Abstract
Plants are under strong evolutionary pressure in developing new and noble R genes to recognize pathogen avirulence (avr) determinants and bring about stable defense for generation after generations. Duplication, sequence variation by mutation, disparity in the length and structure of leucine rich repeats etc., causes tremendous variations within and among R genes in a plant thereby developing diverse recognitional specificity suitable enough for defense against new pathogens. Recent studies on genome sequencing, diversity and population genetics in different plants have thrown new insights on the molecular evolution of these genes. Tandem and segmental duplication are important factors in R gene abundance as inferred from the distribution of major nucleotide binding site-leucine rich repeats (NBS-LRRs) type R-genes in plant genomes. Likewise, R-gene evolution is also thought to be facilitated by cluster formation thereby causing recombination and sequence exchange and resulting in haplotypic diversity. Population studies have further proven that balancing selection is responsible for the maintenance of allelic diversity in R genes. In this review, we emphasize and discuss on improved perspectives towards the molecular mechanisms and selection pressure responsible for the evolution of NBS-LRR class resistance genes in plants.
Keywords: R-genes, NBS-LRRs, Evolution, Diversification, Duplication
Introduction
Plants employ a network of intertwined mechanisms to defend themselves against a vast array of pathogens such as bacteria, fungi, viruses, nematodes and other plants attacking them in their natural ecosystems. In the ongoing battle between plants and pathogens, the latter secrete a large amount of virulence factors called microbe associated molecular patterns (MAMP) that acts as molecular saboteurs. MAMP elicitors are recognized by the plants through specialized MAMP receptors leading to the induction of defense response. However, multiple microorganisms secrete effector proteins into host cells that intercept MAMP-triggered defense signals and thereby attenuate MAMP triggered immunity. Some pathogens may even cause direct suppression of MAMP induced basal defense (Kim et al. 2005). Thus, co-evolution of the virulent pathogens along with the plant hosts has resulted in the establishment of effector triggered immunity (Jones and Dangl 2006). Plants have also evolved many resistance (R) proteins that activate highly efficient defense reactions upon specific recognition of pathogen effectors which include a ‘hypersensitive response (HR)’ of programmed cell death at the infection site. R gene-mediated recognition of pathogen effectors activate a series of defense signaling cascades and induce pathogenesis-related (PR) gene expression to generate systemic acquired resistance (SAR) with a global, durable and broad-spectrum resistance in plants (Durrant and Dong 2004).
Over 70 different R genes has been cloned and characterized in different plants species during the last 15 years (Liu et al. 2007). The known R proteins are grouped into just a few main classes based primarily upon their combination of a limited number of structural motifs (Joshi and Nayak 2011). The most prevalent among them encode proteins that have a putative amino-terminal signaling domain, a nucleotide binding site (NBS) and a series of carboxy-terminal leucine rich repeats (LRRs). These NBS-LRR proteins has been classified as those that encode an amino-terminal putative leucine zipper (LZ) or coiled-coil sequences (LZ/CC-NBS-LRR or CNL proteins) and those with an amino terminal Toll/interleukin receptor domain (known as TIR-NBS-LRR or TNL proteins).
Each domain of NBS-LRR protein is predicted to have a specific function. The NBS domain is suggested to have NTP-hydrolyzing activity and regulating signal transduction through conformational changes (Martin et al. 2003). The LRR domain contains tandemly arrayed repeats in the carboxy-terminal region of R-genes and its predicted biochemical function is to mediate protein-protein interaction. Although, the genetic basis of disease resistance by NBS-LRR genes is well recognized for many plant-pathogen interactions, one of the major points of discussion has been their nature of origin and evolution. The consistent detection of NBS-LRR class of proteins in diverse plant species suggests that these genes are a pillar of plant defenses and their resistance function either evolved by convergence or originated in a common ancestor of plant species. Furthermore, results from various nucleotide polymorphism analyses demonstrate extremely high levels of inter and intraspecific variation of NBS-LRR genes which suggest constant variation in the recognition patterns of plants to pathogen elicitors. Due to this, major effort has been made in the recent times towards understanding how the NBS-LRR R genes evolve. Recent advances in comparative genomics, sequence analysis, population genetic studies and whole genome sequencing has resulted in remarkable progresses in understanding different perspectives towards NBS-LRR R gene evolution. This review highlights the recent insights on the evolutionary perspectives of NBS-LRR class R genes in plants considering global view points with special emphasis on genome sequence and population genetic analysis.
Evolutionary characterization through genome analyses
Complete genome sequence analysis and EST development of model dicot, monocot and tree plants has revealed the genomic organization of NBS-LRR R genes and has paved ways for their evolutionary analyses (Meyers et al. 2003; Zhou et al. 2004; Kohler et al. 2008). Global sequencing projects and PCR-based surveys confirm that all plants maintain large and diverse NBS-LRR families involved in pathogen surveillance or other unknown functions. These studies also corroborate that lineages within the NBS-LRR super family are not equally represented among all plant taxa. The rice genomic sequences contain more than 500 NBS coding sequences all of which encode for CNL (Coiled Coil NBS-LRR) R-genes (Bai et al. 2002; Zhou et al. 2004). In contrast, of the 149 NBS-LRR genes and 58 shorter related genes in Arabidopsis, two third are TIR-NBS-LRR (TNLs) and one third encodes CC-NBS-LRR (CNLs) whereas Populus trichocarpa contains 60 % of CNLs and 40 % of TNLs. Although TNL genes outnumber CNL genes by nearly two to one in the Arabidopsis genome, several lines of evidence suggested that the CNL genes may be the more ancient group. There is a greater degree of diversity among CNL proteins than TNLs across different plant species (Cannon et al. 2002). A large-scale phylogenetic analysis of CNLs grouped them into four distinct lineages some of which marks their existence prior to angiosperm/gymnosperm divergence. Further, the branch length of the CNL tree are reportedly longer and the intron positions are less conserved as compared to TNLs (Cannon et al. 2002).
