Abstract
A widespread leaf deformity disease of mentha (mint), accompanied by whiteflies, the vectors of begomoviruses, was observed in Punjab in the last few years. The presence of begomovirus was indicated by DNA dot-blot analysis using the conserved coat protein and replication-associated protein genes of another begomovirus, Sri Lankan cassava mosaic virus (SLCMV). A DNA fragment (2.0 kb), representing a partial genomic DNA of a begomovirus, amplified from the symptomatic mentha leaves was used to design end-primers and further amplify an additional 0.9 kb fragment, representing the remaining portion of the resident viral DNA. The two sequences, assembled together (2.7 kb), showed that they represented the complete sequence of an isolate of Tomato leaf curl Karnataka virus (ToLCKV) DNA. Using universal betasatellite primers, a 1.4 kb fragment was amplified from the same sample. This cloned DNA fragment showed complete sequence identity with the previously reported Cotton leaf curl Multan betasatellite (CLCuMB). Majority of the symptomatic mentha leaf samples, collected from four districts of Punjab, showed cross-hybridization in DNA dot-blot using cloned SLCMV and CLCuMB DNA, indicating the presence of one or more begomoviruses related to SLCMV and the betasatellite, CLCuMB. The begomovirus and betasatellite could be mechanically transmitted to Nicotiana benthamiana. Whitefly transmission of the resident begomovirus could also be demonstrated on mentha. The evidence indicates the association of ToLCKV and CLCuMB, a hitherto new combination of a begomovirus and a betasatellite associated with a leaf deformity disease in mentha in Punjab.
Keywords: Mentha, Begomovirus, Betasatellite, Whitefly
Introduction
Mentha, popularly known as mint, is a perennial herbaceous flowering plant of the family Lamiaceae, and the genus Mentha and is cultivated in more than 0.15 million hectares in Punjab for the production of menthol is used widely as an additive and a flavouring agent in processed foods such as jellies, syrups, ice creams, chocolates and in cosmetics such as breath fresheners and perfumes. Cultivation of mentha is a relatively recent phenomenon in Punjab and is gaining popularity because of the growing market for its product. In recent years, widespread incidences of leaf deformity and stunting have been observed in mentha plants in Punjab. Three widely cultivated mentha species; Japanese (menthol) mint (Mentha arvensis), peppermint (M. piperita, a cross between watermint, M. aquatica and spearmint, M. spicata) and spearmint were found to be equally affected.
Geminiviruses (Family Geminiviridae) are plant-infecting DNA viruses, having twinned geminate particles encapsidating a single-stranded circular DNA of approximately 2.7 kb [13, 16]. Begomoviruses (Genus Begomovirus) the largest of the four genera in the family Geminiviridae, have members infecting mainly vegetables, legumes and fibre crops and are transmitted by whiteflies (Bemisia tabaci) [32]. Begomoviruses can be either monopartite (encapsidating a single type of DNA molecule) or bipartite (two types of DNA molecules, DNA-A and DNA-B) [32]. The DNA-A component and its counterpart in monopartite begomoviruses encodes proteins which include the coat protein (CP), a replication-associated protein (Rep), a transcriptional activator and a replication enhancing protein; while the DNA-B component encodes proteins involved with movement of viral DNA (nuclear shuttle protein and movement protein) [18].
In the Old World, begomoviruses are often found associated with small circular DNA molecules (satellite DNAs or sub-viral agents) [5], which are completely dependent upon the viral proteins for replication initiation. The most common satellite associated with begomoviruses is known as the betasatellite, a mutant of which was first reported to be associated with Tomato leaf curl virus (ToLCV) [11] and subsequently, with many, mostly monopartite, begomoviruses [3, 8, 19, 29]. The single open reading frame present on the betasatellite encodes a protein (β-C1), which is associated with functions such as symptom production [18], suppression of RNA-interference [15] and viral movement [3, 26]. Here, we report for the first time, the presence of a complex of a begomovirus and betasatellite in field grown mentha plants showing leaf deformity.
Materials and Methods
Collection of Samples
Mentha leaf samples were collected initially from experimental plants, showing symptoms of leaf deformity, and maintained at Punjab Agricultural University, Ludhiana. For a wider survey, approximately 10% of the total area under mentha cultivation in Punjab were surveyed and samples from 30 mentha plants showing leaf deformity were collected from farmers’ fields from the districts of Jalandhar, Kapurthala, Ludhiana and Nawan Shehar in Punjab, representing Japanese mint, peppermint and spearmint, the numbers of samples collected being 14, 7 and 9, respectively.
