Abstract
The Wall-Associated Kinase, one of the receptor-like kinase (RLK) gene families in plant, plays important roles in cell expansion, pathogen resistance and heavy metal stress tolerance in Arabidopsis thaliana. Here, we isolated a cDNA encoding a novel WAK from indica rice and designated as OsiWAK1 (Oryza sativa indica WAK-1). In this study, the RNAi construct with OsiWAK1 gene cloned in sense and antisense orientation separated by a functional intron under constitutive promoter, was introduced through biolistic gene gun method into the rice cultivar “IR-50” to determine the effect of OsiWAK1 transcript silencing on rice plant development. Examination of the transgenic plants reveals that OsiWAK1 transcript silencing in rice results in dwarf plants because of the reduction in the size of leaves, flag-leaves, internodes and panicle. The development of root primordia during germination, root hairs and lateral rooting was also effected. Microscopic analysis revealed that the decrease in size is due to reduction in the cell size but not the number of cells. In addition, the transgenic plants also exhibited sterile phenotype due to anther indehiscence and 40 % reduction in pollen viability. These data suggest that OsiWAK1 may play an important role in rice plant growth and development.
Keywords: Anther indehiscence, Oryza sativa ssp indica, OsiWAK1, RNAi, WAK
Introduction
Plant development is related to the morphological changes of cells and tissue, which is caused by structural modification and reorganization of wall components and the synthesis and insertion of new material into the existing wall. The biochemical and physical factors that regulate wall modification and expansion are not fully understood. It is known that the cell shape and the mechanical forces dictate several processes that directly or indirectly effect the plant development. The integrated cytoskeleton and its dynamic adhesion to the plasma membrane allow these complex interactions between mechanical forces and chemical signals (Geiger and Bershadsky 2001; Sheetz 2001). Thus, efficient communication between the plant cell wall and the cytoplasm is important in plant development and in responding to several external stimuli (Kohorn 2000; Brownlee 2002; Somerville et al. 2004). Understanding the mechanism of this communication is fundamentally important. Transmembrane proteins of the receptor-like kinase (RLK) super family that perceive external stimuli by their extracellular domains and transmit the signals via their cytoplasmic kinase domains (Shiu and Bleecker 2001; Morris and Walker 2003) have the potential to play important role in this communication. Recent studies have suggested that RLKs are ubiquitous throughout the plant kingdom (Verica and He 2002; Shiu et al. 2004), but the precise function for most of the RLKs has not been defined (Becraft et al. 1996; Clark et al. 1997; Yokoyama et al. 1998; Jinn et al. 2000; Takasaki et al. 2000). Out of numerous proteins found in the cell wall (Kohorn 2000; Baluska et al. 2003), only a few have the potential to act as molecular linkers between the elements of the cytoskeleton and components of cell wall. These include proteins like the arabinogalactan proteins (AGPs; Nothnagel 1997), COBRA (Schindelman et al. 2001), wall-associated Kinases (WAKs; Wagner and Kohorn 2001; reviewed in Kanneganti and Gupta 2008), formins (Deeks et al. 2002) and proline rich extension-like receptor kinases (PERKs; Nakhamchik et al. 2004). Among these, only the WAKs and PERKs have the potential to signal directly to the cytoplasm via the protein kinase domain.
Cell Wall-associated kinases (WAKs) belong to unique subfamily of plant RLKs (He et al. 1996, 1999), that in addition to their association with the cell wall, display the typical eukaryotic Ser/Thr kinase signature and an extra-cytoplasmic domain (ectodomain) containing several EGF-like repeats (Kohorn 2000; Lally et al. 2001; Wagner and Kohorn 2001; Kohorn et al. 2006a, b). WAKs are encoded by five tightly linked and highly similar genes (WAK1-WAK5) in Arabidopsis thaliana, and are expressed in leaves, meristems and cells undergoing expansion (He et al. 1996, 1999; Wagner and Kohorn 2001). WAK gene expression is also induced by pathogens, wounding, Aluminum toxicity and numerous other stresses (He et al. 1996; Wagner and Kohorn 2001; Sivaguru et al. 2003). Functional studies of different WAK members in Arabidopsis demonstrated that they are involved in various functions of plants, including pathogen resistance (He et al. 1998), heavy metal tolerance (Sivaguru et al. 2003) cell elongation and plant development (Lally et al. 2001; Wagner and Kohorn 2001).
Biochemical and functional studies demonstrated that the WAK’s are localized in the plasma membrane and are covalently bound to pectin in the cell wall (Wagner and Kohorn 2001) and are required for cell expansion, in part because of their regulation of invertase transcriptional activity, and hence solute concentrations, within the cell (Kohorn 2000; Lally et al. 2001; Wagner and Kohorn 2001; Kohorn et al. 2006a). The extracellular non-EGF domain of WAK1 and WAK2 was shown to bind to pectin in vitro and this is enhanced by a calcium-induced bridging of the pectin (Decreux and Messiaen 2005; Decreux et al. 2006). Also, it has been shown that the assembly and crosslinking of WAKs may begin at an early stage within a cytoplasmic compartment rather than in the cell wall itself, and is coordinated with synthesis of surface cellulose (Kohorn et al. 2006b). Moreover, WAK2 has been recently shown to be required for the activation by pectin of numerous genes in protoplasts, including that encoding a vacuolar invertase (Kohorn et al. 2009). In addition, WAK1 has been shown to be a component of an approximately 500-kDa protein complex that includes a gly rich extracellular protein AtGRP-3 (Park et al. 2002) and a cytoplasmic type 2C protein phosphatase, KAPP (Anderson et al. 2001), that has potential for signaling. Apart from pectin, WAK1 was shown to mediate perception of oligogalacturonides (OGs) (Brutus et al. 2010), that function as danger signals and induce the expression of defense genes and proteins protecting plants against fungal diseases apart from affecting several aspects of plant growth and development. Apart from WAKs, WAK-like gene clusters were also found in Arabidopsis (Verica and He 2002; Verica et al. 2003) and play a role in pathogen defence (Meier et al. 2010).
The existence of WAKs and WAKLs is not unique to Arabidopsis. Studies done using anti-WAK antibody, revealed the presence of immunologically related proteins in pea, tobacco and maize. Expressed sequence tags (ESTs) for WAK/WAKL genes have been identified in tomato, soybean, wheat and rice (Verica and He 2002). WAK gene family is greatly expanded in the rice with the identification of 125 WAKs (OsWAKs) in the sequenced rice genome (Zhang et al. 2005). Such an increase in size of tandemly repeated and clustered gene families is correlated with a role in disease resistance and response to environmental stresses (Richter and Ronald 2000; Lespinet et al. 2002). However, functions of these putative OsWAK genes are yet to be determined. The ubiquitous distribution of WAKs implies their potential roles in plants life processes. However, the biological roles of WAKs are little known in these plants except for Arabidopsis. There is a single report of functional characterization of OsWAK genes. Recently in 2009 Li et al., reported isolation of a novel WAK from rice, named as OsWAK1 that play an important role in rice blast disease resistance. In this paper we report the identification of another novel wall-associated protein kinase, OsiWAK1, from indica rice, which is induced during organogenesis of rice calli. RNAi approach was exploited to understand the role of OsiWAK1 in plant development. Analysis of the transgenic rice plants with either reduced or zero levels of OsiWAK1 transcript accumulation suggested that this gene plays an important role in overall plant development by regulating the cell expansion.
Materials and methods
Plant material and tissue culture conditions
Indica rice cv IR-50 seeds obtained from Farm Aid service, Tamilnadu, India were used throughout the study. Rice tissue culture conditions were as previously described (Anoop and Gupta 2003) with little modifications. In brief, the de-husked seeds were surface sterilized in 0.1 % HgCl2 for 4 min, washed thoroughly in sterile distilled water, followed by 70 % ethanol treatment for 40 s. The seeds were then washed with sterile water and cultured in MS medium supplemented with 2 mg l−1 2,4-D and 0.5 mg l−1 Kinetin (Callus Induction Medium; CIM). The cultures were incubated in dark at 25 ± 2 °C for callusing. Embryogenic calli formed after 28 days were transferred to callus regeneration medium (CRM; MS basal supplemented with 5 mg l−1 benzyl amino purine and 0.5 mg l−1 kinetin) and kept at 16 h photoperiod with light intensity of 110–130 μEm−1 s−1 for different number of days. The calli thus obtained were used for RNA extraction and subsequent analysis.