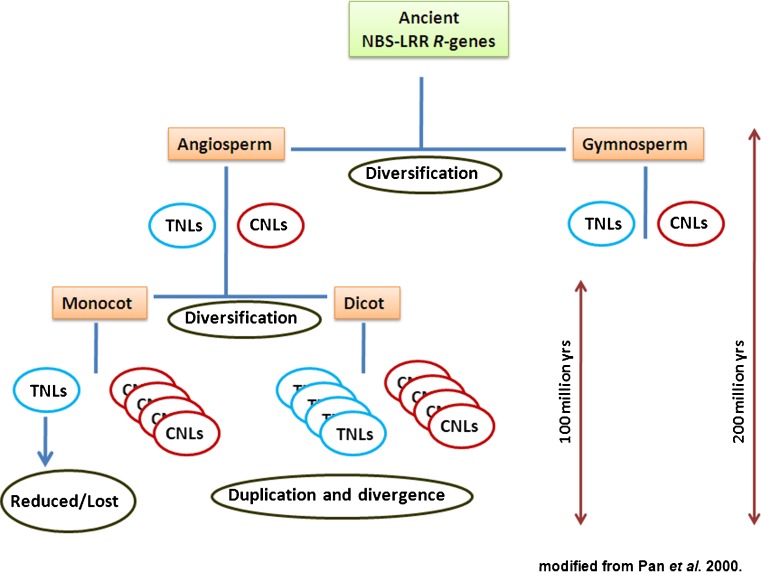
On the other hand, TNLs are largely over expressed in dicot genomes as compared to CNLs. Arabidopsis itself consist of double the number of TNLs than CNLs within its genome (Meyers et al. 2003). The presence of TNLs in pine and moss further indicate that this subfamily of NBS-LRRs have also evolved prior to the angiosperm–gymnosperm split (which occurred at least 200 millions years ago). According to Pan et al. 2000, the evolution of TNLs and CNLs involved two stages (Fig. 1). Stage I exhibited the presence of both CNLs and TNLs with broad spectrum specificity which evolved during the divergence of angiosperm and gymnosperm about 200 million years ago. Stage II was dominated by gene duplication and diversification which characterized the evolution of TNLs and CNLs. This took place after the monocot-dicot separation about 100 million years ago and led to the degeneration of TNL type R-genes in cereals. Although, Pan et al. 2000 suggested that TNLs were lost in cereals, this could be possibly true for the entire monocots in general. So far, the NBS sequences from the Triticum-Thinopyrum line are the only reported monocot TNLs (Jiang et al. 2005). Beside the non existence of TIR-NBS sequences in agriculturally important cereal monocots (Order-Poales), a recent extensive characterization of NBS sequences in four other monocot orders (Zingiberales, Arecales, Asparagales and Alismatales) also resulted in no retrieval of TNLs (Tarr & Alexander 2009). Further, two TNL type sequences isolated from Agrostis species never had true open reading frame steady with NBS domain to qualify them as monocot TNLs (Budak et al. 2006). Thus, it may be concluded that, although TIR-NBS-LRRs were present in early land plants, they either never developed or have been significantly reduced or lost in monocotyledonous plants (Fig. 1). Another interesting point is TIR-NBS sequences are rarely found in magnolids as well. This makes it even more unclear whether TNLs were lost before the divergence of monocots and magnolids or degenerated independently in both lineages. An extensive characterization of NBS gene sequences in additional taxa of monocots, magnolids and other basal angiosperms is needed to further validate the proposed evolutionary nature of the plant R genes.
Fig. 1.
Evolutionary pattern of NBS-LRR class resistance genes in plants. Diversification of TIR-NBS-LRR (group I) and non-TIR-NBS-LRR (group II) took place during differentiation of angiosperms and gymnosperms. The separation of monocot and dicot was followed by extensive gene duplication and diversification resulting in NBS-LRR genes with diverse recognitional specificities
Stahl et al. (1999) provided the first attempt to estimate the age of a functional R gene using a comparative analysis of DNA sequence variation in regions flanking the RPM1 locus. They compared variation in sequence among accessions of two Arabidopsis species, and concluded that the functional resistance allele and the null deletion allele have coexisted at this locus for approximately 10 million years. This estimate further validated the predicted divergence of Brassica and Arabidopsis lineages, in which deletion of RPM1 seem to have occurred independently (Grant 1998). The CNL evolutionary groups are also unevenly distributed in dicot taxa. Cannon et al. (2002) reported 18 sequence representations from a CNL lineage in Arabidopsis but only 4 in soybean and Medicago truncatula while another lineage represented 42 sequences in soybean and Medicago truncatula but only two in Arabidopsis. Nei and Rooney (2005) has categorized this as ‘birth and death hypothesis’ according to which many NBS-LRR lineages has been lost and supplemented with new lineages in the recent times whereas some lineages has been able to retain themselves for a pretty long time period. Further, this also suggests that the NBS-LRR diversity cannot be based on the pattern variability exhibited by any single plant genome. Only a thorough comparison of NBS-LRR sequences from different monocot, dicot and gymnosperms may possibly provide a universally acceptable model to study evolutionary dynamics of NBS-LRR genes.
Apart from the CNLs and TNLs, the existence of modified R gene families such as TX (TIR-X) and TN (TIR-NBS) add up new twist to the NBS-LRR evolutionary pattern. The TIR-NBS proteins lack the LRR domain while the TX protein lacks both the characteristics NBS and LRR domains found in an R gene. TX and TN proteins are reportedly expressive in pines and grasses. Two TN proteins has been reported to be conserved in both Arabidopsis and rice suggesting these are the ancient group of NBS-LRR protein families (Meyers et al. 2002). Although a few TX- and TN-like sequences have been found in cereals, no TNL genes have been identified in cereal genomes (Bai et al. 2002; Meyers et al. 2002). However, the presence of TNL genes in coniferous genomes suggests that the grasses might have lost these types of genes during evolution (Fig. 1) (Bai et al. 2002). More recently, the identification and characterization of a fusion product of TN and TNL proteins in Arabidopsis for resistance against Peronospora parasitica further complicate the situation (Sinapidou et al. 2004). Thus, more comprehensive analysis of TNL and CNL genes in additional plant families is required to infer the evolutionary events leading to the differences in R gene composition.
