Abstract
A rapid and efficient sap inoculation method for tobacco streak virus (TSV) was developed in sunflower. Sap from TSV-infected sunflower plants was freshly extracted in phosphate buffer and diluted serially from 10−1 to 10−8. Two-day old seedlings of sunflower were injured at the meristem and immersed in the sap for 10 min, maintained at 20 °C for 2–3 days and shifted to greenhouse. The surviving seedlings in the respective sap dilution were scored for symptoms of sunflower necrosis disease (SND). SND symptoms were seen in 80 % of the seedlings inoculated with a sap dilution of 10−5. ELISA and RT-PCR analysis of coat protein and movement protein of TSV confirmed SND symptoms. The methodology was also found to be reproducible when the sap from the infected plants was inoculated onto healthy plants. The main aim of the study was to develop a primary screening strategy for the selection of transgenics developed for SND resistance. This methodology can also be extended for the analysis of resistance against other viruses.
Keywords: Illarvirus, Sap inoculation, Sunflower, Sunflower necrosis disease, Tobacco streak virus, Thrips
Introduction
Despite substantial advances in plant disease control strategies, our global food supply is still threatened by a multitude of pathogens and pests (Garelik 2002). Sunflower, a major edible oilseed crop of importance is susceptible to several diseases caused by various agents including virus (Nagaraju 1995). One such virus causing severe reduction in growth leading to yield loss is the tobacco streak virus (TSV).
Tobacco streak virus belonging to the serogroup Illarvirus sub group 1; genus Illarvirus and the family Bromoviridae, was first reported on Nicotiana tabacum (Van vloten-Doting 1981). The virus is seed, pollen and thrips-transmitted (Johnson et al. 1984; Cupertino et al. 1984).
Symptoms of sunflower necrosis disease (SND) comprises of chlorotic and necrotic ringspots, leaf distortion, leaf and stem necrosis and eventual death (Ravi et al. 2001). Identification/development of virus-resistant cultivars has profound importance. Availability of an efficient screening strategy for the identification of resistant plants is very important. Mechanical sap inoculation for the analysis of virus transmission involves manually swabbing the extracted sap added with abrasives onto the leaf surface and scoring for symptoms (Ajith Prasad and Nagaraju 2005). These virus transmission studies depend on various factors like, plant age, temperature, source of the inoculum, buffer with additives like antioxidants and abrasives (carborundum and celite), anti inhibitors of sap transmission etc. The sap inoculation techniques besides being cumbersome and laborious also result in escapes that make them unreliable. Therefore, evolution and standardization of efficient high throughput sap inoculation methodologies for various viruses has received greatest emphasis (Mandal et al. 2002, 2008; Laidlaw 1987). The present study describes the establishment of a novel, high throughput and less labour intensive mechanical sap inoculation method for the transmission of TSV in sunflower.
Material and methods
Plant material and sap extraction
As a source of inoculum, 1 g freshly infected young sunflower leaves from sunflower necrosis diseased plants (preferably before flowering) was extracted in ice cold 0.01 M phosphate buffer (pH 7) with a tissue to buffer ratio of 1:5. The sap was later filtered through muslin cloth and maintained under ice cold condition. To test the efficacy of sap or inoculum, it was diluted serially from 10−1 to 10−8 such that the final volume was 10 ml.