Plant growth conditions and stress treatments
Healthy seeds were surface sterilized with 70 % ethanol, rinsed thoroughly, soaked in double distilled water overnight, and the seedlings were raised on the water saturated cotton beds in a Biotron PH100 growth chamber (Nippon, Japan) maintained at 28 °C, 80 % relative humidity and 16 h photoperiod (180 μEm−1 s−1). After 8 days of growth, they are transferred to pots containing soil and were grown to maturity in the green house. For RNA extraction, the root and shoot samples were collected from 8-day-seedlings and the leaves, leaf sheaths and pre-pollinated inflorescence samples were collected from 90-day-old rice plants grown in green house.
Extraction of nucleic acids
Total RNA was isolated using TRIreagent kit (Sigma Aldrich, Bangalore, India) according to the manufacturer’s protocol. Total plant DNA was extracted from the 2-week old rice seedlings by Dellaporta method (Dellaporta et al. 1983).
Isolation of OsiWAK1
Total RNA was extracted from the embryogenic calli (grown on CIM) and also from the differentiating calli (cultured on CRM) at various time intervals (4, 8, 12 days). To ensure that the samples were free of DNA, the RNA samples were treated with amplification grade RNase- free DNase (Gibco BRL) prior to RT-PCR. First strand cDNA was synthesized from 1 μg of RNA with 1 μM OligodT (15 mer), 1 μM random hexamers, 1X RT buffer, 10 mM dNTPmix (Gibco BRL), 10 U RNase inhibitor (Gibco BRL) and 200 U M-MLV reverse transcriptase (Gibco BRL). The reaction mixture was incubated at 37 °C for 1 h and then M-MLV was heat inactivated at 92 °C for 2 min. PCR was performed in a volume of 25 μl containing the appropriate amount of cDNA template, 20 μM each of dNTP, 2 mM MgCl2, 5 μM of random primer (decamers), 1X PCR buffer and 1 U Taq DNA Polymerase (MBI Fermentas). The PCR was programmed for 45 cycles at 94 °C for 30 s, 35 °C for 30 s, 72 °C for 2 min followed by a final extension at 72 °C for 7 min. Twenty microliter of each PCR product was analyzed by agarose gel electrophoresis on a 1.5 % agarose gel stained with ethidium bromide (0.5 μg/ml). To ensure that approximately equal amounts of cDNA from different samples were used for amplification, we also performed control PCR reactions with the housekeeping gene GAPDH. A differential amplicon of 984 bp (A984) obtained from the RNA extracted from the rice callus incubated for 12 days on the CRM with the random primer 5′-AAGAGCCCGT-3′ was electro-eluted, subcloned into pGEM-T vector (Promega) and was sequenced on an ABI prism automated sequence analyzer at Microsynth (Switzerland) using T7 and SP6 primers.
Rapid amplifiction of cDNA ends (RACE)
Completion of the full-length cDNA sequence was done using First choice RLM-RACE kit (Ambion, 3′ and 5′ Rapid Amplification of 3′ and 5′ ends) according to manufacturer’s instructions. Total RNA from roots of 8-day-old rice seedlings was used as template to generate 5′ and 3′- RACE cDNA. 3′ RACE fragment was obtained by using Ambion’s 3′ RACE adapter specific outer and inner primers with OsiWAK1 gene-specific 3′ outer and inner primers, 3′ ROP (5′-CGCCCACAGCTACTGTGTAA-3′) and 3′ RIP (5′-TAAGGGACTCCACGATGACC-3′) respectively. Same way 5′ RACE fragment was obtained by using Ambion’s 5′ adapter specific outer and inner primers with OsiWAK1 gene specific 5′ outer and inner primers 5′ ROP (5′-AGGTCAGGTCCATGAAGCAC-3′) and 5′ RIP (5′-CGTTGCTAGTGTGGCGACTA -3′). To ensure the specificity of the 5′ and 3′ RACE fragments, care was taken in designing the gene specific primers such that the RACE products have an overlapping region of at least 400 bp with the 984 bp differential amplicon. The PCR conditions used for RACE are as follows: The PCR reaction was set in a final volume of 25 μl with 1X PCR buffer, 200 μM each of dNTP, 10 μM of each primer and 1 U Taq DNA polymerase (MBI Fermantas). The cycling parameters were set to 95 °C for 3 min for initial denaturation followed by 35 cycles of 95 °C for 30 s, 55 °C for 30 s and 72 °C for 90 s followed by a final extension at 72 °C for 7 min. The 5′ and 3′ RACE products were subcloned into pGEM-T vector and sequenced by using T7 and SP6 primers.
Semi-quantitative RT-PCR
Semi-quantitative RT-PCR was conducted with the primers 3′ RIP and 5′ ROP used in RACE analysis to analyze OsiWAK1 mRNA accumulation in different plant organs. These primers amplify a 292 bp region. Another pair of primers, GAF1 (5′-GAAGCACAGCGACATCAAGA-3′) and GAR1 (5′-TTGACACCGCAGACAAACAT-3′) was used to amplify 256 bp region from rice GAPDH that serves as a control. The DNase treated RNA samples were used as templates in RT-PCR. The first strand cDNA was synthesized as described above. PCR reactions were performed in a final volume of 25 μl containing the normalized amounts of c-DNA template, 10 μM each of the gene-specific primers, 0.5 mM each of dNTPs, 1X PCR buffer, 2.5 mM MgCl2 and 1 U TaqDNA polymerase (MBI Fermentas). The PCR cycle conditions were set at 95 °C for 3 min followed by 25 (for GAPDH) or 35 cycles (for OsiWAK1) of 95 °C for 45 s, 55 °C for 45 s, 72 °C for 45 s and a final extension at 72 °C for 7 min. Total volume of the PCR product was loaded and analyzed by electrophoresing on 1.5 % agarose gel.
RNA interference construction and plant transformation
For the expression of small hairpin RNA (shRNA) targeted to the OsiWAK1, a shRNA-encoding plasmid was constructed using pKANNIBAL vector (Wesley et al. 2001). First, using PCR amplification, two 440 nt “arms” were made from the C-terminus of the OsWAK2 gene (nt 1,061–2,042) in opposite orientations. For this, primers WAKSF1 = 5′-CTCGAGTGGACCTGACCTACGGGC-3′ (XhoI site is underlined) and WAKSR1 = 5′- GAATTCGGCTGAAGGCTGTGGGC 3′ (EcoRI site is underlined) were used for preparing the “sense arm” and primers WAKAF2 = 5′-GGATCCTGGACCTGACCTACGGGC (BamHI site is underlined) and WAKAR2 = 5′-AAGCTTGGCTGAAGGCTGTGGGC-3′ (HindIII site is underlined) were used for preparing the “antisense arm”. These arms were then sub-cloned into XcmI digested pXcmKn12 (NIG, Japan). These constructs were denoted as pXcmWAK-‘sense’ and pXcmWAK-‘Antisense’. Then the 440 bp XhoI-EcoRI fragment from pXcmWAK-‘sense’ and the 440 bp BamHI-HindIII fragment from pXcmWAK-‘Antisense’ were inserted upstream and downstream of a pdk (phospho-di-ortho kinase) intron in inverse orientation into pKANNIBAL vector. The resulting RNAi construct, OsiWAK1-RNAi was co-bombarded along with pHX4 (harboring hygromycin phosphotransferase (hph) gene) into rice embryogenic calli using PDS-1000 He-biolistic particle delivery system (Bio-Rad) according to the method previously described (Anoop and Gupta 2003) to generate RNAi plants.
Southern blot hybridization
For analysis of transgenic RNAi plants, 10 μg of total genomic DNA extracted from leaves was digested with HindIII, which cuts once in the OsiWAK1-RNAi vector outside the pdk intron. The blotting was carried out essentially in the same way as described previously (Sambrook and Russell 2001). Hybridization was carried out by ∝ 32P dCTP labeled 700 bp pdk intron fragment obtained by digesting OsiWAK1-RNAi construct with XhoI and HindIII.