NBS-LRR diversification
The diversification of nucleotide substitution patterns in R genes is also a major tool in the evolutionary development and adaptation of selected coding domains such as the leucine rich repeats (LRRs). For an increasing number of R genes, including the NBS-LRR genes, evidence of the selection for diversity of codons encoding residues in the LRR region that are predicted to be solvent exposed, and hence may constitute ligand contact points. Protein variation among NBS-LRR genes can be assessed by comparing base-pair changes in nucleotide sequence from orthologues or paralogues variants of the gene that either alter the amino acids resulting in non-synonymous substitutions (Ka) or leave the amino acid unaltered resulting in synonymous substitutions (Ks) (Kreitman and Akashi 1995). A positive selection for amino acid substitution can be realized when the ratio between Ka and Ks is significantly greater than one (Stahl and Bishop 2000). Adaptive selection experiments in various plant species like rice, tomato, lettuce etc. reveal high rates of amino acid replacement changes in the solvent-exposed residues of the LRR domain. Parniske et al. (1997) were the first to use this comparative method to analyze sequence variation in tandemly repeated genes at the Cf4/Cf9 locus from different subspecies of tomato. Likewise, although Xa21D and Xa21 share about 99 % of sequence similarity, nonsynonymous substitutions occur significantly more frequently than do synonymous substitutions in the LRRs (Wang et al. 1998). It is always expected that the fragments of a protein that binds to ligand will be subjected to stronger adaptive selection than regions of those proteins that play a structural function. The LRR regions of R-genes are receptor domains for recognition of specific pathogen elicitors and may be involved in ligand binding activity. Variability at the LRR domain can provide specific advantage to a NBS-LRR gene in recognizing, binding and defending against a vast array of pathogens. Thus, the LRR domain shows much higher levels of diversity, particularly at the solvent exposed faces in the repeats, than other domains within the NBS encoding genes. Adaptive evolution in the LRR is reliable within an evolutionary race in such way that pathogens should impose selection to continually alter recognition specificity.
Further, R-gene diversification also depends upon variation in the length of the LRR units. For example, the Cf2/5 locus in tomato has undergone extensive deletion and expansion in individual LRR repeat units generating variable paralogues (Dixon et al. 1998). In flax, the L locus is characterized by highly degenerative LLR repeats that determine specificity differences between paralogues (Thomas et al. 1997). Like the initial analysis of Cf gene in tomato, subsequent comparison of DNA sequences within NBS-LRR gene loci has revealed evidence of past exchanges of blocks of sequence by recombination (Ellis et al. 1999 and Noel et al. 1999). Block sequences of LRRs have undergone duplications and these direct repeat sequences undergo unequal exchange events that give rise to cycles of repeat expansion and reduction. However, it is still unclear whether such exchanges occur by sequential crossing over or gene conversion. Mutant R genes due to opening out and closing in of LRRs have been reported in flax and Arabidopsis (Parker et al. 1997). Thus, the combined effect of point mutations and changes in the number of LRR repeats indicate that the variation in the LRR domain may be important for determining the specificity of a given R gene.
Genetic duplication and recombination
NBS-LRR genes are generally organized as genetically defined clusters in most of the plant species which may be simple consisting of homologous R-gene sequences from a single gene family or complex one consisting of R-gene sequences derived from different unrelated families. For example, 76 % of rice NBS-LRRs is represented in 44 gene clusters while the rest occurs as singletons (Zhou et al. 2004). This is true even for other plant species such as Arabidopsis, Maize and barley (Meyers et al. 2003; Bai et al. 2002). Sometimes, gene copy number can vary widely among haplotypes within a species such as that are found in maize Rp1, lettuce Dm3 or potato MLB clusters (Smith et al. 2004; Kuang et al. 2004; Yue et al. 2012; Jupe et al. 2012). Zhang et al. 2010 found that the number of genes in the NBS family not only vary among congeneric species but also among conspecific cultivars. This variation could be attributed to gene duplication, deletion, pseudogenization and functional diversification. Since NBS family is highly crucial to plant defense, rapid variation in their numbers is a necessity because they need to defend themselves from rapidly varying races of pathogens in a surrounding.
In some R gene clusters, unequal recombination occurs frequently (such as Rp1 gene clusters of maize), whereas in others it is rare (such as Dm3 of lettuce) (Michelmore and Meyers 1998). As a consequence, at loci similar to Dm3, orthologues genes from two different lines are more similar to each other than they are to paralogues genes within the same cluster. Similarly, analysis of the Arabidopsis genome also indicate that numerous small-scale genomic duplications have copied or translocated one or several NBS-LRR genes from these clusters to distal and probably random locations in the genome. Molecular studies have demonstrated that this clustering usually results from tandem duplications of paralogues sequences (Meyers et al. 2003; Michelmore and Meyers 1998; Zhu et al. 2002). Duplication and insertion of repetitive sequences such as transposons may result in genic and intergenic sequence repeats within NBS-LRR R genes, which can cause mispairing during recombination events. This will lead to unequal crossovers and interlocus gene conversions resulting in variation in gene copy number in clusters. Gene conversion between paralogs members and inter-genome sequence exchanges has been found to be a major force in the SH3-CNL copy genes in Coffea arabica (Ribas et al. 2011). Thus, R-genes can exhibit altered expressions because of structural modification caused by intergenic unequal crossover. It is also possible that intragenic mispairing will result in the development of chimeric R-genes that can encode novel functions. This can cause change in the NBS-LRR gene copy numbers within a cluster depending upon the positioning of the mispaired recombination sites. Recently, Baumgarten et al. (2003) suggested segmental duplication (duplication and translocation of entire chromosomal segments) as responsible for the genomic dispersion of NBS-LRR genes rather than small scale ectopic duplications. However, when physically separated from their closest relatives, NBS-LRR genes might also adopt and preserve new functions by escaping sequence homogenization occurring because of recombination. Thus, the tandem, segmental and ectopic duplication events have been demonstrated to be an important force for the maintenance of the sequence polymorphism in the R gene loci in plants, such as the generation of Arabidopsis NBS-LRR genes (Meyers et al. 2003).