Detection of Viral and Satellite DNAs Using Dot-Blot
Young mentha leaves were stored frozen at −70°C and total nucleic acid was extracted using a standard method [21]. The extracted DNA was quantified using a Beckman DU 640B spectrophotometer or visually by electrophoresis in a 1% agarose gel using a known standard (λ/HindIII DNA size-marker, Bangalore Genei). For dot-blot hybridization of viral DNA, approx. 100 ng of DNA from each samples were blotted onto a positively charged nylon membrane following the method described earlier [2] using a dot-blot apparatus (Schleicher and Schuell, Germany). Dot-blots for detecting satellite DNAs were performed using ~1 μg DNA. For Southern-blot hybridization for viral DNA, ~1 μg of DNA was resolved in 1% agarose gel and transferred to membrane by capillary transfer overnight. The membranes were air-dried, UV cross-linked twice at 70 J/cm2 in a UV cross-linker (Pharmacia Biotech) and incubated in standard pre-hybridization solution for 4 hours at 65°C in a hybridization oven (Techne, UK). The Rep and CP genes of Sri Lankan cassava mosaic virus, SLCMV-[Adi] (GenBank accession no. AJ579307) [12] were PCR-amplified using the primer pairs RepF/RepR (TCCAATTGACCGAGTC/TTGAGGTCTACTCTCCG, annealing temperature 55°C) and CpF/CpR (TGAGTCCAGACACGATGTGG/ATGACCTTACCTATATGGAC, annealing temperature 50°C) respectively, using Taq DNA-polymerase (Bangalore Genei) in a thermocycler (Techne, UK). Twenty-five microlitre of reaction was performed with 1× reaction buffer containing 1.5 mM MgCl2, 1.25 U Taq DNA-polymerase (Bangalore Genei), 50 mM KCl, 0.2 mM dNTP, 1% Tween-20, and 50 pM of each primer. The amplified DNAs were used to prepare the probe using 32P-labeled dATP (Board of Radiation and Isotope Technology, Govt. of India) with Ready-To-Go DNA labeling beads (GE Healthcare) according to the manufacturer’s instructions. Probes were also prepared using amplified DNA fragments representing the full-length satellite DNAs. Hybridization was carried out according to standard methods [28], followed by autoradiography using Kodak diagnostic film for 8 hours at −70°C.
Detection of Virus by ELISA and Immunocapture PCR
ELISA was performed according to a standard protocol [10] (using 1:5,000 dilutions of primary antibody raised against maltose-binding protein (MBP)-fused CP of Mungbean yellow mosaic India virus (MYMIV), and 1:30,000 dilution of secondary antibody (Anti-Rabbit IgG alkaline phosphatase conjugate, Sigma-Aldrich) and assayed with substrate (Sigma FastTMp-nitrophenyl phosphate tablet sets, Sigma-Aldrich). Immune-capture PCR (IC-PCR) was performed according to a standard method [25], with 1:500 dilution of primary antibody (mentioned above). After the antigen coating, PCR was performed with primers specific for SLCMV, IRf (CTTTGGTGAGAGAACAC)/IRr (GGCGACGAACCTTC), using the conditions described above.
Amplification, Cloning and Sequence Analysis of Viral and Satellite DNAs
Primers were designed based on the SLCMV-[Adi] sequence for amplification of the 2 kb fragment of the viral DNA [M1.8F (TGTGACGCGGACAGTGG)/M1.8R (ACCGTATTAATACATATAATC), annealing temperature: 55°C]. For amplification of the rest of the viral genome, primers [M9F (GTCGAAGCGACCAGCAG)/M9R (ATGTTTCCTAGTCTTCC), annealing temperature 45°C] were designed based on the 2 kb fragment already obtained, with approx. 90 bp overlaps in the ends. Primer was designed using the software Generunner, version 3.05. For amplification of the satellite DNA, universal primers for betasatellite [4] were used. The primers were commercially synthesized from Sigma-Aldrich. PCR for amplification of geminiviral DNA was performed in a thermocycler (Techne, UK) using the following set-up: 30 cycles of denaturation at 94°C for 30 s, annealing at respective temperature for 30 s and extension at 72°C for 1 min; denaturation was carried out for additional 5 mins before the first cycle and extension was carried out for additional 7 mins after the last cycle of the reaction. The reaction was carried out in a 15 μl reaction volume (made up with water), using PhusionTM Hot Start High-Fidelity DNA-polymerase (New England Biolabs), according to the manufacturer’s instructions. PCR for amplification of betasatellite was performed according to a method published earlier [4]. The PCR-products were resolved in a 1% agarose gel in 1× TBE buffer along with a DNA size-marker (λ/HindIII). Gel images were captured using the Quantity-one gel-documentation system (BioRad). The PCR-products were gel purified and visually quantified in 1% agarose gel comparing with a known DNA size-marker (λ/HindIII). The products were then cloned using InsT/A PCR cloning kit (Fermentas) following manufacturer’s instructions. The bacterial colonies were selected using blue–white selection, checked by colony-PCR and sequenced commercially. Analysis of the DNA sequences was performed using the Megalign programme of the DNASTAR software.