Sap inoculation
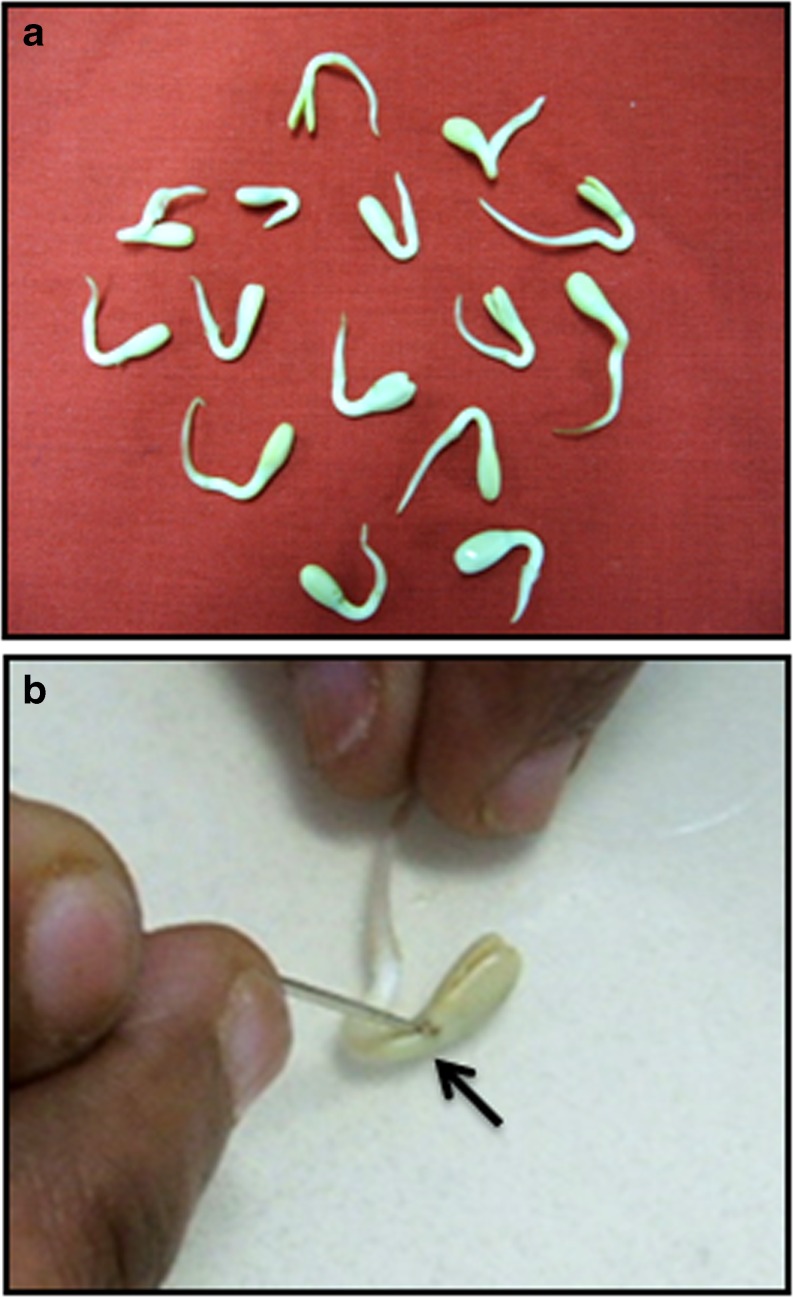
Two day old germinating seedlings of SND-susceptible sunflower cv, RHA 95-C-1 (procured from National Seeds Project, University of Agricultural Sciences, GKVK, Bangalore) (Fig. 1a) were pricked at the meristem with a needle to enhance the entry of viral particles (Fig. 1b). The seedlings were later immersed in different dilutions of viral sap for 10 min. Three replications of 10 seedlings each were inoculated per sap dilution. During the entire process care was taken to maintain the sap on ice. Similarly, a set of absolute control was maintained where only distilled water was used. The seedlings were later transferred to the soil pan which was maintained at 20 °C for 2–3 days. They were later maintained under greenhouse conditions and scored for germination, curling of newly opened leaves, chlorosis and other symptoms of SND. The explants were further confirmed for the systemic infection by analysing the expression and transcript accumulation of TSV proteins.
Fig. 1.
Sap inoculation of tobacco streak virus. a. Two day old seedlings of sunflower cv. 95-c-1 selected for sap inoculation. b. Targeting the viral particles in the sap to the meristem by wounding with a needle
Confirmation of systemic infection by ELISA and RT-PCR
Direct antigen coated ELISA was performed using antibody against coat protein (gift from Dr. R. K. Jain, IARI, New Delhi), following the standardized protocol in our laboratory (Balol 2008). Antibody specific to coat protein was used in 1:4,000 dilution for the expression analysis of the coat protein gene with 5 μg total protein as the antigen source.