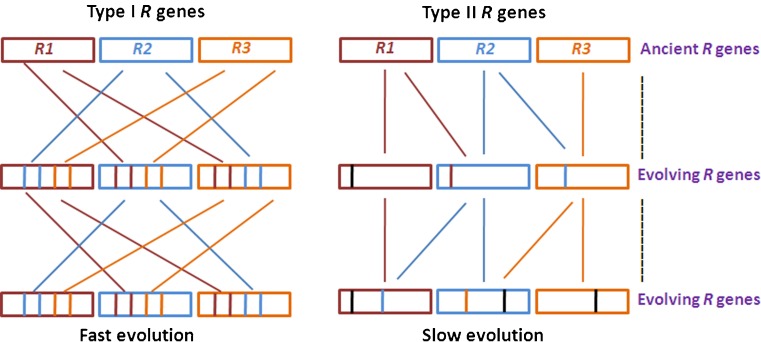
Although sequence exchanges are integral factor in R gene cluster evolution, there also exist a few NBS-LRR R gene lineages that exist in isolation with little or no sequence exchanges (Ellis et al. 2000). This suggests that the frequency of sequence exchange even varies between genes within individual clusters. The three distinct gene subclasses within the N cluster in Linium usitassimum are completely different and never recombine with each other (Dodds et al. 2001). However, two paralogues within a subclass exhibiting high degree of sequence similarity has been reported to undergo high frequency sequence exchange. Kuang et al. (2004) reported that individual clades within the clusters undergo either fast or slow patterns of evolution termed as type I and type II. Type I of the RGC2 genes from lettuce evolve rapidly and display chimeric structures while type II was largely conserved genes with little or no recombination. Luo et al. 2011 investigated the evolution of Rp1 gene family in Poaceae further validating the dual evolutionary pattern of R gene clusters. The extant species of Poaceae has one to five Rp1 loci as compare to two loci in the common ancestor. Zea mays genotypes resulted in a large number of Rp1 homologues through duplication possibly due to sequence exchange among paralogues. A distinct differentiation of type I and type II genes was observed among Oryza species as well. While one member in the Oryza Rp1 cluster did not change sequences with its paralogues, other paralogues had frequent sequence exchanges. Although sequence exchange occurs only between clade members, type I clade paralogs exhibit high degree of sequence exchange thereby resulting in high homology among paralogs and high haplotypic diversity (Fig. 2). On the other hand the type II gene clades exhibit occasional sequence exchanges thereby resulting in high homology among orthologs. It may be assume that structural rearrangement of type I genes may have inhibited mispairing and reduce recombination thereby generating the type II genes. An evolutionary attempt to conserve most of the important resistance specificities of NBS-LRR R genes developed through course of evolution may have resulted in type II gene subclass while most of the on going activities for development of resistance specificities with respect to new pathogen elicitors would be the responsibility of the type I genes. Overall, the type I genes evolve more rapidly that type II genes, reflecting different rates of evolution and selection pressures. There can be several approaches to bring out such modification. It is evident from different experiments that sequence exchanges between divergent sequences are rare while exchange between similar genes appear frequently (Meyers et al. 2003, Baumgarten et al. 2003). Sometimes, considerable divergence in the intergenic regions between haplotypes might have inhibited mispairing thereby stabilizing novel haplotypes. Further, novel resistance specificities might be protected from homogenization due to concerted evolution by the dispersal of novel haplotypes to physically distant sites ether by segmental duplication or ectopic recombination.
Fig. 2.
Fast and slow pattern of evolution exhibited by type I and type II genes in plants. Type I clade paralogs exhibit high degree of sequence exchange thereby resulting in high haplotypic diversity. Type II gene clades exhibit occasional sequence exchanges thereby resulting in high homology among orthologs. Color lines represent the changes accumulated within R genes through evolution. Dotted line represent the evolutionary time
Although differentiation of conserved and variable regions within the R gene cluster may be attributed to random sequence exchanges including mutations and transposon insertions, another possible power guiding the evolution of R gene clusters may be inhibition of recombination. Recent reports suggest that pathogen challenge result in elevated somatic recombination frequency or DNA rearrangements induced by a ‘systemic plant signal’. This systemic recombination signal generating genome instability due to pathogen stress is heritable, resulting in increased recombination in the progeny of stressed plants (Lucht et al. 2002; Molinier et al. 2006). Further, Durant et al. (2007) reported that chromatin modification may also conditionally regulate R gene expression and recombination in response to pathogen attack suggesting that chromatin modification may repress sequence exchange at R gene clusters in absence of pathogen attack. Boyko et al. (2007) observed increased rate of recombination in the homologs of the LRR regions of the N allele in tobacco under Tobacco Mosaic Virus (TMV) infection whereas no such instability was detected in any other loci. Such recombination event resulted in overall increase in DNA methylation while decrease in methylation specifically at R-genes in the progeny of the infected plants. This suggests that the frequency of evolution can be highly heterogenous due to variable pattern of methylation within R gene clusters. This pathogen induced restructuring of R gene cluster make us to assume an episodic evolutionary pattern. Pathogen absence inhibits recombination and transposon insertion due to chromatin modification thereby limiting sequence exchange and increase the conservation of haplotypes. On the contrary, pathogen stress improve the methylation resulting in high recombination frequency from generation after generation causing haplotypic gene duplication and generation of chimeric NBS-LRR R genes.