RNA gel blot analysis and siRNA detection
RNA gel blot analysis was performed using NorthernMax Kit (Ambion, USA) according to manufacturer’s instructions. Ten micrograms of total RNA was electrophoresed on a 1 % formaldehyde gel and blotted onto a positively charged nylon membrane (BrightStar-Plus Membrane, Ambion, USA). The membrane was probed with ∝-32P labeled OsiWAK1 cDNA fragment (440 bp used in RNAi vector construction), using standard procedures (Sambrook and Russell 2001). For siRNA detection, total RNA was extracted from plant tissue using TRI reagent. Total RNA (50 μg) was resuspended in 50 % formamide and denatured for 10 min at 65 °C. After adding a one-third volume of 5X loading solution (4X TBE, 0.08 % (w/v) bromophenol blue), samples were separated by electrophoresis according to the published protocol (Hamilton and Baulcombe 1999). The lower part of the gel was cut and was blotted onto a N+ membrane (BrightStar-Plus Membrane, Ambion, USA) using a semidry blotting apparatus. The membrane was prehybridized with ultrahyb buffer (Ambion, USA) at 47 °C and hybridized in the same solution containing the denatured OsiWAK1 cDNA (440 bp used in RNAi vector construction), labeled with ∝-32P dCTP by use of random primer labeling kit (Bangalore Genei, India). Hybridization was performed overnight at 47 °C followed by post-hybridization washes (each for 30 min) performed sequentially with 2X SSC, 0.5X SSC and 0.2X SSC along with 0.1 % SDS. Signals were visualized by autoradiography on X-ray film at −80 °C.
Investigation of anther dehiscence and pollen viability
Investigations on spikelet opening, filament elongation, anther dehiscence and pollen viability were conducted on the day of anthesis. Pollen viability was examined by staining with 1 % iodine-potassium iodide (I2/KI) solution. From control plants pollen grains were collected on a slide by shaking dehiscing anthers. And pollen grains were manually squeezed onto a slide from indehiscent anthers of OsiWAK1-RNAi transgenic plant. Observations were immediately made by microscopic investigation following the addition of 1 % I2/KI solution. Five spikelets were checked each from control plants and five OsiWAK1-RNAi transgenic rice plants. For pollen germination assay, the pollen were collected from both the wild-type and RNAi plants as described above and were incubated in the solution containing 0.29 M sucrose, 0.4 mM boric acid and 1 mM calcium nitrate at 28 °C for 6 h.
Computer analysis
We performed the BLAST search of the rice genomic database at NCBI (http://www.ncbi.nlm.nih.gov/Blast/Genome/PlantBlast.shtml) for the genomic sequence of this gene. Various tools from Expasy (http://www.expasy.org/tools) were used to deduce the translated product and compute theoretical pI and molecular weight. The putative domains were identified using the interproscan search (http://www.ebi.ac.uk/interproscan/). The potential signal peptide was predicted using SignalP (http://www.cbs.dtu.dk/services/signalP). The transmembrane region was predicted by Sosui analysis at http://bp.nuap.nagoya_u.ac.jp/sosui. The degree of aminoacid sequence identity was determined by the use of Wu-Blast from EBI (http://www.ebi.ac.uk/blast2). Multiple sequence alignments involved use of ClustalW (http://www.ebi.ac.yk/clustalW). The cis-acting regulatory elements in the promoter region were predicted using PLACE database (http://www.dna.affrc.go.jp/PLACE).
Results and discussion
Isolation and sequence analysis of a new wall-associated protein kinase gene, OsiWAK1, from rice
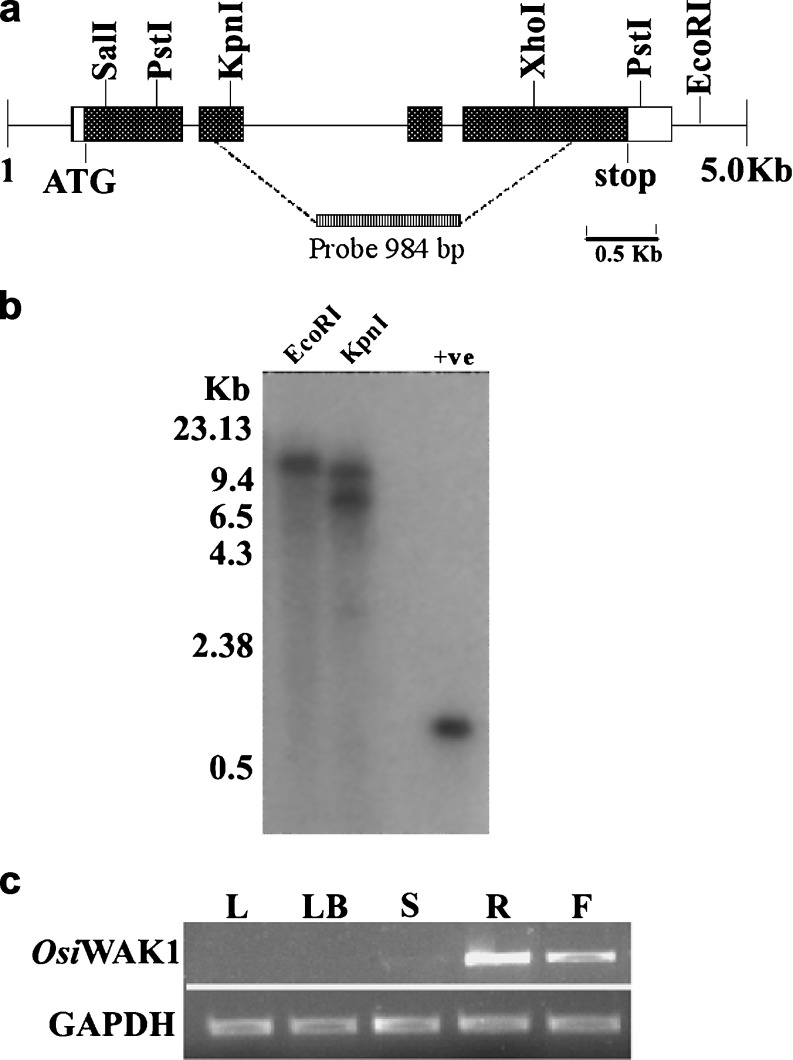
OsiWAK1 was originally isolated as a unique amplification of 984 bp (A984) fragment from differentiating rice calli in a RAPD based RT-PCR experiment (see materials and methods). RT-PCR analysis showed that the steady state level of A984 corresponding mRNAs was induced in differentiating calli (data not shown). Comparison of A984 nucleotide sequence with entries of GenBank and EMBL showed the absence of 5′ and 3′ ends. The cDNA sequence corresponding to the full-length A984 clone was completed by 5′ and 3′ RACE analysis using rice root cDNA as template. One thousand one hundred fifteen basis point and 1,097 bp sequence obtained from 5′ and 3′ RACE analysis respectively showed 100 % alignment at a stretch of 475 bp and 307 bp with the 984 bp fragment obtained by RAPD-RT-PCR analysis. A complete cDNA sequence of 2,414 bp (AF353091) was obtained by overlapping all these sequences. In silico analysis of this cDNA sequence revealed that it contains a single long open reading frame of 2,157 bp capable of encoding a protein with 718 aminoacids (Fig. 1) The protein is predicted to have molecular mass of 78.2 KDa and a theoretical pI of 5.7. BLAST search against the Genbank database indicated that the protein is similar to wall-associated kinases (WAKs) identified in Arabidopsis and to OsWAK1 identified in japonica rice. Thus, this protein was named as OsiWAK1 as it was the first WAK gene to be isolated from indica rice. Also its 100 % homolog was not found in japonica rice. Nucleotide sequence comparison (BlastN) of OsiWAK1 suggests that it is not 100 % identical to any of the 125 OsWAK gene family members from japonica rice reported by Zhang et al., in 2005. However it was 100 % identical to the genomic sequences of contig ctg021131 of chromosome 7 from indica rice. The exact localization of the OsiWAK1 in the rice genome could not be located since 100 % identical genome in the mapped Oryza sativa could not be obtained. The hybridization pattern observed in the autoradiograph of Southern blot analysis revealed it to be a single copy gene (Fig. 2a, b). These observations suggests that OsiWAK1 is a new member of OsWAK gene family apart from the already reported 125 OsWAK members and could be located on chromosome 7 of rice genome as a single copy gene.