Mechanical Transmission
Transmission of the resident begomovirus from symptomatic mentha plants to Nicotiana benthamiana was carried out by preparing an extract from symptomatic leaves (0.2 g) in 1 ml of sterile MQ water. The slurry was centrifuged at RT at 10,000 rpm for 10 min and 10 μl of the supernatant was rubbed onto fully expanded apical leaves of N. benthamiana plants dusted with fine grade carborandum powder at 3–4 leaf stage. Inoculated plants were maintained at 10/14 h photoperiod at 25°C in glasshouse.
Vector Transmission
For inoculation experiments, adult whiteflies (Bemisia tabaci) were collected using a light trap and were released on healthy mentha plants inside wire-mesh cages. They were maintained until they laid eggs, the eggs were collected, released and were allowed to hatch on the infected plants. The adults, after they emerged from the eggs, were fed on the infected plants for 24 h, collected and released onto the healthy plants for inoculation feeding for 24–48 h. Thereafter the plants were kept in insect-free cages in the glasshouse and symptoms were scored daily.
Results
Evidence for the Presence of Geminivirus in Mentha Plants
In a survey comprising four districts of Punjab, where mentha is cultivated in significant acreage, more than 80% of menthol mint plants and about 30% of peppermint and spearmint plants displayed leaf distortion symptoms (Fig. 1). Of the 14 symptomatic menthol mint DNA samples analyzed, ten hybridized with SLCMV cp and SLCMV rep. Similarly, all of the seven peppermint samples and seven out of nine spearmint samples hybridized with the SLCMV cp, indicating the presence of begomovirus(es) in the hybridizing samples. Samples from non-symptomatic plants did not give any signal. Seven representative samples of menthol mint, showing hybridization with SLCMV cp, were further tested by ELISA for the presence of cross-reacting viral proteins, using MYMIV antibodies. All samples showed absorbance values at least three times more than the non-symptomatic controls (data not shown). Out of the seven samples, six were further selected for testing for the presence of geminiviral DNA by IC-PCR. Amplification of a product of the expected size (587 bp) was obtained in all six ELISA-positive samples by IC-PCR, using MYMIV antibody, while no amplification was found in the absence of antigen and in the non-symptomatic sample. This indicated the presence of a begomovirus in the symptomatic mentha plants, which cross-reacted to MYMIV antibody and had sequence identity with SLCMV primers used.
Fig. 1.

Leaf deformity symptoms observed in Japanese mint (top), Pepper mint (middle) and Spear mint (bottom); symptomatic plants shown on the right, compared to non-symptomatic plants on the left
Amplification of Viral and Satellite DNAs and Sequence Analysis
DNA extracted from a symptomatic menthol mint sample, hybridizing with both SLCMV cp and Rep probes in the dot-blot experiment was used to amplify the full-length DNA of the resident begomovirus(es). Amplification of the full-length sequence was attempted with abutting primers derived from cp sequences amplified previously but no product could be obtained. However, amplification was successful with the primers M1.8F and M1.8R resulting in a product of ~2.0 kb. Primers designed from end-sequences (M9F/R) of the above product to amplify the remaining portions of the geminiviral DNA produced a 0.9 kb product. The approximately 90 bp overlap between both the ends of the two sequences were identical and indicated them to be part of a single molecule. The 2.0 kb and the 0.9 kb fragments were thus assembled to obtain the complete sequence, which was submitted to public nucleic acid sequence database (accession no. FJ514798). The size of the complete genome is 2,759 bp, and the gene arrangement typical of begomoviruses, with four open reading frames (ORFs) in the complementary sense and two ORFs in the viral sense strand. Sequence analysis showed the highest (94%) identity with Tomato leaf curl Karnataka virus-Bangalore [India:Bangalore:1993] (ToLCKV-Ban[IN:Ban:93], U38239). Thus, following the ICTV criteria, the virus is a strain of ToLCKV-Ban and can be assigned the strain descriptor of Tomato leaf curl Karnataka virus-Bangalore-[India:Ludhiana:Mentha:2007] ToLCKV-Ban[IN:Lud:Men:2007]. The sequence identities (percent) at the nucleotide level of the different viral ORFs, as compared with ToLCKV-Ban were as follows: V1 87, V2 96, C1 96, C2 96, C3 95 and C4 99. PCR performed using primers for DNA-B did not give any amplification product.