Total RNA was extracted according to the protocol described by Sambrook et al. (1989). For RT-PCR, about 5 μg of total RNA was reverse transcribed to single stranded cDNA using M-MuLV reverse transcriptase (MBI Fermentas). Initially, a 10 μl reaction mixture [consisting of 5 μg of total RNA, 1 μl of Oligo (dT)18 primer (10 pmol/μl) and RNase free sterile distilled water] was prepared in a 0.2 ml PCR tube and incubated at 70 °C for 5 min (to remove any secondary structure) and immediately placed on ice. Subsequently, 10 μl of another reaction mixture [consisting of 2 μl of dNTPs (10 mM), 4 μl of M-MuLV RT buffer (5X), 50 U of M-MuLV Reverse Transcriptase (MBI-Fermentas) and sterile water] was added and briefly spinned. The resulting 20 μl reaction mixture was incubated at 42 °C for 1 h. Further, the enzyme was inactivated by incubating the reaction mixture at 90 °C for 5 min. Subsequently, RT-PCR was carried out using primers for coat protein (forward 5′ ATGAATACTTTGATCCAAGG 3′; reverse 5′ TCAGTCTTGATTCACCAG 3′) and movement protein (forward 5′ GAGACATCTCTGAAGAAGGC 3′; reverse 5′ CGACGGAAGCTTGACCACG 3′) of TSV. The PCR reaction mixture (20 μl) contained 0.3 U Taq DNA polymerase (Bangalore Genei, Bangalore, India) 1X assay buffer (10 mM pH 9.0 Tris–HCl, 50 mM KCl, 1.5 mM MgCl2, 0.01 % gelatin), 150 μM of each dNTP, 1 μl of each forward and reverse primer at a final concentration of 0.25 μM and 100 ng template DNA. The PCR (Eppendorf, Germany) reaction profile comprised of 35 cycles, with strand separation at 94 °C for 30 s, annealing at 60 °C for 30 s and extension at 72 °C for 1 min. The program was extended for 10 min at 72 °C. The products were electrophoresed on a 1 % agarose gel, stained with ethidium bromide and visualized under ultraviolet light (Sambrook et al. 1989). The amplified PCR product was further confirmed for the coat protein gene by cloning the purified PCR product (gel eluted by Gel extraction kit, MBI Fermentas) into T/A cloning vector pTZ57R/T (MBI Fermentas) for sequencing (ABI Prism). The sequence obtained was aligned with the original sequence using CLUSTALW bioinformatics software tool.
Results and discussion
The seedlings could be easily scored for SND, 10 days after sap inoculation. Typical symptoms of puckering and twisting of leaves were observed in the emerging seedlings (Fig. 2a) when compared to the lack of symptoms in the healthy plants maintained as absolute control (Fig. 2b). There was variation in the number of seedlings that developed symptoms with the increase in the dilution of the sap. In the sap dilution of 10−1 to 10−3, 10–15 % of the germinated seedlings showed symptoms. The percentage of infected seedlings increased to 40 % upon dilution to 10−4. It was observed that with the dilution of the sap to 10−5, a maximum of 80 % of the germinated seedlings displayed SND symptoms. The symptoms gradually decreased with further increase in the dilutions (Table 1). The reduction in the appearance of symptoms when the sap dilution was between 10−1 and 10−3 could be due to the agglutination of viral particles in the highly concentrated sap (Hull 2002). Experiments suggested that the optimum dilution to produce high efficiency symptoms was 10−5 which decreased on further dilution, due to the reduction in the density of the viral particles. Further confirmatory analysis was carried out with the infected seedlings raised from 10−5 sap dilution. ELISA for coat protein in the 10 day old seedlings with progressive symptoms indicated the presence of the tobacco streak virus particles in the tissues of the germinated seedlings (Fig. 3d). ELISA (OD) values in the plants with SND symptoms ranged between 1.1 and 1.6, when compared to 0.2 in the seedlings maintained as absolute control. In order to confirm the consistency of the SND symptoms, the seedlings were analysed (Fig. 3a–c) for the persistence of TSV and thereby SND symptoms, 20 days after infection. High ELISA values were observed in the leaves of the seedlings infected with TSV (Fig. 3d) when compared to nearly negligible values in the absolute control. To corroborate the expression analysis, RT-PCR (Fig. 3e) confirmed amplification of two TSV-specific proteins, a 717 bp coat protein (Fig. 3e (i)) and a 792 bp movement protein (Fig. 3e (ii)). To prove the identity of the amplified product, the RT-PCR product from the sap-inoculated seedling was sequenced and compared with the known sequence of the tobacco streak virus coat protein using the multiple sequence alignment CLUSTALW tool. It was found that there was 100 % homology between the sequences confirming that the sap-inoculated seedlings were indeed infected by TSV. This further established the persistence of the virus and therefore efficacy of the methodology.