Fig. 1.
Deduced aminoacid sequences of OsiWAK1 gene. The OsiWAK1 gene encodes for a protein of 718 aa having a single transmembrane region (underlined residues). The conserved cysteines in Epidermal growth factor (EGF) like repeats are shaded
Fig. 2.
Physical map and genomic organization of OsiWAK1 in rice genome and its expression analysis. a Schematic representation of OsiWAK1 gene. The solid boxes indicate the exons and the lines represent introns. The restriction sites are also indicated with in the 5.0 kb region spanning OsiWAK1 gene. b Genomic southern blot analysis of OsiWAK1. λ-DNA digested with HindIII was used as molecular weight marker. Ten micrograms of genomic DNA from leaves was digested with EcoRI and KpnI and hybridized with the α32P labeled OsiWAK1-984 bp cDNA probe indicated in Fig. 1. The 984 bp fragment used for probe preparation was used as positive control. c Expression analysis of OsiWAK1 in rice. PCR was performed with the first strand cDNA synthesized from the total RNA extracted from the leaves (L), leaf base (LB), shoot (S), root (R) and flowers (F) of indica rice cv IR-50. PCR amplification of endogenous rice GAPDH is represented in the lower panel that served as control
Further analysis of this 718 aminoacid sequence of OsiWAK1 using interproscan search (http://www.ebi.acuk/interproscan/) predicted the presence of characteristic domains of WAKs that includes a central hydrophobic region (357–376 aminoacid residues) representing a putative transmembrane helix region (Fig. 1) separating the C-terminal protein kinase domain (415–717 aminoacid residues) from the extracellular EGF-domain (256–349 aminoacid residues). Different EGF repeats like Type I EGF (256–301; 305–349 aminoacid residues), EGF_3 (302–349 aminoacid residues) and EGF_Ca+2 (302–333 aminoacid residues) with conserved cysteines present either adjacent or overlapping with each other (Fig. 1). A potential Asp/Asn hydroxylation site (324–335 aminoacid residues) overlapping with EGF_3 domain was also predicted. Apart from these, a potential ATP binding site (located at the residue 421) and a serine/threonine kinase active site (559–571 aminoacid residues) were predicted, which are essential for carrying out the serine/threonine kinase ctivity. An N-terminal signal peptide of 25 amino acids long (1–25 aminoacid residues) is also predicted which may be incharge of the membrane targetting. OsiWAK1 is closely related to OsWAK1 from rice and WAK5 from Arabidopsis, with 54 % and 33 % identity respectively over the entire amino acid sequence. Though the aminoacid sequence of WAKs from Arabidopsis is relatively more conserved at the protein kinase domain (86 % identity) than extracellular domain (64 % identity) (He et al. 1999), OsWAK1 and OsiWAK1 share 50 % and 60 % identity over the aminoacid sequence of protein kinase domain and extracellular region respectively.
OsiWAK1 is expressed in roots and flowers of rice plant at normal conditions
OsiWAK1 was initially isolated from differentiating rice calli. However, under normal conditions, no OsiWAK1 transcripts were detected in young rice leaves. To investigate whether OsiWAK1 is expressed in other rice tissues, mRNA accumulation of OsiWAK1 was compared by semi-quantitative RT-PCR in different rice tissues including leaves, leaf-base, shoot, root and flowers. As shown in the Fig. 2c, OsiWAK1 transcripts were not detectable in leaves and leaf-bases, but they were accumulated preferentially in roots, weakly in flowers and barely in shoots. The tissue specific expression of OsiWAK1 in roots and flowers was further substantiated by the prediction of root motif and pollen-specific element in the 5′ regulatory region of OsiWAK1 gene. Also AtWAKs were shown to express in organ specific manner. For instance WAK1 and WAK2 are expressed in young leaves and meristems, WAK3 is expressed in leaves and stems, WAK4 and WAK5 are expressed in fruits and roots respectively (He et al. 1999).
Generation and screening of OsiWAK1-RNAi lines
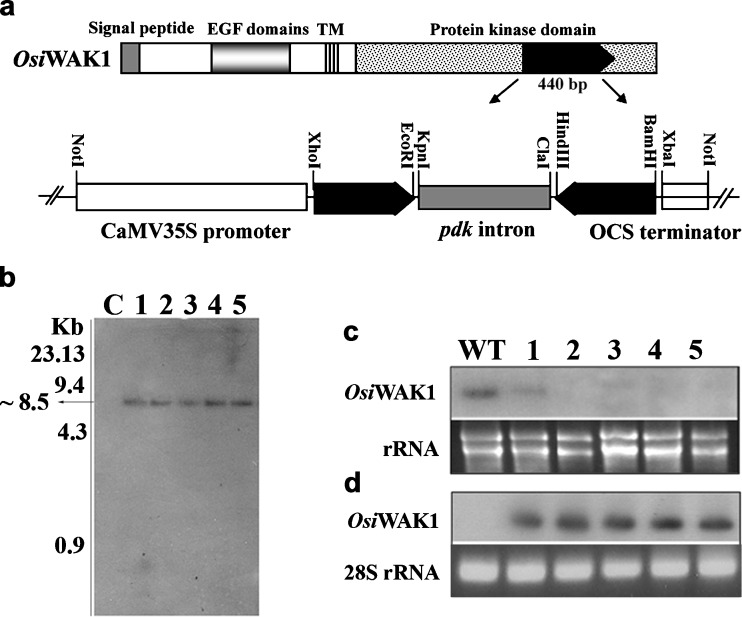
To address the role of OsiWAK1 in rice plant development, we introduced OsiWAK1 cDNA fragment in an RNAi construct (OsiWAK1-RNAi) driven by CaMV 35S promoter (Fig. 3a) into rice calli by biolistic transformation (According to Anoop and Gupta 2003). Out of 70 putative hygromycin resistant transgenic plants obtained, 67 harbored OsiWAK1-RNAi construct as evinced by the DNA dot blot analysis done using radiolabelled 700 bp fragment of pdk intron as probe. Further molecular analysis was performed with five randomly selected southern dot blot positive plants designated as R1, R2, R3, R4 and R5 hereafter. Southern blot analysis of R1–R5 plant genomic DNA revealed that the probe hybridized to ~8.5 kb fragment from the DNA of all the five transgenic lines (R1–R5, Fig. 3b). However, no such hybridization was seen with DNA from the untransformed control suggesting the integration of OsiWAK1-RNAi in the genome of R1, R2, R3, R4 and R5 as a single copy (Fig. 3b). Hybridization to a fragment of same size also suggests that all the five plants (R1–R5) may be product of single transformation event. The transgenic plants harboring OsiWAK1-RNAi showed stunted growth during the vegetative phase. All the transgenic lines essentially showed same phenotypes. Compared to wild-type plants, the T0 generation rice transgenic plants harboring OsiWAK1-RNAi cassette were shorter by 33.14 %. The dwarfness was due to the reduction in the sizes of internodes, leaves, flag-leaves and panicles. All the transgenic plants were almost sterile and the rate of seed set (~8.9 %) was much less compared to that of wild-type plants (~>93.12 %). It is not known whether the silencing of OsiWAK1 would result in the silencing of all the OsWAK gene family members. As noted earlier, a 440 bp region from the protein kinase domain of the OsiWAK1 was used as RNAi trigger. Initial Blast analysis of this 440 bp revealed it to be 100 % specific only to the OsiWAK1. But the recent Blast analysis indicated that apart from OsiWAK1, this region is 89 % similar to the OsWAK (Os.19895) located on chromososme 11. Thus the expression of this OsiWAK1 shRNA might also silence the Os.19895 gene expression. In Arabidopsis it was also reported that the suppression of any one WAK led to the suppression of all the other WAK proteins (Lally et al. 2001). Also, it is reported that all the attempts to express the antisense WAK transcripts by constitutive promoter, CaMV 35S promoter failed to yield any viable transgenic plants in Arabidopsis (He et al. 1998; Lally et al. 2001). Thus, in Arabidopsis subsequent attempts have been made using inducible promoters like PR1 promoter and/ or gluco-corticoid inducible promoter (He et al. 1998; Lally et al. 2001) to study the physiological roles of WAKs. The observations from our study revealed that as in the case of Arabidopsis, the silencing of OsiWAK1 in rice was also sub-lethal and thus frequency of generation of viable transgenic plants from different integration events was very less. Since OsiWAK1 is overexpressed during organogenesis of rice calli, the sub-lethality observed as stated above suggests that OsiWAK1 may play a role in organogenesis. Future work would be done to drive the OsiWAK1-RNAi cassette under an inducible promoter to study the role of OsiWAK1 under various developmental stages.