Although most of the R genes proliferate in clusters, a few of them are even represented by single copy loci such as Arabidopsis RPM1 and RPS5. It has been proposed that these single copy R loci have evolved a very long time ago and now are subject to purifying selection and therefore a reduction in gene diversity (McDowell and Simon 2006). However, strong diversifying selection can also result in many alleles at a single gene locus. This can be seen in case of flax L locus showing resistance to flax rust pathogen Melanospora lini. Frequent interallelic recombination events has resulted the development of at least a dozen of novel alleles at the L locus (Dodds et al. 2006). Most of the recessive resistance genes also occur as single-copy loci distributed at different locations in plant genomes. Although no formal proof of recessive resistance gene clustering has yet been obtained, they are often mapped in the same genetic interval for resistance specificity against the same pathogen. Bauer et al. (1997) mapped all the four recessive resistance genes rym8, rym9, rym12 and rym13 against Barley Bymo Virus within a short genetic interval in the sub-telomeric region on the long arm of the chromosome 4 H in barley. Although it is hypothesized that singleton genes may act as origin for the establishment of new gene clusters, a recent report on the mapping of NBS genes in potato throw new insights into their evolutionary role. Lozano et al. 2012 has reported the occurrence of high rate of pseudogenization (41.6 %) among the total R-genes in Solanum tuberosum as compared to other plant genomes. A similar result has also been obtained in Lotus japonicus (Li et al. 2010). Even if a criterion bias in the characterization of pseudogenes cannot be ruled out, a recent study suggests that pseudogenes can regulate coding gene expression as they compete for microRNA binding (Poliseno et al. 2010). As viral siRNAs has been found to exhibit the possibility of targeting host resistance genes, existence of a large number of pseudogenes could be a defense attribute against such actions. Thus, the NBS pseudogenes may be involved in preventing the degradation of homologues functional R-genes through a restricted silencing approach.
Population genetics and NBS-LRR evolution
Population genetic analysis can also determine the diversity of R alleles in nature where selection forces maintain resistance thereby leading the evolution for new specificities in natural populations. There has been an extensive ‘arm race’ going on between the plants that develop the resistance specificities and pathogen that try to overcome recognition by these plants (Dawkins and Krebs 1979). Thus, higher is the disease pressure, greater is the chance that the older R genes will be replaced with the newer one. The defeated r alleles are supplanted by new R alleles through selective sweeps in which the functionless alleles are removed from the population. This may cause widespread occurrence of younger R genes and monomorphic R gene loci (Bergelson et al. 2001). However, the recent studies on the population genetics with respect to R gene loci show the existence of relatively long-lived polymorphism within it. One recent evolutionary study used a collection of 26 ecotypes of Arabidopsis to investigate allelic variation at the single gene locus RPM1 (Stahl et al. 1999). The authors found that RPM1 is a single copy gene in the resistance ecotypes of Arabidopsis and is completely absent from the susceptible (rpm1) ecotypes. Both alleles are found widely in Arabidopsis throughout its natural distribution. The functional RPM1 locus of Arabidopsis was found to be syntenic with RPM1 loci of Brassica oleracea and Arabidopsis lyrata thereby confirming that the rpm1 allele in Arabidopsis thaliana was created by deletion of a functional RPM1 gene. Studying the molecular evolutionary analysis through DNA sequence polymorphism flanking the locus, Stahl et al. (1999) further established that the two alleles are about a million years old and have been maintained by balancing selection and have fluctuated in frequency. But the transient polymorphism ‘arm race’ model according to which R alleles are replaced in each cycle by new ones was summarily rejected by Stahl et al. (1999) on the basis that firstly, resistance and susceptible alleles have existed at RPM1 locus for more than a million years with its origin close to the time of Arabidopsis speciation and secondly, plant populations in general show considerable variation at R-gene loci. Rather, they proposed a recycling polymorphism ‘trench warfare’ model in which the functional and the susceptible alleles are long lived but their relative frequencies fluctuate dynamically over time due to regular periods of negative frequency dependent selections. Evidences from many other loci of Arabidopsis such as RPP1, RPP8 and RPP13 with common alternative alleles also shows that the polymorphism has been generated, accumulated and maintained at these loci for millions of years (Rose et al. 2004).
Downstream components of R-gene signaling pathways are also co-adapted within species to particular R-gene products. Tai et al. (1999) cloned a non-TIR class NBS-LRR R gene Bs2 from Capsicum annuum and found that it is functional in several solanaceous plants but not in species outside Solanaceae. This suggest that although NBS-LRR R genes are largely ubiquitous, they also exhibit a phenomenon called ‘restricted taxonomic functionality’. Further, no pathogenic strains of the pathogen lacking avrBs2 have emerged in spite of strong selection pressure imposed by the use of this gene in commercial pepper cultivars. This observation underlines the potential role for ‘defeated’ R genes in natural populations. Such host genes prevent the increase in frequency of the corresponding avr genes in the pathogen, thus potentially decreasing the overall fitness of the pathogen population.
The maintenance of the deletion allele suggests that under certain conditions, the active allele imposes a genetic load on the host and is sometimes disadvantageous in the nonexistence of pathogen pressure. Heidel et al. (2004) has demonstrated the association of fitness penalties with generally induced R alleles that confers constitutive resistance. Tian et al. (2003) transformed an rpm1 type null line of Arabidopsis line with the functional RPM1 to study the fitness cost associated with presence and absence of functional R allele. They reported that the transgene carrying the functional RPM1 resulted in 9 % decrease in seed production in field conditions under no pathogen pressure. This further strengthens the view that balancing selection has conserved the null rpm1 allele for such a long period only to counterbalance the highly pricy RPM1 allele. However, this cannot be considered universal as Arabidopsis R genes RPS2 and RPP5 has been reported to be less costly in absence of pathogen pressure (Korves and Bergelson 2003). Thus, the cost-benefits associations for individual R genes may largely vary based on the allelic diversity at the R gene locus, increase response to pathogens, nutritional availability and environmental conditions. It is required to evaluate several NBS-LRR R alleles in varied plant species under different conditions to determine the molecular basis of R gene dependent fitness cost.