The amplification reaction using the betasatellite primers resulted in a fragment of 1.4 kb in size. Sequence analysis of the cloned fragment (deposited in the database under accession no. EU862816) showed that it has a length of 1,351 nucleotides and is completely identical with three entries of betasatellite sequences in the database, namely Cotton leaf curl Multan betasatellite [India:New Delhi 2:2004] (accession no. AY795604), Cotton leaf curl Multan betasatellite [India:Sri Ganganagar:2002] (accession no. AY083590) and Tomato leaf curl betasatellite [India:New Delhi:2003] (accession no. AY438562). Based on the sequence analysis, it was concluded that the amplified DNA represents a new isolate of Cotton leaf curl Multan betasatellite, the first report from mentha and the name, Cotton leaf curl Multan betasatellite [India:Ludhiana:Mentha:2007] CLCuMB-[IN:Lud:Men:07] is proposed, in accordance with the recommendations for naming such molecules [14]. PCR with primers specific for DNA-I, another satellite molecule reported earlier to be associated with some geminiviruses, did not give any amplified product. Interestingly, PCR using the betasatellite primers also amplified a 1.1 kb amplified product along with the CLCuMB-[IN:Lud:Men:07] DNA. The product was cloned and its sequence analysis revealed it to be 1,019 nucleotides in length (deposited in the database under the accession no. EU862815) and showed high nucleotide identity (98%) with a begomovirus-associated satellite-like molecule (DNA-II, AY836366). The molecule does not contain any ORF. The name, Mentha leaf deformity associated DNA-II (MLDA-DNA-II) is proposed for this molecule.
Occurrence of the Virus and Satellites
Of the 30 DNA samples obtained from symptomatic plants from various field locations in Punjab, 24 cross-hybridized with SLCMV-cp gene probe (the cp of SLCMV and ToLCKV-Ban[IN:Lud:Men:2007] share 83% nucleotide sequence identity) comprising 12/14 menthol mint, 7/7 of peppermint and 5/9 spearmint samples. The non-symptomatic negative controls showed no hybridization with the probe. Titers of the cross-hybridizing ToLCKV DNA (as estimated from the intensities of the hybridizing signals) were similar across the three species of mint. All 24 samples hybridizing with SLCMV cp probe also hybridized with CLCuMB-[IN:Lud:Men:07] probe. However, of the 24 samples, only 18 showed cross-hybridization with MLDA-DNA-II probe. There was no noticeable difference in symptoms between the plants which hybridized and which did not hybridize with the MLDA-DNA-II probe. The six samples which did not hybridize with the SLCMV cp probe, showed no hybridization with the CLCuMB-[IN:Lud:Men:07] or MLDA-DNA-II probes.
Transmission of the Virus and Satellites
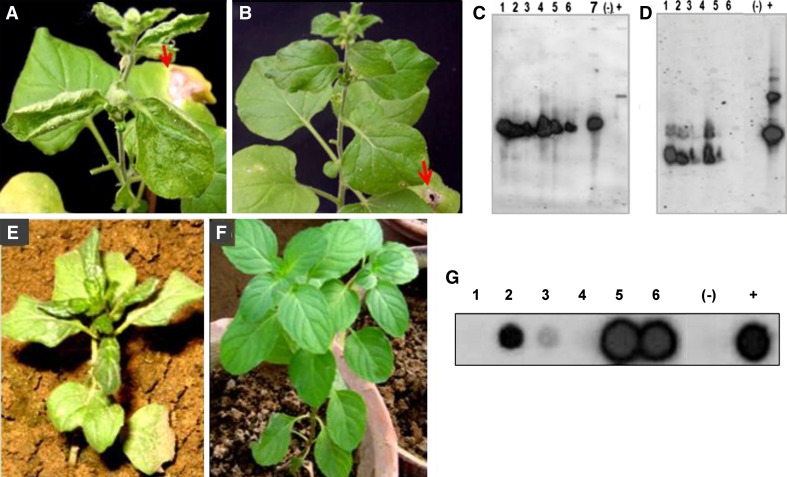
When the ability of the resident virus to be transmitted mechanically to the experimental host N. benthamiana was checked, all (6/6) the rub-inoculated plants (with leaf-extracts from symptomatic mentha plants) showed symptoms of leaf curling and crinkling, 3 weeks post-inoculation (Fig. 2a), while mock-inoculated plants failed to produce any such symptoms (Fig. 2b). Southern analysis of the total DNA isolated from systemic leaves of the above symptomatic N. benthamiana plants using SLCMV cp, cloned DNAs of CLCuMB-[IN:Lud:Men:07] and that of MLDA-DNA-II as probes indicated the accumulation of DNA species cross-hybridizing with the SLCMV cp and the CLCuMB-[IN:Lud:Men:07] probes in 6/6 and 5/6 inoculated plants, respectively (Fig. 2c, d). MLDA-DNA-II probe did not hybridize with any sample tested (data not shown). Transmission of the virus was performed with the natural whitefly vector. Symptoms, shown in Fig. 2e, similar to those seen in the field, appeared after about 1 month of inoculation on 4/10 plants inoculated. For comparison, an un-inoculated plant is shown in Fig. 2f. Of the ten plants, eight were further checked for the accumulation of viral DNA by dot-blot using SLCMV cp as probe. Signals of varying intensities were found in DNA samples from six plants (Fig. 2g), indicating transmission of the resident virus.