Fig. 3.
Generation and analysis of OsiWAK1-RNAi plants. a Schematic representation of the OsiWAK1-RNAi construct. A 440 bp OsiWAK1 gene-specific fragment was used as the RNAi trigger. Transcripts of the RNAi trigger region would be expressed constitutively under the control of CaMV35S promoter. b Southern blot analysis of OsiWAK1-RNAi plants. ~8.5 kb fragment was hybridized to α32P labeled 700 bp pdk intron in all the RNAi plants tested (R1-R5; lanes 1-5) but not in control (C) plants. c RNA gel blot analysis of OsiWAK1 transcripts from roots of wild-type and OsiWAK1-RNAi lines (1-5). The upper panel shows the RNA gel blot probed with the 440 bp OsiWAK1cDNA region used as RNAi trigger. The lower panel shows ethidium bromide stained rRNA as a loading control. d siRNA detection of OsiWAK1-RNAi lines in root RNA. The upper panel shows that siRNA, a molecular marker for double-stranded RNA- based gene silencing was detected in all OsiWAK1-RNAi lines probed with α32P labeled OsiWAK1 440 bp fragment. The lower panel shows ethidium bromide stained 28 s rRNA as loading control
Knockdown of OsiWAK1 mRNA in RNAi plants is related to their pleiotropic phenotypes
It is known that the expression of dsRNAs can knockdown the endogenous transcript level of their target genes (Kusaba 2004; Lu et al. 2004). First we checked the intactness of the introduced OsiWAK1-RNAi construct (sense arm-intron-antisense arm) in transgenic lines by doing PCR analysis using the primers WAKSF1 and WAKAF2, complementary to the 5′ end sequences of sense and antisense arms of OsiWAK1-RNAi construct. An expected amplicon of 1.58 kb fragment was amplified from all the transgenic lines (data not shown). To investigate whether the observed phenotypic changes in transgenic lines correlates to RNAi mediated knockdown of this target mRNA, endogenous OsiWAK1 transcripts were determined by northern analysis using the RNA prepared from the flowers of five transgenic plants (R1–R5) as well as wild-type. Compared to the wild-type flowers, the OsiWAK1 transcript was greatly down-regulated in the flowers of R1 and was undetectable in the flowers of R2–R5 plants (Fig. 3c). To further estimate whether the decrease in endogenous OsiWAK1 mRNA resulted from RNA interference mediated by the introduced RNAi construct, siRNA detection was carried out and the results indicated that the OsiWAK1 cDNA specific small RNAs of ~23 nt was detectable in all the transgenic lines R1–R5 and undetectable in control (Fig. 3d). So the RNAi mediated OsiWAK1 deficiency at mRNA levels is correlated with the observed phenotypic changes in the transgenic lines R1–R5.
Affect of RNAi-mediated silencing of OsiWAK1 expression on vegetative growth
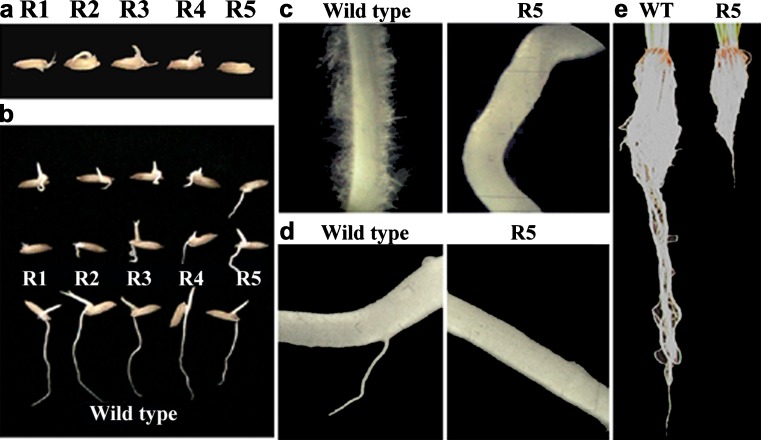
An effort has been made to study the effect of OsiWAK1 silencing on seed germination and root development. For this, all the seeds from the T0 plants (R1–R5) were surface sterilized and allowed to germinate on water soaked cotton for 10 days. Seeds from untransformed plants were used as control. Observations were made periodically every day till 10 days of germination. Emergence of shoot primordial was observed within 2 days after germination (DAG) in all seeds from transgenic lines as well as control plants. Formation and subsequent growth of root primordia was inhibited by 30 % in all transgenic lines as compared to control (Fig. 4a). Further growth of roots was observed in the remaining 70 % of the transgenic lines and in all control seeds. However, the roots of transgenic lines were short, thick and irregular in shape (Fig. 4b) without a single root hair (Fig. 4c) in contrast to the roots from control plants, which were long, smooth and straight (Fig. 4b) with numerous root hairs (Fig. 4c). The formation of lateral roots was delayed by 4 days in the transgenic lines (Fig. 4d). These results suggest that the silencing of OsiWAK1 expression effects rice root development during seedling stage leading to defective root primordial formation, reduced rate of root elongation, absence of root hairs and delayed lateral rooting. After 10 days, the plantlets were transferred to 15 cm pots with vermiculate and soil in 1:3 ratio and were allowed to harden in the growth chamber. Later, the plants were transferred to green house where they grew to maturity with flowering and seed-set. Effect of OsiWAK1 silencing on the root growth was studied in terms of root length, root fresh weight and dry weight. For this, the control and transgenic plants were uprooted after completion of flowering and seed set and the roots were washed thoroughly and the root length, fresh-weight and dry weight were measured (Table 1). It was observed that transgenic lines showed significant reduction in root length (54.34 %), root fresh weight (58.45 %) and root dry weight (64.28 %) as compared to control plants (Fig. 4e). It may be suggested that the silencing of OsiWAK1 affects root biomass production. Also an attempt was made to know if the reduction of root biomass had an impact on shoot growth and development by comparing height of the plants, length of leaf and flag-leaf (Table 1). It was observed that transgenic lines showed 17.5 %, 32.5 %, 58.33 % and 63.66 % reduction in height compared to control (untransformed) plant at 30, 60, 90 and 120 days respectively. Therefore, effect of silencing of OsiWAK1 was more pronounced during later stages of development (Fig. 5a). In addition, length of leaf decreased by 61.8 % and that of flag-leaf by 52.8 % in 120 days old transgenic lines as compared to control (Fig. 5b). Since all the transgenic lines showed similar phenotypes, only the phenotype of T1 generation from R5 were shown for representation. From the above results, it may be suggested that silencing of OsiWAK1 effects shoot growth also. In Arabidopsis also suppression of WAKs effected the root and shoot growth. Suppression of AtWAK4 resulted in dwarf plants with small rosette leaves, condensed inflorescence stem, short siliques and short primary roots (Lally et al. 2001). Suppression of WAK2 resulted in small leaves (Wagner and Kohorn 2001) and affected the rate of root hair elongation (Kohorn et al. 2006a).
Fig. 4.