Concluding remarks
There has been considerable progress during the last few years in understanding NBS-LRR R gene systematics and evolution. Recombinational analysis of RPM1 gene in Arabidopsis thaliana and L alleles in Linium usitassimum especially has made a huge difference in our philosophy towards the evolutionary perspectives of R-genes. The distribution of R genes within plant genome is resulted through elaborate exchange involving recombination, selection, mutation and is influenced by the reproductive behavior and ecological limitations. The diversification and separation of NBS-LRR sequences into distinct ancient lineages explains about the structural features and pragmatic sequence diversity within this family. Large-scale sequence analysis of NBS-LRR haplotypes shows extensive clustering pattern of R genes and their diversification through duplication and recombination. Infact, RGC genes within the same cluster exhibit heterogeneity in the evolutionary rates with one being rapidly evolving between sequence exchange between paralogs while the other one evolving slowly independent of the paralogs. The contraction/expansion of LRR domains allows for the speedy development of altered resistance specificities. Molecular analysis of population genetics of R-genes suggest the nature of selection processes acting on the complex NBS-LRR R gene loci involved in plant-pathogen interactions. The molecular evolutionary analysis also suggest that the resistance and susceptible alleles of NBS-LRR gene are about a million years old and have been maintained by balancing selection and have fluctuated in frequency. However, the cost benefit associated with an individual R gene depends upon the response to pathogen, ecological conditions and availability of food and nutrition. Although several hypotheses have been tested about R gene evolution, numerous important questions are still there to be addressed. Why dicot has two NBS-LRR R proteins, TNLs and CNLs while monocots have only one? What is the nature of relationship between plant and animal with respect to resistance evolution? How does a locus residing in only one of the genomes evolve to confer resistance in polyploids? Why the NBS-LRR R genes exhibit duality in recognizing pathogens? Complete structural details of NBS-LRR protein complexes will only show the evolutionary interpretation of nucleotide substitutions and recombination in R gene sequences. Thorough elucidation of the available plant genomic resources in future and their utilization in focus experiments in different plant models will enable us to assess the exact nature of the various NBS-LRR R gene propositions.
Acknowledgements
The authors are grateful to Prof. Manoj Ranjan Nayak, President, Siksha O Anusandhan University for his encouragement and support.
Abbreviations
- R-genes
Resistance genes
- NBS
Nucleotide binding site
- LRR
Leucine rich repeats
- CNL
Coiled coil NBS-LRR
- TNL
Toll interleukin-1 receptor NBS-LRR
References
- Bai J, Pennill LA, Ning J, Lee SW, Ramalingam J, Webb CA, Zhao B, Sun Q, Nelson JC, Leach JE, Hulbert SH. Diversity in nucleotide binding site-leucine-rich repeat genes in cereals. Genome Res. 2002;12:1871–1884. doi: 10.1101/gr.454902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer E, Weyen J, Schiemann A, Graner A, Ordon F. Molecular mapping of novel resistance genes against barley mild mosaic virus (BaMMV) Theor Appl Genet. 1997;95:1263–1269. doi: 10.1007/s001220050691. [DOI] [Google Scholar]
- Baumgarten A, Cannon S, Spangler R, May G. Genome level evolution of resistance genes in Arabidopsis thaliana. Genetics. 2003;165:309–319. doi: 10.1093/genetics/165.1.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergelson J, Kreitman M, Stahl EA, Tian D. Evolutionary dynamics of plant R-genes. Science. 2001;292:2281–2285. doi: 10.1126/science.1061337. [DOI] [PubMed] [Google Scholar]
- Boyko A, Kathiria P, Zemp FJ, Yao Y, Pogribny I, Kovalchuk I. Transgenerational changes in the genome stability and methylation in pathogen-infected plants. Nuc Acids Res. 2007;35:1714–1725. doi: 10.1093/nar/gkm029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budak H, Su S, Ergen N. Revealing constitutively expressed resistance genes Agrostis species using PCR based motif directed RNA fingerprinting. Gen Res. 2006;88:165–175. doi: 10.1017/S0016672307008518. [DOI] [PubMed] [Google Scholar]
- Cannon SB, Zhu H, Baumgarten AM, Spangler R, May G, Cook DR, Young ND. Diversity, distribution, and ancient taxonomic relationships within the TIR and non-TIR NBS-LRR resistance gene subfamilies. J Mol Evol. 2002;54:548–562. doi: 10.1007/s00239-001-0057-2. [DOI] [PubMed] [Google Scholar]
- Dawkins R, Krebs J (1979) Arm races between and within species. Proceedings of the royal society of London 205:480-511 [DOI] [PubMed]
- Dixon MS, Hatzixanthis K, Jones DA, Harrison K, Jones JDG. The tomato Cf-5 disease resistance gene and six homologs show pronounced allelic variation in leucine-rich repeat copy number. Plant Cell. 1998;10:1915–1925. doi: 10.1105/tpc.10.11.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds PN, Lawrence G, Catanzariti AM, Teh T, Wang CIA, Ayliffe MBK, Ellis J. Direct protein interaction underlies gene-for-gene specificity and co-evolution of the flax L5/L6/L7 resistance genes and flax rust AvrL567 avirulence genes. Proc Natl Acad Sci USA. 2006;103:8888–8893. doi: 10.1073/pnas.0602577103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds PN, Lawrence GJ, Ellis JG. Contrasting modes of evolution acting on the complex N locus for rust resistance in flax. Plant J. 2001;27:439–453. doi: 10.1046/j.1365-313X.2001.01114.x. [DOI] [PubMed] [Google Scholar]