Fig. 2.
a Systemic symptoms produced in N. benthamiana plants rub-inoculated with symptomatic mentha extract 21 days post-inoculation and b mock-inoculated plants. Arrows show the inoculated leaves. c Southern-blot probed with radioactively labeled DNA fragment encoding SLCMV-CP, Lanes 1–6 total DNA extracted from N. benthamiana plants inoculated with leaf-extracts of symptomatic mentha plants, lane 7 DNA from cassava plant naturally infected with SLCMV as positive control; (−) Un-inoculated N. benthamiana plants (+) Cloned SLCMV DNA. d Southern-blot probed with radioactively labeled CLCuMB-[IN:Lud:Men:07], lanes 1–6: total DNA extracted from N. benthamiana plants inoculated with leaf-extracts of symptomatic mentha plants (−) Asymptomatic mentha plant; (+) Cloned DNA of CLCuMB-[IN:Lud:Men:07]. e Symptoms produced in whitefly-inoculated mentha plants and f un-inoculated control mentha plant and g Dot-blot analysis of the whitefly-inoculated mentha plants, probed with DNA encoding SLCMV-CP. Lane 1–6 inoculated plants (+) Virus source plant (−) Negative control (non-symptomatic plant)
Discussion
During this investigation, widespread leaf deformity symptoms in the mentha crop were seen in the districts surveyed in Punjab and were generally associated with the presence of whiteflies. Nucleic acid-based evidence, in the form of strong hybridization signals upon DNA dot-blot analysis, with symptomatic mentha plants using genes from a representative begomovirus (SLCMV), and serological evidence, with ELISA and IC-PCR indicated that the plants were infected with one or several begomoviruses. Although full-length amplification of the resident begomoviral DNA was unsuccessful, the amplification of DNA fragments representing about 2.0 kb was successful from symptomatic plants. Primers designed from the ends of the above fragment, facing outside, gave rise to the remaining approximately 0.9 kb amplified fragment, representing the remaining part of the begomoviral DNA. According to the proposed classification and naming schemes for geminiviruses [14], since overall nucleotide sequence identity of the new begomoviral isolate is 94% with ToLCKV-Ban it can be considered its new variant from mentha. Earlier, in mentha, reports have indicated the presence of various classes of viruses, including a hordeivirus [1], Strawberry latent ringspot virus [33], Mint virus X [34] and a vitivirus [35], all reports emanating from the USA. Recently, a partial DNA sequence of a geminivirus related to Tomato leaf curl Pakistan virus was reported from mentha from Uttar Pradesh [27]. This investigation has provided evidence for the presence of a second tomato-infecting begomovirus (ToLCKV-Ban) in mentha. In addition, a betasatellite (CLCuMB), which showed near complete identity (99%) with betasatellites reported earlier from cotton, was also detected in the same mentha plants. The betasatellite is considered a variant of CLCuMB according to the proposed classification and naming conventions [6]. Both ToLCKV and CLCuMB could be efficiently transmitted by rub-inoculation to N. benthamiana, based on symptom development and Southern analysis. Mechanical transmission of betasatellites has not been reported yet and this is the first report of such a transmission. DNA dot-blot analysis of symptomatic mentha plants in the field gave a strong indication that infection with ToLCKV or similar begomoviruses and CLCuMB or related molecules was fairly widespread in Punjab.
