants subjected to 200 mM NaCl (Rosa et al. 2009). In the developing fruits of tomato (Solanum lycopersicum L. cv. Micro-Tom), AGP gene expression (isoforms, AgpL1, AgpL2 and AgpS1) and AGPase activity were up-regulated when plants were subjected to 160 mM NaCl, causing a decrease in starch content in the fruit tissues. Also, the expression of GBSS gene in rice was up-regulated when plants were subjected to low temperature (15/10 °C; day/night temperature), whereas it was down-regulated in plants exposed to high temperatures (>30/25 °C; day/night temperature) and drought stress (Wang et al. 2006). Starch degradation in salt-stressed plants may play a role as a major source of carbon, indicated by enriched soluble sugar (3 times that of the control), including glucose, fructose and sucrose (Yin et al. 2010). In Pokkali, soluble starch increased early during the salt exposure period (1–4 d), then dropped. The functional role of soluble sugar in the salt defense mechanism is as an osmoregulation system, controlling the osmotic pressure in the plant cells in Na+-enriched conditions (Cha-um et al. 2009), and starch content may perform as a chelating agent, namely “Na+-starch granule bound” (Kanai et al. 2007). In the present study, novel transcriptional genes involved in starch metabolism, in both salt-tolerant and salt-sensitive genotypes of rice, were discovered in seedlings grown in salt-stress conditions. The major route of soluble sugar in Pokali, salt-tolerant, was found to be that from the photosynthetic system and starch degradation, while in IR29, salt–susceptible, it only flowed through starch metabolism when plants were exposed to salt stress.
In conclusion, photosynthetic abilities, including Fv/Fm, ΦPSII, qP, WUE and Pn in the IR29 salt-sensitive cultivar dropped significantly and were lower than those in the Pokkali salt–tolerant cultivar, leading to inhibited growth characters in plants exposed to 200 mM NaCl. Endogenous soluble sugar contents (glucose and fructose) in both salt-tolerant and salt-sensitive genotypes of rice, increased in plants subjected to salt stress. The accumulation of soluble sugars in seedlings of the salt-sensitive cultivar grown under salt-stress may be from the starch metabolism, as identified by the transcriptional expression of genes involving starch, whereas sugar accumulation in the salt-tolerant cultivar may be generated from photosynthesis and starch degradation.
References
- Balibrea ME, Amico JD, Balarín MC, Pérez-Afocea FP. Carbon partitioning and sucrose metabolism in tomato plants growing under salinity. Physiol Plant. 2000;110:503–511. doi: 10.1111/j.1399-3054.2000.1100412.x. [DOI] [Google Scholar]
- Ballicora MA, Iglesias AA, Preiss J. ADP-glucose pyrophosphorylase: a regulatory enzyme for plant starch synthesis. Photosyn Res. 2004;79:1–24. doi: 10.1023/B:PRES.0000011916.67519.58. [DOI] [PubMed] [Google Scholar]
- Brumós J, Colmenero-Flores JM, Conesa A, Izquierdo P, Sánchez G, Iglesias DJ, López-Climent MF, Gómez-Cadenas A, Talón M. Membrane transporters and carbon metabolism implicated in chloride homeostasis differentiate salt stress responses in tolerant and sensitive Citrus rootstocks. Funct Integr Genom. 2009;9:293–309. doi: 10.1007/s10142-008-0107-6. [DOI] [PubMed] [Google Scholar]
- Cha-um S, Supaibulwatana K, Kirdmanee C. Water relation, photosynthetic ability, and growth of Thai jasmine rice (Oryza sativa L. ssp. indica cv. KDML105) to salt stress by application of exogenous glycinebetaine and choline. J Agron Crop Sci. 2006;192:25–36. doi: 10.1111/j.1439-037X.2006.00186.x. [DOI] [Google Scholar]