- Durrant WE, Dong X. Systemic acquired resistance. Annu Rev Phytopathol. 2004;42:185–209. doi: 10.1146/annurev.phyto.42.040803.140421. [DOI] [PubMed] [Google Scholar]
- Durant WE, Wang S, Dong X. Arabidopsis SNI1 and RAD51D regulate both gene transcription and DNA recombination during the defense response. Proc Natl Acad Sci USA. 2007;104:4223–4227. doi: 10.1073/pnas.0609357104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis JG, Lawrence GJ, Luck JE, Dodds PN. Identification of regions in alleles of the flax rust resistance gene L that determine differences in gene-for-gene specificity. Plant Cell. 1999;11:495–506. doi: 10.1105/tpc.11.3.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis J, Dodds P, Pryor T. Structure, function and evolution of plant disease resistance genes. Curr Opin Plant Biol. 2000;3:278–284. doi: 10.1016/S1369-5266(00)00080-7. [DOI] [PubMed] [Google Scholar]
- Grant M. Independent deletion of a pathogen resistance gene in Brassica and Arabidopsis. Proc Natl Acad Sci USA. 1998;95:15843–15848. doi: 10.1073/pnas.95.26.15843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidel AJ, Clarke JD, Antonovics J, Dong X. Fitness costs of mutations affecting the systemic acquired resistance pathway in Arabidopsis thaliana. Genetics. 2004;168:2197–2206. doi: 10.1534/genetics.104.032193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JDG, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- Joshi RK, Nayak S. Functional characterization and signal transduction ability of nucleotide-binding site leucine rich repeats resistance genes in plants. Genet Mol Res. 2011;10(4):2637–2652. doi: 10.4238/2011.October.25.10. [DOI] [PubMed] [Google Scholar]
- Jiang SM, Hu J, Yin WB, Chen YH, Wang RRC, Hu ZM. Cloning of resistance gene analogs located on the alien chromosome in an addition line of wheat-Thinopyrum intermedium. Theor Appl Genet. 2005;111:923–931. doi: 10.1007/s00122-005-0022-3. [DOI] [PubMed] [Google Scholar]
- Jupe F, Pritchard L, Etherington GJ, Mackenzie K, Cock PJA, Wright F, Sharma SK, Bolser D, Bryan GJ, Jones JDG, Ingo H. Identification and localization of the NB-LRR gene family within the potato genome. BMC Genomics. 2012;13:75. doi: 10.1186/1471-2164-13-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MG, da Cunha L, McFall AJ, Belkhadir Y, Debroy S, Dangl JL, Mackey D. Two Pseudomonas syringae type III effectors inhibit RIN4-regulated basal defense in Arabidopsis. Cell. 2005;121:749–759. doi: 10.1016/j.cell.2005.03.025. [DOI] [PubMed] [Google Scholar]
- Kohler A, Rinaldi C, Duplessis S, Baucher M, Geelan D, Duchaussoy F, Meyers BC, Boerjan W, Martin F. Genome-wide identification of NBS resistance genes in Populus trichocarpa. Plant Mol Biol. 2008;66:619–636. doi: 10.1007/s11103-008-9293-9. [DOI] [PubMed] [Google Scholar]
- Korves TM, Bergelson J. A developmental response to pathogen infection in Arabidopsis. Plant Physiol. 2003;133:339–347. doi: 10.1104/pp.103.027094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreitman M, Akashi H (1995) Molecular evidence for natural selection. Annu Rev Ecol Syst 26:403–422. doi: http://www.jstor.org/stable/2097213
- Kuang H, Woo SS, Meyers BC, Nevo E, Michelmore RW. Multiple genetic processes result in heterogeneous rates of evolution within the major cluster disease resistance genes in lettuce. Plant Cell. 2004;16:2870–2894. doi: 10.1105/tpc.104.025502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Cheng Y, Ma W, Zhao Y, Jiang H, Zhang M. Identification and characterization of NBS-encoding disease resistance genes in Lotus japonicus. Plant Syst Evol. 2010;289:101–110. doi: 10.1007/s00606-010-0331-0. [DOI] [Google Scholar]
- Liu J, Liu X, Dai L, Wang G. Recent progress in elucidating the structure, function and evolution of disease resistance genes in plants. J Genet Genom. 2007;34(9):765–776. doi: 10.1016/S1673-8527(07)60087-3. [DOI] [PubMed] [Google Scholar]
- Lozano R, Ponce O, Ramirez M, Mostajo N, Orjeda G. Genome wide identification and mapping of NBS encoding resistance genes in Solanum tuberosum Group Phureja. PLoS One. 2012;7(4):e34775. doi: 10.1371/journal.pone.0034775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucht JM, Mauch-Mani B, Steiner HY, Metraux JP, Ryals J, Hohn B. Pathogen stress increases somatic recombination frequency in Arabidopsis. Nature Genet. 2002;30:311–314. doi: 10.1038/ng846. [DOI] [PubMed] [Google Scholar]
- Luo S, Peng J, Li K, Wang M, Kuang H. Contrasting evolutionary patterns of the Rp1 resistance gene family in different species of Poaceae. Mol Biol Evol. 2011;28(1):313–325. doi: 10.1093/molbev/msq216. [DOI] [PubMed] [Google Scholar]
- Martin GB, Bogdanove AJ, Sessa G. Understanding the functions of plant disease resistance proteins. Annual Rev Plant Biol. 2003;54:23–61. doi: 10.1146/annurev.arplant.54.031902.135035. [DOI] [PubMed] [Google Scholar]
- McDowell JM, Simon SA. Recent insights into R gene evolution. Mol Plant Pathol. 2006;7:437–448. doi: 10.1111/j.1364-3703.2006.00342.x. [DOI] [PubMed] [Google Scholar]
- Meyers BC, Kozik A, Griego A, Kuang HH, Michelmore RW. Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell. 2003;15:809–834. doi: 10.1105/tpc.009308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers BC, Morgante M, Michelmore RW. TIR-X and TIR-NBS proteins: two new families related to disease resistance TIR NBS-LRR proteins encoded in Arabidopsis and other plant genomes. Plant J. 2002;32:77–92. doi: 10.1046/j.1365-313X.2002.01404.x. [DOI] [PubMed] [Google Scholar]
- Michelmore RW, Meyers BC. Clusters of resistance genes in plants evolve by divergent selection and a birth-and-death process. Genome Res. 1998;8:1113–1130. doi: 10.1101/gr.8.11.1113. [DOI] [PubMed] [Google Scholar]