Tomato leaf curl viruses with bipartite genomes are present mainly in northern India and those with monopartite genomes in southern India [17, 23, 24, 31]. Tomato leaf curl Gujarat virus was the first tomato-infecting monopartite begomovirus reported from northern India [30] and subsequently monopartite tomato begomoviruses have been reported in hosts other than tomato (cotton) in northern India [20]. Although ToLCKV has been reported mainly from southern India, there exists one report of a tomato isolate from the north-western part of the country, based on partial genome sequences (CP-region) [9]. It is possible that the isolate ToLCKV-[men] may have had its origin from such infected tomato plants or other ToLCKV-infected hosts.
Cotton leaf curl disease is prevalent in Pakistan and mainly in the north-western states of India including Punjab, Haryana and Rajasthan, apart from southern India. Several begomoviruses causing Cotton leaf curl disease have been reported from these areas [22, 30, 36], all of which are found to be associated with betasatellites. Artificial inoculation experiments have shown that the betasatellites are necessary for producing typical symptoms of Cotton leaf curl disease [3]. Betasatellites have also been reported to be associated with begomoviruses causing Tomato leaf curl disease [7, 9]. It is difficult to speculate the origin of CLCuMB-[IN:Lud:Men:07], but it has most likely originated in either tomato or cotton plants, both crops being cultivated close to mentha-growing areas.
No published information is available on DNA-II. MLDA-DNA-II accumulated in a majority, but not all symptomatic mentha plants examined, and could not be transmitted to N. benthamiana by mechanical inoculation. Thus, the possibility exists that MLDA-DNA-II is restricted to mentha. Further work is necessary on this interesting satellite-like molecule to determine its role, if any, in the development of leaf deformity in mentha.
Based on this investigation, we conclude that a begomovirus (ToLCKV) and a betasatellite (CLCuMB) are strongly associated with a widespread leaf deformity disease of mentha in Punjab. This is the first report of simultaneous occurrence of begomovirus and betasatellite in mentha and illustrates the invasiveness and promiscuity of such infectious agents triggered by rapid changes in agricultural practices in India.
Acknowledgments
BKB is grateful to Council of Scientific and Industrial Research, Govt. of India for Research fellowship. Authors are grateful to Dr V. G. Malathi, Indian Agricultural Research Institute, New Delhi, for providing the antisera against MYMIV.
Footnotes
References
- 1.Beczner L, Hamilton RI, Rochon DM. Properties of the Mentha strain of lychnis ringspot virus. Intervirology. 1992;33:49–56. doi: 10.1159/000150230. [DOI] [PubMed] [Google Scholar]
- 2.Borah BK, Johnson AMA, Sai Gopal DVR, Dasgupta I. A comparison of four DNA extraction methods for the detection of Citrus yellow mosaic badna virus from two species of citrus using PCR and dot-blot hybridization. Virus Res. 2008;151:321–324. doi: 10.1016/j.jviromet.2008.05.022. [DOI] [PubMed] [Google Scholar]
- 3.Briddon RW, Mansoor S, Bedford ID, Pinner MS, Saunders K, Stanley J, Zafar Y, Malik KA, Markham PG. Identification of DNA components required for induction of cotton leaf curl disease. Virology. 2001;285:234–243. doi: 10.1006/viro.2001.0949. [DOI] [PubMed] [Google Scholar]
- 4.Briddon RW, Bull SE, Mansoor S, Amin I, Markham PG. Universal primers for PCR-mediated amplification of DNA-β. Mol Biotechnol. 2002;20:315–318. doi: 10.1385/MB:20:3:315. [DOI] [PubMed] [Google Scholar]
- 5.Briddon RW, Stanley J. Subviral agents associated with plant single-stranded DNA viruses. Virology. 2006;344:198–210. doi: 10.1016/j.virol.2005.09.042. [DOI] [PubMed] [Google Scholar]