- Cha-um S, Supaibulwatana K, Kirdmanee C. Glycinebetaine accumulation, physiological characterizations, and growth efficiency in salt tolerant and salt sensitive lines of indica rice (Oryza sativa L. spp. indica) response to salt stress. J Agron Crop Sci. 2007;193:157–166. doi: 10.1111/j.1439-037X.2007.00251.x. [DOI] [Google Scholar]
- Cha-um S, Charoenpanich A, Roytrakul S, Kirdmanee C. Sugar accumulation, photosynthesis and growth of two indica rice varieties in response to salt stress. Acta Physiol Plant. 2009;31:477–486. doi: 10.1007/s11738-008-0256-1. [DOI] [Google Scholar]
- Cha-um S, Ashraf M, Kirdmanee C. Screening upland rice (Oryza sativa L. spp. indica) genotypes for salt-tolerance using multivariate cluster analysis. Afri J Biotechnol. 2010;9:4731–4740. [Google Scholar]
- Chen HJ, Chen JY, Wang SJ. Molecular regulation of starch accumulation in rice seedling leaves in response to salt stress. Acta Physiol Plant. 2008;30:135–142. doi: 10.1007/s11738-007-0101-y. [DOI] [Google Scholar]
- Chinnusamy V, Jagendorf A, Zhu JK. Understanding and improving salt tolerance in plants. Crop Sci. 2005;45:437–448. doi: 10.2135/cropsci2005.0437. [DOI] [Google Scholar]
- Djanaguiraman M, Sheeba JA, Shanker AK, Devi DD, Bangarusamy U. Rice can acclimate to lethal level of salinity by pretreatment with sublethal level of salinity through osmotic adjustment. Plant Soil. 2006;284:363–373. doi: 10.1007/s11104-006-0043-y. [DOI] [Google Scholar]
- Ferdose J, Kawasaki M, Taniguchi M, Miyake H. Differential sensitivity of rice cultivars to salinity and its relation to ion accumulation and root tip structure. Plant Prod Sci. 2009;12:453–461. doi: 10.1626/pps.12.453. [DOI] [Google Scholar]
- Ghosh N, Adak MK, Ghosh PD, Gupta S, Gupta DNS, Mandal C. Differential responses of two rice varieties to salt stress. Plant Biotechnol Rep. 2011;5:89–103. doi: 10.1007/s11816-010-0163-y. [DOI] [Google Scholar]
- Golldack D, Quigley F, Michalowski CB, Kamasani UR, Bohnert HJ. Salinity stress-tolerant and –sensitive rice (Oryza sativa L.) regulate AKT1-type potassium channel transcripts differently. Plant Mol Biol. 2003;51:71–81. doi: 10.1023/A:1020763218045. [DOI] [PubMed] [Google Scholar]
- Gupta AK, Kaur N. Sugar signalling and gene expression in relation to carbohydrate metabolism under abiotic stresses in plants. J Biosci. 2005;30:761–776. doi: 10.1007/BF02703574. [DOI] [PubMed] [Google Scholar]
- Hasegawa PM, Bressan RA, Zhu JK, Bohnert HJ. Plant cellular and molecular responses to high salinity. Annu Rev Plant Physiol Mol Biol. 2000;51:463–499. doi: 10.1146/annurev.arplant.51.1.463. [DOI] [PubMed] [Google Scholar]
- James MG, Denyer K, Myers AM. Starch synthesis in the cereal endosperm. Curr Opin Plant Biol. 2003;6:215–222. doi: 10.1016/S1369-5266(03)00042-6. [DOI] [PubMed] [Google Scholar]
- Kader MA, Lindberg S. Uptake of sodium in protoplasts of salt-sensitive and salt tolerant cultivars of rice, Oryza sativa L. determined by the fluorescent dye SBFI. J Exp Bot. 2005;56:3149–3158. doi: 10.1093/jxb/eri312. [DOI] [PubMed] [Google Scholar]
- Kafi M, Stewart WS, Borland AM. Carbohydrate and proline contents in leaves, roots and apices of salt-tolerant and salt sensitive wheat cultivars. Russ J Plant Physiol. 2003;50:174–182. doi: 10.1023/A:1022956727141. [DOI] [Google Scholar]