Fig. 2.
Response of the sap-inoculated seedlings to TSV 10 days after inoculation. a. Sap inoculated seedlings with SND symptoms b. Absolute control
Table 1.
Variation in infection with sap dilution (t1-t8 are sap dilutions 10−1 to 10−8)
| Sap dilutions | Germinated seeds | Infected seeds |
|---|---|---|
| t1 | 9.33 | 1 |
| t2 | 9.33 | 1.33 |
| t3 | 8.66 | 1.33 |
| t4 | 9.33 | 4 |
| t5 | 9.33 | 8 |
| t6 | 9.66 | 2 |
| t7 | 10 | 0 |
| t8 | 9 | 0 |
| SEm± | 0.35 | 0.412 |
| CV % | 6.56 | 54.74 |
Fig. 3.
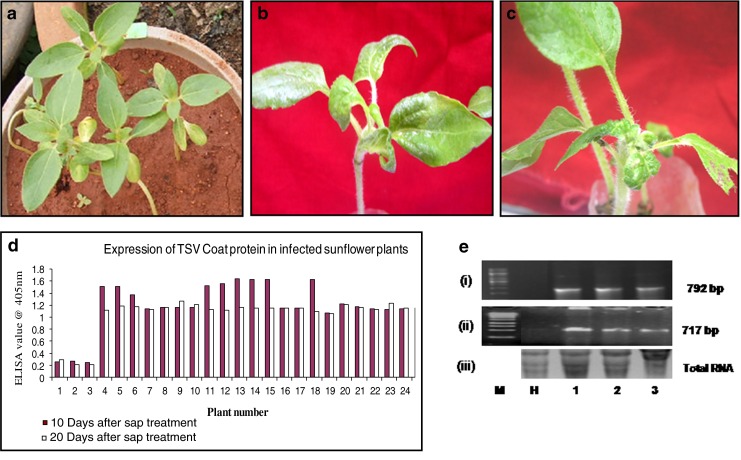
a–c: Response of the sap inoculated seedlings to TSV 20 days after inoculation. a: absolute control; b & c: sap inoculated seedlings with SND symptoms. d: Expression analysis by ELISA for coat protein 10 days and 20 days after sap inoculation in the inoculated and uninoculated (absolute control) seedlings. Bars 1–3: absolute control; Bars 4–25: sap inoculated seedlings showing SND symptoms. e: RT-PCR analysis. (i) RT-PCR analysis using movement protein gene specific primers (ii) RT-PCR analysis using coat protein gene specific primers; (iii) Total RNA as loading control. Lane M: marker; Lane H: healthy absolute control; Lanes 1–3: amplified products from sap inoculated seedlings
The authenticity and reproducibility of the methodology was further confirmed when the sap from the infected seedlings induced TSV symptoms in 20 days old healthy sunflower plants (Fig. 4a). The unswabbed controls however did not show any symptoms (Fig. 4b). PCR analysis confirmed the presence of the tobacco streak virus in the tissues when a 717 bp product of the coat protein gene amplified in the tissue which exhibited the symptoms whereas the band was absent in the tissue without TSV symptoms (Fig. 4c). A similar amplified product was also obtained from a TSV-infected plant from the field further confirming TSV infection. This proved unequivocally the credibility of the sap inoculation methodology.
Fig. 4.
Reproducibility of the sap-inoculation method. a Appearance of the symptoms of sunflower necrosis disease in 20-days old healthy sunflower seedlings following inoculation of the sap from the infected seedlings; b Absence of the symptoms in healthy uninoculated seedlings; c PCR analysis of the coat protein gene to confirm the presence of TSV. Lanes 1: marker; lanes 2: DNA from a TSV-infected plant from the field; Lane 3: DNA from the sap-inoculated seedlings with TSV symptoms; lane 4: DNA from the seedlings with no symptoms
The study has demonstrated that a high throughput methodology for sap inoculation of TSV has been developed and the symptoms confirmed as SND by both expression and transcript accumulation of the viral proteins. Conventionally, sap inoculation of TSV has been through the swabbing of leaf lamina along with an abrasive that makes it a very tedious and cumbersome procedure. This is a very first report which allows rapid screening of a large number of seedlings in sunflower for resistance to tobacco streak virus. Such non conventional methods have been developed earlier (Hoffmann et al. 2001; Helleco-Kervarrec et al. 2002; Aebig et al. 2005). The present methodology was designed in such a way that the dilution of sap should be enough to induce detectable symptoms of SND. The utility of the methodology is seen in transmission studies and in rapid screening for resistance against TSV in sunflower germplasm as well as a primary screening of transgenics.