- 6.Briddon RW, Brown JK, Moriones E, Stanley J, Zerbini M, Zhou X, Fauquet CM. Recommendations for the classification and nomenclature of the DNA-β satellites of begomoviruses. Arch Virol. 2008;153:763–781. doi: 10.1007/s00705-007-0013-6. [DOI] [PubMed] [Google Scholar]
- 7.Chakraborty S, Pandey PK, Banerjee MK, Kalloo G, Fauquet CM. Tomato leaf curl Gujarat virus, a new Begomovirus species causing a severe leaf curl disease of tomato in Varanasi, India. Phytopathology. 2003;93:1485–1495. doi: 10.1094/PHYTO.2003.93.12.1485. [DOI] [PubMed] [Google Scholar]
- 8.Chatterjee A, Ghosh SK. A new monopartite begomovirus isolated from Hibiscus cannabinus L. in India. Arch Virol. 2007;152:2113–2118. doi: 10.1007/s00705-007-1029-7. [DOI] [PubMed] [Google Scholar]
- 9.Chowda Reddy RV, Colvin J, Muniyappa V, Seal S. Diversity and distribution of begomoviruses infecting tomato in India. Arch Virol. 2005;150:845–867. doi: 10.1007/s00705-004-0486-5. [DOI] [PubMed] [Google Scholar]
- 10.Clark MF, Adams AN. Characteristics of the microplate method of enzyme-linked immunosorbent assay for the detection of plant viruses. J Gen Virol. 1977;34:475–483. doi: 10.1099/0022-1317-34-3-475. [DOI] [PubMed] [Google Scholar]
- 11.Dry IB, Krake LR, Rigden JE, Rezaian MA. A novel subviral agent associated with a geminivirus: the first report of a DNA β satellite. Proc Natl Acad Sci USA. 1999;4:7088–7093. doi: 10.1073/pnas.94.13.7088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dutt N, Briddon RW, Dasgupta I. Identification of a second begomovirus, Sri Lankan cassava mosaic virus, causing cassava mosaic disease in India. Arch Virol. 2005;150:2101–2108. doi: 10.1007/s00705-005-0579-9. [DOI] [PubMed] [Google Scholar]
- 13.Fauquet CM, Stanley J. Geminivirus classification and nomenclature: progress and problems. Ann. Appl. Biol. 2003;142:165–189. doi: 10.1111/j.1744-7348.2003.tb00241.x. [DOI] [Google Scholar]
- 14.Fauquet CM, Briddon RW, Brown JK, Moriones E, Stanley J, Zerbini M, Zhou X. Geminivirus strain demarcation and nomenclature. Arch Virol. 2008;153:783–821. doi: 10.1007/s00705-008-0037-6. [DOI] [PubMed] [Google Scholar]
- 15.Gopal P, Kumar PP, Sinilal B, Jose J, Yadunandam AK, Usha R. Differential roles of C4 and β-C1 in mediating suppression of post-transcriptional gene silencing: evidence for transactivation by the C2 of Bhendi yellow vein mosaic virus, a monopartite begomovirus. Virus Res. 2007;23:8–9. doi: 10.1016/j.virusres.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 16.Harrison BD. Advances in Geminivirus research. Annu Rev Phytopathol. 1985;23:55–82. doi: 10.1146/annurev.py.23.090185.000415. [DOI] [Google Scholar]
- 17.Hong YG, Harrison BD. Nucleotide sequences from tomato leaf curl viruses from different countries: evidence for three geographically separate branches in evolution of coat protein of whitefly transmitted geminiviruses. J Gen Virol. 1995;76:2043–2049. doi: 10.1099/0022-1317-76-8-2043. [DOI] [PubMed] [Google Scholar]
- 18.Hull R. Matthews’ plant virology. San Diego: Academic Press; 2002. p. 180. [Google Scholar]
- 19.Jose J, Usha R. Bhendi yellow vein mosaic disease in India is caused by association of a DNA beta satellite with a begomovirus. Virology. 2003;305:310–317. doi: 10.1006/viro.2002.1768. [DOI] [PubMed] [Google Scholar]
- 20.Kirthi N, Priyadharshini CGP, Sharma P, Maiya SP, Hemalatha V, Sivaraman P, Dhawan P, Rishi N, Savithri HS. Genetic variability of begomoviruses associated with cotton leaf curl disease originating from India. Arch Virol. 2004;149:2047–2057. doi: 10.1007/s00705-004-0352-5. [DOI] [PubMed] [Google Scholar]
- 21.Kobayashi N, Horikishi T, Katsuyama H, Handa T, Takayanagi K. A simple and efficient DNA extraction method for plants, especially woody plants. Plant Tissue Culture Biotechnol. 1998;4:76–80. [Google Scholar]