- Kanai M, Higuchi K, Hagihara T, Konishi T, Ishii T, Fujita N, Nakamura Y, Maeda Y, Tadano T. Common reed produces starch granules at the shoot base in response to salt stress. New Phytol. 2007;176:572–580. doi: 10.1111/j.1469-8137.2007.02188.x. [DOI] [PubMed] [Google Scholar]
- Karkacier M, Erbas M, Usiu MK, Aksu M. Comparison of different extraction and detection methods for sugar using amino-bonded phase HPLC. J Chromatogr Sci. 2003;41:331–333. doi: 10.1093/chromsci/41.6.331. [DOI] [PubMed] [Google Scholar]
- Kerepesi I, Galiba G. Osmotic and salt stress-induced alteration in soluble carbohydrate content in wheat seedlings. Crop Sci. 2000;40:482–487. doi: 10.2135/cropsci2000.402482x. [DOI] [Google Scholar]
- Khelil A, Menu T, Ricard B. Adaptive response to salt involving carbohydrate metabolism in leaves of a salt-sensitive tomato cultivar. Plant Physiol Biochem. 2007;45:551–559. doi: 10.1016/j.plaphy.2007.05.003. [DOI] [PubMed] [Google Scholar]
- Kötting O, Kossmann J, Zeeman SC, Lloyd JR. Regulation of starch metabolism: the age of enlightenment. Curr Opin Plant Biol. 2010;13:321–329. doi: 10.1016/j.pbi.2010.01.003. [DOI] [PubMed] [Google Scholar]
- Lee KS, Choi WY, Ko JC, Kim TS, Gregorio GB. Salinity tolerance of japonica and indica rice (Oryza sativa L.) at seedling stage. Planta. 2003;216:1043–1046. doi: 10.1007/s00425-002-0958-3. [DOI] [PubMed] [Google Scholar]
- Lee SK, Hwang SK, Han M, Eom JS, Kang HG, Han Y, Choi SB, Cho MH, Bhoo SH, An G, Hahn TR, Okita TW, Jeon JS. Identification of the ADP-glucose pyrophosphorylase isoforms essential for starch synthesis in the leaf and seed endosperm of rice (Oryza sativa L.) Plant Mol Biol. 2007;65:531–546. doi: 10.1007/s11103-007-9153-z. [DOI] [PubMed] [Google Scholar]
- Liu T, van Staden J. Partitioning of carbohydrate in salt-sensitive and salt-tolerant soybean callus cultures under salinity stress and its subsequent relief. Plant Growth Regul. 2001;33:13–17. doi: 10.1023/A:1010687711334. [DOI] [Google Scholar]
- Loggini B, Scartazza A, Brugnoli E, Navari-Izzo F. Antioxidant defense system, pigment composition, and photosynthetic efficiency in two wheat cultivars subjected to drought. Plant Physiol. 1999;119:1091–1099. doi: 10.1104/pp.119.3.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malagoli P, Britto DT, Schulze LM, Kronzucker HJ. Futile Na+ cycling at the root plasma membrane in rice (Oryza sativa L.): kinetic, energetics, and relationship to salinity tolerance. J Exp Bot. 2008;59:4109–4117. doi: 10.1093/jxb/ern249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour MMF, Salama KHA. Cellular basis of salinity tolerance in plants. Environ Exp Bot. 2004;52:113–122. doi: 10.1016/j.envexpbot.2004.01.009. [DOI] [Google Scholar]
- Maxwell K, Johnson GN. Chlorophyll fluorescence – a practical guide. J Exp Bot. 2000;51:659–668. doi: 10.1093/jexbot/51.345.659. [DOI] [PubMed] [Google Scholar]
- McCleary BV, Monaghan DA. Measurement of resistant starch. J AOAC Inter. 2002;85:665–675. [PubMed] [Google Scholar]
- Morsy MR, Jouve L, Hausman JF, Hoffmann SJM. Alteration of oxidative and carbohydrate metabolism under abiotic stress in two rice (Oryza sativa L.) genotypes contrasting in chilling tolerance. J Plant Physiol. 2007;164:157–167. doi: 10.1016/j.jplph.2005.12.004. [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant. 1962;15:473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x. [DOI] [Google Scholar]
- Orzechowski S. Starch metabolism in leaves. Acta Biochim Polonic. 2008;55:435–445. [PubMed] [Google Scholar]