- Molinier J, Ries G, Zipfel C, Hohn B. Tran generation memory of stress in plants. Nature. 2006;442:1046–1049. doi: 10.1038/nature05022. [DOI] [PubMed] [Google Scholar]
- Nei M, Rooney AP. Concerted and birth-and-death evolution of multigene families. Annu Rev Genet. 2005;39:121–152. doi: 10.1146/annurev.genet.39.073003.112240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noel L, Moores TL, van Der Biezen EA, Parniske M, Daniels MJ, Parker JE, Jones JDG. Pronounced intraspecific haplotypes divergence at the RPP5 complex disease resistance locus of Arabidopsis. Plant Cell. 1999;11:2099–2112. [PMC free article] [PubMed] [Google Scholar]
- Pan Q, Wendel J, Fluhr R. Divergent evolution of plant NBS-LRR resistance gene homologues in dicot and cereal genomes. J Mol Evol. 2000;50:203–213. doi: 10.1007/s002399910023. [DOI] [PubMed] [Google Scholar]
- Parker JE, Coleman MJ, Szabo V, Frost LN, Schmidt R, van der Biezen EA, Moores T, Dean C, Daniels MJ, Jones JDG. The Arabidopsis downy mildew resistance gene RPP5 shares similarity to the toll and interleukin-1 receptors with N and L6. Plant Cell. 1997;9:879–894. doi: 10.1105/tpc.9.6.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parniske M, Hammond-Kosack KE, Golstein C, Thomas CM, Jones DA, Harrison K, Wulff BBH, Jones JDG. Novel disease resistance specificities result from sequence exchange between tandemly repeated genes at the Cf-4/9 locus of tomato. Cell. 1997;91:821–832. doi: 10.1016/S0092-8674(00)80470-5. [DOI] [PubMed] [Google Scholar]
- Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–1038. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribas AF, Cenci A, Combes MC, Etienne H, Lashermes P. Organization and molecular evolution of a disease resistance gene cluster in coffee trees. BMC Genomics. 2012;12:240. doi: 10.1186/1471-2164-12-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose LE, Bittner-Eddy PD, Langley CH, Holub EB, Michelmore RW, Beynon JL. The maintenance of extreme amino acid diversity at the disease resistance gene, RPP13, in Arabidopsis thaliana. Genetics. 2004;166:1517–1527. doi: 10.1534/genetics.166.3.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinapidou E, Williams K, Nott L, Bahkt S, Tor M, Crute I, Bittner-Eddy P, Beynon J. Two TIR-NB-LRR genes are required to specify resistance to Perenospora parasitica isolate cala2 in Arabidopsis. Plant J. 2004;38:898–909. doi: 10.1111/j.1365-313X.2004.02099.x. [DOI] [PubMed] [Google Scholar]
- Smith SM, Pryor AJ, Hulbert SH. Allelic and haplotypic diversity at the rp1 rust resistance locus of maize. Genetics. 2004;167:1939–1947. doi: 10.1534/genetics.104.029371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl EA, Bishop JG. Plant–pathogen arms races at the molecular level. Curr Opin Plant Biol. 2000;3:299–304. doi: 10.1016/S1369-5266(00)00083-2. [DOI] [PubMed] [Google Scholar]
- Stahl EA, Dwyer G, Mauricio R, Kreitman M, Bergelson J. Dynamics of disease resistance polymorphism at the Rpm1 locus of Arabidopsis. Nature. 1999;400:667–671. doi: 10.1038/23260. [DOI] [PubMed] [Google Scholar]
- Tarr DEK, Alexander HM. TIR-NBS genes are rare in monocots: evidence from diverse monocot orders. BMC Research Notes. 2009;2:197. doi: 10.1186/1756-0500-2-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai TH, Dahlbeck D, Clark ET, Gajiwala P, Pasion R, Whalen MC, Stall RE, Staskawicz BJ. Expression of the Bs2 pepper gene confers resistance to bacterial spot disease in tomato. Proc Natl Acad Sci USA. 1999;96:14153–14158. doi: 10.1073/pnas.96.24.14153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas CM, Jones DA, Parniske M, Harrison K, Balint-Kurti PJ, Hatzixanthis K, Jones JDG. Characterization of the tomato Cf-4 gene for resistance to Cladosporium fulvum identifies sequences that determine recognitional specificity in Cf-4 and Cf-9. Plant Cell. 1997;9:2209–2224. doi: 10.1105/tpc.9.12.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian D, Traw MB, Chen JQ, Kreitman M, Bergelson J. Fitness costs of R-gene-mediated resistance in Arabidopsis thaliana. Nature. 2003;423:74–77. doi: 10.1038/nature01588. [DOI] [PubMed] [Google Scholar]
- Wang GL, Ruan DL, Song WY, Sideris S, Chen LL, Pi LY, Zhang S, Zhang Z, Fauquet C, Gaut B, Ronald P. Xa21D encodes a receptor–like molecule with a leucine-rich repeat domain that determines race specific recognition and is subject to adaptive evolution. Plant Cell. 1998;10:765–779. doi: 10.1105/tpc.10.5.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue JX, Meyers BC, Chen JQ, Tian D, Yang S. Tracing the origin and evolutionary history of plant nucleotide binding site leucine rich repeat (NBS-LRR) genes. New Phytol. 2012;193(4):1049–1063. doi: 10.1111/j.1469-8137.2011.04006.x. [DOI] [PubMed] [Google Scholar]
- Zhang M, Wu YH, Lee MK, Liu YH, Rong Y, Santos TS, Wu C, Xie F, Nelson RL, Zhang HB. Number of genes in NBS and RLK families vary by more than four fold within a plant species and are regulated by multiple factors. Nuc Acids Res. 2010;30:1–13. doi: 10.1093/nar/gkp822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Wang Y, Chen JQ, Araki H, Jing Z, Jiang K, Shen J, Tian D. Genome-wide identification of NBS genes in japonica rice reveals significant expansion of divergent non-TIR NBS-LRR genes. Mol Genet Genom. 2004;271:402–415. doi: 10.1007/s00438-004-0990-z. [DOI] [PubMed] [Google Scholar]
- Zhu HY, Cannon SB, Young ND, Cook DR. Phylogeny and genomic organization of the TIR and non-TIR NBSLRR resistance gene family in Medicago truncatula. Mol Plant Microbe Interact. 2002;15:529–539. doi: 10.1094/MPMI.2002.15.6.529. [DOI] [PubMed] [Google Scholar]