Silencing of OsiWAK1 affects root morphology and growth in rice. a Root primordial formation was inhibited in 30 % of the seeds from OsiWAK1-RNAi lines. b The roots of OsiWAK1-RNAi seedlings were short, thick irregular and c without root hairs when compared to wild-type roots on 4th day after germination (DAG). d The lateral root formation was not seen in OsiWAK1-RNAi plants on 6th DAG. e The length and the fresh-weight of the OsiWAK1-RNAi lines are less compared to that of wild-type roots in mature plants. R1-R5 represents the seeds from OsiWAK1-RNAi lines 1–5. Observations for R5 are represented in the picture. Similar observations were obtained for all five lines (R1–R5)
Table 1.
Comparison of the growth parameters of wild type rice (Ssp indica cv IR-50) with that of Osiwak1-RNAi lines in T1 generation
| Sl.No | Parameter | Wild-type (Avg ± StDev) | Osiwak1-RNAi (T1 generation) (Avg ± StDev) |
|---|---|---|---|
| 1 | Height of the Plant (cm) | 90.4 ± 4.24 | 33 ± 3.53 |
| 2 | Length of the leaf (cm) | 51.4 ± 1.14 | 19.6 ± 1.14 |
| 3 | Length of the flag leaf (cm) | 20.8 ± 0.836 | 9.8 ± 0.28 |
| 4 | Length of the inflorescence stem (cm) | 40.2 ± 1.41 | 11.3 ± 0.15 |
| 5 | Number of spikes per plant | 8 ± 1 | 11 ± 1 |
| 6 | Length of the spike (cm) | 24.6 ± 1.14 | 7 ± 0.212 |
| 7 | No. of flowers per spike | 84 ± 5 | 23 ± 5 |
| 8 | No. of matured seeds per plant | 632 ± 20 | 0 |
| 9 | No. of pollen per anther | 1,286 ± 40 | 1,124 ± 20 |
| 10 | % of pollen fertility | 93.5 % | 31 % |
| 11 | Length of the root (cm) | 46 ± 1.41 | 21 ± 1.41 |
| 12 | Fresh weight of root (gm per plant) | 5.825 ± 0.233 | 2.42 ± 0.212 |
| 13 | Dry weight of root (gm per plant) | 1.82 ± 0.098 | 0.65 ± 0.127 |
All the recordings were made from mature plants. The recordings were done at least three times. The values represent the mean of different samples with standard deviation. In all cases n = 5
Fig. 5.
Silencing of OsiWAK1 affects vegetative growth of rice plants. a The OsiWAK1-RNAi lines showed stunted growth. b The size of leaf (L), Inflorescence stem (IS), Inflorescence (I) and Flagleaf (FL) of WT and OsiWAK1-RNAi plants are compared. All the organs in OsiWAK1-RNAi plants are stunted. Bar = 20 cm
Knockdown of OsWAK2 mRNA in rice plants effects cell elongation
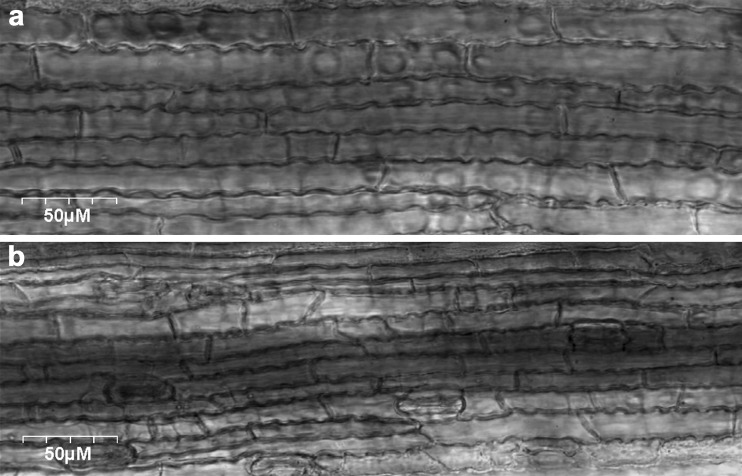
In the present study, we also made an effort to understand the reason for development of dwarf rice plants when OsiWAK1 transcript was suppressed. For this, we analyzed the leaf epidermal cells of both control rice plants and transgenic rice plants harboring OsiWAK1-RNAi construct using confocal microscopy. It is observed that the cell size was reduced and not the cell number in the OsiWAK1 silenced plants (Fig. 6b) compared to that of control plants (Fig. 6a). So, OsiWAK1 may be playing an important role in the cell elongation or expansion. In Arabidopsis also suppression of WAKs affected the leaf expansion (Lally et al. 2001) and root cell elongation (Kohorn et al. 2006a). The exact mechanism as to how the WAKs are involved in cell expansion is not clear. Recently the WAK2 of Arabidopsis was shown to affect either directly or indirectly the transcription and activity of vacuolar invertase thus, may be modulating solute concentrations (Kohorn et al. 2006a). Changes in the concentration of solutes are known to be involved in turgor regulation (Koch 2004). It is hypothesized that WAKs may sense cell wall expansion by their attachment to pectin, and provides mechanism for transducing these signals to systems regulating solute changes. This in turn can influence the maintenance of turgor as the cell enlarges (Kohorn et al. 2006a).
Fig. 6.
Silencing of OsiWAK1 affects cell elongation in rice. a, b, Confocal microscopy pictures of leaves from either a wild-type plant (a) or OsiWAK1-RNAi (R5) plant (b). Bars = 50 μm
Affect of RNAi-mediated silencing of OsiWAK1 expression on flowering and productivity
In Arabidopsis silencing of WAK4 affected flowering and seed set and thus led to sterile plants. The transgenic Arabidopsis plants with suppressed WAK4 transcript accumulation led to the development of plants with short siliques and unopened miniature flowers (Lally et al. 2001). In the present study we analyzed the effect of OsiWAK1 silencing on flowering and seed set in terms of productivity of the transgenic lines (T1 generation) compared to that of control plants. For this, length of the inflorescence stem and spike was measured and also number of spikes per plant, number of flowers per spike and number of mature seeds per plant was counted (Table 1). In the transgenic plants, the length of inflorescence stem and panicle was reduced by ~71.8 % and 71.5 % respectively compared to that of control plants (Fig. 5b). Further, transgenic lines showed 37.5 % more spikes compared to control plant. However, number of flowers per spike in transgenic lines was reduced by ~73 % compared to control plants. Apart from this, all the T1 transgenic lines showed no seed set. Therefore, it may be suggested that OsiWAK1 silencing led to reduction in inflorescence size, increase in number of spikes per plant but decrease in number of flowers per spike with no seed set. An attempt was made to understand if the inability of transgenic plants to set seeds was due to defect in flower or floral organ development. For this, floral organs from flowers of both wild-type and T1 transgenic plants were compared. No major differences or defects were observed in the floral organ arrangement or structure or size except for the shorter anther filament in the transgenic lines compared to that of wild type (Fig. 7a). Because male gametophyte development is one of the key regulators of successful fertilization in sexual plants, the anther development in the flowers of transgenic plants was compared with that of wild-type plants. In wild type plants, the anther dehiscence occurred soon after flower opening (Fig. 7b), pollens were deposited on the stigma, fertilization occurred and endosperm formation was seen within a day post anthesis. Whereas, in case of transgenic plants anthers fail to dehisce even after flower opening and thereafter (Fig. 7c). Hence the pollen grains were not released and no fertilization occurred, no endosperm formation and hence no seed set. Both the stigma and anthers withered away in these plants. These observations indicate that the OsiWAK1 silencing affected the anther dehiscence mechanism in rice either directly or indirectly.
Fig. 7.