- Praxedes SC, de Lacerda CF, Ferreira TM, Prisco JT, DaMatta FM, Gomes-Filho E. Salt tolerance is unrelated to carbohydrate metabolism in cowpea cultivars. Acta Physiol Plant. 2011;33:887–896. doi: 10.1007/s11738-010-0615-6. [DOI] [Google Scholar]
- Qadir M, Tubeileh A, Akhtar J, Larbi A, Minhas PS, Khan MA. Productivity enhancement of salt–affected environments through crop diversification. Land Degrad Develop. 2008;19:429–453. doi: 10.1002/ldr.853. [DOI] [Google Scholar]
- Rosa M, Hilal M, González JA, Prado FE. Low-temperature effect on enzyme activities involved in sucrose-starch partitioning in salt-stressed and salt-acclimated cotyledons of quinoa (Chenopodium quinoa Willd.) seedlings. Plant Physiol Biochem. 2009;47:300–307. doi: 10.1016/j.plaphy.2008.12.001. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- Senadheera P, Singh RK, Maathuis FJM. Differentially expressed membrane transporters in rice roots may contribute to cultivar dependent salt tolerance. J Exp Bot. 2009;60:2553–2563. doi: 10.1093/jxb/erp099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanji KK. Salinity in the soil environment. In: Lauchli A, Luttge U, editors. Salinity environment–plant–molecules. Dordrecht: Kluwer Academic; 2002. pp. 21–51. [Google Scholar]
- Tetlow IJ. Understanding storage starch biosynthesis in plants: a means to quality improvement. Can J Bot. 2006;84:1167–1185. doi: 10.1139/b06-089. [DOI] [Google Scholar]
- Tiwari BS, Bose A, Giiosii B. Photosynthesis in rice under a salt stress. Photosynthetica. 1997;34:303–306. doi: 10.1023/A:1006857027398. [DOI] [Google Scholar]
- Voigt EL, Almeida TD, Chagas RM, Ponte LFA, Viégas RA, Silveira JAG. Source-sink regulation of cotyledonary reserve mobilization during cashew (Anacadium occidentale) seedling establishment under NaCl salinity. J Plant Physiol. 2009;166:80–89. doi: 10.1016/j.jplph.2008.02.008. [DOI] [PubMed] [Google Scholar]
- Walia H, Wilson C, Condamine P, Liu X, Ismail AM, Zeng L, Wanamaker SI, Mandal J, Xu J, Cui X, Close TJ. Comparative transcriptional profiling of two contrasting rice genotypes under salinity stress during the vegetative growth stage. Plant Physiol. 2005;139:822–835. doi: 10.1104/pp.105.065961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SJ, Liu LF, Chen CK, Chen LW. Regulation of granule-bound starch synthase I gene expression in rice leaves by temperature and drought stress. Biol Plant. 2006;50:537–541. doi: 10.1007/s10535-006-0085-2. [DOI] [Google Scholar]
- Wang RL, Hua C, Zhou F, Zhou QC. Effects of NaCl stress on photochemical activity and thylakoid membrane polypeptide composition of a salt-tolerant and a salt-sensitive rice cultivar. Photosynthetica. 2009;47:125–127. doi: 10.1007/s11099-009-0019-2. [DOI] [Google Scholar]
- Yin YG, Kobayashi Y, Sanuki A, Kondo S, Fukuda N, Ezura H, Sugaya S, Matsukura C. Salinity induces carbohydrate accumulation and sugar-regulated starch biosynthetic genes in tomato (Solanum lycopersicum L. cv. ‘Micro-Tom’) fruits in an ABA- and osmotic stress-independent manner. J Exp Bot. 2010;61:563–574. doi: 10.1093/jxb/erp333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeeman SC, Smith SM, Smith AM. The diurnal metabolism of leaf starch. Biochem J. 2007;401:13–28. doi: 10.1042/BJ20061393. [DOI] [PubMed] [Google Scholar]
- Zeeman SC, Kossmann J, Smith AM. Starch its metabolism, evolution, and biotechnological modification in plants. Annu Rev Plant Biol. 2010;61:209–234. doi: 10.1146/annurev-arplant-042809-112301. [DOI] [PubMed] [Google Scholar]