- 22.Mansoor S, Briddon RW, Bull SE, Bedford ID, Bashir A, Hussain M, Saeed, Zafar MY, Malik KA, Fauquet C, Markham PG. Cotton leaf curl disease is associated with multiple monopartite begomoviruses supported by single DNA beta. Arch Virol. 2003;148:1969–1986. doi: 10.1007/s00705-003-0149-y. [DOI] [PubMed] [Google Scholar]
- 23.Muniyappa V, Venkatesh HM, Ramappa HK, Kulkarni RS, Zeiden M, Tarba CY, Ghanim M, Czosnek H. Tomato leaf curl virus from Bangalore (ToLCV-Ban4): sequence comparison with Indian ToLCV isolates, detection in plants and insects, and vector relationships. Arch Virol. 2000;145:1583–1598. doi: 10.1007/s007050070078. [DOI] [PubMed] [Google Scholar]
- 24.Padidam M, Beachy RN, Fauquet CM. Tomato leaf curl geminivirus from India has a bipartite genome and coat protein is not essential for infectivity. J Gen Virol. 1995;76:25–35. doi: 10.1099/0022-1317-76-1-25. [DOI] [PubMed] [Google Scholar]
- 25.Rampersad SN, Umaharan P. Detection of begomoviruses in clarified plant extracts: a comparison of standard, direct-binding, and immunocapture polymerase chain reaction techniques. Phytopathology. 2003;93:1153–1157. doi: 10.1094/PHYTO.2003.93.9.1153. [DOI] [PubMed] [Google Scholar]
- 26.Saeed SY, Zafar Y, Randles JW, Rezaian MA. A monopartite begomovirus-associated DNA beta satellite substitutes for the DNA B of a bipartite begomovirus to permit systemic infection. J Gen Virol. 2007;88:2881–2889. doi: 10.1099/vir.0.83049-0. [DOI] [PubMed] [Google Scholar]
- 27.Samad A, Gupta MK, Shasany AK, Ajayakumar PV and Alam M. Begomovirus related to Tomato leaf curl Pakistan virus newly reported in Mentha crops in India. New Dis. Rep. 2008;18.
- 28.Sambrook J, Russell DW. Molecular cloning: a laboratory manual. 3. NY: Cold Spring Harbor Press; 2001. [Google Scholar]
- 29.Saunders K, Stanley J. A nanovirus-like DNA component associated with yellow vein disease of Ageratum conyzoides: evidence for interfamilial recombination between plant DNA viruses. Virology. 1999;264:142–152. doi: 10.1006/viro.1999.9948. [DOI] [PubMed] [Google Scholar]
- 30.Sharma P, Rishi N, Malathi VG. Molecular cloning of coat protein gene of an Indian cotton leaf curl virus (CLCuV-HS2) isolate and its phylogenetic relationship with others members of Geminiviridae. Virus Genes. 2005;30:85–91. doi: 10.1007/s11262-004-4585-x. [DOI] [PubMed] [Google Scholar]
- 31.Shih SL, Tsai WS, Green SK, Hanson PM, Valand G, Kalloo G, Shrestha SK, Joshi S. Molecular characterisation of a new tomato begomovirus infecting from India. Plant Dis. 2003;87:598. doi: 10.1094/PDIS.2003.87.5.598A. [DOI] [PubMed] [Google Scholar]
- 32.Stanley J, Bisaro DM, Briddon RW, Brown JK, Fauquet CM, Harrison BD, Rybicki EP, Stenger DC. Geminiviridae. In: Ball LA, editor. Virus taxonomy. 8th report of the international committee on the taxonomy of viruses. London: Elsevier/Academic Press; 2005. pp. 301–326. [Google Scholar]
- 33.Tzanetakis IE, Postman JD, Gergerich RC, Martin RR. A virus between families: nucleotide sequence and evolution of Strawberry latent ringspot virus. Virus Res. 2006;121:199–204. doi: 10.1016/j.virusres.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 34.Tzanetakis IE, Postman JD, Martin RR. Mint virus X: a novel potexvirus associated with symptoms in ‘Variegata’ mint. Arch Virol. 2006;151:143–153. doi: 10.1007/s00705-005-0586-x. [DOI] [PubMed] [Google Scholar]
- 35.Tzanetakis IE, Postman JD, Martin RR. Identification, detection and transmission of a new vitivirus from Mentha. Arch Virol. 2007;152:2027–2033. doi: 10.1007/s00705-007-1030-1. [DOI] [PubMed] [Google Scholar]
- 36.Zhou X, Liu Y, Robinson DJ, Harrison BD. Four DNA-A variants among Pakistan isolates of cotton leaf curl virus and their affinities to DNA-A of geminivirus isolates from okra. J Gen Virol. 1998;79:915–923. doi: 10.1099/0022-1317-79-4-915. [DOI] [PubMed] [Google Scholar]