Silencing of OsiWAK1 affects anther dehiscence and pollen viability in rice. a Flowers of wild type and OsiWAK1-RNAi plants. The length of the anther filament is reduced in OsiWAK1-RNAi plants. b, c Mature and dehiscent anthers of wild type plants (b) and mature and nondehiscent anthers of OsiWAK1-RNAi plants (c). Anthers from opened flowers (left) and closed flowers after anthesis (right) are shown. d Pollen of the wild-type and OsiWAK1-RNAi plants stained with I2/KI. Pollen grains filled with starch granules showed dark blue stain but those with little or no starch stained light yellow with I2/KI
There are reports indicating the association of anther dehiscence and reduced pollen viability and/or fertility. Therefore, in this study, an attempt has been made to study the effect of OsiWAK1 silencing on the pollen grain development in terms of pollen grain number, pollen viability/fertility and pollen tube formation. The results indicated that the anthers from both the OsiWAK1 deficient lines and wild-type plants have approximately equal number of pollen grains but the pollen viability was reduced by around 70 % in RNAi lines as shown by the I2/KI staining assay (starch filling). The deposition of starch and other secondary metabolites in the pollen grain is crucial for its viability (Raghavan 1988). The I2/KI staining assay showed that only ~30 % of pollen grains (n = 1,260) from plant R5 were round and had a uniform size with a dark blue black reaction. Remaining pollen grains were not filled and were much smaller, empty, shrunken and variable in shape. Whereas ~93 % of pollen grains (n = 1,286) from the wild type were round and are of uniform size with a dark blue black reaction (Fig. 7d). Further, pollen tube assay revealed that none of the pollen grains from transgenic lines show pollen tube formation and elongation (data not shown). These results indicate that OsiWAK1 silencing affects pollen fertility and also pollen germination. These results clearly suggest that anther indehiscence, reduced pollen viability and defective pollen germination may be the main reasons of sterility in the OsiWAK1-RNAi lines. The extracellular domain of WAK1 from Arabidopsis was shown to interact with cell wall pectins in a calcium-induced conformation (Decreux and Messiaen 2005). Also, pectin methyl esterases (PMEs) act as potential key regulators of pollen tube growth by determining the rheological properties of the apical pollen tube wall (Bosch et al. 2005; Jiang et al. 2005). WAKs may be regulating the pollen tube elongation probably by its interaction with pectins.
Further investigations into the role of WAKs in the signaling pathway potentially linking input from the cell wall to the adjustments in the endogenous sugar metabolism would help us to further understand the WAKs function in cell elongation and ultimately their role in regulating plant growth and development.
In conclusion, we have isolated OsiWAK1, a new member of OsWAK gene family from indica rice. It is found to be present as a single copy in the rice genome on chromosome 7. Knockout plants developed using RNAi showed pleiotropic phenotypes like defective root development and male sterility (due to anther indehiscence and reduced pollen viability and germination), indicating the critical role played by OsiWAK1 in rice plant growth and development. To our knowledge, this work constitutes the second report of functional characterization of OsWAK gene family member in rice (Oryza sativa).
Acknowledgements
We would like to thank Dr. Waterhouse (CSIRO, Australia) for providing us the construct pKANNIBAL. This work was supported partially by grants from Department of Biotechnology (DBT)-India, Centre for potential for genomic sciences-University grants commission (CPSGS-UGC) and UGC-SAP programmes. K.V is the recipient of a research fellowship from the Council for Scientific and Industrial Research, India.
Footnotes
OsiWAK1 is deposited in the GenBank with Accession Number: AF353091
References
- Anderson CM, Wagner TA, Perret M, He ZH, He D, Kohorn BD. WAKs: cell wall-associated kinases linking the cytoplasm to the extracellular matrix. Plant Mol Biol. 2001;47:197–206. doi: 10.1023/A:1010691701578. [DOI] [PubMed] [Google Scholar]
- Anoop N, Gupta AK. Transgenic indica rice cv IR-50 over-expressing vigna aconitifolia delta-1-pyrroline-5-carboxylase synthetase cDNA shows tolerance to high salt. J Plant Biochem Biotechnol. 2003;12:109–116. [Google Scholar]
- Baluska F, Samaj J, Wojtaszek P, Volkmann D, Menzel D. Cytoskeleton–plasma membrane–cell wall continuum in plants. Emerging links revisited. Plant Physiol. 2003;133:482–491. doi: 10.1104/pp.103.027250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becraft PW, Stinard PS, McCarty DR. CRINKLY4: a TNFR-like receptor kinase involved in maize epidermal differentiation. Science. 1996;273:1406–1409. doi: 10.1126/science.273.5280.1406. [DOI] [PubMed] [Google Scholar]
- Bosch M, Cheung AY, Hepler PK. Pectin methylesterase, a regulator of pollen tube growth. Plant Physiol. 2005;138:1334–1346. doi: 10.1104/pp.105.059865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownlee C. Role of the extracellular matrix in cell-cell signalling: paracrine paradigms. Curr Opin Plant Biol. 2002;5:396–401. doi: 10.1016/S1369-5266(02)00286-8. [DOI] [PubMed] [Google Scholar]
- Brutus A, Sicilia F, Macone A, Cervone F, De Lorenzo G. A domain swap approach reveals a role of the plant wall-associated kinase 1 (WAK1) as a receptor of oligogalacturonides. Proc Natl Acad Sci USA. 2010;107:9452–9457. doi: 10.1073/pnas.1000675107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SE, Williams RW, Meyerowitz EM. The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell. 1997;89:575–585. doi: 10.1016/S0092-8674(00)80239-1. [DOI] [PubMed] [Google Scholar]
- Decreux A, Messiaen J. Wall-associated Kinase WAK1 interacts with cell wall pectins in a calcium-induced conformation. Plant Cell Physiol. 2005;46:268–278. doi: 10.1093/pcp/pci026. [DOI] [PubMed] [Google Scholar]
- Decreux A, Thomas A, Spies B, Brasseur R, Van Cutsem P, Messiaen J. In vitro characterization of the homogalacturonan-binding domain of the wall-associated kinase WAK1 using site-directed mutagenesis. Phytochemistry. 2006;67:1068–1079. doi: 10.1016/j.phytochem.2006.03.009. [DOI] [PubMed] [Google Scholar]
- Deeks MJ, Hussey PJ, Davies B. Formins: intermediates in signal-transduction cascades that affect cytoskeletal reorganization. Trends Plant Sci. 2002;7:492–498. doi: 10.1016/S1360-1385(02)02341-5. [DOI] [PubMed] [Google Scholar]
- Dellaporta SI, Wood J, Hicks JB. A plant DNA minipreparation version II. Plant Mol Biol Rep. 1983;1:19–21. doi: 10.1007/BF02712670. [DOI] [Google Scholar]
- Geiger B, Bershadsky A. Assembly and mechanosensory function of focal contacts. Curr Opin Cell Biol. 2001;13:584–592. doi: 10.1016/S0955-0674(00)00255-6. [DOI] [PubMed] [Google Scholar]
- Hamilton AJ, Baulcombe DC. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- He ZH, Fujiki M, Kohorn BD. A cell wall-associated receptor-like protein kinase. J Biol Chem. 1996;271:19789–19793. doi: 10.1074/jbc.271.33.19789. [DOI] [PubMed] [Google Scholar]
- He ZH, He D, Kohorn BD. Requirement for the induced expression of a cell wall associated receptor kinase for survival during the pathogen response. Plant J. 1998;14:55–63. doi: 10.1046/j.1365-313X.1998.00092.x. [DOI] [PubMed] [Google Scholar]
- He ZH, Cheeseman I, He D, Kohorn BD. A cluster of five cell wall-associated receptor kinase genes, WAK1–5, are expressed in specific organs of Arabidopsis. Plant Mol Biol. 1999;39:1189–1196. doi: 10.1023/A:1006197318246. [DOI] [PubMed] [Google Scholar]