References
- Aebig JA, Kamo K, Hsu H. Biolistic inoculation of gladiolus with cucumber mosaic cucumovirus. J Virol Meth. 2005;123:89–94. doi: 10.1016/j.jviromet.2004.09.010. [DOI] [PubMed] [Google Scholar]
- Ajith Prasad HN, Nagaraju Transmission of sunflower necrosis virus disease through sap, seed, insect vector and pollen. Environ Econ. 2005;23:125–128. [Google Scholar]
- Balol GB (2008) Development of transgenic sunflower expressing coat protein gene their characterisation and evaluation. M.Sc. (Agri.) Thesis, University of Agricultural Sciences, Bangalore, 121 pp
- Cupertino FP, Grogan RG, Petersen LJ, Kimble KA. Tobacco streak virus infection of tomato and some natural weed hosts in California. Plant Dis. 1984;68:331–333. doi: 10.1094/PD-68-331. [DOI] [Google Scholar]
- Doting VV. Tobacco streak virus (nucleic acid) Acta Horticult. 1981;76:456. [Google Scholar]
- Garelik G. Taking the bite out of potato blight. Sci. 2002;298:1702–1704. doi: 10.1126/science.298.5599.1702. [DOI] [PubMed] [Google Scholar]
- Helleco-kervarrec C, Riault G, Jacquot E. Biolistic–mediated inoculation off immature wheat embryos with barley yellow dwarf virus-PAV. J Virol Meth. 2002;102:161–166. doi: 10.1016/S0166-0934(01)00446-3. [DOI] [PubMed] [Google Scholar]
- Hoffmann K, Verbeek M, Romano A, Dullemans AM, Van den Heuvel JFJM, Van de Walk F. Mechanical transmission of Poleroviruses. J Virol Meth. 2001;91:197–201. doi: 10.1016/S0166-0934(00)00256-1. [DOI] [PubMed] [Google Scholar]
- Hull R (eds) (2002) Transmission 2: Mechanical, seed, pollen and epidemiology. In Mathew’s Plant Virology. Academic Press, New York, pp 533–581
- Johnson HA, Jr, Converse RH, Amorao A, Esperjo JI, Fruzier NW. Seed transmission of tobacco streak virus in strawberry. Plant Dis. 1984;68:390–392. doi: 10.1094/PD-68-390. [DOI] [Google Scholar]
- Laidlaw WMR. A new method for mechanical transmission and factors affecting its sensitivity. EPPO Bulletin. 1987;17:81–89. doi: 10.1111/j.1365-2338.1987.tb00011.x. [DOI] [Google Scholar]
- Mandal B, Pappu HR, Culbreath AK, Holbrook DCC, Gorbet JW, Todd W. Differential response of selected peanut (Arachis hypogaea) genotypes to mechanical inoculation by tomato. Plant Dis. 2002;9:939–944. doi: 10.1094/PDIS.2002.86.9.939. [DOI] [PubMed] [Google Scholar]
- Mandal B, Csinos AS, Martinez-Ochoa N, Pappu HR. A rapid and efficient inoculation method for tomato spotted wilt tospovirus. J Virol Meth. 2008;149:195–198. doi: 10.1016/j.jviromet.2007.12.007. [DOI] [PubMed] [Google Scholar]
- Nagaraju (1995) Studies on sunflower mosaic virus disease. Ph.D. Thesis, University of Agricultural Sciences, Bangalore, 157 pp
- Ravi KS, Buttgereitt A, Kitkaru AS, Deshmukh S, Lesemann DE, Winter S. Sunflower necrosis disease from India is caused by an ilarvirus related to Tobacco streak virus. New Dis Rep. 2001;3:141. [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. 2. Cold Spring Harbour: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]