- Jiang LX, Yang SL, Xie LF, Puah CS, Zhang XQ, Yang WC, Sundaresan V, Ye D. VANGUARD1 encodes a pectin methylesterase that enhances pollen tube growth in the Arabidopsis style and transmitting tract. Plant Cell. 2005;17:584–596. doi: 10.1105/tpc.104.027631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinn TL, Stone JM, Walker JC. HAESA, an Arabidopsis leucine-rich repeat receptor kinase, control floral organ abscission. Genes Dev. 2000;14:108–117. [PMC free article] [PubMed] [Google Scholar]
- Kanneganti V, Gupta AK. Wall associated kinases from plants – an overview. Physiol Mol Biol Plants. 2008;14:109–118. doi: 10.1007/s12298-008-0010-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch K. Sucrose metabolism: regulatory mechanisms and pivotal roles in sugar sensing and plant development. Curr Opin Plant Biol. 2004;7:235–246. doi: 10.1016/j.pbi.2004.03.014. [DOI] [PubMed] [Google Scholar]
- Kohorn BD. Plasma membrane-cell wall contacts. Plant Physiol. 2000;124:31–38. doi: 10.1104/pp.124.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohorn BD, Kobayashi M, Johansen S, Riese J, Huan LF, Koch K, Fu S, Dotson A, Byers N. An Arabidopsis cell wall-associated kinase required for invertase activity and cell growth. Plant J. 2006;46:307–316. doi: 10.1111/j.1365-313X.2006.02695.x. [DOI] [PubMed] [Google Scholar]
- Kohorn BD, Kobayashi M, Johansen S, Friedman HP, Fischer A, Byers N. Wall-associated kinase 1 (WAK1) is crosslinked in endomembranes, and transport to the cell surface requires correct cell-wall synthesis. J Cell Sci. 2006;119:2282–2290. doi: 10.1242/jcs.02968. [DOI] [PubMed] [Google Scholar]
- Kohorn BD, Johansen S, Shishido A, Todorova T, Martinez R, Defeo E, Obregon P. Pectin activation of MAP kinase and gene expression is WAK2 dependent. Plant J. 2009;60:974–982. doi: 10.1111/j.1365-313X.2009.04016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusaba M. RNA interference in crop plants. Curr Opin Biotechnol. 2004;15:139–143. doi: 10.1016/j.copbio.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Lally D, Ingmire P, Tong HY, He ZH. Antisense expression of a cell wall-associated protein kinase, WAK4, inhibits cell elongation and alters morphology. Plant Cell. 2001;13:1317–1333. doi: 10.1105/tpc.13.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lespinet O, Wolf YI, Koonin EV, Aravind L. The role of lineage-specific gene family expansion in the evolution of eukaryotes. Genome Res. 2002;12:1048–1059. doi: 10.1101/gr.174302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Zhou SY, Zhao WS, Su SC, Peng YL. A novel wall-associated receptor-like protein kinase gene, OsWAK1, plays important roles in rice blast disease resistance. Plant Mol Biol. 2009;69:337–346. doi: 10.1007/s11103-008-9430-5. [DOI] [PubMed] [Google Scholar]
- Lu S, Shi R, Tsao CC, Yi X, Li L, Chiang VL. RNA silencing in plants by the expression of siRNA duplexes. Nucleic Acids Res. 2004;32:e171. doi: 10.1093/nar/gnh170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier S, Ruzvidzo O, Morse M, Donaldson L, Kwezi L, Gehring C. The Arabidopsis Wall Associated Kinase-Like 10 gene encodes a functional guanylyl cyclase and is co-expressed with pathogen defense related genes. PLoS ONE. 2010;5(1):e8904. doi: 10.1371/journal.pone.0008904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris ER, Walker JC. Receptor-like protein kinases: the key to response. Curr Opin Plant Biol. 2003;6:339–342. doi: 10.1016/S1369-5266(03)00055-4. [DOI] [PubMed] [Google Scholar]
- Nakhamchik A, Zhao Z, Provart NJ, Shiu SH, Keatley SK, Cameron RK, Goring DR. A comprehensive expression analysis of the Arabidopsis proline-rich extensin-like receptor kinase gene family using bioinformatic and experimental approaches. Plant Cell Physiol. 2004;45:1875–1881. doi: 10.1093/pcp/pch206. [DOI] [PubMed] [Google Scholar]
- Nothnagel EA. Proteoglycans and related components in plant cells. Int Rev Cytol. 1997;174:195–291. doi: 10.1016/S0074-7696(08)62118-X. [DOI] [PubMed] [Google Scholar]
- Park JH, Halitschke R, Kim HB, Baldwin IT, Feldmann KA, Feyereisen R. A knock-out mutation in allene oxide synthase results in male sterility and defective wound signal transduction in Arabidopsis due to a block in jasmonic acid biosynthesis. Plant J. 2002;31:1–12. doi: 10.1046/j.1365-313X.2002.01328.x. [DOI] [PubMed] [Google Scholar]
- Raghavan V. Anther and pollen development in rice. Am J Bot. 1988;75:183–196. doi: 10.2307/2443885. [DOI] [Google Scholar]
- Richter TE, Ronald PC. The evolution of disease resistance genes. Plant Mol Biol. 2000;42:195–204. doi: 10.1023/A:1006388223475. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular cloning: a laboratory manual. 3. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Schindelman G, Morikami A, Jung J, Baskin TI, Carpita NC, Derbyshire P, McCann MC, Benfey PN. COBRA encodes a putative GPI-anchored protein, which is polarly localized and necessary for oriented cell expansion in Arabidopsis. Genes Dev. 2001;15:1115–1127. doi: 10.1101/gad.879101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheetz MP. Cell control by membrane-cytoskeleton adhesion. Nat Rev Mol Cell Biol. 2001;2:392–396. doi: 10.1038/35073095. [DOI] [PubMed] [Google Scholar]
- Shiu SH, Bleecker AB. Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc Natl Acad Sci USA. 2001;98:10763–10768. doi: 10.1073/pnas.181141598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu SH, Karlowski WM, Pan R, Tzeng YH, Mayer KF, Li WH. Comparative analysis of the receptor-like kinase family in Arabidopsis and Rice. Plant Cell. 2004;16:1220–1234. doi: 10.1105/tpc.020834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivaguru M, Ezaki B, He ZH, Tong H, Osawa H, Baluska F, Volkmann D, Matsumoto H. Aluminum-induced gene expression and pro-tein localization of a cell wall-associated receptor kinase in Arabidopsis. Plant Physiol. 2003;132:2256–2266. doi: 10.1104/pp.103.022129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville C, Bauer S, Brininstool G, Facette M, Hamann T, Milne J, Osborne E, Paredez A, Persson S, Raab T, Vorwerk S, Youngs H. Towards a systems approach to understanding plant cell walls. Science. 2004;306:2206–2211. doi: 10.1126/science.1102765. [DOI] [PubMed] [Google Scholar]
- Takasaki T, Hatakeyama K, Suzuki G, Watanabe M, Isogai A, Hinata K. The S receptor kinase determines self-incompatibility in Brassica stigma. Nature. 2000;403:913–916. doi: 10.1038/35002628. [DOI] [PubMed] [Google Scholar]
- Verica JA, He ZH. The cell wall-associated kinase (WAK) and WAK-like kinase gene family. Plant Physiol. 2002;129:455–459. doi: 10.1104/pp.011028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verica JA, Chae L, Tong H, Ingmire P, He ZH. Tissue-specific and developmentally regulated expression of a cluster of tandemly arrayed cell wall-associated kinase-like kinase genes in Arabidopsis. Plant Physiol. 2003;133:1732–1746. doi: 10.1104/pp.103.028530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner TA, Kohorn BD. Wall-associated kinases are expressed throughout plant development and are required for cell expansion. Plant Cell. 2001;13:303–318. doi: 10.1105/tpc.13.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesley SV, Helliwell C, Smith NA, Wang MB, Rouse D, Liu Q, Gooding P, Singh S, Abbott D, Stoutjesdijk P, Robinson S, Gleave A, Green A, Waterhouse PM. Constructs for efficient, effective and high Throughput gene silencing in plants. Plant J. 2001;27:581–590. doi: 10.1046/j.1365-313X.2001.01105.x. [DOI] [PubMed] [Google Scholar]
- Yokoyama R, Takahashi T, Kato A, Torii KU, Komeda Y. The Arabidopsis ERECTA gene is expressed in the shoot apical meristem and organ primordial. Plant J. 1998;15:301–310. doi: 10.1046/j.1365-313X.1998.00203.x. [DOI] [PubMed] [Google Scholar]
- Zhang S, Chen C, Li L, Meng L, Singh J, Jiang N, Deng XW, He ZH, Lemaux PG. Evolutionary expansion, gene structure, and expression of the rice wall-associated kinase gene family. Plant Physiol. 2005;139:1107–1124. doi: 10.1104/pp.105.069005. [DOI] [PMC free article] [PubMed] [Google Scholar]